Abstract
Light signals perceived by photoreceptors are transduced to negatively regulate COP1, a key repressor of photomorphogenic development. To identify genes involved in light inactivation of COP1, a genetic screen was employed to identify extragenic modifier mutations of a temperature-sensitive cop1 allele. One suppressor mutation isolated also exhibited a far-red-specific long hypocotyl phenotype in a wild-type background. Further phenotypic analyses of this new mutation, named fin219, suggested that it defines a novel phytochrome A signaling component. Genetic analysis indicated that FIN219 interacts closely with another phytochrome A signaling component, FHY1. Molecular characterization of FIN219 indicated that it encodes a cytoplasmic localized protein highly similar to the GH3 family of proteins and its expression is rapidly induced by auxin. In contrast to its loss-of-function mutant phenotype, overexpression of FIN219 results in a far-red-specific hyperphotomorphogenic response. Our data suggest that FIN219 may define a critical link for phytochrome A-mediated far-red inactivation of COP1 and a possible cross-talk juncture between auxin regulation and phytochrome signaling.
Keywords: Arabidopsis development, COP1 repressor, FIN219 gene, light control, auxin regulation, phytochrome signaling
Being fixed in space, plants have evolved highly plastic developmental programs to adapt to the changing environment. Light is one of the most influential environmental factors that govern plant growth and development (Kendrick and Kronenberg 1994). Besides providing the energy source for plants via photosynthesis, light also serves as an information signal about their surroundings. Two major groups of photoreceptors help plants to sense the changes in the quality, quantity, direction, and duration of the light environment. They are the red/far-red light-absorbing phytochromes and blue/UV-A-absorbing cryptochromes and phototropin (for review, see Deng and Quail 1999; Neff et al. 2000). Phytochromes consist of a common chromophore covalently attached to apoproteins, which are encoded by a gene family with five members (PHYA–PHYE) in Arabidopsis. Different phytochromes have distinct yet partially overlapping photosensory functions. For example, phytochrome A is primarily responsible for mediating far-red light-mediated inhibition of hypocotyl elongation during seedling development, whereas phytochrome B is the major photoreceptor for red light-mediated inhibition of hypocotyl elongation.
In the light, phytochromes are capable of interconversion between two distinct conformational states and initiate signal transduction events. Recent studies have revealed a number of potential signaling components that are either specific to an individual phytochrome or shared by more than one phytochrome. For instance, genetic analyses have identified FHY1, FHY3 (Whitelam et al. 1993), SPA1 (Hoecker et al. 1998), FIN2 (Soh et al. 1998), and FAR1 (Hudson et al. 1999) as phytochrome A-specific signaling components, whereas several other genes, RED1 (Wagner et al. 1997), PEF2, and PEF3 (Ahmad and Cashmore 1996), have been defined as potential phytochrome B-specific signaling intermediates. At the same time, genetic and molecular approaches have also identified potential signaling components that act downstream of both phytochrome A and phytochrome B. This group includes PEF1 (Ahmad and Cashmore 1996), PSI2 (Genoud et al. 1998), PIF3 (Ni et al. 1998), PKS1 (Fankhauser et al. 1999), and NDPK2 (Choi et al. 1999). On the other hand, biochemical approaches have implied the involvement of trimeric G protein, calmodulin, and cGMP in phytochrome signaling (Romero and Lam 1993; Bowler et al. 1994; Neuhaus et al. 1997). It is evident that each phytochrome may have both unique signaling components and shared ones. However, it remains unclear as to how those unique and shared signaling components work together to transduce the light signal.
At the cellular level, light activation of both phytochrome A and phytochrome B in the cytoplasm initiates their nuclear import and accumulation (Sakamoto et al. 1996; Kircher et al. 1999; Yamaguchi et al. 1999). This nuclear accumulation of phytochromes in the light would indicate that phytochrome signaling entails both cytoplasmic and nuclear events. Molecular identification of four phytochrome signaling components included both nuclear [PIF3 (Ni et al. 1998); SPA1 (Hoecker et al. 1999); FAR1 (Hudson et al. 1999)] and cytoplasmic [PKS1 (Fankhauser et al. 1999)] localized factors. Furthermore, recent evidence supports a direct interaction and regulation of a nuclear transcription factor (Ni et al. 1999) or an enzyme (Choi et al. 1999) by the activated phytochromes within the nucleus.
Genetic screens for mutants that display photomorphogenic development in darkness resulted in the identification of 11 pleiotropic COP/DET/FUS loci (Wei and Deng 1999). The recessive nature of all these mutations indicated that the pleiotropic COP/DET/FUS gene products act as negative regulators of photomorphogenesis. Molecular and biochemical analyses of this group of genes suggested that they might form four functional entities: COP1, DET1, COP10, and COP9 signalosome, which consists of eight distinct subunits (Wei and Deng 1999). Among these entities, COP1 was defined as a rate-limiting or regulatory component in mediating the suppression of photomorphogenesis in the dark and its activity is negatively regulated by light (McNellis et al. 1994b; Osterlund et al. 1999). COP1 is a 76-kD protein with a RING finger motif, a putative coiled–coil domain, and multiple WD-40 repeats characteristic of the β-subunit of the trimeric G protein (Deng et al. 1992; McNellis et al. 1994a). In contrast to phytochromes, COP1 is enriched in the nuclei of the hypocotyl cells of dark-grown seedlings and is excluded from these nuclei upon exposure to light (von Arnim and Deng 1994). Recent studies have suggested that COP1 acts within the nucleus to directly interact with and negatively regulate multiple transcription factors, which act to promote light-regulated gene expression and development (Ang et al. 1998; Yamamoto et al. 1998). This negative regulation of target transcription factors by COP1 may involve targeted degradation via the 26S proteasome (Osterlund et al. 2000).
It has been demonstrated that phytochrome A, phytochrome B, and cryptochromes are primarily responsible for mediating far-red, red, and blue light inactivation of COP1 (Ang and Deng 1994; Ang et al. 1998; Osterlund and Deng 1998). However, little is known regarding how light signals that are perceived by individual photoreceptors are transduced to regulate COP1 activity and its cellular localization. To identify the players involved, we have devised a genetic screen for extragenic suppressors by employing the temperature-sensitive cop1-6 mutant. Among a large number of extragenic suppressor loci recovered, one modifier mutant, fin219 (far-red insensitive 219), exhibited a long hypocotyl phenotype specific to the far-red high irradiance response (FR-HIR) condition. Further characterization of this mutation indicated that it defined a new signaling component that has a role in the phytochrome A-mediated light inactivation of COP1. Molecular cloning of this locus revealed that FIN219 is an auxin-inducible gene and may represent a cross-talk juncture between auxin response and light regulation.
Results
The suppressor screen using a temperature-sensitive cop1 allele
A screen to identify extragenic modifier mutations of a given gene is most sensitive if using a mutation that displays a level of gene activity at or close to the threshold necessary for wild-type function (Simon et al. 1991). Analyses of all available cop1 mutant alleles revealed that cop1-6, a weak mutation encoding a COP1 protein with five additional amino acids inserted between codon 301 and 302 (McNellis et al. 1994a), exhibits a temperature-dependent phenotype. When grown under ambient temperatures below 28°C in darkness, cop1-6 exhibits constitutively photomorphogenic development (Fig. 1A), whereas it appears as the wild type when grown above 28°C. Besides the elongated hypocotyl and unexpanded cotyledons in wild type-like phenotype, expression of light-inducible genes such as CAB and RBCS was also repressed in cop1-6 seedlings that were grown above 28°C (data not shown). Hence, cop1-6 is clearly capable of providing levels of COP1 activity over the threshold when the temperature is above 28°C.
