Abstract
The Rad51 protein of Saccharomyces cerevisiae is a eukaryotic homolog of the RecA protein, the prototypic DNA strand-exchange protein of Escherichia coli. RAD51 gene function is required for efficient genetic recombination and for DNA double-strand break repair. Recently, we demonstrated that RecA protein has a preferential affinity for GT-rich DNA sequences—several of which exhibit enhanced RecA protein-promoted homologous pairing activity. The fundamental similarity between the RecA and Rad51 proteins suggests that Rad51 might display an analogous bias. Using in vitro selection, here we show that the yeast Rad51 protein shares the same preference for GT-rich sequences as its prokaryotic counterpart. This bias is also manifest as an increased ability of Rad51 protein to promote the invasion of supercoiled DNA by homologous GT-rich single-stranded DNA, an activity not previously described for the eukaryotic pairing protein. We propose that the preferred utilization of GT-rich sequences is a conserved feature among all homologs of RecA protein, and that GT-rich regions are loci for increased genetic exchange in both prokaryotes and eukaryotes.
Keywords: Rad51 protein, in vitro selection, genetic recombination, homologous pairing, genetic instability
In the yeast Saccharomyces cerevisiae, genes belonging to the RAD52 epistasis group are required for homologous recombination and for DNA double-strand break repair (for reviews, see Resnick 1987; Petes et al. 1991; Game 1993). The RAD52 group includes the RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, MRE11, and XRS2 genes. Genetic characterization of this group has identified RAD51 as one of the most important members of this group. Mutations in the RAD51 gene result in a myriad of defects including sensitivity to ionizing radiation and alkylating agents, a deficiency in mating-type switching, and deficiencies in mitotic and meiotic recombination (Resnick 1987; Petes et al. 1991; Shinohara et al. 1992; Game 1993). The Rad51 protein shares significant sequence as well as structural homology with the prototypic and most well-studied DNA strand exchange protein RecA from Escherichia coli (Aboussekhra et al. 1992; Basile et al. 1992; Shinohara et al. 1992). The RecA protein is absolutely essential for homologous genetic recombination and DNA repair in E. coli (for reviews, see Clark 1973; Radding 1988; West 1992; Cox 1993; Kowalczykowski et al. 1994).
The recent purification and biochemical characterization of the yeast Rad51 protein has provided an opportunity to compare it more thoroughly to RecA protein. Similar to RecA protein, Rad51 protein has DNA-dependent ATPase activity (Sung 1994; Sugiyama et al. 1997), and possesses the ability to promote homologous pairing and DNA strand exchange in vitro (Sung 1994; Sung and Robberson 1995; Sugiyama et al. 1997). Also, Rad51 protein forms a nucleoprotein filament that is indistinguishable from the filament formed by the RecA protein (Ogawa et al. 1993b; Sung and Robberson 1995). However, although these activities establish Rad51 protein as the yeast analog of the RecA protein, some differences do exist. First, the Rad51 protein catalyzes the hydrolysis of ATP at a rate that is ∼35- to 40-fold lower than for RecA protein (Sung 1994; Sugiyama et al. 1997). Second, the Rad51 protein does not require ATP hydrolysis at any step during DNA strand exchange (Sung and Stratton 1996). Finally, the Rad51 protein catalyzes DNA strand exchange with a polarity (3′ → 5′), which is opposite to that of RecA protein (Sung and Robberson 1995).
These DNA strand exchange proteins are characterized by their ability to bind DNA without regard to sequence and to pair any homologous DNA sequence to its partner (for reviews, see West 1992; Ogawa et al. 1993a; Kowalczykowski and Eggleston 1994; Kowalczykowski et al. 1994; Camerini-Otero and Hsieh 1995). This permits genetic recombination to occur anywhere along the length of homologous chromosomes. However, although binding and pairing of DNA are generally regarded as being nonspecific, loci exist within both prokaryotes and eukaryotes that are either enhanced or depressed for homologous recombination. Previously, we showed that a component of this variance could be attributed directly to the DNA strand exchange process. Using in vitro selection and homologous pairing assays, we showed that (1) the E. coli RecA protein has a preferential affinity for GT-rich DNA sequences; (2) these selected sequences display enhanced RecA protein-dependent homologous pairing activity; and (3) that these sequences are found at genomic loci within prokaryotes and eukaryotes that display enhanced recombinational activity, such as the E. coli recombination hot spot Chi (χ; 5′-GCTGGTGG-3′) (Lam et al. 1974; Dixon and Kowalczykowski 1991), microsatellite DNA from humans, the constant region of heavy chains of immunoglobulins from several mammals, and Alu repetitive elements from humans (Tracy and Kowalczykowski 1996). These findings suggested that the genetic instability observed at these sequences is attributable, at least in part, to the recombinational activity of the respective DNA strand exchange protein. This suggestion proved accurate for the χ sequence, as DNA substrates containing χ exhibited elevated levels of homologous pairing in RecA protein-dependent joint molecule formation assays (Tracy and Kowalczykowski 1996).
Because of the genetic and biochemical similarities between the RecA and Rad51 proteins, we wanted to determine whether Rad51 protein displays an analogous preference for binding and homologously pairing GT-rich DNA. Using in vitro selection, we show that the yeast Rad51 protein has a similar bias for GT-rich DNA sequences. This preference is also displayed as an increased rate and extent of homologous pairing activity. Confirming our supposition above, many of the sequences share significant homology with eukaryotic genomic sequences that are genetically unstable. Therefore, we suggest that such GT-rich DNA sequences are universally important loci for genetic exchange.
