Abstract
BACKGROUND:
The most common type of congenital heart disease is the cardiac septal defects, which has reported to be caused by a missense mutation (G296S) in exon 3 of the GATA4 gene.
AIMS:
The present study was undertaken to find out whether GATA4 gene is the prime cause of the septal defects in Mysore population.
MATERIALS AND METHODS:
GATA4 gene analyses were undertaken on 21 confirmed CHD cases by PCR and DNA sequencing.
RESULTS AND CONCLUSION:
Analysis of this particular mutation in 21 septal defect patients revealed that none of the patients had the mutation, indicating that this mutation is population specific or septal defect in Mysore population is caused due to mutations in other regions of the GATA4 gene.
Keywords: Cardiac septal defects, congenital heart disease, GATA4, missense mutation
Introduction
Division of a common atrium and ventricle into right and left sided chambers represents an essential evolutionary milestone in the development of a four-chambered heart and is necessary for the separation of oxygenated and deoxygenated blood.[1] However, this process of separation fails to occur resulting in septal defects, which accounts for about 50% of all the congenital heart disease (CHD). Recently, molecular and developmental biologists have elucidated the molecular pathways that regulate cardiac development.[1] One of the important genes, which have an active role during the process of heart development is GATA4 located on chromosome 8p23.1-p22 with six exons. It codes for a 3372 bp long transcript with 443 amino acid residues. This gene is a member of the GATA family of zinc-finger transcription factors, which are a group of structurally related transcription factors that control gene expression and differentiation in a variety of cell types. Members of this family of DNA-binding proteins recognize a consensus sequence known as the ‘GATA’ motif, which is an important cis-element in the promoters of GATA genes.[2] Garg et al [3] have reported a missense mutation (G296S) in exon 3 of the GATA4 gene as a prime cause of cardiac septal defect in a large family with 16 individuals having atrial septal defect, of which, eight had additional types of defect like ventricular septal defect (VSD), atrioventricular septal defect (AVSD) and pulmonary valve thickening. Studies in 3000 unrelated individuals without any septal defect of diverse ethnicity did not have this mutation. This heterozygous missense mutation was found to cause a G to A transition at nucleotide 886 which results in a glycine to serine substitution at codon 296 (G296S) disrupting a highly conserved glycine residue adjacent to the second zinc finger of GATA4, which is critical for protein-protein interactions. This mutation results in diminished DNA binding affinity and transcriptional activity of GATA4. Furthermore, the G296S mutation abrogated a physical interaction between GATA4 and TBX5, T-box protein responsible for a subset of syndromic cardiac Septal Defects.[3]
In view of this, in the present investigation this GATA4 mutation was screened in 21 septal defect patients from Mysore (South India) to know whether it is the prime cause of the septal defects in our population.
Materials and Methods
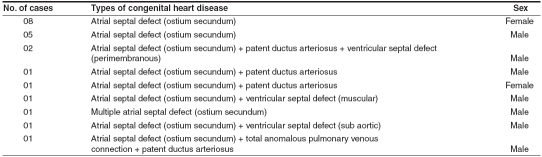
The present investigation was conducted in Mysore city (Karnataka state), South India from September 2003 to August 2005 in three major hospitals: K. R. Hospital, CSI Holdsworth Memorial Hospital and J.S.S Hospital. The suspected CHD patients had been subjected by the pediatricians for extensive X-ray analysis, electrocardiogram and echocardiography examination for confirmation of the defect. A total of 21 confirmed CHD cases were considered for the present study, which included ostium secundum type of atrial septal defect (ASD), ventricular septal defect (VSD), patent ductus arteriosus (PDA) and total anomalous pulmonary venous connection (TAPVC) [Table 1]. After informed consent was obtained, DNA was extracted from peripheral blood leukocytes of 21 patients using standard procedure of Lahiri et al[4] with suitable modifications. A pair of intronic primers (Forward primer-CCGAGTGGGCCTCTCCTGTGC; Reverse primer- CTACTTTGCTGGCCTCTTCCGTCCT) were designed for the complete exon 3 of GATA4 (GenBank accession number NM_002052) which amplified a 276bp fragment. PCR products were purified using the Gel Extraction Kit (Sigma, St Louis, USA] and the purified products were sequenced on an automated DNA sequencer (Applied Biosystems, Foster City, USA).