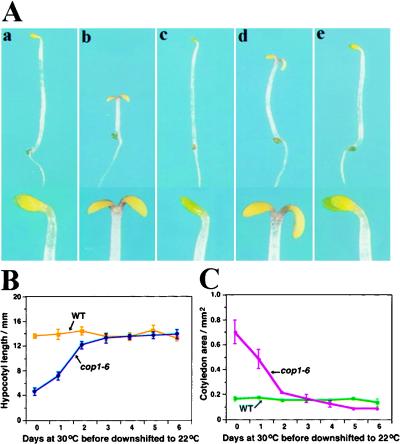
Figure 1.
Seedling growth characteristics of the cop1-6 mutant and its suppressor. (A) Wild-type Columbia (a), cop1-6 (b), suppressor line 2A9 (c), double homozygous mutant fin219/cop1-6 (d), and fin219 (e) were grown in the dark for 4 days. Panels a–e were taken under the same magnification; close-ups of their cotyledons are represented at the bottom of each panel. (B,C) Seedlings of cop1-6 and wild type were grown in the dark at 30°C for 0-to-6 days before they were transferred to 22°C. The hypocotyl lengths (B) and cotyledon area (C) of 25 seedlings from each treatment were measured at 10 days postgermination. The error bars represent standard deviations.
To define the critical window during seedling development in which COP1 activity is required, we germinated the seeds of cop1-6 mutants in darkness at 30°C for 0 to 6 days before they were transferred to 22°C. To assess the morphological changes of cop1-6, the hypocotyl lengths and cotyledon sizes of 10-day-old cop1-6 seedlings were recorded. As shown in Figure 1, B and C, cop1-6 seedlings that were grown at the permissive temperature (30°C) for <2 days before they were transferred to the nonpermissive temperature (22°C) had clearly assumed different degrees of photomorphogenic development. In contrast, seedlings that had been grown at the permissive temperature for 2 days or more before the transfer were largely committed to a normal (etiolation) mode. The seedlings showed little expansion of their cotyledons and their hypocotyls were elongated. These data indicated that COP1 activity was critically required during the initial 2 days during seedling development to make the commitment between the developmental programs.
On the basis of the observation above, we designed a screen at the 3-day-old seedling stage for extragenic mutations, which suppress the photomorphogenic phenotype of cop1-6 at 22°C. Twenty-six inheritable suppressor mutations of cop1-6 were recovered and further genetic complementation tests indicated that they defined 19 extragenic suppressor loci (see Materials and Methods). As a first step in analyzing these new mutants, we decided to focus on the extragenic mutations that also exhibited specific light-regulatory defects in a wild-type background. To this end, the extragenic suppressors were outcrossed to wild-type plants and the resulting F2 progenies were examined for the segregation of novel phenotypes. An extensive analysis of possible phenotypes under various light conditions revealed two suppressor loci that exhibited their own phenotypes in a wild-type background. One was the HY5 locus (Ang et al. 1998), and the other exhibited a far-red light-specific long hypocotyl phenotype. The latter mutation identified a new locus, FIN219, which is the focus of this report (see below).
Genetic interaction between cop1 and fin219
The original suppressor line (2A9) that segregated the fin219 mutation exhibited a complete wild-type phenotype in dark-grown seedlings at 22°C (Fig. 1A, c). In the cop1-6 background, the suppressor mutation behaved in a dominant manner. However, after three generations of backcrosses to the parent cop1-6 mutant line, the suppressor phenotype weakened and lost its ability to suppress the cotyledon opening and enlargement (Fig. 1A, d). Therefore, it is most likely that the initial suppressor line contains background mutations that enhance the suppressor mutation phenotype. To confirm that the fin219 mutation segregating from the initial suppressor line is the suppressor mutation, it was reintroduced into a cop1-6 mutant background. The double mutant of fin219 and cop1-6 developed similarly to the backcrossed suppressor mutant shown in Figure 1A (d).
The fact that the fin219 mutation can suppress the phenotype of cop1-6 in a dominant fashion is consistent with the notion that fin219 normally regulates COP1 activity negatively and its loss of function (even a 50% reduction in the heterozygote) would effectively result in an increase in COP1 activity over its threshold requirement in the cop1-6 mutant background. To support the notion that COP1 acts downstream of FIN219, fin219 was introduced into cop1-5, a null allele of cop1, by genetic crosses. Double mutants of fin219/cop1-5 were grown in the dark and light and compared with the parental strains. In both dark and light growth conditions, fin219/cop1-5 double mutants resembled the cop1-5 mutant, indicating that cop1 is epistatic to fin219 (data not shown). Hence, COP1 likely acts downstream of FIN219.
FIN219 is a novel locus involved in far-red-mediated inhibition of hypocotyl elongation
In a wild-type background, the far-red long hypocotyl phenotype of fin219 is largely a loss-of-function mutation, with a slightly semidominant effect in heterozygotic seedlings. As shown in Figure 2, homozygous fin219 mutant seedlings exhibited a long hypocotyl phenotype only under continuous far-red light and no obvious defect under red, blue, or white light or in complete darkness (Fig. 2A,C). By comparing the seedling phenotypes of fin219 mutants that were grown under far-red light for 2–6 days, it is clear that far-red induction of cotyledon development is delayed in fin219 mutants as well (data not shown). Hence, the fin219 mutant seems to be defective in phytochrome A-mediated FR-HIR. This phenotype resembles that of the phyA mutants and those mutants (fhy1, fhy3, fin2, far1) defective in PHYA-specific signaling processes (Parks and Quail 1993; Nagatani et al. 1993; Whitelam et al. 1993; Soh et al. 1998; Hudson et al. 1999).
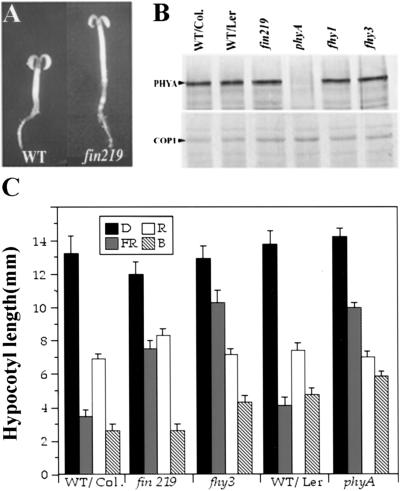
Figure 2.
Phenotypic characterization of the fin219 mutant. (A) Three-day-old wild-type and fin219 mutant seedlings grown under continuous far-red light. (WT) Wild type Columbia. (B) Western blot analysis of wild type and fin219 for PHYA and COP1 protein levels. The protein extracts were prepared from 6-day-old far-red light-grown seedlings, and equal amounts of total protein were loaded onto each lane. PHYA and COP1 protein bands detected by the corresponding antibodies are indicated. (C) The average hypocotyl lengths of 6-day-old seedlings grown under blue (B), red (R), far-red (FR) light, or in darkness (D) (n = 25). (Col.) Columbia; (Ler) Landsberg. Error bars represent standard deviations.
To determine if fin219 defines a new locus, genetic complementation tests among fin219 and fhy1, fhy3, and phyA were carried out. The F2 progenies of all three pair-wise crosses segregated wild type and long hypocotyl mutants under continuous far-right light, indicating that fin219 is not allelic to phyA, fhy1, or fhy3. Furthermore, genetic mapping of fin219 also indicated that it is located in a chromosomal region distinct from fin2 (Soh et al. 1998) and far1 (Hudson et al. 1999). Hence, fin219 defines a new locus distinct from all previously reported far-red-specific long hypocotyl mutations.
