Abstract
The yeast HPR1 gene plays an important role in genome stability, as indicated by the observation that hpr1 mutants have high frequencies of DNA repeat recombination and chromosome loss. Here we report that HPR1 is required for transcriptional elongation. Transcription driven from constitutive and regulated yeast promoters cannot elongate through the bacterial lacZ coding region in hpr1Δ cells, but progresses efficiently through other sequences such as yeast PHO5. We show that HPR1 is not required for transcription activation and that the previously reported effects of hpr1Δ on the activation of different promoters is a consequence of the incapacity of hpr1Δ cells to elongate transcription through lacZ, used as reporter. Transcriptional defects are also observed in yeast DNA sequences of hpr1Δ cells in the presence of the transcription elongation inhibitor 6-azauracil. In all cases, the blockage of transcription elongation in hpr1Δ is associated with both the high frequency of deletions and the increase in plasmid instability that we report here. Therefore, in addition to the identification of a new element involved in transcriptional elongation, our work provides evidence for a new source of genomic instability.
Keywords: HPR1, transcription elongation, transcription-induced recombination, genomic instability, DNA repeats, lacZ reporter
Genetic recombination is required for DNA repair in mitosis, for the generation of genetic diversity and for proper reductional division in meiosis. In addition, it is responsible for chromosomal aberrations associated with some types of cancer and hereditary diseases, many of which occur by recombination between DNA repeats. The identification of the genes and functions that control initiation of recombination between repeats is therefore essential to understand the origin of chromosome aberrations and genome instability.
One important factor affecting the frequency of initiation of recombination is transcriptional activity. Recombination is stimulated by transcription as first shown by the identification of HOT1, a sequence from the rDNA region of Saccharomyces cerevisiae required for RNA polymerase I (Pol I)-dependent transcription (Keil and Roeder 1984; Voelkel-Meiman et al. 1987). Deletions between direct repeats have also been shown to be stimulated by RNA Pol II-mediated transcription in yeast (Thomas and Rothstein 1989). The relevance of transcription in homologous recombination has also been reported in mammalian cells (Nickoloff 1992; Thyagarajam et al. 1995). Probably the most significant connection between transcription and recombination is the one observed in immunoglobulin gene rearrangements (Blackwell et al. 1986; Lauster et al. 1993). Other examples of transcription-stimulated recombination have been provided in S. cerevisiae (Klar et al. 1981; Nevo-Caspi and Kupiec 1994), Schizosaccharomyces pombe (Grimm et al. 1991) and phages and bacteria (Dul and Drexler 1988; Vilette et al. 1992).
Transcriptional activity may induce recombination because unwinding of the DNA duplex, changes in the local supercoiling or disruption of chromatin structure associated with transcription could provide a better accessibility of recombination proteins to the transcribed DNA, could lead to DNA structures susceptible to DNA breaks, whether or not mediated by nucleases, or could facilitate the pairing reaction (McCormack and Thompson 1990; Dröge 1993; Kotani and Kmiec 1994). Other explanations cannot be excluded, however, such as a role of the transcription machinery in recruiting recombination proteins to transcribed genes that could be responsible for an increase in recombination, as is the case of transcription-coupled nucleotide excision repair (for review, see Basthia et al. 1996; Hoeijmakers et al. 1996). In this sense, the recent identification of the Rad51p recombinational repair protein in the human RNA Pol II holoenzyme is noteworthy (Maldonado et al. 1996). Despite our poor understanding of the molecular basis of transcription-induced recombination, it seems clear that transcription is an important potential source of spontaneous recombination between repeats leading to genomic instability and needs to be investigated.
