Abstract
Channa striata, locally known as “haruan”, is economically important in fisheries and aquaculture industries in several Asian countries. DNA sequencing, based on a partial segment of the Cytochrome oxidase c subunit 1 (CO1) gene, was used to determine genetic variation in C. striata samples from four different populations on the west coast of Peninsular Malaysia. The highest nucleotide and haplotype diversities were observed in the Linggi population (π = 0.0067, h = 0.835), and the lowest in the Timah Tasoh population (π = 0.0008, h = 0.286). Apart from Kajang-Linggi, which was insignificant, FST values were significant (p < 0.05) in all pairwise-population comparisons. Consequently, it is inferred that genetic structuring C. striata populations in this region was largely shaped by a common origin, with secondary influences from geographical factors and isolation.
Keywords: Channa striata, snakehead, mtDNA, CO1, isolation by distance
Channa striata, locally known as “haruan”, is a carnivorous freshwater air-breather (Jianguang and Fast, 1997). A popular food-fish in Malaysia (Ali, 1999; Mat Jais, 2007), Thailand and Indo-China, it is also cultivated in India, Pakistan and Thailand (Hossain et al., 2008). Very hardy, it can stay alive out of water for a long time, whereby is the normal marketing procedure (Hossain et al., 2008). It is also widely used in traditional medicine for wound-healing (Zakaria et al., 2004). Studies by Ng et al. (2004) showed that a C. striata extract is useful as an alternative treatment in, osteoarthritis, a joint-disorder involving the softening and disintegration of articular cartilage, with subsequent changes in the underlying bones.
Due to its higher mutation rate of base substitution compared to nuclear DNA (Qiongying et al., 2006), maternally inherited mitochondrial DNA is an efficient genetic marker in genetic-differentiation studies. Mitochondrial cytochrome c oxidase 1 (CO1) being the prime DNA bar-coding region for taxonomically identifying animals, such as fishes, insects, amphibians and sponges (Erpenbeck et al., 2005; Seifert et al,. 2007; Alessandrini et al., 2008; Rock et al., 2008; Smith et al., 2008), is sometimes useful for investigating phylogeographic groups within a single species (Yu et al., 2009). The objective herein was to determine the genetic variation in C. striata populations from four different locations on the west coast of Peninsular Malaysia, through DNA sequencing of CO1 of mtDNA.
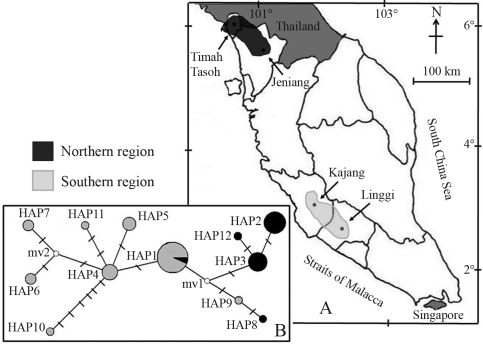
Forty five samples were collected from two northern sites, Timah Tasoh (Perlis) and Jeniang (Kedah) and two southern Linggi (Negeri Sembilan) and Kajang (Selangor) (Figure 1). With the exception of Kajang, a cultured species of unknown origin, but believed to be from the southern region itself, all were wild populations. Prior to DNA extraction, fin tissues were preserved in TNES-Urea (100 mM Tris-HCL pH 7.5, 125 mM NaCl, 10 mM EDTA pH 7.5, 1% SDS, 3 M Urea), modified according to Valles-Jimenez et al. (2004).
Figure 1.
A. Sampling localities of the four Channa striata populations analyzed in the study. B. The minimum spanning-network relationships among the twelve haplotypes of mtDNA CO1 in the four populations of Channa striata. Each bar with a crossed connection between haplotypes represents one mutation site. The size of each circle is an approximate indication of haplotype frequency [black circle found in northern region populations; grey circle found in southern region populations; open circle is the intermediate (mv)].
DNA extraction was with an Aquagenomic Kit (Bio-Syntech, Salt Lake City, Utah, USA), according to manufacturer’s instructions. A segment of the CO1 mtDNA gene was amplified using a pair of primers - forward primer L6154 (5′-AYC ARC AYY TRT TYT GRT TCT-3′) and reverse primer H6556 (5′ TGR AAR TGI CGI ACW ACR TA-3′) (Telechea et al., 2006). PCR was done in a Peltier thermal cycler (MJ Research Waltham, MA, USA), with the following profile: pre-denaturation at 95 °C for 5 min, 30 cycles of denaturation at 94 °C, annealing at 50 °C and extension at 70 °C for 1 min each, followed by final extension at 72 °C for 5 min. The PCR products were then purified using Wizard® SV Gel and a PCR Clean-Up System by Promega (Promega Madison, WI) and sequenced on an ABI3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Sequences were edited using MEGA 4.0 (Tamura et al., 2007) and aligned with Clustal W 1.6, implemented in same software. Arlequin version 3.1 (Schneider et al., 2000) was used for calculating nucleotide and haplotype diversities, haplotype frequency and FST values. Bonferroni correction was applied, with global significance level at 0.05 to correct for multiple comparison. The relationships among all haplotypes were constructed using the Network program (Bandelt et al., 1999).