Table 1.
Number of patients and their cardiac phenotypes of the present study
Results and Discussion
In the present study, the sequence analysis of 276bp fragments from all the 21 patients showed the absence of G296S missense mutation. Similarly, Okubo et al[5] in Japan have screened GATA4 gene, for the missense mutation (G296S), in a large family of 22 members with 11 ASD patients. But they did not find this mutation in any of the patients. However, they found a novel 1bp deletion (c.1074delC) in exon 6 in nine individuals with ASD. In another study, Hirayama-Yamada et al[6] again in Japan have screened 16 ASD patients and found a frameshift mutation (E359del) in exon 5 in one patient with ASD as reported by Garg et al.[3] In another patient, a novel missense mutation (c.155C>T) was found in exon 1. However, both the groups failed to find the G296S missense mutation. On the contrary, Sarkozy et al[7] have found G296S missense mutation in 5 of 29 ASD from Italy.
The present investigation suggests that, the G296S missense mutation does not cause septal defect in patients from Mysore city. This indicates that the G296S missense mutation may be a population specific. Hence, the mutations in other regions of the GATA4 gene may be responsible for the formation of septal defect in Mysore population. However, this possibility needs to be further investigated.
Acknowledgments
We thank all the family members who have taken part in the study, the Department of Pediatrics, Cheluvamba Hospital, CSI Holdsworth Memorial Hospital and J.S.S. Hospital; Doctors and PG students who have provided with the necessary information to conduct the present study; University of Mysore for giving us (SR) an opportunity to carry out our research activities; Dr. Vidu Garg, USA, Mr. Harshavardhan M. Gawde, Ms. Pushpa Saviour, Ms. Shoma, Dr. Mikhil M. Bamne and Dr. Nagaraj for their help during the study. We also thank Prof. H. A. Ranganath; Chairman, DOS in Zoology for their help and encouragement.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Garg V. Insights into the genetic basis of congenital heart disease. Cell Mol Life Sci. 2006;63:5532–2. doi: 10.1007/s00018-005-5532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arceci RJ, King AA, Simon MC, Orkin SH, Wilson DB. Mouse GATA-4: A retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993;13:2235–46. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, et al. GATA4 mutations cause human congenital heart disease and reveal an interaction with TBX5. Nature. 2003;424:443–7. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 4.Lahiri DK, Bye S, Nurnberger JI, Jr, Hodes ME, Crisp M. A non-organic and non-enzymatic extraction method gives higher yields of genomic DNA from whole-blood samples than do nine other methods tested. J Biochem Biophys Met. 1992;25:193–205. doi: 10.1016/0165-022x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 5.Okubo A, Miyoshi O, Baba K, Takagi M, Tsukamato K, Kinoshita A, et al. A novel GATA4 mutation completely segregated with Atrial Septal Defect in a large Japanese family. J Med Genet. 2004;41:e97. doi: 10.1136/jmg.2004.018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirayama-Yamada K, Kamisago M, Akimoto K, Aotsuka H, Nakamura Y, Tomita H, et al. Phenotypes with GATA4 or NKX2.5 mutations in familial atrial septal defect. Am J Med Genet A. 2005;135:47–52. doi: 10.1002/ajmg.a.30684. [DOI] [PubMed] [Google Scholar]
- 7.Sarkozy A, Conti E, Neri C, D’Agostino R, Digilio MC, Esposito G, et al. Spectrum of atrial septal defects associated with mutations of NKX2.5 and GATA4 transcription factors. J Med Genet. 2005;42:e16. doi: 10.1136/jmg.2004.026740. [DOI] [PMC free article] [PubMed] [Google Scholar]