One gene particularly relevant for the understanding of the putative connection between transcription and recombination as well as for the control of direct repeat recombination in yeast is HPR1. The importance of Hpr1p in genome stability is supported by the high frequencies of recombination and chromosome loss observed in null hpr1Δ mutants (Aguilera and Klein 1990; Santos-Rosa and Aguilera 1994). Hpr1p has been suggested to be required for transcription activation of regulated promoters (Fan and Klein 1994; Zhu et al. 1995). The identification of mutations in different transcription factors as suppressors of either the thermosensitivity (Fan et al. 1996; Uemura et al. 1996) or the hyper-recombination phenotype of hpr1Δ (Piruat and Aguilera 1996; Santos-Rosa et al. 1996) is consistent with a functional role of Hpr1p in transcription. Our recent observations that high rates of deletions in hpr1Δ cells only occurred between direct repeats in which defined regions of the intervening sequence were transcribed (Prado et al. 1997), suggest that Hpr1p might have a functional role in transcriptional elongation. To elucidate the function of Hpr1p on transcription, whether at the initiation or the elongation stage, and to understand the causes of induction of genomic instability by hpr1Δ, we undertook an in vivo molecular analysis that has allowed us to show that: (1) Hpr1p functions in transcriptional elongation, (2) previous results suggesting that Hpr1p is a general transcriptional activator (Fan and Klein 1994; Zhu et al. 1995) are explained by the incapacity of hpr1Δ cells to elongate transcription through the lacZ sequence used as reporter of gene expression and not by an incapacity of hpr1Δ cells to activate transcription at the initiation stage, and (3) the increased levels of direct repeat recombination and chromosome loss observed in hpr1Δ cells is a direct consequence of the blockage of transcription elongation. Our work not only shows a functional role for Hpr1p in transcriptional elongation, but also suggests a new model for the initiation of genomic instability.
Results
HPR1 is required for lacZ transcription
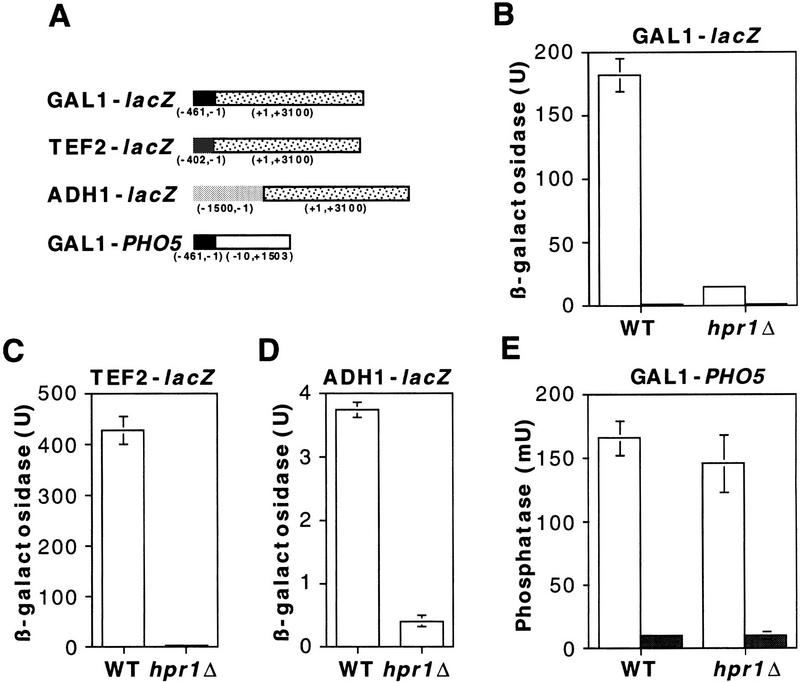
Expression of lacZ under the control of the GAL1 promoter strongly depends on HPR1 (Fan and Klein 1994; Zhu et al. 1995) (Fig. 1B). To assess whether or not lacZ could be expressed in hpr1Δ cells, the lacZ coding region was placed under the control of two different constitutive promoters, TEF2 and ADH1, the latter of which had been shown to promote transcription in the absence of Hpr1p (Zhu et al. 1995). In contrast to the expected result, lacZ was not expressed from either of the two promoters (Fig. 1C,D). The lack of β-galactosidase expression driven from the GAL1 promoter in hpr1Δ, therefore, may not be caused by a defect in transcriptional activation at the promoter. One possibility is that the lack of β-gal expression was caused by a defect in elongation through lacZ. To evaluate such a possibility, the yeast PHO5 gene was placed under the control of the GAL1 promoter. In this case, acid phosphatase was activated to wild-type levels in hpr1Δ cells (Fig. 1E). Therefore, hpr1Δ has no effect on activation of the GAL1 promoter. This fits with the observation that induction of the endogenous GAL1 gene was not affected by hpr1Δ (Fan and Klein 1994).
