Abstract
Alu repeats or Line-1-ORF2 (ORF2) inhibit expression of the green fluorescent protein (GFP) gene when inserted downstream of this gene in the vector pEGFP-C1. In this work, we studied cis-acting elements that eliminated the repression of GFP gene expression induced by Alu and ORF2 and sequence characteristics of these elements. We found that sense and antisense PolyA of simian virus 40 (SV40PolyA, 240 bp) eliminated the repression of GFP gene expression when inserted between the GFP gene and the Alu (283 bp) repeats or ORF2 (3825 bp) in pAlu14 (14 tandem Alu repeats were inserted downstream of the GFP gene in the vector pEGFP-C1) or pORF2. Antisense SV40PolyA (PolyAas) induced stronger gene expression than its sense orientation (PolyA). Of four 60-bp segments of PolyAas (1F1R, 2F2R, 3F3R and 4F4R) inserted independently into pAlu14, only two (2F2R and 3F3R) eliminated the inhibition of GFP gene expression induced by Alu repeats. Deletion analysis revealed that a 17 nucleotide AT repeat (17ntAT; 5′-AAAAAAATGCTTTATTT-3′) in 2F2R and the fragment 3F38d9 (5′-ATAAACAAGTTAACAACA ACAATTGCATT-3′) in 3F3R were critical sequences for activating the GFP gene. Sequence and structural analyses showed that 17ntAT and 3F38d9 included imperfect palindromes and may form a variety of unstable stem-loops. We suggest that the presence of imperfect palindromes and unstable stem-loops in DNA enhancer elements plays an important role in GFP gene activation.
Keywords: Alu, enhancer, GFP, stem-loop structures, SV40PolyA
Introduction
Historically, considerable attention has been given to proteins and their encoding genes. However, with completion of the human and mouse genomes and a better understanding of eukaryotic gene expression, the noncoding sequences of genes have attracted increasing attention. Noncoding sequences are widespread in eukaryotic genomes and contain important genetic information (Eggleston, 2005; Maeshima and Eltsov, 2007; Satzinger, 2008; Depken and Schiessel, 2009) that includes promoters, enhancers and insulators (Tour and Laemmli, 1988), noncoding RNA that directs DNA methylation (Furey and Haussler, 2003), the regulation of axon formation (Dietzel and Belmont, 2001), and small RNA genes (Eggleston, 2005).
Alu and Line-1 repeat elements represent about 10% and 17% of the whole human genome, respectively, and are the most important noncoding sequences (Belgnaoui et al., 2006; Polak and Domany, 2006). Alu elements were initially considered to have no role in gene stability and expression, but recent work has shown that these elements can extensively influence gene expression. In previous work, we have shown that Alu tandem repeats and Line-1-ORF2 (ORF2) inhibited green fluorescent protein (GFP) gene expression when inserted downstream of this gene in the pEGFP-C1 vector (Wang et al., 2009a,b). Downstream noncoding gene sequences are highly structured and contain important regulatory elements such as 3′ UTRs, transcription termination signals (Andreassi and Riccio, 2009) and enhancers (Mao et al., 2010).
In this study, we examined the ability of sense and antisense SV40PolyA to eliminate the repression of GFP gene expression when inserted between the GFP gene and Alu repeats or ORF2 in pAlu14 or pORF2. Antisense SV40PolyA (PolyAas) caused stronger gene expression than its sense orientation (PolyA). We also examined the effects of small fragments of PolyAas on GFP gene expression to identify which PolyAas sequences activated this gene and found that two fragments were critical for activating GFP gene expression. The two fragments both include imperfect palindromes and may form incomplete stem-loop structures that are described as a mechanism for activating GFP gene expression.
Materials and Methods
Construction of expression vectors
The pAlu14 and pORF2 expression vectors were constructed as described elsewhere (Wang et al., 2009a,b) by inserting 14 head-to-tail tandem Alu (283 bp) elements or an ORF2 (3825 bp) downstream of the GFP gene in the pEGFP-C1 vector.
Primers were designed with sites for restriction enzymes (EcoR I or Hind III/Xba I; Kpn I/Nhe I) and the polymerase chain reaction (PCR) was used to amplify the synthetic DNA sequences (as templates) that contained mutated sites and fragments of PolyAas DNA. The PCR products were digested with restriction enzymes and inserted between the GFP gene and Alu repeats in pAlu14 or between the GFP gene and ORF2 in pORF2. When the compatible ends of the DNA fragments digested with Xba I and Nhe I restriction enzymes were ligated by T4 DNA ligase both of the recognition sites for Xba I and Nhe I were destroyed. Using this approach, the expression vectors of two tandem insertion sequences were obtained. The primers and templates used for construction of the expression vectors are shown in Tables 1 and 2, respectively.
Table 1.
Primers used to construct the expression vectors.