The fin219 mutant is not defective in phytochrome A and COP1 accumulation
Because the fin219 mutant is specifically defective in FR-HIR, it is important to test whether fin219 specifically affects PHYA abundance or its downstream signalings. To examine a possible effect of fin219 on the accumulation of PHYA, we examined PHYA levels by Western blot. As shown in Figure 2B, fin219 expresses wild-type levels of PHYA protein, similar to fhy1 and fhy3. This result indicates that fin219 does not affect the accumulation of PHYA apoprotein. Furthermore, because fin219 is specifically defective in response to far-red but not red or white light, it is unlikely that fin219 would affect chromophore biogenesis. This is because a general defect to far-red, red, and white light responses would be anticipated for chromophore biogenesis mutants, as in the case of hy1 and hy2 (Koornneef et al. 1980; Parks and Quail 1991).
As moderate elevation of COP1 would result in a long hypocotyl phenotype (McNellis et al. 1994b), a possible far-red-specific COP1 protein level elevation in fin219 was examined. As shown in Figure 2B, fin219 mutants accumulated wild-type levels of COP1 protein in far-red light. Thus, the long hypocotyl in far-red light observed in fin219 is not likely a result of far-red-specific COP1 elevation.
Far-red-dependent germination and greening block are not affected by fin219
To further determine the role of FIN219, the effect of the fin219 mutation on several other PHYA-mediated responses was examined. It was reported that phytochrome A is responsible for far-red light-dependent germination after extended cold treatment (Shinomura et al. 1994) and far-red greening block (Barnes et al. 1996). As shown in Figure 3A (a), fin219 mutants largely retain normal far-red light-dependent germination, and their germination rate is even enhanced by far-red light in comparison to their wild-type control. This result indicated that the fin219 mutant is not defective in phytochrome A-mediated germination in far-red light. This property of fin219 is similar to fhy1, fhy3 (Barnes et al. 1996), and fin2 (Soh et al. 1998) mutants. On the other hand, the result shown in Figure 3A (b) indicates that the greening of far-red light-grown fin219 seedlings is completely blocked, similar to that of wild type and contrasting with that of phyA and fhy1 mutations.
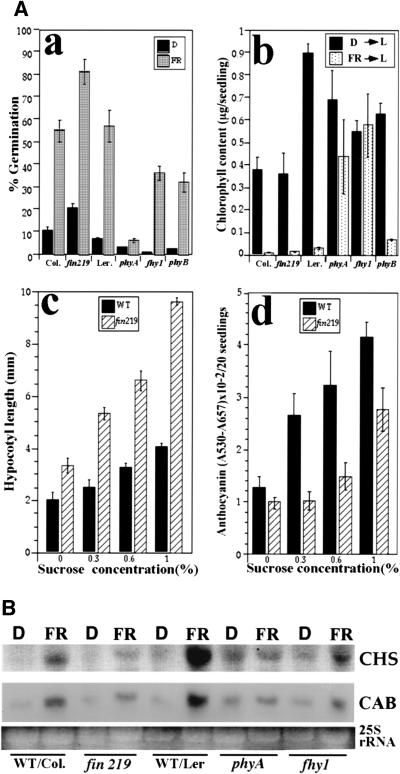
Figure 3.
The effect of fin219 on light-regulated development and gene expression (A). Effect of the fin219 mutation on selected light-regulated responses. (a) Far-red-light-inducible germination. The germination ratio was determined for the seeds that were irradiated with far-red intensity light for 15 min and kept in darkness at 22°C for 7 days (FR) or seeds kept in complete darkness without exposure to any light for 7 days (D) (n = 200). Error bars represent standard deviations. (b) Far-red block of greening. Seedlings grown in darkness for 4 days (D) or under far-red light for 4 days (FR) were transferred into white light (L, ∼30 μmoles/m2 per sec) for 4 days before extraction of chlorophyll. (c,d) Sucrose dependency of the elongation of hypocotyls and anthocyanin accumulation. Seeds were grown on GM plates containing various concentrations of sucrose from 0% to 1% for 3 days under continuous far-red light. (WT) Wild-type Columbia. (B) fin219 affects far-red induction of the CHS and CAB genes. Ten micrograms of total RNA was isolated from 3-day-old dark-grown seedlings that had been exposed to far-red light or kept in darkness for 2 hr. The membrane was hybridized with CHS and CAB3 probes. The stained RNA gel was photographed and the 25S rRNA bands are shown as an equal loading control.
The fin219 phenotype is modified by sucrose
Compared to other far-red-specific long hypocotyl mutants, the fin219 phenotype exhibits unique sucrose dependence. As shown in Figure 3A (c), exogenously applied sucrose enhances hypocotyl elongation in fin219 more than in wild type, thus leading to a large difference in hypocotyl lengths between fin219 and wild type at moderate sucrose concentrations (1% sucrose). Similar effects of sucrose on anthocyanin accumulation were also observed (Fig. 3A, d). These characteristics of fin219 mutants are different from those of fhy1 and fhy3, whose phenotypes are not significantly modified by exogenous sucrose (Whitelam et al. 1993).
The fin219 mutation affects far-red light induction of gene expression
Phytochrome A is also involved in far-red light induction of gene expression. This response has been used successfully to characterize the putative phytochrome A signaling component mutant fhy1 (Barnes et al. 1996). To further define the role of FIN219 in phytochrome A signaling, far-red light induction of CHS and CAB gene expression in fin219 and wild type was examined. Similar to fhy1 and phyA mutants, induction of CHS and CAB gene expression in fin219 was notably compromised (Fig. 3B). Therefore, fin219 has a role in phytochrome A-mediated far-red light induction of gene expression. It is interesting to note that compared to the Columbia ecotype, the Landsberg ecotype exhibited enhanced chlorophyll accumulation and far-red light induction of both CAB and CHS genes. This may reflect some ecotype-specific fine-tuning of light signaling.
fin219 and fhy1 show nonallelic noncomplementation
Complementation results between fin219 and fhy1 revealed an interesting genetic interaction between the two nonallelic mutations. Whereas both are loss-of-function mutations, fin219 exhibits a slightly semidominant phenotype under our conditions. However, the fin219/fhy1 transheterozygotes (fin219/+; fhy1/+) (Fig. 4A, d) exhibited a severe deficiency in far-red light inhibition of hypocotyl elongation similar to their homozygous parental mutants (Fig. 4A, e,f). This effect is not due to allelic complementation, as wild-type seedlings segregated in the F2 progeny. Hence, fin219 and fhy1 mutations appear to show nonallelic noncomplementation. To rule out the possibility that the long hypocotyl in F1 progeny might be due to hybrid vigor, as the fin219 and the initial fhy1 mutations used are from different ecotypes, similar crosses were done with fin219 and two other fhy1 alleles derived from the same Columbia ecotype. The same nonallelic noncomplementation was observed in both crosses (data not shown). Hence, the noncomplementation between fin219 and fhy1 mutations is more likely a result of genetic interactions between these two mutations. As FHY1 is a well-defined component in the phytochrome A-mediated FR-HIR pathway, this observation further supports the notion that FIN219 may provide the bridge between the photosensing phytochrome A and the downstream negative regulator of photomorphogenic development, COP1.
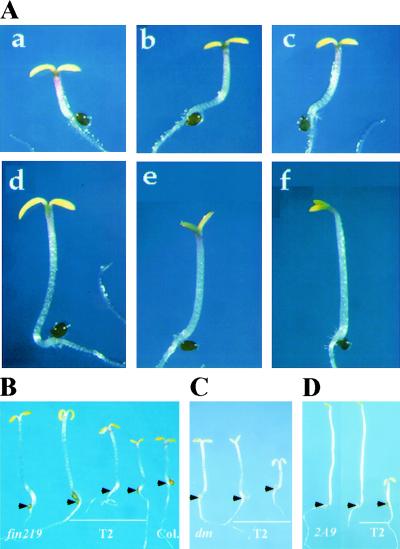
Figure 4.