Figure 1.
Expression of lacZ fusion constructs placed in CEN plasmids in wild-type and hpr1Δ cells. The hpr1Δ mutation abolished transcription through the bacterial lacZ coding region, regardless of the yeast promoter from which it was transcribed, whereas it did not affect transcription through the yeast PHO5 coding region. (A) The lacZ coding region was fused to either the regulated GAL1 promoter or the constitutive TEF2 and ADH1 promoters. The PHO5 coding region was fused to the GAL1 promoter. Numbers below each construct refer to the translation start of each DNA sequence. Expression of lacZ and PHO5 was determined by β-galactosidase (B–D) or acid phosphatase (E) activities, respectively. For the GAL1-driven expression of either lacZ (B) or PHO5 (C), either 2% glucose (Glu, shaded bars, B,C,E) or 2% galactose (Gal, open bars, B,C,E) was added to 16-hr mid-log phase cultures in glycerol–lactate synthetic medium, and enzymatic activities were assayed 8 hr later.
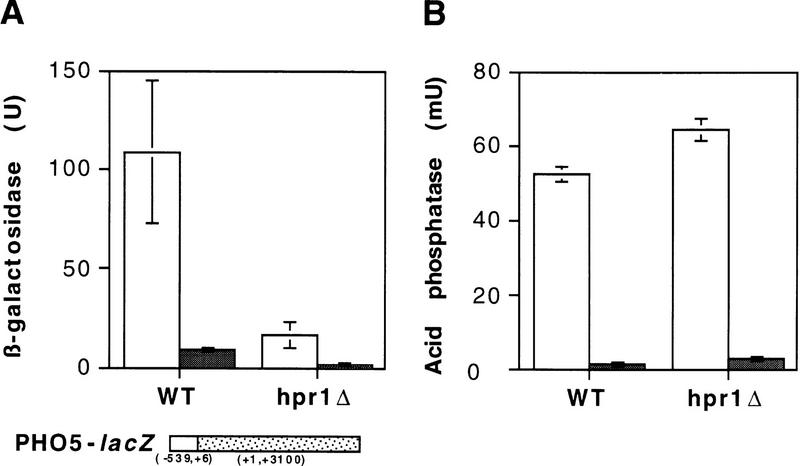
The previous work was performed on fusion constructs located in centromeric plasmids. Nevertheless, we have confirmed the same results in fusion constructs located in chromosomes. Thus, Figure 2 shows that whereas acid phosphatase mainly expressed by the endogenous PHO5 gene reached similar levels of activation in both wild-type and hpr1Δ cells, expression of lacZ under the same PHO5 promoter was seriously impaired in hpr1Δ cells.
Figure 2.
Expression of the chromosomally located lacZ (A) and PHO5 (B) coding sequences under the control of the PHO5 promoter in wild-type and hpr1Δ cells. For repression (+Pi, shaded bars) and induction (−Pi, open bars) conditions see Materials and Methods. Acid phosphatase and β-galactosidase were assayed in the same culture for each sample. Other details as in Fig. 1.