| Primer identification | Sequence | Annotation |
|---|---|---|
| 1F (forward primer) |
EcoR I Xba I 5′-ATCGGAATTCTTAATCTAGATAATGCTTACAATTTACGCGTTA-3′ |
Amplifying 1F1R, PolyAas |
| 1R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCTGCATTCTAGTTGTGGTT T -3′ |
Amplifying 1F1R |
| Poly60-2F (forward primer) |
EcoR I Xba I 5′-ATCGGAATTCTTAATCTAGATAAGTGAAAAAAATGCTTTATT -3′ |
Amplifying 2F2R,45R,30R |
| Poly60-2R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCATAATGGTTACAAATAAAG -3′ |
Amplifying 2F2R, Poly4 |
| 3F (forward primer) |
EcoR I Xba I 5′-ATCGGAATTCTTAATCTAGATAAAAGC TGCAATAAACAAGTT-3′ |
Amplifying 3F3R |
| 3R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCCCCTGA ACCTGAAACATAA-3′ |
Amplifying 3F3R, 3R49, 3F235 |
| 4F (forward primer) |
EcoR I Xba I 5′-ATCGGAATTCTTAATCTAGATAAGGAGGTGTGGGAGGTTTTT-3′ |
Amplifying 4F4R |
| 4R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCTAATCAGCCATACCACATT-3′ |
Amplifying 4F4R, PolyAas |
| PolyAasF (forward primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCTGCTAATGCTTACAATTTACGCGTTA-3′ |
Amplifying PolyA |
| PolyAasR (reverse primer) |
EcoR I Xba I 5′-ATCGGAATTCTTAATCTAGATAATCAGCCATACCACATT-3′ |
Amplifying PolyA |
| FirLoopF (forward primer) |
EcoR I Xba I 5′-ATCGGAATTCTTAATCTAGATAATGTGAAAAAA |
Amplifying 22R, 19R, 16R, 0 nt, 1 nt, 2 nt, 4 nt, 5 nt, 6 nt, TCC, GTC, GCA, CTC, GGC |
| FirLoopR (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCACAAATAA |
Amplifying 0 nt, 1nt, 2 nt, 4 nt, 5 nt, 6 nt, TCC, GTC, GCA, CTC, GGC |
| 1619MR (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCAC-3′ |
Amplifying 19R, 16R |
| Poly45R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCAAAGCAATAGCATCA-3′ |
Amplifying 45R |
| Poly30R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCCAAATTTCACAAATA-3′ |
Amplifying 30R |
| SecloopF (forward primer) |
EcoR I Xba I 5′-ATCGGAATTCTTAATCTAGATAATGCTTTATTTGT-3′ |
Amplifying Secloop |
| SecloopR (reverse primer) |
Kpn I Nhe I 5′ATCGGGTACCATGCTAGCCACAAATTTCAC-3′ |
Amplifying Secloop |
| Poly4F (forward primer) |
EcoR I Xba I 5′-ATCGGAATTCTTAATCTAGATAATGATGCTATTG-3′ |
Amplifying Poly4 |
| EcoXba (forward primer) |
EcoR I Xba I 5′-ATCGGAATTCTT AATCTAGA-3′ |
Amplifying 17ntAT |
| KpnNhe (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGC-3′ |
Amplifying 17ntAT |
| 3F46F(forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGAAAGCTGCAATAAACAAG |
Amplifying 3F46 fragment with reverse primer 3F135R |
| 3R49F (forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGAAACAAGTTAACAACAA |
Amplifying 3R49 fragment with reverse primer 3R; Amplifying 3F135 fragment with 3F135R |
| 3F135R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCCATAAAATGAATG -3′ |
Amplifying 3F135, 3F46 |
| 3F235F(forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGAAACAATTGCATTC-3′ |
Amplifying 3F235 fragment with reverse primer 3R |
| 3F46d2F (forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGAGCTGCAATAAACAAG |
Amplifying 3F46d2 fragment with reverse primer 3F135R |
| 3F46d3F (forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGACTGCAATAAACAAGT-3′ |
Amplifying 3F46d3 fragment with reverse primer 3F135R |
| 3F46d4F (forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGATGCAATAAACAAGTT-3′ |
Amplifying 3F46d4 fragment with reverse primer 3F135R |
| 3F46d5F (forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGAGCAATA AACAAGTTA-3′ |
Amplifying 3F46d5 fragment with reverse primer 3F135R |
| 3F46d6F (forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGACAATAAACAAGTTA-3′ |
Amplifying 3F46d6 fragment with reverse primer 3F135R |
| 3F46d7F (forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGAAATAAACAAGTTA-3′ |
Amplifying 3F46d7 fragment with reverse primer 3F135R |
| 3F46d8F (forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGAATAAACAAGTTA AC-3′ |
Amplifying 3F46d8 fragment with reverse primer 3F135R; Amplifying 3F38d1, 3F38d2, 3F38d3, 3F38d4, 3F38d5, 3F38d6, 3F38d8, 3F38d9, 3F38d10, 3F38d11, 3F38d12, 3F38 d13 with corresponding reverse primers |
| 3F46d9F (forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGATAAACAAGTTAACA-3′ |
Amplifying 3F46d9 fragment with reverse primer 3F135R |
| 3F46d10F (forward primer) |
HindIII Xba I 5′-ATCGAAGCTTAATCTAGAAAACAAGTTA-3′ |
Amplifying 3F46d10 fragment with reverse primer 3F135R |
| 3F38d1R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCATAAAATGAATGCA-3′ |
Amplifying 3F38d1 fragment with forward primer 3F46d8F |
| 3F38d2R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCTAAAATGAATGCAA-3′ |
Amplifying 3F38d2 fragment with forward primer 3F46d8F |
| 3F38d3R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCAAAATGAATGCAAT-3′ |
Amplifying 3F38d3 fragment with forward primer 3F46d8F |
| 3F38d4R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCAAATGA TGCAATT-3′ |
Amplifying 3F38d4 fragment with forward primer 3F46d8F |
| 3F38d5R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCAATGAATGCAATTG-3′ |
Amplifying 3F38d5 fragment with forward primer 3F46d8F |
| 3F38d6R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCATGAATGCAATTG-3′ |
Amplifying 3F38d6 fragment with forward primer 3F46d8F |
| 3F38d8R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCGAATGCAATTG-3′ |
Amplifying 3F38d8 fragment with forward primer 3F46d8F |
| 3F38d9R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCAATGCAATTG-3′ |
Amplifying 3F38d9 segment with forward primer 3F46d8F |
| 3F38d10R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCATGCAATTG-3′ |
Amplifying 3F38d10 fragment with forward primer 3F46d8F |
| 3F38d11R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCTGCAATTG-3′ |
Amplifying 3F38d11 fragment with forward primer 3F46d8F |
| 3F38d12R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCGCAATTGTTGTTGTTAACTT-3′ |
Amplifying 3F38d12 segment with forward primer 3F46d8F |
| 3F38d13R (reverse primer) |
Kpn I Nhe I 5′-ATCGGGTACCATGCTAGCCAATTGTTGTTGTTAACTT-3′ |
Amplifying 3F38d13 fragment with forward primer 3F46d8F |
Underlined sequences indicate restriction enzyme cleavage sites.