Results
In vitro selection of GT-rich DNA sequences by Rad51 protein
To determine whether the Rad51 protein has a preference for a certain DNA composition, we performed in vitro selection starting with a pool of 6 × 1013 54-mers (SKBT18; Table 1). The 54-mers are composed of a random internal region of 18 nucleotides, and two flanking defined regions of 18 nucleotides (Tracy and Kowalczykowski 1996). To ensure significant competition between all of the 54-mers, we used limiting concentrations of purified Rad51 protein during five cycles of selection and amplification. The selected 54-mers were cloned and 40 randomly chosen clones were sequenced. Figure 1 depicts the sequences of the internal 18-mer regions that were selected by Rad51 protein. The average base composition of these sequences is highly over-represented for G and T residues (45% and 31%, respectively), and is significantly under-represented for A and C residues (10% and 15%, respectively). In addition to the 32 different sequences shown, the sequence displayed for clone 1 (selected sequence 1) appeared an additional eight times (which accounts for the missing clones in Fig. 1). Clone 39 is identical to clone 1, except for single-base changes at positions 2 (C → G) and 18 (C → G). Remarkably, the sequence of clone 1 is exactly the same sequence that appeared 16 times in the RecA protein selection experiments (Tracy and Kowalczykowski 1996). The frequent selection of this sequence is not a consequence of it being over-represented in the initial pool, as sequencing results showed that the pool was completely random (data not shown).
Table 1.
Oligodeoxyribonucleotides
| Designation and description
|
Length
|
Sequence
|
|---|---|---|
| SKBT16—Contains selected sequence 1 | 54 | 5′-AGCTTGCATGCCTGCAGGGGCGTGTGTGGTGGTGTGCTAGGATCC CCGGGTACC-3′ |
| SKBT17—Complement of SKBT16 | 54 | 5′-GGTACCCGGGGATCCTAGCACACCACCACACACGCCCCTGCAGGC ATGCAAGCT-3′ |
| SKBT18—Random pool of 54-mers | 54 | 5′-AGGCAAGCTTGGCTGCAG(N18)TAGGATCCGAGTCCTCCT-3′ |
| SKBT19 —Homologous to pBT54CN1 —opposite side of plasmid | 54 | 5′-TGAACGAAATAGACAGATCGCTGAGATAGGTGCCTCACTGATTAA GCATTGGTA-3′ |
| SKBT20 —Homologous to pBT54CN1 —adjacent to SKBT16 | 54 | 5′-TCCCAGTCACGACGTTGTAAAACGACGGCCAGTGCCAAGCTTGCA TGCCTGCAG-3′ |
Figure 1.
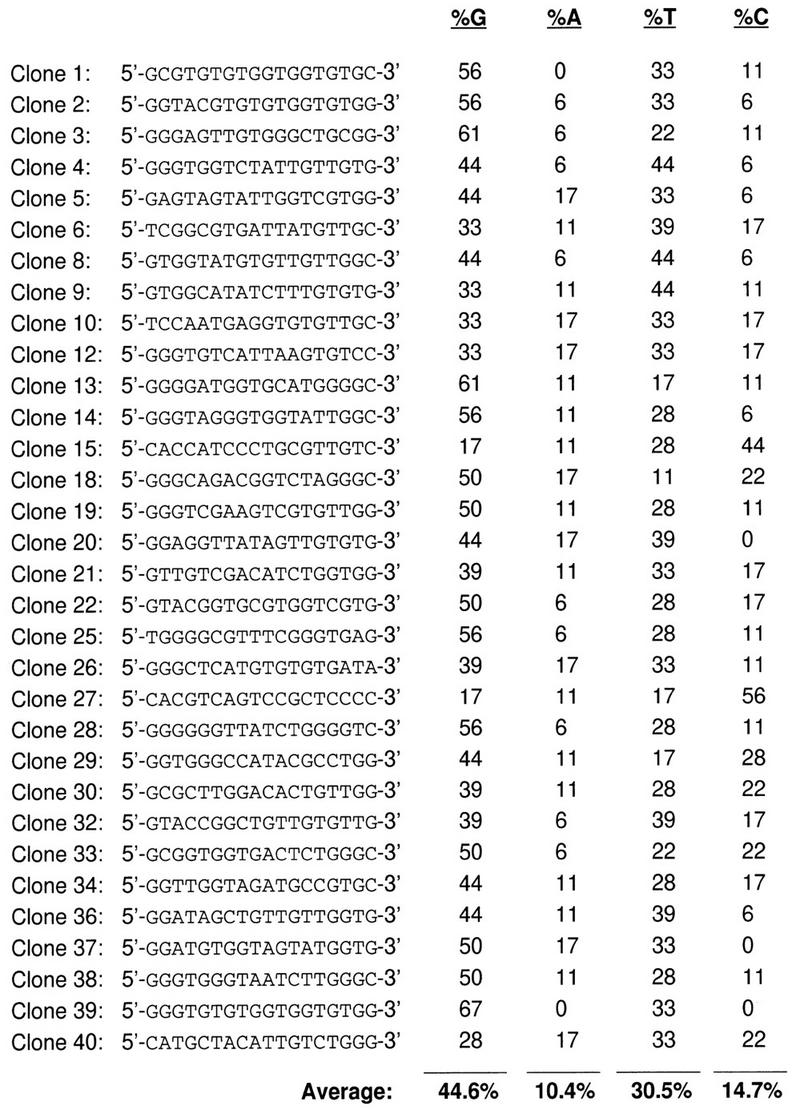
Rad51 protein selects for GT-rich DNA sequences from a random pool of oligonucleotides. In vitro selection was performed using the 54-mers containing a random internal region of 18 nucleotides. The sequences of the 18-nucleotide region after five cycles of selection and amplification are shown. In addition, the frequency of occurrence for each base is shown, along with the average base composition for all of the sequences.
Similarly, control reactions established that the observed preference does not arise from the in vitro selection methodology (Tracy and Kowalczykowski 1996), demonstrating that the bias shown by RecA protein and Rad51 protein is intrinsic to these proteins. These results demonstrate that Rad51 protein has a preference for DNA sequences that are GT-rich, and that it has a particular bias for the sequence found in clone 1. Strikingly, this is nearly identical to the results obtained with RecA protein, except that Rad51 protein appears to have an even greater preference for G’s and a slightly decreased bias for T’s.