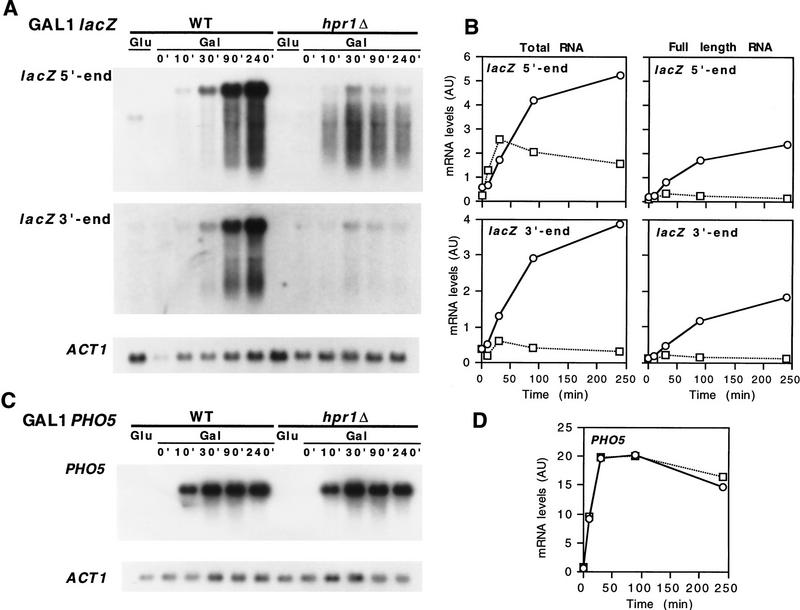
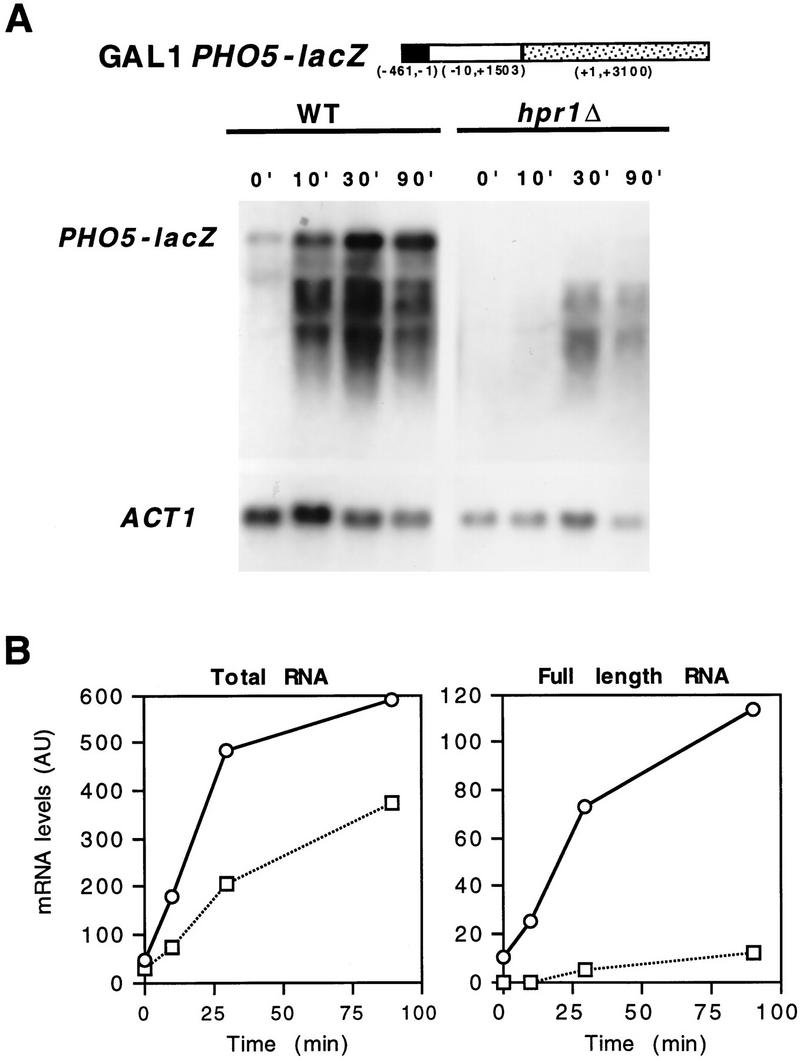
To confirm that the absence of β-galactosidase activity of hpr1Δ cells was caused by transcriptional, rather than post-transcriptional, defects of lacZ expression, we performed Northern analysis of lacZ under the GAL1 and PHO5 promoters. Figure 3 shows that lacZ mRNA driven from the GAL1 promoter was accumulated at high levels in wild type after induction, whereas it did not accumulate in hpr1Δ (Fig. 3A,B). The Northern analysis of hpr1Δ cells (Fig. 3A) shows a band corresponding to the full length lacZ mRNA, which appeared with both the 5′- and 3′-end probes of lacZ, and a smear which presumably contained incomplete 5′ lacZ mRNAs, as it appeared with the 5′-end but not with the 3′-end lacZ probe. At 30 min of induction, the total lacZ mRNA in hpr1Δ (mainly incomplete 5′-end mRNA) was produced in similar amounts as in wild-type (mainly full-length mRNA). This was consistent with the idea that the GAL1 promoter initiated transcription equally well in both wild-type and hpr1Δ, but transcription did not proceed further down from the 5′-end region of lacZ in hpr1Δ. The decrease of total lacZ mRNA observed after 30 min could be explained by the low stability of such incomplete