Table 2.
Synthetic templates used to construct expression vectors.
| Identification | Sequence |
|---|---|
| 19RM | 5′-AATGTGAAAAAAATGCTTTATTGCTAGC-3′ |
| 16RM | 5′-AATGTGAAAAAAATGCTTTGCTAGC-3′ |
| 17ntAT | 5′-CTAGATAATAAAAAAATGCTTTATTTGCTAGCAT-3′ |
| Loop0nt | 5′-GTGAAAAAAATTTATTTGT-3′ |
| Loop1nt | 5′-GTGAAAAAAAGTTTATTTGT-3′ |
| Loop2nt | 5′-GTGAAAAAAATGTTTATTTGT-3′ |
| Loop4nt | 5′-GTGAAAAAAACTGCTTTATTTGT-3′ |
| Loop5nt | 5′-GTGAAAAAAACGTGCTTTATTTGT-3′ |
| Loop6nt | 5′-GTGAAAAAAATCGTGCTTTATTTGT-3′ |
| TCC | 5′-GTGAAAAAAATCCTTTATTTGT-3′ |
| GTC | 5′-GTGAAAAAAAGTCTTTATTTGT-3′ |
| GCA | 5′-GTGAAAAAAAGCATTTATTTGT-3′ |
| CTC | 5′-GTGAAAAAAACTCTTTATTTGT-3′ |
| GGC | 5′-GTGAAAAAAAGGCTTTATTTGT-3′ |
Cell culture and transfection
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum. Cells were plated in each well of a 24-well plate at 0.9 × 105 cells/well and cultured at 37 °C in 5% CO2 for 30–36 h. The cells were transiently transfected with 0.4 μg of expression vector DNA using 2 μL of Lipofectamine2000 reagent (Invitrogen, USA), according to the manufacturer’s instructions, and subsequently cultured for an additional 30–36 h. The transfected cells were used for RNA extraction and fluorescence assays.
Assessment of GFP fluorescence
Transfected HeLa cells were fixed in 4% paraformaldehyde and the expression of GFP protein was assessed by using fluorescence microscopy (Nikon TE2000-U, Japan). Images were obtained under normal and fluorescent illumination.
Northern blotting
Total RNA from transfected cells was extracted with Trizol® reagent (Invitrogen, USA). RNA was electrophoresed in 1.2% agarose gels containing 0.4 M formaldehyde and then transferred to nylon membranes (pore diameter 0.45 μm; Osmonics, USA). A 590-bp fragment from the GFP gene in the pEGFP-C1 vector was amplified by PCR using the forward primer 5′-GGGCGAGGGCGATG-3′ and the reverse primer 5′-CTTGTACAGCTCGTCCAT GC-3′. The PCR product was purified by agarose gel electrophoresis and radiolabeled with [α-32P]-dCTP (Furui, China) using the random primer labeling system (TaKaRa, Japan). The nylon membranes blotted with RNA were hybridized with α-32P-radiolabeled DNA probes at 42 °C in 50% formamide containing 5x SSC (saline sodium citrate), 5x Denhardt’s solution and 100 μg of salmon sperm DNA/mL for 24 h in a UL2000 hybriLinker (UVP, USA). The membranes were washed twice at room temperature with a solution of 1x SSC-0.1% SDS and then washed three times with a solution of 0.1x SSC-0.1% SDS at 68 °C prior to autoradiography. The membranes were subsequently stripped by washing twice at 80 °C for 1 h in a solution containing 50% formamide-5% SDS-50 mM Tris (pH 7.4), and then hybridized with α-32P-radiolabeled probe for neoRNA (containing the cassette for neomycin resistance). A 671-bp fragment from the neo gene in the pEGFP-C1 vector was amplified by PCR using the forward primer 5′-CACAACA GACAATCGGCTGCT-3′ and the reverse primer 5′-AGC GGCGATACCGTAAAAGCAC-3′. The probe for neoRNA was prepared using the random primer labeling system and the 671-bp neo fragment as the template.
Results
PolyA and PolyAas eliminate the repression of GFP gene expression induced by Alu repeats or ORF2
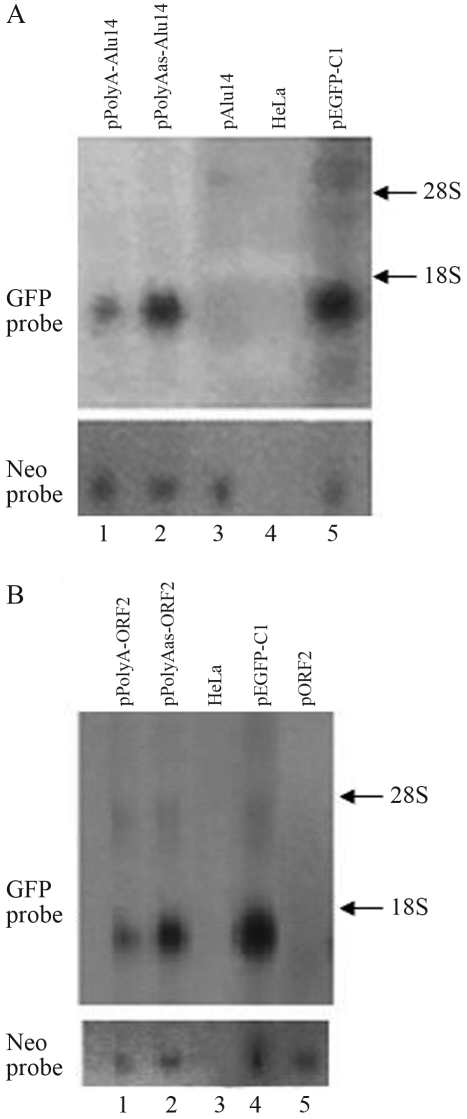
Northern blotting showed that there was almost complete repression of GFP expression in HeLa cells transfected with the expression vectors pAlu14 and pORF2 (Figure 1A, lane 3 vs. lane 5; Figure 1B, lane 5 vs. lane 4). PolyA and PolyAas sequences inserted between the GFP gene and Alu repeats or ORF2 partly abolished the repression of GFP expression caused by Alu repeats or ORF2 (Figure 1A, lanes 1 and 2 vs. lane 3; Figure 1B, lanes 1 and 2 vs. lane 5). PolyAas reversed the repression of GFP expression to a greater extent than its sense orientation in pAlu14 and pORF2 (Figure 1A, lane 2 vs. lane 1; Figure 1B, lane 2 vs. lane 1).