Genetic interaction of fin219 and fhy1 and phenotypic rescue of the mutant phenotype. (A) Genetic interaction between fin219 and fhy1. (a) Wild-type Columbia; (b) fin219 heterozygote in Columbia background; (c) fhy1 heterozygote in Landsberg background; (d) fin219/+;fhy1/+ transheterozygote; (e) fin219 homozygous in Columbia background; (f) fhy1 homozygous in Landsberg background. All the seedlings were grown under continuous far-red light for 3 days. (B) Functional rescue of fin219. The fin219 mutants were transformed with a 5.1-kb BamHI DNA fragment containing the FIN219 gene. The transgenic T2 seedlings grown under far-red light for 3 days segregated long, intermediate, and short hypocotyls in the T2 generation with the expected ratio (1:2:1 for single-locus lines). The representative transgenic seedlings (middle three), wild type (right), and fin219 parental mutant (left) are shown. For each seedling, the junction between the hypocotyl and the root is marked by an arrowhead. Complementation tests of the backcrossed suppressor mutant (C) and the original suppressor line (D) were transformed with the 5.1-kb genomic DNA fragment containing the FIN219 gene. In both cases, two representative segregating transgenic T2 seedlings (middle and right) are shown together with the suppressor seedlings (dm in C; 2A9 in D). All seedlings were grown in the dark for 4 days. Only ∼5%–25% of the T2 progeny in the single-locus transgenic lines exhibited a short hypocotyl (rescued) phenotype (C,D,right), which shows indistinguishable phenotype from the cop1-6 mutant seedlings. For each seedling, the position of hypocotyl and root junction is marked by an arrowhead.
Positional cloning of FIN219
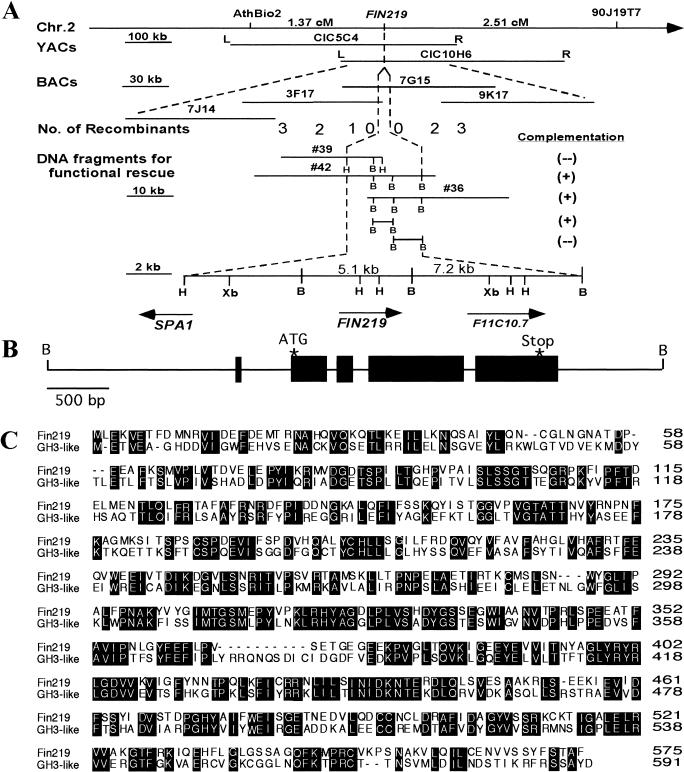
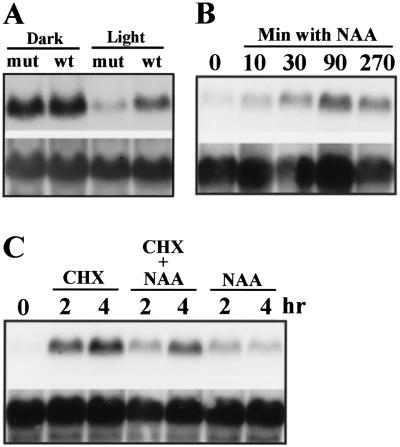
To clone FIN219, PCR-based simple sequence length polymorphism (SSLP) (Bell and Ecker 1994) markers were used to map FIN219 to the lower arm of chromosome 2, below the SSLP marker nga168. After examining 1154 recombinant chromosomes, FIN219 was mapped to one BAC clone (T7G15) located within the region bordered by AthBio2 and 90J19T7 markers (Fig. 5A; Materials and Methods). An overlapping set of binary cosmid clones were constructed from T7G15 and used to transform fin219 mutants for functional rescue. Transformation of two cosmid clones (nos. 36 and 42), but not clone 39 (which contains the SPA1 gene), complemented the mutant fin219, suggesting that the FIN219 gene must reside in their overlapping region (see Materials and Methods). This overlapping region was divided further into two BamHI DNA fragments of 5.1 and 7.2 kb. Both fragments were used to rescue the fin219 far-red-specific long hypocotyl phenotype and only the 5.1-kb BamHI fragment was able to do so (Fig. 4B; see Materials and Methods). This 5.1-kb BamHI fragment was used to screen a cDNA library and recovered a total of 15 cDNA clones encoding the same gene. One cDNA clone, M11-2, containing the longest insert (∼1.8 kb), and the 5.1-kb BamHI genomic fragment were sequenced completely. Comparison of these sequences revealed that this fragment contains only one transcribed region, which contains five exons and four introns (Fig. 5B). Therefore, this gene is most likely the FIN219 locus. A detailed analysis of the fin219 mutant indicated that it is an epigenetic mutation due to an altered methylation pattern (data not shown) and reduced expression in the light (Fig. 6A). Furthermore, the fin219 mutation has no detectable effect on the expression pattern of an adjacent gene SPA1 (Hoecker et al. 1998, 1999), a negative regulator of the phytochrome A pathway (data not shown).
Figure 5.
Positional cloning and molecular characterization of FIN219. (A) Positional cloning of FIN219. Genetic mapping first located the gene on the lower arm of chromosome 2 between SSLP marker AthBio2 and CAPS marker 90J19T7. The arrow points to the telomere on the bottom arm of chromosome 2. First-round analysis of recombinant lines positioned the gene within the overlapping region of CIC10H6 and CIC5C4. Further mapping of the gene using overlapping BAC clones covering the region located the gene within a single BAC clone (TAM7G15). Overlapping binary cosmid clones were constructed using this BAC clone to transform fin219 mutants. Two cosmid clones (nos. 36 and 42) rescued the mutant phenotype. The overlapping region between clones 36 and 42 was dissected further into 5.1- and 7.2-kb BamHI fragments and used for functional rescue again. Only the 5.1-kb BamHI fragment complemented the mutant fin219. (B) BamHI; (H) HindIII; (Xb) XbaI. FIN219 is located between SPA1 and an unknown gene, F11C10.7. (B) Diagram of the 5.1-kb BamHI fragment, which contains one transcribed region with five predicted exons and four introns. The locations of the initiation codon (ATG) and stop codon are indicated by stars. (C) Sequence alignment of FIN219 (accession no. AF279129) and a GH3-like protein (accession no. AF071527.1) from Arabidopsis. Identical residues of these two proteins are shown in shaded boxes.
Figure 6.