transcripts (for review, see Ross 1995) and an eventual reduction in the efficiency of reinitiation of transcription from the GAL1 promoter as a consequence of a block of transcription in lacZ. Instead, PHO5 mRNA was accumulated in both wild-type and hpr1Δ, and only full-length mRNA was observed (Fig. 3C,D).
Figure 3.
Northern analysis of GAL1–lacZ and GAL1–PHO5. The Northern analysis and kinetics of induction of lacZ (A,B) and PHO5 (C,D) mRNAs driven from the GAL1 promoter is shown. Wild-type (○, B,D) and hpr1Δ (□, B,D) transformants were obtained from overnight cultures in glycerol–lactate synthetic media lacking uracil and diluted in identical fresh media to an OD600 of 0.5. Galactose (Gal) was then added and samples were taken for Northern analysis after different times, as specified. For repression conditions (Glu) total RNA was isolated from mid-log phase cultures in 2% glucose synthetic media lacking uracil. The DNA probes used were (lacZ 5′ end) the 0.5-kb BamHI–HpaI fragment of pLGZ containing the 5′ end of lacZ; (lacZ 3′ end) the 0.4-kb PvuII fragment of pLGZ containing the 5′- end of lacZ; (PHO5) the 1.5-kb EcoRI–PstI internal PHO5 fragment of pJDB207–PHO5 (Eco); (ACT1) the 0.6-kb ClaI internal ACT1 fragment of plasmid pYA301. (AU) arbitrary units.