Figure 1.
PolyA and PolyAas inserted downstream of the GFP gene in pAlu14 (A) and pORF2 (B) eliminated the repression of GFP gene expression induced by Alu repeats or ORF2. HeLa cells were transfected with the expression vectors and GFP RNA was detected by northern blotting.
The neo gene was used as a control to assess the efficiency of transfection with the GFP gene. The occurrence of both genes on the same expression vector eliminated the possibility that variation in the efficiency of transfection contributed to the differences observed in the experimental results.
The effects of PolyAas segments on GFP gene expression
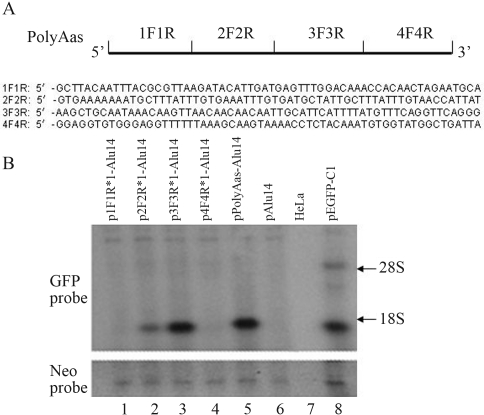
To determine which segments in PolyAas eliminated the repression of GFP gene expression caused by Alu repeats we produced four 60-bp segments of PolyAas (1F1R, 2F2R, 3F3R and 4F4R; Figure 2A) that were then inserted between the GFP gene and Alu repeats in the pAlu14 vector used to transiently transfect HeLa cells. Northern blotting showed that 1F1R and 4F4R did not stimulate GFP gene expression (Figure 2B, lanes 1 and 4 vs. lane 6) whereas 2F2R and 3F3R did (Figure 2B, lanes 2 and 3 vs. lane 6).
Figure 2.
The effect of 60-bp segments of PolyAas (1F1R, 2F2R, 3F3R and 4F4R) on GFP gene expression. (A) Positions and sequences of the four segments. (B) 1F1R, 2F2R, 3F3R, 4F4R and PolyAas were inserted downstream of the GFP gene in pAlu14. HeLa cells were transfected with the expression vectors and GFP RNA was detected by northern blotting.
The effects of 2F2R and its deleted fragments on GFP gene expression
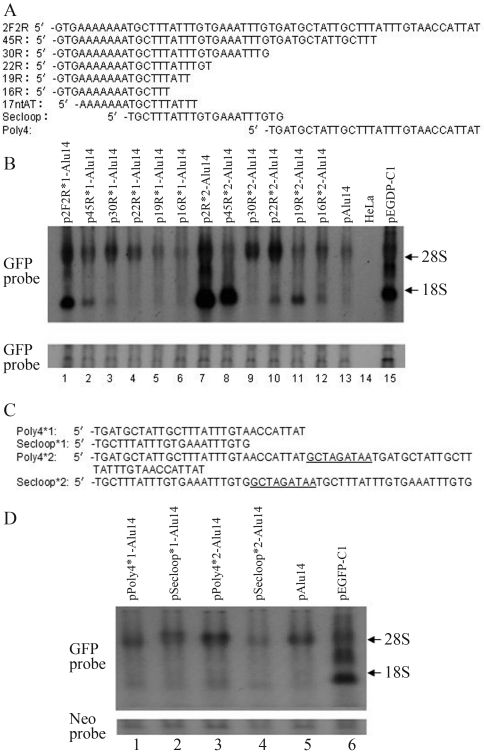
To determine which fragments of 2F2R were responsible for the activation of GFP gene expression we deleted selected regions of the 2F2R DNA (Figure 3A). The bases in the 3′ end of 2F2R were deleted and the single sequence or double tandem sequences of deleted 2F2R (45R, 30R, 22R, 19R and 16R) were inserted into pAlu14. Fragments 45R, 30R and 22R activated GFP gene expression (Figure 3B, lanes 2, 3 and 4 vs. lane 13, and lanes 8, 9 and 10 vs. lane 13), whereas 19R and 16R induced weaker GFP gene expression (Figure 3B, lanes 5, 6, 11 and 12 vs. lane 13). The double tandem sequences of 2F2R and their deleted sequences induced stronger GFP gene expression than the corresponding single sequences (Figure 3B, lanes 7–12 vs. lanes 1–6, respectively). Although 45R, 30R and 22R all enhanced GFP gene expression, the activation of 45R was weaker than that of 2F2R (Figure 3B, lane 2 vs. lane 1; lane 8 vs. lane 7), and the activation by 30R and 22R was weaker than that of 45R (Figure 3B, lanes 3 and 4 vs. lane 2; lanes 9 and 10 vs. lane 8). These results indicated that the 3′ deleted sequences in 2F2R contributed to GFP gene expression. The base deletions influenced the termination of transcription, with the double tandem sequences of 2F2R and 45R resulting mainly in low molecular mass transcripts (Figure 3B, lanes 7 and 8), whereas double tandems of 30R and 22R yielded mainly high molecular mass transcripts (Figure 3B, lanes 9 and 10).