FIN219 mRNA expression and its auxin regulation. (A) FIN219 expression is reduced in light-grown fin219 mutants. Equal amounts of total RNA from 7-day-old dark- and light-grown wild-type (wt) and fin219 mutant (mut) seedlings were loaded in each lane. (B) Time course of expression of FIN219 mRNA induction by auxin. Seven-day-old seedlings grown in light were transferred to a liquid medium in the presence of 10 μm NAA. (C) Similar auxin induction test as in B with 2 or 4 hr after addition of auxin. The effect of 70 μg/ml cycloheximide and NAA alone, or together, was tested.
fin219 is a suppressor mutation of cop1-6
To further confirm that the fin219 mutation was largely responsible for suppression of the cop1-6 phenotype, the same 5.1-kb BamHI DNA fragment covering the FIN219 locus was introduced into the original suppressor line (2A9), as well as the backcrossed suppressor line (see Fig. 1A, c,d). When germinated in darkness, T2 progenies from multiple single-insertion locus lines in both complementation tests segregated up to one-fourth cop1-6-like mutant phenotypes (Fig. 4C; see Materials and Methods). The fact that introduction of the FIN219 locus resulted in rescue of the suppressor phenotype confirms that fin219 is the responsible suppressor mutation in this line.
FIN219 is a GH3-like protein and its expression is rapidly auxin inducible
The cDNA sequence revealed a 575-amino-acid ORF with a predicted protein of ∼64.3 kD in size. Comparison with known sequences in the database revealed that FIN219 is most closely related to a GH3-like protein, with an overall 47% identity and 66% similarity (Fig. 5C). The GH3 protein was first identified from soybean as an early auxin-responsive gene (Hagen et al. 1984, 1991). However, the function of GH3 is not known. In Arabidopsis, a family of at least six other GH3-like proteins share 37%–39% identity with FIN219. Computer analysis also revealed putative coiled–coil domains in both amino- and carboxy-terminal regions of the predicted FIN219 protein (COILS, v. 2.1, A.N. Lupas, Human Genome Center, Baylor College of Medicine, Houston, TX).
As the FIN219 protein shows strong homology with GH3 proteins, which were defined as early auxin-inducible genes in soybean (Hagen et al. 1984), we examined whether auxin regulates its expression. Three biologically active auxin compounds (IAA, NAA, and 2,4-D) have the same effect on FIN219 expression, and the result of NAA is shown in Figure 6, B and C. Furthermore, FIN219 expression was rapidly auxin-inducible and increased only 10 min after addition of auxin. It takes ∼90 min to 2 hr to reach the maximal level. Also similar to early auxin-inducible genes (Franco et al. 1990; Roux and Perrot-Rechenmann 1997), FIN219 is inducible by cycloheximide. However, the effects of auxin and cycloheximide are not additive. This result suggests that a highly labile repressor whose turnover rate is controlled by auxin might regulate FIN219 expression.
FIN219 is a cytoplasmic protein
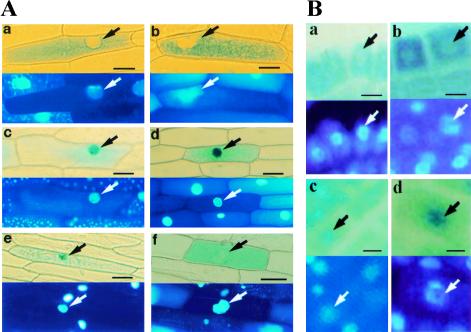
To determine the subcellular localization of FIN219, a FIN219 full-length cDNA was fused in frame to the carboxyl terminus of GUS (GUS–FIN219) and introduced into plant cells via both transient expression and stable transformation. When introduced into onion cells by particle bombardment, GUS–FIN219 was located in the cytoplasm both in darkness and in the light (Fig. 7A, a,b). As reported previously (von Arnim and Deng 1994), under the same conditions, GUS–COP1 was nuclear localized in darkness and cytoplasmically localized in the light (Fig. 7A, e,f). The control, GUS–NIa, was constitutively nuclear localized (Fig. 7A, c,d). Supporting the transient result, examination of stable transgenic lines carrying GUS–FIN219 also revealed cytoplasmic localization in both light- and dark-grown seedlings (Fig. 7B, a,b). Again, stable transgenic GUS–NIa seedlings exhibited constitutive nuclear localization (Fig. 7B, c,d). Taken together, our results suggest a constitutive cytoplasmic localization of the FIN219 protein.
Figure 7.
FIN219 is a cytoplasmic protein. (A) Localization of FIN219 in onion cells in a transient assay. Constructs encoding either GUS–FIN219 (a,b), GUS–NIa (c,d), or GUS–COP1 (e,f) were introduced into onion cells by particle bombardment. The onion cells were incubated in complete darkness (a,c,e) or under continuous white light (b,d,f) after bombardment for 2 days, stained for GUS activity, and then mounted whole in the presence of DAPI. The bottom each panel shows DAPI staining of the same cells to visualize the position of the nucleus (arrow). Scale bars, 100 μm in a–f. (B) Localization of FIN219 in stable transgenic plants. GUS–FIN219 (a,b) and GUS–NIa (c,d) staining patterns in root tips are shown. (a,c) Dark-grown seedlings; (b,d) light-grown seedlings. The bottom of each panel shows DAPI staining of the same field to visualize the position of the nucleus (arrow). Scale bars, 100 μm in a–d.
FIN219 overexpression results in far-red hyperphotomorphogenic response
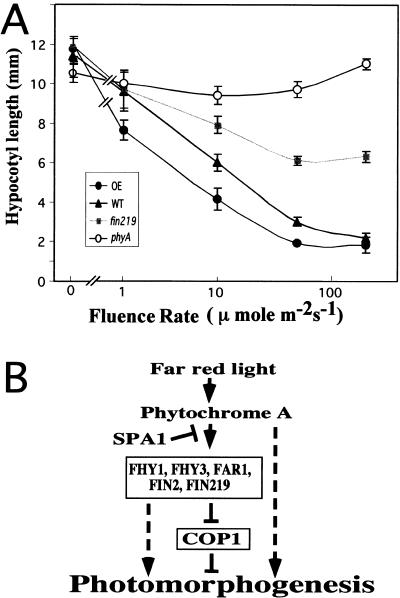
To further substantiate our conclusion that FIN219 acts in phytochrome A-mediated FR-HIR, an overexpression construct driven by the 35S promoter was made and introduced into wild-type Columbia. Homozygous lines from multiple single-locus transgenic lines were selected and their phenotypes analyzed. Although no obvious phenotype was found in high-intensity continuous far-red, red, and blue light, the overexpressor lines exhibited significantly shorter hypocotyls only under low-intensity continuous far-red light. Figure 8 illustrates this effect with a representative FIN219 overexpressor line in comparison to wild type, phyA, and fin219. In the light intensity range we examined, phyA mutants exhibit the same hypocotyl length, indicating that phytochrome A is an essential photoreceptor mediating FR-HIR. We also noted that FIN219–GUS transgenic lines exhibited the same overexpressor phenotype under low-intensity far-red light, further indicating that its cytoplasmic localization is biologically relevant. As these FIN219–GUS lines are clearly overexpressing the fusion protein due to their high GUS staining activity, the observed far-red-specific hyperphotomorphogenic response is likely a gain-of-function phenotype. Therefore, the overexpression of FIN219 results in the anticipated opposite effect of the loss-of-function fin219 mutation.
Figure 8.