Transcription is blocked at the lacZ 5′ end in hpr1Δ cells
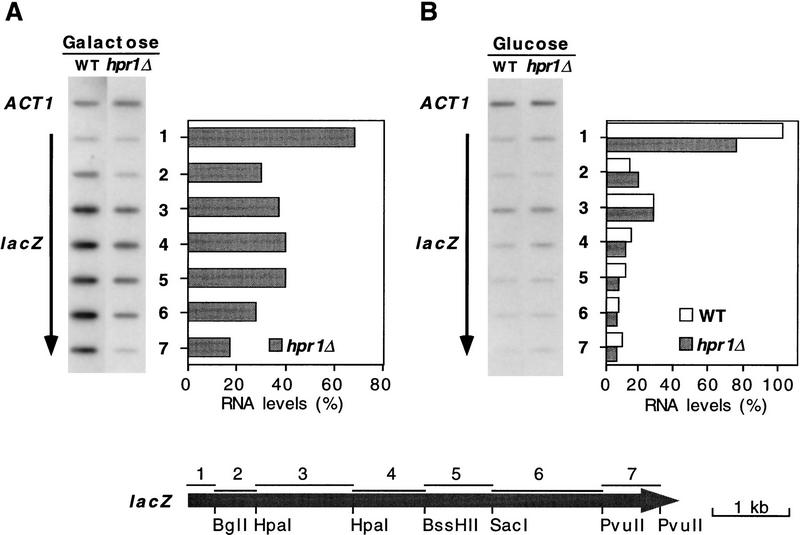
To confirm that transcription could be paused or blocked at the lacZ 5′-end region we performed transcriptional run-on analysis of GAL1–lacZ in permeabilized cells (Fig. 4). RNA Pol II was paused or blocked in the first 170 bp of the lacZ 5′ end, as deduced from the observation that under induction conditions (Gal) the amount of radiolabeled mRNA hybridizing with the first 170 bp of lacZ in hpr1Δ was ∼70% of the wild-type levels (Fig. 4A), but 17%–39% for the next six downstream fragments. As expected, the levels of radiolabeled lacZ mRNA bound to each DNA fragment under repression conditions (Glu) was significantly lower for all DNA fragments. The only exception was the 5′-end 170-bp fragment (Fig. 4B), which bound to similar amounts of radiolabeled mRNA under repression and induction conditions in both wild-type (101%) and hpr1Δ cells (74%). This implies that the polymerase is normally paused at the 5′ end of lacZ under repression conditions as recently reported (Akhtar et al. 1996), and that the initiation of transcription is equally efficient in both wild-type and hpr1Δ cells. According to our results, Hpr1p would be required to allow the RNA Pol II to travel farther down from the lacZ 5′-end region under activation conditions, but not for the establishment of the RNA Pol II at the 5′ end, as is also the case for Kin28p, Srb2p, or the carboxy-terminal domain (CTD) of RNA Pol II (Akhtar et al. 1996).
Figure 4.
Transcriptional run-on analysis in wild-type and hpr1Δ cells. Total RNA was isolated from wild-type and hpr1Δ cells transformed with single-copy p416GAL1–lacZ plasmid under induction (A) and repression (B) conditions. Two percent-galactose or 2% glucose was added to yeast cultures in glycerol–lactate synthetic medium at an OD600 of 0.05, 5 hr prior to the run-on analysis. The 0.6-kb internal ACT1 fragment and seven different DNA fragments (1–7) from the lacZ coding region were immobilized in hybond-N+ filters. The lacZ region covering each of the seven DNA fragments used is shown at the bottom. In all cases, the percentage of radiolabeled mRNA bound to each lacZ fragment was normalized with respect to their corresponding levels in galactose-grown wild-type cells, taken as 100% for each. The orientation of lacZ arrows indicates the direction of transcription. As negative control, we used DNA from Salmonella typhimurium (not shown).