Aligned sequences of 364 bp in the mtDNA CO1 gene were obtained. Twelve unique haplotypes were identified from the four populations of 45 individuals. The sequences were all submitted to the Genbank under accession numbers GQ204314, GQ204319, GQ204321, GQ204323, GQ204326, GQ204330, HM192913 and HM192914. The highest nucleotide and haplotype diversities were observed in Linggi (π = 0.007, h = 0.835) and the lowest in Timah Tasoh (π = 0.0008, h = 0.286). Linggi presented 13 polymorphic sites, Jeniang and Kajang 7 each, and Timah Tasoh only one (Table 1).
Table 1.
Summary of variable sites and the distribution of haplotypes, nucleotide diversity, number of haplotypes and number of polymorphic sites among the 4 populations of Channa striata (n = sample size).
| Haplotypes | Variable sites
|
Timah Tasoh n = 7 | Jeniang n = 10 | Kajang n = 14 | Linggi n = 14 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coordinate | 6° 35′ N 100° 13′ E | 5° 49’ N 100° 38’ E | 2° 59’ N 101° 47’ E | 2° 28’ N 101° 03′ E | |||||||||||||||||||
| 5 | 6 | 21 | 66 | 109 | 117 | 123 | 186 | 188 | 219 | 243 | 250 | 258 | 261 | 297 | 330 | 333 | 353 | 358 | |||||
| Hap1 | T | G | G | G | T | T | T | T | A | A | T | T | G | A | A | T | C | T | C | 0.1 | 0.714 | 0.286 | |
| Hap2 | . | . | . | . | . | . | . | C | . | . | . | . | A | . | G | . | . | . | . | 0.8 | |||
| Hap3 | . | . | . | . | . | . | . | C | . | . | . | . | . | . | G | . | . | . | . | 0.857 | |||
| Hap4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | 0.286 | |||
| Hap5 | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | C | . | . | . | 0.214 | |||
| Hap6 | . | . | . | . | . | . | C | . | . | G | . | . | . | . | . | C | . | . | . | 0.142 | |||
| Hap7 | . | . | . | A | . | . | C | . | . | . | . | . | . | . | . | C | . | . | . | 0.143 | |||
| Hap8 | . | . | . | . | G | . | . | . | G | G | . | . | . | . | G | . | . | . | . | 0.1 | |||
| Hap9 | . | . | . | . | . | . | . | . | . | G | . | . | . | . | G | . | . | . | . | 0.071 | |||
| Hap10 | A | A | T | . | . | . | . | . | . | . | C | . | . | . | . | C | A | C | . | 0.071 | |||
| Hap11 | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | C | . | . | A | 0.071 | |||
| Hap12 | . | . | . | . | . | C | . | C | . | . | . | . | . | . | G | . | . | . | . | 0.143 | |||
| Nucleotide diversity | 0.0008 | 0.0047 | 0.0039 | 0.0067 | |||||||||||||||||||
| No. of haplotypes | 2 | 3 | 3 | 6 | |||||||||||||||||||
| Haplotype diversity | 0.286 | 0.378 | 0.483 | 0.835 | |||||||||||||||||||
| No. of oolymorphic sites | 1 | 7 | 7 | 13 | |||||||||||||||||||
The minimum spanning-network relationships among all the haplotypes are represented in Figure 1. Except for Timah Tasoh, Hap1, the most common haplotype, was found in the remaining three populations. From this common haplotype, two clades emerged, the northern and the southern. The spanning network showed that geographically close populations, i.e. Timah Tasoh and Jeniang (north) and Kajang and Linggi (south), were also genetically close.
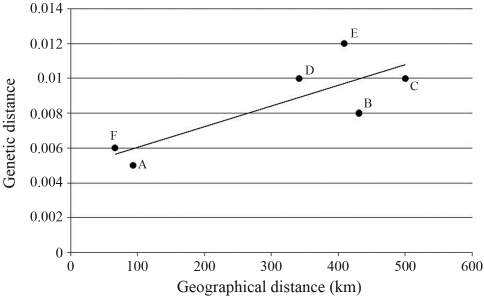
The highest inter-population genetic distance occurred between Linggi and Jeniang, at 0.011, and the lowest between Timah Tasoh and Jeniang, at 0.004 (Table 2). Genetic differentiation, as revealed by pairwise FST analysis, was insignificant between the Kajang and Linggi populations, and significant between the remaining two (Table 2). The scatterplot revealed a highly positive correlation of genetic isolation versus geographical distance (Figure 2) thereby fitting the ‘isolation by distance’ model (Sun et al., 2004). Nevertheless, caution is called for, due to the small sample sizes. Due to geological events, such as river formation (Tzeng et al., 2006), the distribution of freshwater fishes is usually restricted, thereby lowering the chances of free-gene-flow.