(A) Overexpression of FIN219 results in hypersensitivity to far-red light. Wild-type (WT), phyA, fhy1, and fin219 mutant, and FIN219 overexpressor (OE) seedlings grown under 0, 1.5, 10, 50, and 200 μmoles/m2 per sec of continuos far-red light were used to measure hypocotyl length. Each data point represents the average of 20 seedlings. Error bars represent standard deviations. (B) Diagram indicating the hypothetical relationship of FIN219 and other known components involved in phytochrome A-mediated far-red light inactivation of COP1 and photomorphogenesis. Phytochrome A can promote photomorphogenesis through other pathways independent of COP1 (broken lines). (See text for details.)
Discussion
This report describes the identification and characterization of a new light regulatory component, FIN219. FIN219 was isolated as a suppressor of the weak cop1-6 mutation and was shown to be a phytochrome A signaling component. Thus, it is possible that FIN219 has a crucial role in phytochrome A-mediated FR inactivation of COP1 and in promoting photomorphogenesis. The fact that FIN219 expression is auxin inducible implies molecular cross talk between auxin response and light regulation.
A critical window of COP1 function during seedling development
Because cop1-6 is a temperature-sensitive mutation and in darkness develops photomorphogenically at 22°C but etiolates normally at 30°C, it was used to test the temporal requirement of COP1 during seedling developmental switch. By performing temperature shift tests, we were able to conclude that the first 2 days during Arabidopsis seedling development are critical for COP1 activity. The seedling developmental pattern is essentially determined by COP1 activity during that 2-day period. Alteration of COP1 activity after this 2-day period has little effect on the predetermined developmental program. This insight has facilitated our design of an extragenic modifier mutant screen on 3-day-old dark-grown seedlings, a stage at which dark-grown cop1-6 mutants can still green and survive after transferring to white light (Ang and Deng 1994). This finding also provided an important basis for the future understanding of the molecular mechanism of COP1 action.
FIN219 is involved in light inactivation of COP1 activity
The fact that the FIN219 locus introduced into the suppressor strain resulted in rescue of the suppressor phenotype indicated that fin219 is the suppressor mutation. As fin219 behaved dominantly in suppressing the cop1-6 mutation but was unable to suppress other cop1 mutant alleles (data not shown), it is reasoned that FIN219 may be involved in negatively regulating COP1 activity. In the cop1-6 background, the partial reduction of FIN219 activity in heterozygotes results in a sufficient increase of COP1 activity above a necessary threshold level, thus inducing a wild-type-like phenotype with respect to hypocotyl elongation. In addition, the null cop1-5 mutation is completely epistatic to the fin219 mutation, indicating that COP1 acts downstream of FIN219. The fact that the fin219 mutation can cause suppression of the cop1-6 phenotype in darkness would indicate that FIN219 possesses basal level activity toward COP1 even in darkness or absence of phytochrome activation. Presumably light activation of phytochrome would lead to higher FIN219 activity. However, our present data could not exclude other possible explanations.
FIN219 is a new component of the phytochrome A signaling pathway
FIN219 represents a novel locus whose mutation results in a long hypocotyl only in far-red light. So far, it has been reported that four other mutants, fhy1, fhy3 (Whitelam et al. 1993), far1 (Hudson et al. 1999), and fin2 (Soh et al. 1998), also exhibit a far-red-specific phenotype similar to that of phyA mutants. It is possible, therefore, that those genes somehow work together to inactivate COP1 and mediate far-red light inhibition of hypocotyl elongation (Fig. 8B). Although all of these mutants exhibit a far-red-specific long hypocotyl phenotype, there are subtle distinctions in other phytochrome A-regulated responses. For example, unlike FHY1 and FHY3, FIN219 is not involved in the far-red block of greening. Therefore, it is possible that different combinations of signaling components may be used for specific phytochrome A-mediated processes.
Strong nonallelic noncomplementation between fin219 and fhy1 further supports the notion that FIN219 is a phytochrome A signaling component. Nonallelic noncomplementation is considered a hallmark for genes whose products act closely within the same pathway and most likely interact directly. For example, in Drosophila, mutations in α- and β-tubulin genes have been described as nonallelic noncomplementing (Fuller et al. 1989). In that case, α- and β-tubulin were shown to interact physically to assemble into functional microtubules. However, mutations in α- and β-tubulin genes are strictly recessive. As our fin219 mutation is slightly semidominant and results from altered methylation in the promoter region (H.-L. Hsieh and X.-W. Deng, unpubl.), this interpretation of our genetic interaction remains to be tested once the FHY1 gene is cloned. Nevertheless, this result suggests that FIN219 and FHY1 may act in the same pathway.
Among far-red-specific long hypocotyl loci, FAR1 is the only downstream regulator whose molecular identity has been reported and shown to encode a novel nuclear protein. Interestingly, our molecular study of FIN219 indicates that it is a cytoplasmic protein. The fact that the two cloned phytochrome A-specific signaling components defined by similar far-red-specific long hypocotyl phenotypes are nuclear in one case (FAR1) and cytoplasmic in the other (FIN219) suggests that phytochrome A signaling entails specific components that exist in both the nucleus and the cytoplasm. Further molecular characterization of other far-red-specific long hypocotyl loci should provide a foundation for which to examine the molecular and biochemical interaction and regulation of those components.
A possible link between auxin response and photomorphogenesis
FIN219 is highly similar to a family of proteins defined by the soybean early auxin-inducible gene GH3. The identity of FIN219 to Arabidopsis GH3-like proteins ranged from 37% to 47%. More importantly, similar to GH3, FIN219 expression is also rapidly inducible by auxin. It is possible, therefore, that through regulating FIN219 expression, auxin may affect the light regulation of development. Similar to a recent report documenting cross talk between phytochrome signaling and auxin response (Tian and Reed 1999), our data provided yet another case. Auxin is capable of regulating many aspects of plant development, including inhibition of root elongation, promotion of hypocotyl and stem elongation, root and shoot gravitropism, promotion of lateral root, adventitious root and root hair formation, and maintenance of apical dominance (Estelle and Klee 1994). A careful examination of the fin219 mutant in the presence and absence of auxin did not reveal notable alteration in its auxin-regulated processes when compared to wild type; thus, it is unlikely that FIN219 is itself an auxin-signaling component. Therefore, this is a likely case in which auxin was used to regulate a light signaling component expression level. It is feasible that molecular cross talk such as this has been developed during evolution to closely coordinate phytochrome signaling and auxin regulation. With this sort of cross talk, it is possible that cell- or tissue-specific alterations in auxin may be reflected in changes of FIN219 expression, which in turn result in altered COP1 activity and, thus, developmental pattern regulation.
Materials and methods
Plant materials and growth conditions
cop1-6, the derived fin219 and the fhy3 mutant used in this study are in the Arabidopsis thaliana Columbia ecotype. Three other mutants, phyA-1 and fhy1 (Whitelam et al. 1993), and the putative null phyB-1 (Koornneef et al. 1980; Reed et al. 1993) are in the Landsberg erecta ecotype. Surface sterilization and cold treatment of the seeds and seedling growth under different light sources were described previously (Ang and Deng 1994). Continuous far-red light of ∼200 μmole/m2 per sec was obtained from far-red fluorescent tubes (FL20S.FR-74; Toshiba, Japan), filtered through a one-fourth-inch-thick acrylic plate (FRF700; Westlake, PA1). The remaining light sources used for the experiments were described by McNellis et al. (1994a), and the light intensities of red or blue light used for the experiments were ∼30 and 50 μmole/m2 per sec, respectively. Arabidopsis seedlings were grown on GM agar plates containing 0.3% sucrose for mutant screening and hypocotyl length measurement studies. Chlorophyll determination and RNA induction analyses were performed with seedlings grown on GM plates without sucrose, and germination experiments were performed on aqueous medium as described by Shinomura et al. (1994). For hypocotyl length measurement, an average of 30 seedlings were placed on the surface of GM plates and measured under the light microscope. Chlorophyll and anthocyanin content were determined based on methods described previously (Deng and Quail 1992).