Natural regulatory blocks of transcription described in several eukaryotic genes (for review, see Eick et al. 1994; Bentley 1995) are near their 5′ ends and require transcriptional activators to be bypassed by RNA Pol II. In contrast, the lack of lacZ expression from ADH1 and TEF2 promoters (Fig. 1) suggests that the block of transcription at lacZ in hpr1Δ cells is promoter-independent. To confirm this, we decided to place the lacZ coding sequence in the UTR of the GAL1–PHO5 construct right after the PHO5 stop codon (see Materials and Methods). Thus, the RNA Pol II transcription machinery has to transcribe the complete 1.4-kb PHO5 coding sequence before entering the lacZ sequence. As can be seen in Figure 5, full PHO5–lacZ message is produced in wild-type cells but not in hpr1Δ cells. A smear corresponding to incomplete messages was observed in hpr1Δ cells. Such smear is also observed in wild-type cells, as in the GAL1–lacZ construct (see Fig. 3) suggesting that, indeed, elongation through lacZ is inefficient in yeast. A new run-on analysis of this GAL1–PHO5–lacZ fusion construct showed that elongation was mostly blocked at the 5′-end of the lacZ coding sequence in hpr1Δ cells: The RNA Pol II engaged at the 5′ end of lacZ was 37%–53% of the RNA Pol II engaged at the PHO5 coding sequence (data not shown). This result is consistent with the run-on analysis of the GAL1–lacZ construct (see Fig. 4) and confirms that the incapacity of hpr1Δ cells to make complete PHO5–lacZ mRNA (Fig. 5) and lacZ mRNA (Fig. 3) is caused by the incapacity to elongate transcription through lacZ, regardless of its position relative to the promoter from which it is transcribed. Therefore, HPR1 has a functional role in elongation, and not in initiation or activation of transcription.
Figure 5.
(A) Northern analysis of PHO5 mRNA. The DNA probe used for Northern analysis was the 1.5-kb EcoRI–PstI internal PHO5 fragment of pJDB207–PHO5 (Eco). Arbitrary units of mRNA were calculated according to the same standards for all experiments. (B) The mRNA values are given with respect to rRNA levels (see Materials and Methods). (○) Wild type; (□) hpr1Δ. Other details as in Fig. 3.
Transcriptional elongation blocks as a cause of genome instability
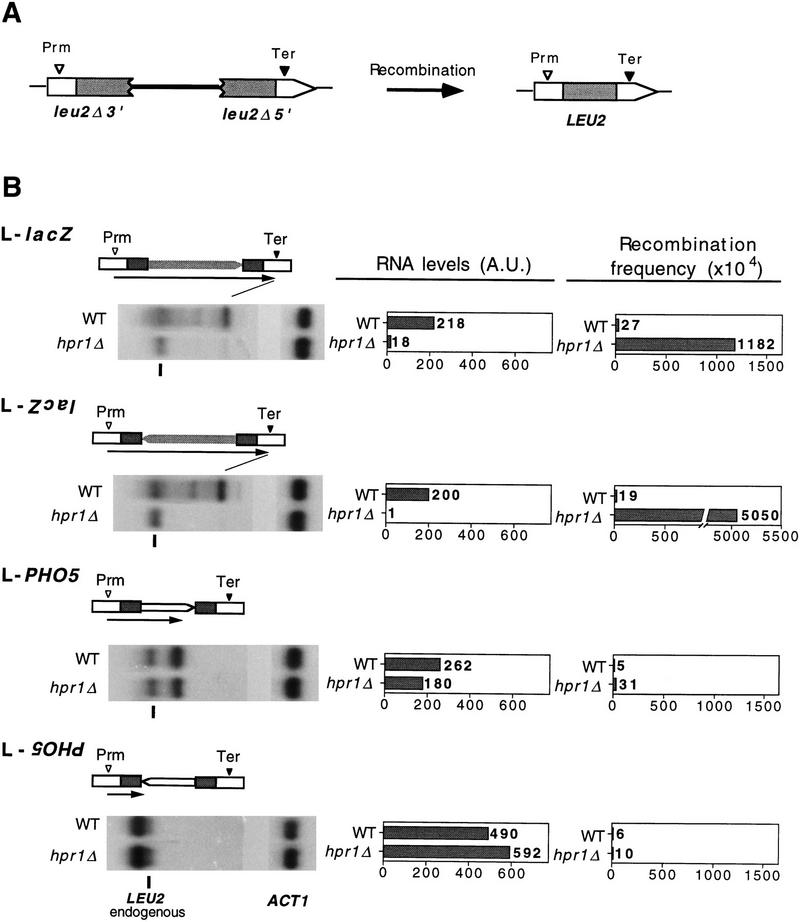
To confirm that the blockage of transcription elongation was responsible for the hyper-rec phenotype of hpr1Δ, we inserted the lacZ and PHO5 coding sequences between two 0.6-kb direct repeats, immediately downstream of a 3′-end truncated copy of LEU2 and immediately upstream of a 5′-truncated copy of LEU2. In these constructs, transcription of both lacZ and PHO5 was initiated at the unique LEU2 promoter, as determined by RNAse A protection (data not shown). Consequently, RNA Pol II must transcribe 760 bp of LEU2 before entering either the lacZ or the PHO5 coding sequences. According to our expectations, lacZ should cause a reduction in the level of full mRNA initiated at the LEU2 promoter and a strong hyper-recombination phenotype in hpr1Δ cells, which are both consequences of blockage or stalling of transcription elongation at lacZ (L–lacZ constructs), but none of these phenotypes should be caused by PHO5 (L–PHO5 constructs).