Figure 3.
The effects of 2F2R deletions on GFP gene expression. (A) Positions and sequences of the deletion mutations in 2F2R. (B) Single fragments or double tandem fragments of 2F2R and deletions (45R, 30R, 22R, 19R and 16R) were inserted downstream of the GFP gene in pAlu14. HeLa cells were transfected with the expression vectors and GFP RNA was detected by northern blotting. (C) Nucleotide sequences of Poly4 and Secloop and their double tandems. The nucleotides linking two fragments are underlined. (D) Poly4 and Secloop and their double tandems were inserted downstream of the GFP gene in pAlu14. HeLa cells were transfected with the expression vectors and the GFP RNA was detected by northern blotting.
The single or double tandem sequences of Poly4 and Secloop (the sequences and their positions are shown in Figure 3A and Figure 3C) in 2F2R were inserted downstream of the GFP gene. HeLa cells were transiently transfected with the expression vectors. Northern blotting showed that neither Poly4 nor Secloop significantly activated GFP gene expression (Figure 3D, lanes 1–4 vs. lane 5).
The effects of the 22R fragment and its deleted sequences on GFP gene expression
The deletion of three upstream bases (5′ -GTG) and two downstream bases (GT-3′) of fragment 22R yielded a 17 nucleotide repeat of AT (17ntAT; sequence and position shown in Figure 3A). 17ntAT activated GFP gene expression to the same extent as 22R (Figure 4B, lane 2 vs. lane 5), indicating that the five deleted bases were not important for GFP gene activation. The 19R fragment, i.e., 22R from which three downstream bases (TGT-3′) had been deleted, caused much lower GFP gene expression (Figure 4B, lane 3 vs. lane 5), indicating an important role for these bases in GFP gene activation. Double tandems of 17ntAT produced more transcripts than the corresponding single sequence (Figure 4B, lane 2 vs. lane 4). Figure 4A shows the sequences inserted into pAlu14.
Figure 4.
The effects of 22R and its deleted sequences on GFP gene expression. (A) Nucleotide sequences of double tandems of 16R, 17ntAT, 19R, 22R and single 17ntAT. The nucleotides linking two fragments are underlined. (B) 22R and its deleted sequences were inserted downstream of the GFP gene in pAlu14. HeLa cells were transfected with the expression vectors and GFP RNA was detected by northern blotting.
The effects of 3F3R fragments on GFP gene expression
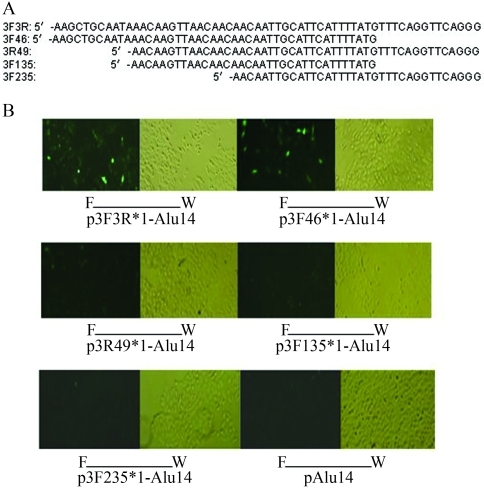
To identify which fragments in 3F3R enhanced GFP gene expression, we deleted sections of 3F3R DNA (Figure 5A). The single sequences of deleted 3F3R (3F46, 3R49, 3F135 and 3F235) were inserted into pAlu14. Figure 5B shows that 3F46 activated GFP gene expression, whereas 3R49, 3F135 and 3F235 did not.
Figure 5.
The effect of different fragments of 3F3R on GFP protein expression. (A) Positions and nucleotide sequences of 3F3R fragments. (B) The sequences of 3F46, 3R49, 3F135 and 3F235 were inserted downstream of the GFP gene in pAlu14. HeLa cells were transfected with the expression vectors and GFP protein was detected by fluorescence microscopy (W: white light, F: fluorescent light, x 100 times).
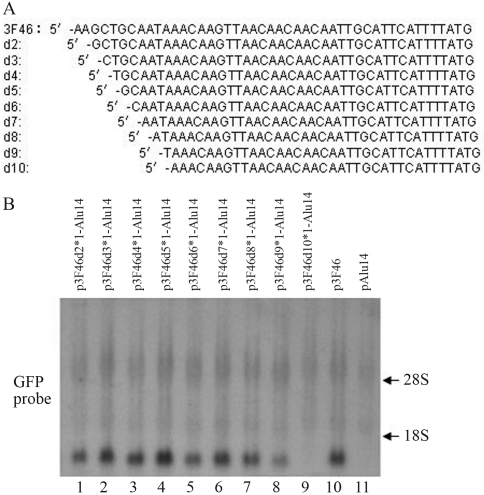
The effects of 3F46 deletions on GFP gene expression
To identify the 3F46 cis-element responsible for gene activation we deleted the nucleotides upstream of 3F46 (Figure 6A) and constructed expression vectors. Northern blotting showed that 3F46d2–3F46d8 (deletion of 2–8 bases upstream of 3F46) still activated GFP gene expression (Figure 6B, lanes 1–7 vs. lane 11), whereas 3F46d9 caused only weak activation (Figure 6B, lane 8 vs. lane 11) and 3F46d10 produced hardly any activation (Figure 6B, lane 9 vs. lane 11).
Figure 6.
The effects of 3F46 deletions on GFP gene expression. (A) Nucleotide sequences of 3F46 and its deleted fragments. (B) The sequences of 3F46 and deleted sequences were inserted downstream of the GFP gene in pAlu14. HeLa cells were transfected with the expression vectors and GFP RNA was detected by northern blotting.