Table 2.
Pairwise FST values (below diagonal), and genetic distances (above diagonal) between the four populations of Channa striata.
| Population | Jeniang | Kajang | Timah Tasoh | Linggi |
|---|---|---|---|---|
| Jeniang | 0.010 | 0.004 | 0.011 | |
| Kajang | 0.000 (0.01) | 0.008 | 0.006 | |
| Timah Tasoh | 0.001 (0.03) | 0.000 (0.01) | 0.010 | |
| Linggi | 0 000 (0.01) | 0.005 (0.05) | 0.000 (0.02) |
Values in parentheses indicate significant FST value (p < 0.05).
Figure 2.
Relationships between the geographical (km) and the genetic distances. (A) Timah Tasoh and Jeniang; (B) Timah Tasoh and Kajang; (C) Timah Tasoh and Linggi; (D) Jeniang and Kajang; (E) Jeniang and Linggi; (F) Kajang and Linggi.
Thus, fairly low intra-population genetic diversity (π = 0.0008–0.0047, h = 0.286–0.483) was the case, except in the Linggi population (π = 0.0067, h = 0.835). Esa et al. (2008), also through CO1-gene analysis, reported even lower variability indices (π = 0–0.001 and h = 0.186–0.450) in an endangered fish, Tor tambroides, from Sarawak (Malaysian Borneo). Furthermore, although low (0.4%–1.1%), the inter-population distances in C. striata were higher (0.1%–0.3%) than in Tor tambroides. Thus, the indications of prevailing genetic variability advocates initiating management procedures. Nonetheless, as a consequence of marker characteristics, an underestimation of the true levels of genetic variability is possible. Several recent studies employing the bar-coding approach have shown that the CO1 gene is a more useful marker for unambiguous-species identification, due to its genetically conserved within-species characteristics (Dasmahapatra and Mallet, 2006; Lara et al., 2009; Valdez-Moreno et al., 2009). Notwithstanding, other markers for population studies, such as microsatellites, or more variable mtDNA markers, such as the D-loop, should also be investigated.
The absence of the common haplotype Hap1 in the Timah Tasoh population is the evidence of its isolation. The presence of a dam could be a likely factor, through its construction possibly changing the original riverine environment into a lacustrine. Notwithstanding, although Hap1 is missing, the main Timah Tasoh haplotype (Hap3) only differed from it by two bases. Thus, the most probable scenario for this population would be a shared origin which evolved independently, due to its isolation. Wang et al. (2004) described the common haplotype as having immense potentiality for producing mutational derivatives. Through being more recently derived, the other haplotypes were unique.
Based on this preliminary analysis, it can be inferred that the genetic structuring of the Peninsular Malaysia C. striata population was largely shaped by a common origin, with secondary influences from geographical factors and isolation. Further studies, with more efficient markers and larger populations, especially from the northern and southern regions, are required to corroborate the findings.
Acknowledgments
We would like to thank all the colleagues involved in sample collection and laboratory work. This study was funded by a Universiti Sains Malaysia Research University Grant (1001/PBiologi/8150123) received by the corresponding author, and a Universiti Sains Malaysia Fellowship by the first.