Significant over-representation of GT-rich dinucleotides and trinucleotides in the selected DNA sequences
Because both the Rad51 and RecA proteins bind two to three nucleotides per protein monomer (Kowalczykowski 1991; Shinohara et al. 1992; Ogawa et al. 1993b), the distribution of dinucleotide and trinucleotide frequencies in the protein-selected sequences serves to quantify the intrinsic pattern of preferences displayed by a protein monomer for a given sequence. Table 2 shows that a group of trinucleotides (GTG, TGT, TGG, GGT, GGG, and TTG) are significantly over-represented. Except for one case (GGG), all of these triplets were also highly over-represented in the RecA protein-selected sequences (Tracy and Kowalczykowski 1996). Also, as observed for RecA protein, many triplets containing A and C residues are significantly under-represented. An analysis of the dinucleotide frequencies showed that three (GT, TG, and GG) are significantly over-represented relative to the expected frequency and to the other dinucleotide frequencies (Table 2). As observed for the trinucleotides, dinucleotides containing A and C residues are significantly avoided; the RecA protein-selected sequences displayed an identical bias. A χ2 test demonstrated that the frequency pattern for both the trinucleotide and dinucleotide distributions is not the result of random chance (data not shown). Interestingly, the most over-represented triplet in the S. cerevisiae genome is TGG/CCA, and the second most over-represented dinucleotide is TG/CA (Burge et al. 1992), two of the most highly over-represented in the selected sequences. A similar correlation was observed between the RecA protein-selected sequences and the E. coli genome (Tracy and Kowalczykowski 1996).
Table 2.
Frequency of trinucleotide and dinucleotide distribution in the 18-nucleotide sequences selected by Rad51 protein
| Trinucleotides
|
Frequency (%)a
|
Trinucleotides
|
Frequency (%)a
|
|---|---|---|---|
| GTG | 10.4 | CAC | 0.6 |
| TGT | 7.4 | CTA | 0.6 |
| TGG | 7.2 | CTT | 0.6 |
| GGT | 7.0 | CTC | 0.6 |
| GGG | 5.6 | CCG | 0.6 |
| TTG | 4.5 | CCA | 0.6 |
| GTT | 3.7 | CCC | 0.6 |
| GGC | 2.7 | AGA | 0.4 |
| GTC | 2.7 | AAG | 0.4 |
| GTA | 2.3 | AAT | 0.4 |
| CGT | 2.1 | ACT | 0.4 |
| TGC | 2.0 | ACC | 0.4 |
| ATG | 2.0 | TAA | 0.4 |
| CTG | 2.0 | TTT | 0.4 |
| TAT | 1.8 | CGA | 0.4 |
| CAT | 1.8 | CAG | 0.4 |
| AGT | 1.6 | CCT | 0.4 |
| GCG | 1.6 | GAA | 0.2 |
| TCT | 1.6 | AGC | 0.2 |
| GCT | 1.4 | TTC | 0.2 |
| TAG | 1.4 | CAA | 0.2 |
| TCG | 1.4 | AAA | 0.0 |
| CGG | 1.4 | AAC | 0.0 |
| GGA | 1.2 | ||
| GAT | 1.2 | Dinucleotides | Frequency (%)b |
| ATT | 1.2 | GT | 18.0 |
| GAG | 1.0 | TG | 17.8 |
| ATA | 1.0 | GG | 17.5 |
| ATC | 1.0 | GC | 5.7 |
| ACG | 1.0 | TT | 5.5 |
| TGA | 1.0 | AT | 4.8 |
| TAC | 1.0 | TC | 4.6 |
| TCC | 1.0 | TA | 4.4 |
| GAC | 0.8 | CG | 4.2 |
| AGG | 0.8 | CT | 3.5 |
| TTA | 0.8 | GA | 2.9 |
| GCA | 0.6 | AG | 2.9 |
| GCC | 0.6 | CA | 2.8 |
| ACA | 0.6 | CC | 2.4 |
| TCA | 0.6 | AC | 2.2 |
| CGC | 0.6 | AA | 0.7 |
Frequency was calculated by determining the number of times a certain trinucleotide occurred in all 32 sequences and dividing this number by the total number of trinucleotide occurences (512). The expected frequency for a trinucleotide is 1.6% (1/43 = 1/64).
Frequency was calculated by determining the number of times a certain dinucleotide occurred in all 32 sequences and dividing this number by the total number of dinucleotide occurrences (544). The expected frequency for a dinucleotide is 6.3% (1/42 = 1/16).
Selected sequence 1 displays enhanced Rad51 protein-dependent joint molecule formation
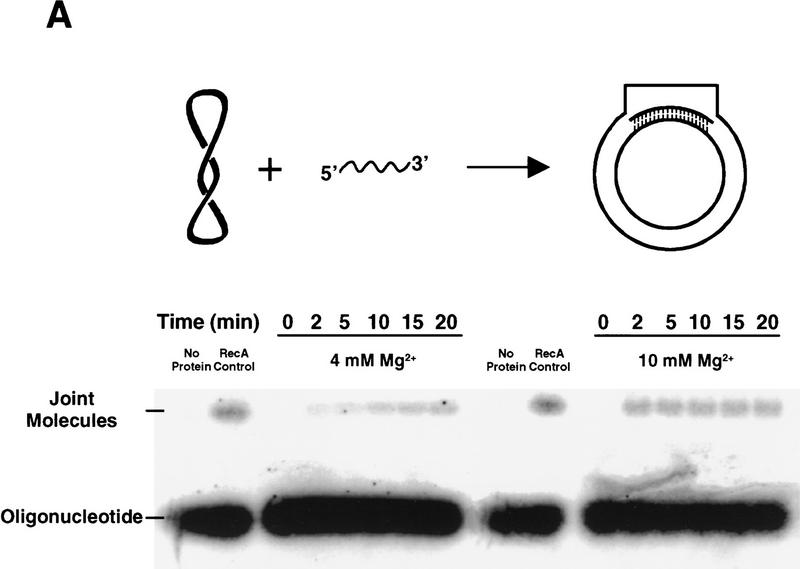
Although Rad51 protein can promote DNA strand exchange between circular single-stranded DNA (ssDNA) and linear double-stranded DNA (dsDNA), its ability to promote invasion of supercoiled DNA by homologous ssDNA is unknown. Hence, we examined this aspect of Rad51 protein in the context of these selected DNA sequences. Because both RecA protein and Rad51 protein selected the sequence shown for clone 1, and as this sequence displayed an enhanced rate and extent of joint molecule formation (homologous pairing) in RecA protein-dependent pairing assays (3.5- and 3.4-fold increases, respectively) (Tracy and Kowalczykowski 1996), this sequence was used with Rad51 protein. Joint molecule formation was examined using the plasmid pBT54CN1, which contains selected sequence 1 as dsDNA in the multiple cloning site, and one of the following ssDNAs, (1) selected sequence 1 (SKBT16; Table 1); (2) the complement of SKBT16 (SKBT17; Table 1), which pairs in exactly the same region of the plasmid and thus eliminates the possibility of plasmid position-dependent effects; or (3) sequences (SKBT19 and SKBT20; Table 1) that are homologous to pBT54CN1 outside the vicinity of the selected sequence. In addition, because both the absolute level of homologous pairing and the pairing bias displayed by selected sequence 1 in the RecA protein-dependent reactions are sensitive to the magnesium ion (Mg2+) concentration, and as the optimal Mg2+ concentration for in vitro pairing assays with Rad51 protein is unknown, we examined joint molecule formation at several different Mg2+ concentrations.