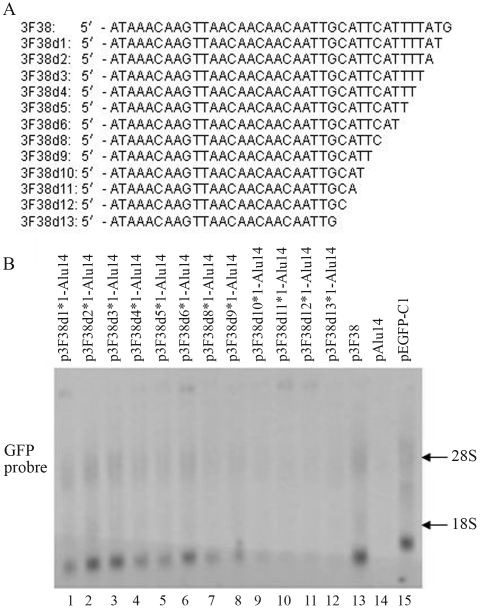
The effects of base deletions downstream of 3F38 (3F46d8) on GFP gene expression
The deletion of selected nucleotides was used to establish the downstream boundary for GFP gene activation by fragment 3F38 (Figure 7A). Northern blotting showed that 3F38d1–3F38D6, 3F38d8 and 3F38d9 activated the GFP gene (Figure 7B, lanes 1–8 vs. lane 14) whereas 3F38d10–3F38d13 did not (Figure 7B, lanes 9–12 vs. lane 14). These results identified 3F38d9 as the critical sequence of 3F38 for GFP gene activation.
Figure 7.
The effect of 3F38 (3F46d8) and its deleted fragments on GFP gene expression. (A) Nucleotide sequences of 3F38 and its deleted fragments. (B) The sequences of 3F38 and its deleted sequences were inserted downstream of the GFP gene in pAlu14. HeLa cells were transfected with the expression vectors and GFP RNA was detected by northern blotting.
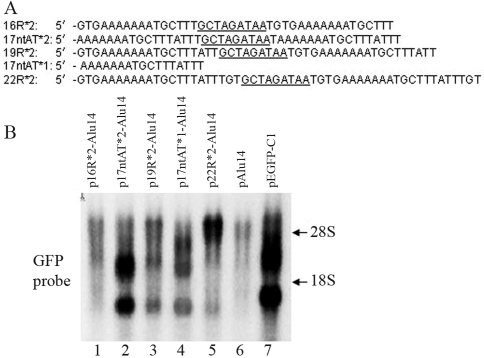
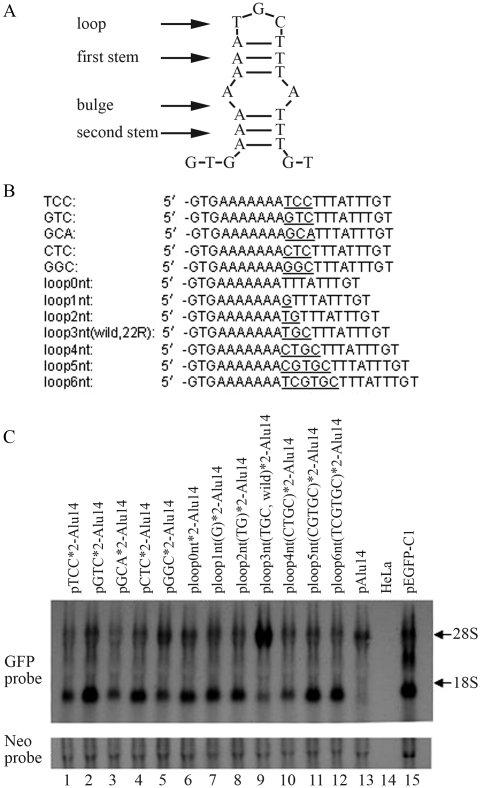
The effects of mutations in 22R DNA on GFP gene expression
Analysis of the DNA sequence of 22R indicated that this fragment may form an incomplete stem-loop structure that included a loop (3 nt), an initial stem (3 bp), a bulge (2 nt) and a second stem (3 bp) (Figure 8A). We examined the influence of loop base type and loop length on the ability of 22R to influence GFP gene activation by introducing mutations in these regions and inserting the fragment into the vector pAlu14 for transfection in HeLa cells. The loop base combination TGC (22R*2, wild type) induced the strongest GFP gene expression (Figure 8C, lane 9 vs. lanes 1–5), whereas changing the loop base number from 0 nt to 6 nt showed that a 3 nt loop (22R*2, wild type) produced the strongest GFP gene expression (Figure 8C, lane 9 vs. lanes 6, 7, 8, 10, 11 and 12). Although most of the loop mutants were able to enhance GFP gene expression they were generally less effective than the wild type fragment (22R) (Figure 8C, lanes 2, 4, 5, 6, 7, 8, 11 and 12 vs. lane 13). This finding indicates that if there are no changes in the palindromes flanking the loop then many types of loops can enhance GFP gene expression.
Figure 8.
The effects of 22R mutants on GFP gene activation. (A) The 22R DNA sequence is predicted to form an imperfect stem-loop structure that includes a loop (3 nt), an initial stem (3 bp), a bulge (2 nt) and a second stem (3 bp). (B) Nucleotide sequences of 22R and its mutants. Predicted loop bases are underlined. (C) The double tandems of 22R and mutants of this fragment were inserted downstream of the GFP gene in pAlu14. HeLa cells were transfected with the expression vectors and GFP RNA was detected by northern blotting.
Discussion
SV40PolyA activates luciferase reporter gene expression in HeLa cells (Zhi-Li et al., 2001). In this study, the insertion of sense or antisense PolyA between the GFP gene and Alu repeats or ORF2 in the vectors pAlu14 or pORF2 resulted in partial recovery of GFP gene expression repressed by Alu repeats or ORF2. This finding indicated that sense and antisense PolyA enhanced GFP gene expression, with PolyAas causing greater induction than PolyA. Nolan et al. (1996) found that reversing the orientation of DRE (a 27-bp enhancer) dramatically decreased growth hormone gene expression, indicating that the binding of transfactors to the DRE and the interaction of this complex with the TATA region are directional.