Footnotes
Associate Editor: Louis Bernard Klaczko
References
- Alessandrini F, Mazzanti M, Onofri V, Turchi C, Tagliabracci A. MtDNA analysis for genetic identification of forensically important insects. Forensic Sci Int Genet Suppl Ser. 2008;1:584–585. [Google Scholar]
- Ali A. Aspects of the reproductive biology of female snakehead (Channa striata Bloch) obtained from irrigated rice agroecosystem, Malaysia. Hydrobiologia. 1999;411:71–77. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra KK, Mallet J. DNA barcodes: Recent successes and future prospects. Heredity. 2006;97:254–255. doi: 10.1038/sj.hdy.6800858. [DOI] [PubMed] [Google Scholar]
- Erpenbeck D, Hooper JNA, Worheide G. CO1 phylogenies in diploblasts and the ‘Barcoding of Life’-are we sequencing a suboptimal partition? Mol Ecol Notes. 2005;6:550–553. [Google Scholar]
- Esa Y, Siraj SS, Daud SK, Rahim KAA, Rovie J, Japning R, Guan SG. Mitochondrial DNA diversity of Tor tambroides Valenciennes (Cyprinidae) from five natural populations in Malaysia. Zool Stud. 2008;47:360–367. [Google Scholar]
- Hossain MK, Latifa GA, Rahman MM. Observations on induced breeding of snakehead murrel, Channa striatus (Bloch, 1793) Int J Sustain Crop Prod. 2008;3:65–68. [Google Scholar]
- Jianguang XH, Fast AW. A bioenergetics model for an air-breathing fish, Channa striatus. Environ Biol Fishes. 1997;50:309–318. [Google Scholar]
- Lara A, Ponce De Leo NJL, Rodríguez R, Casane D, Côté G, Bernatchez L, García-Machado E. DNA bar-coding of Cuban freshwater fishes: Evidence for cryptic species and taxonomic conflicts. Mol Ecol Resour. 2009;10:421–430. doi: 10.1111/j.1755-0998.2009.02785.x. [DOI] [PubMed] [Google Scholar]
- Mat Jais AM. Molecular size of the bio-active component from haruan Channa striatus extract. J Appl Sci. 2007;7:2198–2199. [Google Scholar]
- Ng YTM, Shanthi G, Loqman MY. Effect of orally administered Channa striatus extract against experimentally-induced osteoarthritis in rabbits. Intern J Appl Res Vet Med. 2004;2:171–175. [Google Scholar]
- Qiongying T, Huanzhang L, Mayden R, Bangxi X. Comparison of evolutionary rates in the mitochondrial DNA cytochrome b gene and control region and their implications for phylogeny of the Cobitoidea (Teleostei, Cypriniformes) Mol Phylogenet Evol. 2006;39:347–357. doi: 10.1016/j.ympev.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Rock J, Costa FO, Walker DI, North AW, Hutchinson WF, Carvalho GR. DNA barcodes of fish of the Scotia Sea, Antarctica, indicate priority groups for taxonomic and systematics. Antarct Sci. 2008;20:253–262. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin v. 2000: A software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva; Switzerland: 2000. [Google Scholar]
- Seifert KA, Samson RM, deWaard JR, Houbraken J, Lévesque CA, Moncalvo GLS, Hebert PDN. Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proc Natl Acad Sci USA. 2007;104:3901–3906. doi: 10.1073/pnas.0611691104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Poyarkov JNA, Hebert PDN. CO1 DNA barcoding amphibians: Take the chance, meet the challenge. Mol Ecol Resour. 2008;8:235–246. doi: 10.1111/j.1471-8286.2007.01964.x. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu S, Zhao G, He S, Wu Q, Taniguchi N, Yu Q. Genetic structure of Chinese sucker population Myxocyprinus asiaticus in Yangtze River based on mitochondrial DNA marker. Fish Sc. 2004;70:412–420. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software v. 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Telechea F, Laudet V, Hanni C. Phylogeny of the Gadidae (sensu Svetovidov, 1948) based on their morphology and two mitochondrial genes. Mol Phylogenet Evol. 2006;38:189–199. doi: 10.1016/j.ympev.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Tzeng CS, Lin YS, Lin SM, Wang TY, Feng FY. The phylogeography and population demographics of selected freshwater fishes in Taiwan. Zool Stud. 2006;45:285–297. [Google Scholar]
- Wang JP, Lin HD, Huang S, Pan CH, Chen XL, Chiang TY. Phylogeography of Varocorhinus barbatulus (Cyprinidae) in Taiwan based on nucleotide variation of mtDNA and allozymes. Mol Phylogenet Evol. 2004;31:1143–1156. doi: 10.1016/j.ympev.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Valdez-Moreno M, Ivanova NV, Elías -Gutierez M, Contreras-Balderas S, Hebert PDN. Probing diversity in freshwater fishes from Mexico and Guatemala with DNA barcodes. J Fish Biol. 2009;74:377–402. doi: 10.1111/j.1095-8649.2008.02077.x. [DOI] [PubMed] [Google Scholar]
- Valles-Jimenez R, Cruz P, Perez-Enriquez R. Population genetic structure of Pacific White Shrimp (Litopenaeus vannamei) from Mexico to Panama: Microsatellite DNA variation. Mar Biotechnol. 2004;6:475–484. doi: 10.1007/s10126-004-3138-6. [DOI] [PubMed] [Google Scholar]
- Yu LL, Xiao YK, Zi NY, Jie K, Shen M, Li MC. Genetic diversity and historical demography of Chinese shrimp Feneropenaeus chinensis in Yellow Sea and Bohai Sea based on mitochondrial DNA Analysis. Afr J Biotechnol. 2009;8:1193–1202. [Google Scholar]
- Zakaria ZA, Somchit MN, Sulaiman MR, Mat Jais AM. Preliminary investigation properties of Haruan (Channa striatus) fillet extracted with various solvent systems. Pakistan J Biol Sc. 2004;7:1706–1710. [Google Scholar]