Mutagenesis and genetic screen
Dry cop1-6 seeds were mutagenized with 0.3% EMS for 16 hr. After extensive washing, the mutagenized seeds were sown in soil pots and grown in isolation in Conviron growth chambers to avoid contamination by stray seeds. A total of 4670 M1 plants survived the mutagenesis treatment and matured to set seeds. M2 seeds were harvested in batches of ∼50 M1 plants each. M2 seeds were grown in darkness at 22°C for 3 days on GM plates and screened for suppressor phenotype. At 3 days, the cop1-6 parental lines developed open cotyledons and short hypocotyls. The suppressors of cop1-6 stood out from the cop1-6 seedlings with their elongated hypocotyls, closed cotyledons, and apical hooks resembling wild-type seedlings. The putative suppressor seedlings were transferred to white light for greening before they were potted and grown for setting seeds. Only those suppressor lines that exhibited the same phenotype in subsequent generations were subjected to further genetic tests outlined below.
Genetic analyses
To determine if the suppressor mutations were intragenic or extragenic, the double homozygous mutants were outcrossed to wild type. The segregation ratio of wild type to cop mutants in the F2 generation was scored after growing in darkness for 6 days. For extragenic and unlinked recessive suppressor mutations, the expected ratio of wild type to cop mutants is 15:1. A lower ratio would be expected if the suppressor mutation and cop1 were linked, with an extreme 3:1 ratio for an intragenic suppressor mutation. For dominant suppressors, a 3:1 ratio would be observed for unlinked mutations and a lower ratio for linked mutations, with an extreme of 1:3 for an intragenic suppressor.
To verify if the suppressor mutations were monogenic or multigenic, the double homozygous mutants were backcrossed to cop1-6 homozygotes. For monogenic mutations, the expected segregation ratios of wild-type to cop seedlings in the F2 generation are 3:1 and 1:3 for dominant and recessive suppressors, respectively. Alternatively, if two unlinked genes were involved, the expected ratios of wild type to mutant would be 15:1, 13:3, and 7:9 for two recessive mutations, a dominant and a recessive mutation, and two dominant mutations, respectively. The dominance or recessiveness of the suppressor mutations can be determined in F1 progeny of the same crosses depending if the heterozygous suppressor mutations can suppress the cop1-6 homozygote phenotype.
To discover the number of loci the 26 mutations define, pair-wise complementation tests among the 26 extragenic suppressor mutations were performed. The F1 progeny of these complementation crosses were grown in darkness for 3 days before their seedling phenotypes were scored to determine allelism. Allelism between two recessive suppressor mutations can be determined in the F1 generation depending on whether or not they can complement each other. In cases where one or both suppressor mutations are dominant, allelism can only be determined in the F2 generation.
RNA and protein blot analyses
In most cases, total RNA was isolated from 3- to 6-day-old seedlings grown under desirable conditions using the Qiagen RNeasy Plant Mini prep kit. For auxin-induced gene expression study, 1-week-old white light-grown seedlings were transferred to a liquid medium consisting of 4.3 mg/ml MS basal salt mixture (Sigma) and 10 mm PIPES (pH 6.0) in the presence of 10 μm NAA and/or 70 μm cyclohexamide. The seedlings were incubated at room temperature with gentle shaking for different time points as indicated, harvested, and frozen immediately with liquid nitrogen. Ten micrograms of total RNA was loaded on the gel and blotted to the membrane. For FIN219 expression in the fin219 mutant, 7-day-old dark and white light-grown wild-type (Columbia) and mutant seedlings were used for RNA extraction.
The FIN219 probe was made by in vitro transcription using MAXIscript T7/T3 kits from Ambion. The CHS riboprobe was derived from a 0.9-kb EcoRI fragment containing the Arabidopsis CHS coding region (Deng et al. 1991) cloned into Topo 2.1 vector (Invitrogen). The CAB probe was a 0.5-kb BamHI–SacI fragment of the CAB3 coding region (Deng et al. 1991), labeled by random priming. All hybridizations and washing conditions were done according to a standard method (Sambrook et al. 1989). Equal loading of the RNA was verified by ethidium bromide staining as well as by rehybridizing the blots with an 18S rDNA probe (Deng et al. 1991).
The protein blot analysis was done according to procedures described (Parks and Quail 1993; Ang et al. 1998).
Positional cloning of FIN219 and sequence analysis
For generating the mapping population, the derived fin219 mutant (in Columbia background) was crossed to the Landsberg ecotype. Long hypocotyl seedlings under far-red light were selected in the F2 generation and selfed for seeds. A total of 1154 recombinant chromosomes were used for fine-mapping analyses by using SSLP, CAPS, cosmids, BACs, and YAC end fragment markers available for chromosome II. After the FIN219 gene was located to BAC clone T7G15, a cosmid transformation library was constructed by introducing partially digested DNA fragments (15–40 kb) from T7G15 into the ClaI site of a binary cosmid vector, p04541 (Jones et al. 1992). A cosmid contig over T7G15 was established, and three cosmid clones (nos. 36, 39, and 42) covering the predicted FIN219 locus (based on the recombination breakpoints) were transformed into Agrobacterium strain GV3101 by electroporation. The cosmid DNAs from the transformed Agrobacterium strains were subjected to restriction digestion analysis to confirm that no rearrangement of cosmids had occurred in Agrobacteria. Subsequently, the agrobacteria with constructs 36, 39, and 42 were used for stable transformation and functional rescue of the fin219 mutants by vacuum infiltration. In addition, BamHI fragments (36B3 and 36B2) of the overlapped region of cosmid clones 36 and 42 were cloned further into the BamHI site of the binary transformation vector pPZP221 (Hajdukiewicz et al. 1994) and then transformed into fin219 mutants. Cosmid transgenic seedlings were selected with kanamycin (50 μg/ml), whereas transgenic seedlings for constructs 36B3 (5.1 kb) and 36B2 (7.2 kb) were selected with gentamycin (100 μg/ml). A total of 10, 12, and 32 independent lines were examined for phenotypic rescue using constructs 36, 39, and 42, and 7, 0, and 21 of those lines displayed complementation of the fin219 phenotype, respectively. For constructs 36B3 and 36B2, 53 and 43 independent transgenic lines were obtained. Examination of the T2 generation progeny indicated that 25 and 0 of them show functional rescue of the fin219 mutant phenotype, respectively. The same 36B2 construct was also used to transform into the original suppressor line (2A9) and the backcrossed suppressor lines. In each case, at least five independent transgenic lines were analyzed and confirmed the phenotype rescue (Fig. 4C,D). All DNA isolation and Southern hybridization during the process of chromosome walking were carried out according to standard methods (Sambrook et al. 1989).
The 5.1-kb BamHI fragment that complemented the mutant fin219 was used to screen the cDNA libraries (CD4-13 through CD4-16) obtained from Arabidopsis Biological Resource Center (ABRC). Fifteen clones were selected using this 5.1-kb BamHI fragment and derived from the same gene. The longest cDNA insert (∼1.8 kb) was used for detailed sequence analysis. At the same time, the 5.1-kb genomic DNA fragment that was capable of rescuing the fin219 phenotype was also sequenced.