The wild-type SV40 enhancer contains a double tandem duplication (72-bp repeat). The single 72-bp repeat contains three functional elements (A, B and C) that range in size from 15 to 22 bp (Shepard et al., 1988). Although PolyA is a short sequence (240 bp) it contains various regions that may differ in their ability to activate genes. To examine this hypothesis, we produced four segments of PolyAas (1F1R, 2F2R, 3F3R and 4F4R) (Figure 2A) and inserted them separately downstream of the GFP gene in pAlu14. 2F2R and 3F3R abolished the inhibition of GFP gene expression induced by tandem Alu repeats. To determine which portions of 2F2R activated the GFP gene, we deleted bases from the 3′ end of this segment and found that fragment 22R activated the GFP gene, with double tandem sequences having a stronger effect than the corresponding single sequences. None of the other 2F2R fragments (19R, 16R, Secloop and Poly4) significantly activated the GFP gene.
The 5′ and 3′ UTRs of viral genomes are highly structured and are critical for controlling viral biological processes. The stem-loop structure is important for gene activation (Dai et al., 1997) and Bio-software predicts various stem-loop structures in these regions. Most of the stem-loop structures in viral genomes show bulge sequences in their stems (Yu and Markoff, 2005; Rosskopf et al., 2010; Nickens and Hardy, 2008). To explain the results obtained with the 2F2R fragments, we hypothesized that 22R contained an imperfect palindrome and formed an incomplete stem-loop structure that included a loop (3 nt), an initial stem (3 bp), a bulge (2 nt) and a second stem (3 bp). Fragment 19R [22R with three downstream bases (TGT) deleted] produced fewer transcripts (Figure 4B, lane 3 vs. lane 5), whereas 17ntAT [22R with three upstream bases (GTG) and two downstream bases (GT) deleted] activated the GFP gene when inserted into pAlu14, indicating that the third base (T) downstream of 22R is important for GFP gene activation. 17ntAT was the smallest sequence in 22R to form an incomplete stem-loop structure. The stem-loop structures were destroyed in 19R and 16R, and neither fragment activated the GFP gene significantly, which suggested that an incomplete stem-loop structure (Figure 8A) was important for GFP gene activation. Examination of the 17ntAT sequence (5′-AAAAAAATGCTTTATTT) suggested that it was capable of forming a variety of incomplete, unstable stem-loops. Figure 8A shows one of the presumed stem-loop structures.
To determine the 3F3R sequences involved in GFP gene activation we produced four overlapping fragments (3F46, 3R49, 3F135 and 3F235) of this segment (Figure 5A). The four fragments were inserted separately between the GFP gene and the Alu repeats in the pAlu14 vector that was then used to transfect HeLa cells. Only 3F46 activated the GFP gene. Sequential (one by one) deletion of bases upstream of 3F46 showed that removal of the first eight bases (3F46d8) had little influence on GFP gene activation, whereas elimination of the ninth base (3F46d9) markedly attenuated this activation, indicating a critical role for this base (Figure 6B, lane 8). Sequential (one by one) deletion of the downstream bases of 3F38 (fragment 3F46 in which eight bases were deleted) showed that removal of the first nine bases did not markedly affect GFP gene activation whereas the removal of bases 10–13 eliminated the activation of this gene. Together, these findings indicated that the critical sequence in 3F3R for GFP gene activation was 3F38d9 (5′-ATAAACAAGTTAACAACAACAATTGC ATT-3′). This sequence contained 29 bases (A = 15, C = 5, G = 2, T = 7), with the fragment from A12 to C20 containing three AAC repeats that were flanked by GTT and TTG sequences which formed stem-loop structures with the AAC repeats. Fragment 3F38d9 was thus similar to 17ntAT in that both of them formed unstable stem-loop structures.
Base mutations and variations in the number of bases (from 0 nt to 6 nt) in the 22R loop showed that the TGC loop (22R, wild type) induced the strongest gene expression, although most of the loop mutants showed some ability to induce this gene (Figure 8C). This finding suggested that an unstable stem-loop structure was required for GFP gene activation by 22R, with many loops partly satisfying this criterion. Changes in loop bases influence the stability of stem-loop structures (Lamoureux et al., 2006), and stem-loop structures with 3–4 base loops may be specifically stabilized and have lower folding times (Kuznetsov et al., 2001, 2008). These findings may help to explain the importance of 3nt loops in GFP gene activation.
DNA cruciform structures can be formed when intra-strand pairing occurs between complementary bases of inverted repeat sequences in double-stranded DNA. Cruciform formation is energetically less favorable than B-form DNA so that the extrusion of these structures from duplex DNA requires the driving energy provided by negative supercoiling (Sean et al., 2009). Hairpin structures in the cruciform promoter for the bacteriophage N4 virion RNA polymerase are extruded at physiological superhelical density (Chou et al., 1999). The palindromes in double-stranded DNA may form incomplete stem-loop structures within small scope (Darlow and Leach, 1998). For this reason, the structures formed by these palindrome sequences may play an important role in regulating gene expression in cells.
The critical sequences of 2F2R and 3F3R involved in gene activation have two characteristics in common, namely, (1) they can form various stem-loop structures that increase the probability of creating stem-loops by random impact and (2) the stem-loop structures are incomplete and unstable, which ensures that stem-loops promptly revert to a double helix state. Based on these findings, we propose that sequences containing suitably imperfect palindromes activate gene expression by dynamic fluctuations between unstable stem-loop structures and double-strand forms. Additional experiments are required to confirm this hypothesis.
Acknowledgments
This work was supported by grants from the Hebei Province Natural Science Foundation of China (C2008001065 and C2011206043) and the Key Project of Hebei Province (08276101D-90).