Figure 2 shows Rad51 protein-dependent formation of joint molecules at two different Mg2+ concentrations, 4 and 10 mm. Figure 2A demonstrates that Rad51 protein is able to promote the uptake of ssDNA (SKBT16) into a supercoiled molecule (pBT54CN1) at both 4 and 10 mm Mg2+. At both concentrations, the initial rate and maximal extent of joint molecule formation are significantly increased for the selected sequence (SKBT16) relative to the control oligonucleotides. At 4 mm Mg2+, the initial rate and maximal extent are increased on average ∼4.4- and 4.0-fold, respectively (Fig. 2B). When the Mg2+ concentration was increased to 10 mm, the difference in the rate and extent between SKBT16 and the controls is still significant, although slightly reduced (∼2.0- and 1.7-fold, respectively; Fig. 2C). Pairing was also examined at higher Mg2+ concentrations (15 and 20 mm) for both SKBT16 and SKBT17. Even at these relatively high Mg2+ concentrations, the selected sequence still demonstrated enhanced joint molecule formation (data not shown). Control reactions in the absence of Rad51 protein, using a heterologous plasmid (pBR322), or lacking pBT54CN1 establish that this is an entirely homology-dependent and Rad51 protein-dependent pairing reaction, and for the first time, demonstrate that Rad51 protein is capable of promoting DNA strand invasion (Fig. 2). Similar to the results obtained for the RecA protein-dependent pairing assays, we find that the intrinsic difference in pairing activity between selected sequence 1 and the controls is most apparent at suboptimal in vitro reaction conditions (4 mm Mg2+).
Figure 2.
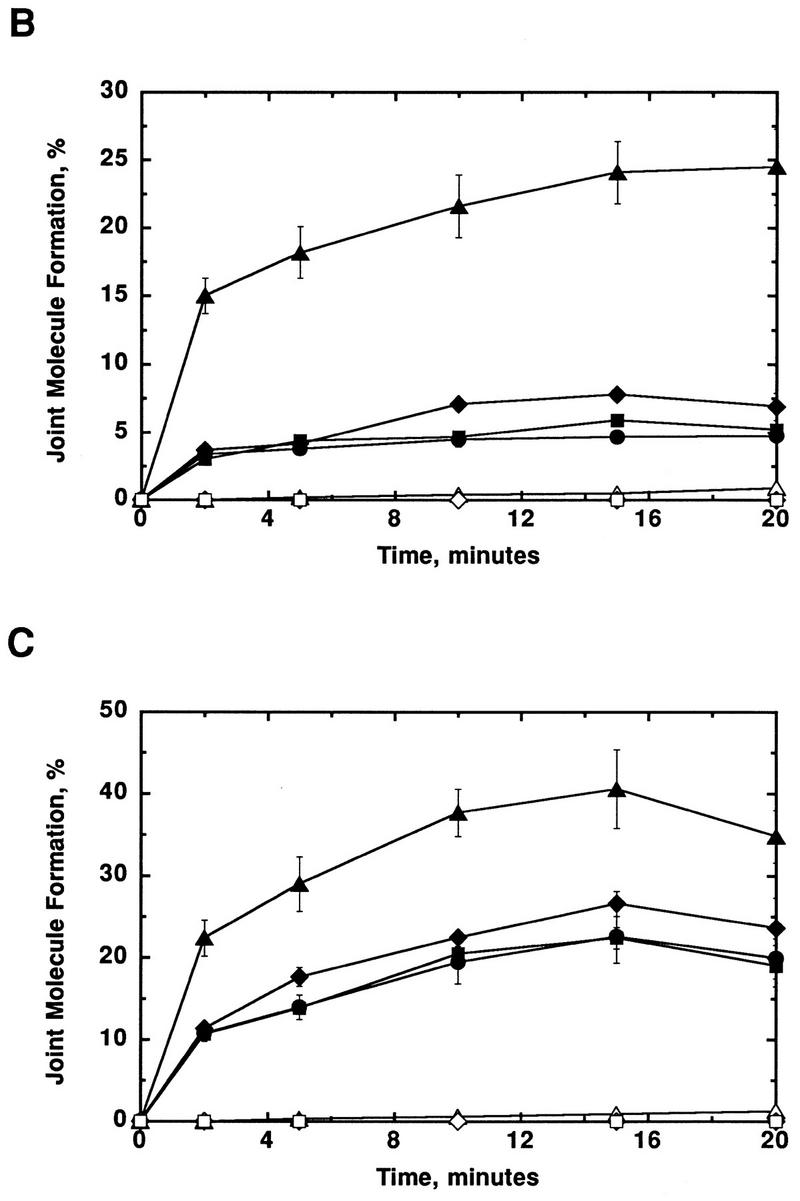
Rad51 protein-promoted joint molecule formation with selected DNA. Formation of joint molecules was performed as described in Materials and Methods. (A) Joint molecule formation with pBT54CN1 and SKBT16 in the presence of either 4 mm or 10 mm magnesium acetate. Joint molecule formation was conducted with a 6-fold molar (molecule) excess of oligonucleotide SKBT16 relative to pBT54CN1; the percentage of joint molecule formation was calculated relative to the limiting amount of plasmid DNA. As a positive control, RecA protein was substituted for Rad51 protein in joint molecule formation; this reaction was conducted at 10 mm Mg2+ for 2 min. As a negative control, protein was omitted. The formation of joint molecules is indicated by the presence of 32P label migrating at the position of supercoiled pBT54CN1. The pairing reaction is depicted at the top. (B) Joint molecule formation with pBT54CN1 and either SKBT16 (▴, contains selected sequence 1); SKBT17 (•, complement of SKBT16); SKBT19 (♦, opposite side of plasmid); or SKBT20 (▪, adjacent to SKBT16). Control reactions included absence of Rad51 protein (▵); use of a heterologous plasmid (pBR322) (⋄); or absence of pBT54CN1 (□). Reactions were performed in the presence of 4 mm magnesium acetate. (C) Joint molecule formation in the presence of 10 mm magnesium acetate. In both B and C, error bars indicate the standard deviation of three experiments.