Subcellular localization of the GUS–FIN219 fusion protein
To construct the GUS–FIN219 fusion protein, FIN219 cDNA was amplified by use of primers 5′-GGATCCAGATCTATGTTGGAGAAGGTTGA-3′ and 5′-GGATCCAGATCTAAAGACAACAACGACGGA-3′ and subcloned into the BglII site of pRTL2–GUS/NIa plasmid to replace NIa. The constructs containing either GUS fused to FIN219 cDNA or GUS sequence alone were used directly for transient assays in onion cells by particle bombard (von Arnim and Deng 1994). GUS–COP1 and GUS–NIa were described previously (von Arnim and Deng 1994). For stable transformations, the PstI DNA fragments were released from pRTL2–GUS/NIa or pRTL2–GUS–FIN219 constructs and ligated into the binary vector pPZP221, which had been digested with PstI. The resulting constructs were used to transform Arabidopsis (Columbia) via agrobacteria. A total of 13 GUS–FIN219 transgenic lines were generated, and three single-locus lines were examined in detail for GUS localization and hypocotyl phenotype. The same results were obtained from all lines examined. GUS staining of the homozygous T2 transgenic seedlings was performed as described (von Arnim and Deng 1994).
Construction of FIN219 overexpressor lines
The full-length cDNA clone was released as a BamHI–SacI fragment from the original vector (M11-2) and inserted into the binary transformation vector pPZPY122. Thus, the FIN219 ORF was driven by the strong 35S promoter. The construct was transformed into wild type (Columbia) plants via agrobacteria. A total of 20 lines were generated and 3 representative single-locus lines were selected for detailed phenotypic studies. Homozygous T2 or T3 transgenic seedlings for those selected lines were used for phenotype study under different light conditions.
Acknowledgments
We thank Christian Hardtke, Mark Osterlund, and Haiyang Wang for critical comments on the manuscript, Dr. Garry Whitelam for mutant seeds, and Dr. Peter Quail for anti-PHYA antibodies. This work was supported by National Institutes of Health grant GM47850 (to X.W.D.) and a Human Frontier Science Program grant to (X.W.D. and M.M.). X.W.D. is a NSF Presidential Faculty Fellow.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
While this paper was under review, another gene, PAT1, also involved in phytochrome A signal transduction was cloned from Arabidopsis by Cordelia Bolle, Csaba Koncz, and Nam-Hai Chua. This paper is published in Genes & Development 14: 1269–1278 (2000).
Footnotes
E-MAIL xingwang.deng@yale.edu; FAX (203) 432-3854.
References
- Ahmad M, Cashmore AR. The pef mutants of Arabidopsis thaliana define lesions early in the phytochrome signaling pathway. Plant J. 1996;10:1103–1110. doi: 10.1046/j.1365-313x.1996.10061103.x. [DOI] [PubMed] [Google Scholar]
- Ang L-H, Deng X-W. Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell. 1994;6:613–628. doi: 10.1105/tpc.6.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L-H, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng X-W. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua N-H. Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell. 1996;8:601–615. doi: 10.1105/tpc.8.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignments of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua N-H. Cyclic GMP and calcium mediate phytochrome phototransduction (published erratum appears in Cell 79: 743) Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS. Phytochrome signaling is mediated through nucleoside diphosphate kinase 2. Nature. 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Quail PH. Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 1992;2:83–95. [Google Scholar]
- ————— Signaling in light-controlled development. Semin Cell Dev Biol. 1999;10:121–129. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Casper T, Quail PH. cop1: A regulatory locus involved in the light-controlled development and gene expression in Arabidopsis. Genes & Dev. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Matsui M, Wei N, Wagner D, Chu AM, et al. COP1, an Arabidopsis regulatory gene, encodes a novel protein with both a Zn-binding motif and a Gβ-protein homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- Estelle M, Klee HJ. Auxin and cytokinin in Arabidopsis. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 555–578. [Google Scholar]
- Fankhauser C, Yeh K-C, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Franco AR, Gee MA, Guilfoyle TJ. Induction and superinduction of auxin-responsive mRNAs with auxin and protein synthesis inhibitors. J Biol Chem. 1990;265:15845–15849. [PubMed] [Google Scholar]
- Fuller MT, Regan CL, Green LL, Robertson B, Deuring R, Hay TS. Interacting genes identify interacting proteins involved in microtubule function in Drosophila. Cell Motil Cytoskeleton. 1989;14:128–135. doi: 10.1002/cm.970140122. [DOI] [PubMed] [Google Scholar]
- Genoud T, Millar AJ, Nishizawa N, Kay SA, Schäfer E, Nagatani A, Chua NH. An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell. 1998;10:889–904. doi: 10.1105/tpc.10.6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Kleinschmidt A, Guilfoyle T. Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta. 1984;162:147–153. doi: 10.1007/BF00410211. [DOI] [PubMed] [Google Scholar]
- Hagen G, Martin G, Li Y, Guifoyle TJ. Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol Biol. 1991;17:567–579. doi: 10.1007/BF00040658. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH. SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes & Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JG, Shlumukov L, Carland F, English J, Scofield SR, Bishop GJ, Harrison K. Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1992;1:285–297. doi: 10.1007/BF02525170. [DOI] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM. Photomorphogenesis in plants. 2nd ed. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schäfer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Miséra S, Deng X-W. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional role for the multiple protein domains. Plant Cell. 1994a;6:487–500. doi: 10.1105/tpc.6.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Deng X-W. Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: Evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell. 1994b;6:1391–1400. doi: 10.1105/tpc.6.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A, Reed JW, Chory J. Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 1993;102:269–277. doi: 10.1104/pp.102.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J. Light: An indicator of time and place. Genes & Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Neuhaus G, Bowler C, Hiratsuka K, Yamagata H, Chua N-H. Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J. 1997;16:2554–2564. doi: 10.1093/emboj/16.10.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- ————— Binding of phytochrome B to its nuclear signaling partner PIF3 is reversibly by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Deng X-W. Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 1998;16:201–208. doi: 10.1046/j.1365-313x.1998.00290.x. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Ang L-H, Deng X-W. The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol. 1999;9:113–118. doi: 10.1016/s0962-8924(99)01499-3. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke C, Wei N, Deng XW. Targeted destabilization of HY5 during light regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Parks BM, Quail PH. Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell. 1991;3:1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell. 1993;5:39–48. doi: 10.1105/tpc.5.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JM, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far red light receptor phytochrome B alter cell elongation and physiological responses through Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero LC, Lam E. Guanine nucleotide binding protein involvement in early steps of phytochrome-regulated gene expression. Proc Natl Acad Sci. 1993;90:1465–1469. doi: 10.1073/pnas.90.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Perrot-Rechenmann C. Isolation by differential display and characterization of a tobacco auxin-responsive cDNA Nt-gh3, related to GH3. FEBS Lett. 1997;419:131–136. doi: 10.1016/s0014-5793(97)01447-6. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shinomura T, Nagatani A, Chory J, Furuya M. The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol. 1994;104:363–371. doi: 10.1104/pp.104.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MA, Bowtell DL, Dodson GS, Laverty TR, Rubin GM. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG. Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J. 1998;16:411–419. doi: 10.1046/j.1365-313x.1998.00307.x. [DOI] [PubMed] [Google Scholar]
- Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126:711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng X-W. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Wagner D, Hoecker U, Quail PH. Red1 is necessary for phytochrome B-mediated red light-specific signal transduction in Arabidopsis. Plant Cell. 1997;9:731–743. doi: 10.1105/tpc.9.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng X-W. Making sense of the COP9 signalosome. A regulatory protein complex conserved from Arabidopsis to human. Trends Genet. 1999;15:98–103. doi: 10.1016/s0168-9525(98)01670-9. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Hardberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Matsui M, Ang L-H, Deng X-W. Role of a COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell. 1998;10:1083–1094. doi: 10.1105/tpc.10.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]