Footnotes
Associate Editor: Carlos F.M. Menck
References
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:1–10. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SH, Tseng YY, Chu BY. Stable formation of a pyrimidine-rich loop hairpin in a cruciform promoter. J Mol Biol. 1999;292:309–320. doi: 10.1006/jmbi.1999.3066. [DOI] [PubMed] [Google Scholar]
- Dai X, Greizerstein MB, Nadas-Chinni K, Rothman-Denes LB. Supercoil-induced extrusion of a regulatory DNA hairpin. Proc Natl Acad Sci USA. 1997;94:2174–2179. doi: 10.1073/pnas.94.6.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlow JM, Leach DR. Evidence for two preferred hairpin folding patterns in d(CGG) d(CCG) repeat tracts in vivo. J Mol Biol. 1998;275:17–23. doi: 10.1006/jmbi.1997.1452. [DOI] [PubMed] [Google Scholar]
- Depken M, Schiessel H. Nucleosome shape dictates chromatin fiber structure. Biophys J. 2009;96:777–784. doi: 10.1016/j.bpj.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel S, Belmont AS. Reproducible but dynamic positioning of DNA in chromosomes during mitosis. Nat Cell Biol. 2001;3:767–770. doi: 10.1038/35087089. [DOI] [PubMed] [Google Scholar]
- Eggleston AK. Unraveling chromatin organization. Nat Struct Mol Biol. 2005;12:6. doi: 10.1038/nsmb0105-6. [DOI] [PubMed] [Google Scholar]
- Furey TS, Haussler D. Integration of the cytogenetic map with the draft human genome sequence. Hum Mol Genet. 2003;12:1037–1044. doi: 10.1093/hmg/ddg113. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SV, Shen Y, Benight AS, Ansari A. A semiflexible polymer model applied to loop formation in DNA hairpins. Biophys J. 2001;81:2864–2875. doi: 10.1016/S0006-3495(01)75927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SV, Ren C, Woodson SA, Ansari A. Loop dependence of the stability and dynamics of nucleic acid hairpins. Nucleic Acids Res. 2008;36:1098–1112. doi: 10.1093/nar/gkm1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux M, Patard L, Hernandez B, Couesnon T, Santini GP, Cognet JA, Gouyette C, Cordier C. Spectroscopic and structural impact of a stem base-pair change in DNA hairpins: GTTC-ACA-GAAC versus GTAC-ACA-GTAC. Spectrochim Acta Part A. 2006;65:84–94. doi: 10.1016/j.saa.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Maeshima K, Eltsov M. Packaging the genome: The structure of mitotic chromosomes. J Biochem. 2007;143:145–153. doi: 10.1093/jb/mvm214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Li C, Zhang Y, Li Y, Zhao Y. Human with-no-lysine kinase-4 3′-UTR acting as the enhancer and being targeted by miR-296. Int J Biochem Cell Biol. 2010;42:1536–1543. doi: 10.1016/j.biocel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Nickens DG, Hardy RW. Structural and functional analyses of stem-loop 1 of the Sindbis virus genome. Virology. 2008;370:158–172. doi: 10.1016/j.virol.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Nolan EM, Cheung TC, Burton DW, Deftos LJ. Transcriptional regulation of the human chromoranin A gene by its 5′ distal regulatory element: Novel effects of orientation, structure, flanking sequences, and position on expression. Mol Cell Endocrinol. 1996;124:51–62. doi: 10.1016/s0303-7207(96)03927-5. [DOI] [PubMed] [Google Scholar]
- Polak P, Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006;7:1–15. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosskopf JJ, Upton JH, 3rd, Rodarte L, Romero TA, Leung MY, Taufer M, Johnson KL. A 3′ terminal stem-loop structure in Nodamura virus RNA2 forms an essential cis-acting signal for RNA replication. Virus Res. 2010;150:12–21. doi: 10.1016/j.virusres.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satzinger H. Theodor and Marcella Boveri: Chromosomes and cytoplasm in heredity and development. Nat Rev Genet. 2008;9:231–238. doi: 10.1038/nrg2311. [DOI] [PubMed] [Google Scholar]
- Sean P, Nguyen JH, Semler BL. Altered interactions between stem-loop IV within the 5′ noncoding region of coxsackievirus RNA and poly(rC) binding protein 2: Effects on IRES-mediated translation and viral infectivity. Virology. 2009;389:45–58. doi: 10.1016/j.virol.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard A, Clarke J, Herr W. Simian virus 40 revertant enhancers exhibit restricted host ranges for enhancer function. J Virol. 1988;62:3364–3370. doi: 10.1128/jvi.62.9.3364-3370.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tour BE, Laemmli UK. The metaphase scaffold is helically folded: Sister chromatids have predominantly opposite helical handedness. Cell. 1988;55:937–944. doi: 10.1016/0092-8674(88)90239-5. [DOI] [PubMed] [Google Scholar]
- Wang XF, Wang XY, Liu J, Feng JJ, Mu WL, Shi XJ, Yang QQ, Duan XC, Xie Y. Alu tandem sequences inhibit GFP gene expression by triggering chromatin wrapping. Genes Genom. 2009a;31:209–215. [Google Scholar]
- Wang XF, Jin X, Wang XY, Liu J, Feng JJ, Yang QQ, Mu WL, Shi XJ, Lu ZJ. Effects of L1-ORF2 fragments on green fluorescent protein gene expression. Genet Mol Biol. 2009b;32:688–696. doi: 10.1590/S1415-47572009005000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Markoff L. The topology of bulges in the long stem of the flavivirus 3′ stem-loop is a major determinant of RNA replication competence. J Virol. 2005;79:2309–2324. doi: 10.1128/JVI.79.4.2309-2324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi-Li Xu, Mizuguchi H, Ishii AW, Uchida E, Mayumi T, Hayakawa T. Optimization of transcriptional regulatory elements for constructing plasmid vectors. Gene. 2001;272:149–156. doi: 10.1016/s0378-1119(01)00550-9. [DOI] [PubMed] [Google Scholar]