Discussion
Rad51 protein-selected sequences are similar to genetically unstable eukaryotic genomic sequences
Because the set of GT-rich sequences selected by RecA protein are similar to genetically unstable DNA sequences in both prokaryotes and eukaryotes, we wanted to determine whether the Rad51 protein-selected sequences might also represent attractive genomic targets for Rad51 protein action. When these sequences were compared to DNA sequences within the National Center for Biotechnology Information (NCBI) databases using the BLAST network service (Altschul et al. 1990), and the GenBank database using FastA, several (clones 1, 2, 3, 8, 9, 10, 13, 14, 20, 25, 26, 28, 30, 32, 33, 36, and 39) were found to share significant homology with sequences from eukaryotic organisms that are recombinationally active, such as microsatellite DNA, the constant region of the heavy chains of immunoglobulins, Alu repetitive elements, and telomeric repeat sequences (Table 3). Several studies showed that microsatellite sequences from mammalian genomes are hot spots for homologous recombination (Jeffreys et al. 1995; Steinmetz et al. 1986; Wahls et al. 1990). Wahls et al. (1990) indicated that their data were most consistent with a specific enzyme recognizing the consensus minisatellite sequence, and subsequently stimulating, either directly or indirectly, its ability to recombine. It is known that the constant region of the heavy chains of immunoglobulins from several mammals undergo rearrangement by homologous recombination at DNA sequences (called switch sequences), which are generally GT-rich (Singer and Berg 1991); the Rad51 protein is expressed and localized in B cells that carry out immunoglobulin heavy chain class switch recombination (Li et al. 1996). Alu repetitive elements are short repetitive DNA sequences (∼300 bp long) that are dispersed throughout the genomes of vertebrates. They are of interest from a physiological point of view because they have been shown to cause genomic rearrangements in humans as a result of homologous recombination (Vanin et al. 1983; Henthorn et al. 1986; Hobbs et al. 1986; Nicholls et al. 1987; Rouyer et al. 1987; Rudiger et al. 1991). Finally, there is mounting evidence that both interstitial and terminal telomeric repeat sequences are hot spots for recombination in several eukaryotic organisms, including S. cerevisiae (Pluta and Zakian 1989), mice (Ashley et al. 1993; Ashley and Ward 1993; Nachman and Churchill 1996), and humans (Brown et al. 1990; Park et al. 1992; Rossi et al. 1993). In Schizosaccharomyces pombe, homologous pairing of chromosomes was shown to be most frequent near centromeres and telomeres (Scherthan et al. 1994). Considering that practically all telomere repeat sequences are GT-rich (Henderson 1995), our results can potentially explain their high recombination frequencies. When the Rad51 protein-selected sequences are compared to the telomere repeat sequences, clones 1 (the most prevalent sequence) and 39 show a close resemblance to the repeat sequence from S. cerevisiae [5′-TGTGGGTGTGGTGTGG-3′ (Singer and Gottschling 1994)]. Previously, it was shown that the maintenance of telomeric repeat arrays in the yeasts S. cerevisiae and Kluyveromyces lactis is strongly dependent on the RAD52 pathway, which involves the Rad51 protein, in strains lacking an active telomerase (McEachern and Blackburn 1996). This demonstrates that homologous recombination can functionally substitute for telomerase, at least in these organisms. In addition, it was proposed that telomere–telomere recombination in S. cerevisiae could potentially be promoted by a RecA-like protein in a RAD52 independent manner (Pluta and Zakian 1989); Rad51 protein is a candidate for this role.
Table 3.
Rad51 protein-selected sequences share significant homology with genetically unstable eukaryotic genomic sequences
| Eukaryotic genomic sequences
|
Organism
|
Clones that display similarity
|
|---|---|---|
| Microsatellites | humans and bovine | 1, 2, 9, 20, 26, 33, 36, and 39 |
| Constant region of heavy chains of immunoglobulins | humans, bovine, sheep, rat, and mouse | 1, 2, 8, 10, 25, 26, 28, 30, 32, 33, and 39 |
| Alu repetitive elements | humans | 1, 2, 3, 13, 14, 20, 32, and 39 |
| Telomere repeat sequences | S. cerevisiae | 1 and 39 |
| Huntington’s disease region | humans | 1, 2, 20, 25, 28, 37, and 39 |
| Fragile X syndrome gene | humans | 2 and 38 |
Similarity was determined by using the VAX computer programs BLAST and FASTA (Genetics Computer Group, Inc.). Homology is considered to be significant when 14 of the 18 bases are identical.
Many of the selected sequences (clones 1, 2, 20, 25, 28, 37, 38, and 39) also share similarity with DNA segments associated with triplet expansion diseases in humans, such as Fragile X syndrome and Huntington’s disease (Table 3). Fragile X syndrome is an X-linked dominant mental retardation disorder that is inherited (Richards and Sutherland 1992). The disorder occurs as a result of significant expansion of the (CGG)n trinucleotide repeat (from 2 to 50 copies in normal individuals to >200–2000 copies in affected individuals) (Kremer et al. 1991; Oberle et al. 1991; Verkerk et al. 1991). Huntington’s disease is an inherited neurodegenerative disorder that is progressive and ultimately results in cognitive loss, motor disturbances, and psychiatric problems (Martin and Gusella 1986). Similar to Fragile X syndrome, Huntington’s disease is characterized by expansion of a trinucleotide repeat (CAGn). The repeat expands from 11–34 copies in the normal population to 42–66 copies in those affected by the disease (Huntington’s Disease Collaborative Research Group 1993). One mechanism that was proposed to account for the variablity and instability associated with these tandem repeat sequences is unequal crossing over during homologous recombination (Smith 1974; Jeffreys et al. 1985). If Rad51 protein were targeted to regions directly adjacent to the triplet repeat sequences, then potentially this preference could offer one explanation for the observed instability of these sequences.
Although we advance the idea that increased genetic exchange by members of the Rad51 protein family contributes to the genetic instability observed at these sequences, we also wish to make it clear that other mechanisms can contribute to the observed genomic instability. In this regard, genetic variation has been attributed to slippage of DNA polymerase during replication (Levinson and Gutman 1987; Wells 1996), whereby the presence of DNA structures (e.g., hairpins) or proteins (e.g., histones) blocks the progression of DNA polymerase, causing it to continually slip. In addition, mismatch DNA repair has been implicated as a factor that contributes to the genetic variance (Strand et al. 1993; Jaworski et al. 1995; Wells 1996).
Because recombination in vivo is dependent on not only Rad51 protein, but also several other proteins from the RAD52 epistasis group, it remains possible that these other proteins could further enhance or reduce the preferential binding and homologous pairing of GT-rich DNA sequences by Rad51 protein. Therefore, if increased genetic exchange mediated by Rad51 protein is indeed at least partially responsible for the observed genetic instability at these GT-rich genomic loci, then an additional consideration is whether other proteins of the RAD52 epistasis group are influencing the ability of Rad51 protein to act preferentially at these sites. Because several members of this group have been purified, it is now possible to ascertain whether they affect Rad51 protein function at GT-rich sequences.
Implications
Taken together, these findings argue that the preferential binding of GT-rich sequences is a universal characteristic of a class of recombination enzymes, which is conserved from bacteria to eukaryotes, and that these GT-rich regions invite homologous recombination in their vicinity. A limitation of this argument is that we have examined only two members of this enzyme family and therefore we cannot exclude the possibility that other RecA protein homologs have preferences for DNA compositions that reflect that organism’s genomic content. However, studies are in progress with a RecA protein homolog from the Archaea Domain, the RadA protein from Sulfolobus solfataricus (E.M. Seitz, R.B. Tracy, and S.C. Kowalczykowski, unpubl.). On the basis of these preliminary data, we favor the universality argument advanced here. The coincidence of GT-rich sequences at recombination hot spots in the chromosomes of a variety of organisms, however, indicates that these particular regions are more attractive for binding and subsequent genetic exchange by several different recombination enzymes. In E. coli, the recombination hot spot χ coordinates the efficient recombinational repair of double-stranded ends that form from direct damage or as a consequence of stalled replication forks (Kuzminov et al. 1994). This coordination results from the ability of χ to focus the efforts of the RecA and RecBCD proteins at “recombination islands” that are rich in GT composition, rather than at random dsDNA breaks (Dixon and Kowalczykowski 1991, 1995; Tracy and Kowalczykowski 1996; Anderson and Kowalczykowski 1997; Tracy et al. 1997). Consequently, this coordination permits a far more efficient maintenance of genomic integrity than any random, uncontrolled process. In eukaryotes, a similarly directed process is likely to be in place, with recombination hot spots playing a key role in both genetic diversity and genomic integrity.
Materials and methods
Chemicals
Chemicals were reagent grade, and solutions were prepared using Barnsted NANOpure water. ATP (Pharmacia LKB Biotechnology, Inc.) was dissolved as a concentrated stock solution at pH 7.5. The concentration of ATP was determined spectrophotometrically by using an extinction coefficient of 1.54 × 104 m/cm at 260 nm.
Proteins
Rad51 protein was purified from E. coli using a protocol that was modified from that described by Sung (1994) (E. Zaitseva, T. Sugiyama, J. New, and S.C. Kowalczykowski, unpubl.). T4 polynucleotide kinase was purchased from New England BioLabs. Taq DNA polymerase was purchased from Pharmacia. T7 sequenase, version 2.0, was purchased from U.S. Biochemical Corp. The enzymes were used as directed by the specific vendor.
DNA substrates
The oligodeoxyribonucleotides used and their sequences are shown in Table 1. All oligonucleotides were synthesized and purified as described (Tracy and Kowalczykowski 1996).
The plasmid pBT54CN1, which contains selected sequence 1 was constructed as described (Tracy and Kowalczykowski 1996). pBT54CN1 was purified by CsCl density gradient centrifugation followed by use of a Qiagen Maxi Kit (Qiagen, Inc., Chatsworth, CA).
In vitro selection assay
The oligodeoxyribonucleotide used for in vitro selection (SKBT18) is shown in Table 1. The random region contained an equimolar mixture of the four bases. The defined regions were used for PCR and cloning. The in vitro selection assay was performed as described (Tracy and Kowalczykowski 1996), except for the following changes. First, the radiolabeled oligonucleotides (100 μm nucleotides) were mixed with selection buffer [30 mm Tris acetate (pH 7.5), 20 mm magnesium acetate, 1 mm DTT, 50 mm potassium acetate, and 2.5 mm ATP] and incubated at 37°C for 2 min; 1 μm Rad51 protein was then added. The reaction mixture (100 μl) was incubated at 37°C for 10 min, immediately applied to a nitrocellulose filter (HAWP 0025 from Millipore Corp.), and washed as described previously (Tracy and Kowalczykowski 1996). Second, after each step of PCR, the amplified DNA was purified through a Microcon-10 column (Amicon).
Determination of dinucleotide and trinucleotide composition within the selected sequences
The VAX computer program Composition (Genetics Computer Group, Inc.) was used to determine the composition of dinucleotides or trinucleotides within the Rad51 protein-selected sequences. Once Composition had tabulated the occurrence of each dinucleotide or trinucleotide, the frequency (percentage) was determined.
Joint molecule formation assay
Oligonucleotides were 5′-end labeled with [γ-32P]ATP and T4 polynucleotide kinase. Joint molecule formation was carried out in a reaction mixture (250 μl) consisting of 30 mm Tris-acetate (pH 7.5), 1 mm DTT, 50 mm potassium acetate, 1 mm ATP, 1 μm nucleotides of oligonucleotides, 0.33 μm Rad51 protein, 18 μm nucleotides of pBT54CN1, and the indicated magnesium acetate concentration. The reactions were initiated by the addition of plasmid DNA, after preincubation of the other components for 5 min at 37°C. At 0, 2, 5, 10, 15, and 20 min, 40-μl aliquots were removed and immediately added to 5 μl of a 10% SDS/0.5 m EDTA mixture and 2 μl of proteinase K (14.4 mg/ml) (Boehringer Mannheim) to deproteinize the samples. Samples were incubated at 37°C for 10 min, followed by the addition of 5 μl of DNA loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol, and 25% Ficoll). Reactions were analyzed by electrophoresis through a 1% agarose gel at 30 V for 14 hr. The gels were dried and quantitated using a Betagen Betascope 603 radioisotopic analyzer. All control reactions were conducted with SKBT16. The percentage of joint molecules formed was determined relative to the total amount of plasmid DNA (which was limiting) present in a reaction.
Acknowledgments
We thank the following members of our laboratory for their helpful discussions and critical comments on the manuscript: Dan Anderson, Joel Brockman, Frederic Chedin, Jason Churchill, Deana Haddox, Frank Harmon, Noriko Kantake, Julie Kleiman, Sindy Law, Alex Mazin, Jim New, Erica Seitz, Tomohiko Sugiyama, and Eugene Zaitsev. This work was supported by a National Institutes of Health (NIH) grant (AI-18987) to S.C.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sckowalczykowski@ucdavis.edu; FAX (916) 752-5939.
References
- Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarites to procaryotic RecA proteins. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson DG, Kowalczykowski SC. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- Ashley T, Ward DC. A “hot spot” of recombination coincides with an interstitial telomeric sequence in the Armenian hamster. Cytogenet Cell Genet. 1993;62:169–171. doi: 10.1159/000133464. [DOI] [PubMed] [Google Scholar]
- Ashley T, Cacheiro NL, Russell LB, Ward DC. Molecular characterization of a pericentric inversion in mouse chromosome 8 implicates telomeres as promoters of meiotic recombination. Chromosoma. 1993;102:112–120. doi: 10.1007/BF00356028. [DOI] [PubMed] [Google Scholar]
- Basile G, Aker M, Mortimer RK. Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol Cell Biol. 1992;12:3235–3246. doi: 10.1128/mcb.12.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WR, MacKinnon PJ, Villasante A, Spurr N, Buckle VJ, Dobson MJ. Structure and polymorphism of human telomere-associated DNA. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- Burge C, Campbell AM, Karlin S. Over- and under-representation of short oligonucleotides in DNA sequences. Proc Natl Acad Sci. 1992;89:1358–1362. doi: 10.1073/pnas.89.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero RD, Hsieh P. Homologous recombination proteins in prokaryotes and eukaryotes. Annu Rev Genet. 1995;29:509–552. doi: 10.1146/annurev.ge.29.120195.002453. [DOI] [PubMed] [Google Scholar]
- Clark AJ. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Cox MM. Relating biochemistry to biology: How the recombinational repair function of RecA protein is manifested in its molecular properties. BioEssays. 1993;15:617–623. doi: 10.1002/bies.950150908. [DOI] [PubMed] [Google Scholar]
- Dixon DA, Kowalczykowski SC. Homologous pairing in vitro stimulated by the recombination hotspot, Chi. Cell. 1991;66:361–371. doi: 10.1016/0092-8674(91)90625-9. [DOI] [PubMed] [Google Scholar]
- ————— Role of the Escherichia coli recombination hotspot, chi, in RecABCD-dependent homologous pairing. J Biol Chem. 1995;270:16360–16370. doi: 10.1074/jbc.270.27.16360. [DOI] [PubMed] [Google Scholar]
- Game JC. DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces. Semin Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- Henderson E. Telomere DNA Structure. In: Blackburn EH, Greider CW, editors. Telomeres. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 11–34. [Google Scholar]
- Henthorn PS, Mager DL, Huisman TH, Smithies O. A gene deletion ending within a complex array of repeated sequences 3′ to the human beta-globin gene cluster. Proc Natl Acad Sci. 1986;83:5194–5198. doi: 10.1073/pnas.83.14.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs HH, Brown MS, Golstein JL, Russell DW. Deletion of exon encoding cysteine-rich repeat of low density lipoprotein receptor alters its binding specificity in a subject with familial hypercholesterolemia. J Biol Chem. 1986;261:13114–13120. [PubMed] [Google Scholar]
- Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s Disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL. Hypervariable “minisatellite” regions in human DNA. Nature. 1985;314:67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Jaworski A, Rosche WA, Gellibolian R, Kang S, Shimizu M, Sinden RR, Wells RD. Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. Proc Natl Acad Sci. 1995;92:11019–11023. doi: 10.1073/pnas.92.24.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC. Biochemistry of genetic recombination: Energetics and mechanism of DNA strand exchange. Annu Rev Biophys Biophys Chem. 1991;20:539–575. doi: 10.1146/annurev.bb.20.060191.002543. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Eggleston AK. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer EJ, Pritchard M, Lynch M, Yu S, Holman K, Baker E, Warren ST, Schlessinger D, Sutherland GR, Richards RI. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- Kuzminov A, Schabtach E, Stahl FW. Chi sites in combination with RecA protein increase the survival of linear DNA in Escherichia coli by inactivating exoV activity of RecBCD nuclease. EMBO J. 1994;13:2764–2776. doi: 10.1002/j.1460-2075.1994.tb06570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam ST, Stahl MM, McMilin KD, Stahl FW. Rec-mediated recombinatorial hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974;77:425–433. doi: 10.1093/genetics/77.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Li MJ, Peakman MC, Golub EI, Reddy G, Ward DC, Radding CM, Maizels N. Rad51 expression and localization in B cells carrying out class switch recombination. Proc Natl Acad Sci. 1996;93:10222–10227. doi: 10.1073/pnas.93.19.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JB, Gusella JF. Huntington’s disease. Pathogenesis and management. N Engl J Med. 1986;315:1267–1276. doi: 10.1056/NEJM198611133152006. [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes & Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- Nachman MW, Churchill GA. Heterogeneity in rates of recombination across the mouse genome. Genetics. 1996;142:537–548. doi: 10.1093/genetics/142.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Fischel-Ghodsian N, Higgs DR. Recombination at the human alpha-globin gene cluster: Sequence features and topological constraints. Cell. 1987;49:369–378. doi: 10.1016/0092-8674(87)90289-3. [DOI] [PubMed] [Google Scholar]
- Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas MF, Mandel JL. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Shinohara A, Nabetani A, Ikeya T, Yu X, Egelman EH, Ogawa H. RecA-like recombination proteins in eukaryotes: Function and structures of RAD51 genes. Cold Spring Harbor Symp Quant Biol. 1993a;58:567–576. doi: 10.1101/sqb.1993.058.01.063. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993b;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Park VM, Gustashaw KM, Wathen TM. The presence of interstitial telomeric sequences in constitutional chromosome abnormalities. Am J Hum Genet. 1992;50:914–923. [PMC free article] [PubMed] [Google Scholar]
- Petes TD, Malone RE, Symington LS. Recombination in yeast. In: Broach JR, Jones E, Pringle J, editors. The molecular and cellular biology of the yeast Saccharomyces: Genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- Pluta AF, Zakian VA. Recombination occurs during telomere formation in yeast. Nature. 1989;337:429–433. doi: 10.1038/337429a0. [DOI] [PubMed] [Google Scholar]
- Radding CM. Homologous pairing and strand exchange promoted by Escherichia recA protein. In: Kucherlapati R, Smith GR, editors. Genetic recombination. Washington, DC.: American Society for Microbiology; 1988. pp. 193–229. [Google Scholar]
- Resnick MA. Investigating the genetic control of biochemical events in meiotic recombination. In: Moens PB, editor. Meiosis. New York, NY: Academic Press; 1987. pp. 157–210. [Google Scholar]
- Richards RI, Sutherland GR. Fragile X syndrome: The molecular picture comes into focus. Trends Genet. 1992;8:249–254. [Google Scholar]
- Rossi E, Floridia G, Casali M, Danesino C, Chiumello G, Bernardi F, Magnani I, Papi L, Mura M, Zuffardi O. Types, stability, and phenotypic consequences of chromosome rearrangements leading to interstitial telomeric sequences. J Med Genet. 1993;30:926–931. doi: 10.1136/jmg.30.11.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouyer F, Simmler MC, Page DC, Weissenbach J. A sex chromosome rearrangement in a human XX male caused by Alu-Alu recombination. Cell. 1987;51:417–425. doi: 10.1016/0092-8674(87)90637-4. [DOI] [PubMed] [Google Scholar]
- Rudiger NS, Hansen PS, Jorgensen M, Faergeman O, Bolund L, Gregersen N. Repetitive sequences involved in the recombination leading to deletion of exon 5 of the low-density-lipoprotein receptor gene in a patient with familial hypercholesterolemia. Eur J Biochem. 1991;198:107–111. doi: 10.1111/j.1432-1033.1991.tb15992.x. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Bahler J, Kohli J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J Cell Biol. 1994;127:273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Singer M, Berg P. Genes & Genomes. Mill Valley, CA: University Science Books; 1991. [Google Scholar]
- Singer MS, Gottschling DE. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase [see comments] Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Smith GP. Unequal crossover and the evolution of multigene families. Cold Spring Harbor Symp Quant Biol. 1974;38:507–513. doi: 10.1101/sqb.1974.038.01.055. [DOI] [PubMed] [Google Scholar]
- Steinmetz M, Stephan D, Fischer Lindahl K. Gene organization and recombinational hotspots in the murine major histocompatibility complex. Cell. 1986;44:895–904. doi: 10.1016/0092-8674(86)90012-7. [DOI] [PubMed] [Google Scholar]
- Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Sung P, Stratton SA. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J Biol Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- Tracy RB, Kowalczykowski SC. In vitro selection of preferred DNA pairing sequences by the Escherichia coli RecA protein. Genes & Dev. 1996;10:1890–1903. doi: 10.1101/gad.10.15.1890. [DOI] [PubMed] [Google Scholar]
- Tracy RB, Chedin F, Kowalczykowski SC. The recombination hot spot chi is embedded within islands of preferred DNA pairing sequences in the E. coli genome. Cell. 1997;90:205–206. doi: 10.1016/s0092-8674(00)80328-1. [DOI] [PubMed] [Google Scholar]
- Vanin EF, Henthorn PS, Kioussis D, Grosveld F, Smithies O. Unexpected relationships between four large deletions in the human beta-globin gene cluster. Cell. 1983;35:701–709. doi: 10.1016/0092-8674(83)90103-4. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Wahls WP, Wallace LJ, Moore PD. Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell. 1990;60:95–103. doi: 10.1016/0092-8674(90)90719-u. [DOI] [PubMed] [Google Scholar]
- Wells RD. Molecular basis of genetic instability of triplet repeats. J Biol Chem. 1996;271:2875–2878. doi: 10.1074/jbc.271.6.2875. [DOI] [PubMed] [Google Scholar]
- West SC. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]