The results confirmed our predictions (Fig. 6). Transcription in L–lacZ constructs initiated at the LEU2 promoter upstream of the first repeat, traversed lacZ and terminated at the LEU2 terminator downstream of the second repeat in both wild type and hpr1Δ. Transcript levels in hpr1Δ were 12 times lower than in wild type if lacZ was transcribed in its natural direction, and 200 times if transcribed in the opposite direction, whereas recombination frequencies were increased 44- and 266-fold, respectively. In the L–PHO5 constructs, both the pattern and the level of transcripts were identical in wild type and hpr1Δ, and no significant difference was observed in recombination. In one orientation, transcription initiated at the LEU2 promoter, traversed PHO5 and terminated at the PHO5 terminator with similar efficiency in both wild-type and hpr1Δ cells (Fig. 6). A weak effect was observed on recombination (sixfold increase). In the opposite orientation, transcription terminated immediately downstream of the first LEU2 copy once it encountered the PHO5 transcription terminator, with similar efficiency in both wild-type and hpr1Δ. No effect was observed on recombination.
Figure 6.
Transcription and recombination analysis of direct repeat systems carrying lacZ (3 kb) or PHO5 (1.5 kb) coding regions. A scheme of the deletion product formed by recombination between the direct repeats used is shown (A). The diagram of each direct repeat system indicates the 0.6-kb repeated sequences (shaded boxes), the orientation of the lacZ and PHO5 coding regions, the LEU2 promoter (Prm) and transcription terminators (Ter), and the transcripts driven from the LEU2 promoter (arrow), whose 3′ ends have been made to coincide with the position of the corresponding band in each gel (B). Total RNA was isolated from overnight cultures in synthetic media lacking tryptophan. The DNA probes used in the hybridization experiments were the 598-bp ClaI–EcoRV LEU2 repeat, and the 581-bp ClaI internal ACT1 fragment. The transcript corresponding to the LEU2 endogenous chromosomal band is indicated. No transcript initiates in the internal lacZ and PHO5 internal sequences as determined with specific lacZ and PHO5 DNA probes (data not shown).
We have also observed that the blockage of transcription at lacZ is associated with an increase in plasmid loss. The replicative pRS416 plasmids containing either GAL1–lacZ (p416GAL1–lacZ) or GAL1–PHO5 (pSCh202) (see Figs. 1 and 2) had similar stability in hpr1Δ as in wild-type cells under repression conditions of transcription (55%–62% of the cells contained the plasmids after 23–25 generations in nonselective glucose-based media). Under induction conditions, however, p416GAL1–lacZ was clearly unstable in hpr1Δ cells. The proportion of hpr1Δ cells that contained p416GAL1–lacZ after 23–25 generations in nonselective galactose-based media was 37 times lower than that of wild-type cells, whereas only a threefold reduction was found for pSCh202 (data not shown).
Genome instability and transcription defects of hpr1Δ cells are observed in yeast DNA sequences in the presence of 6-azauracil
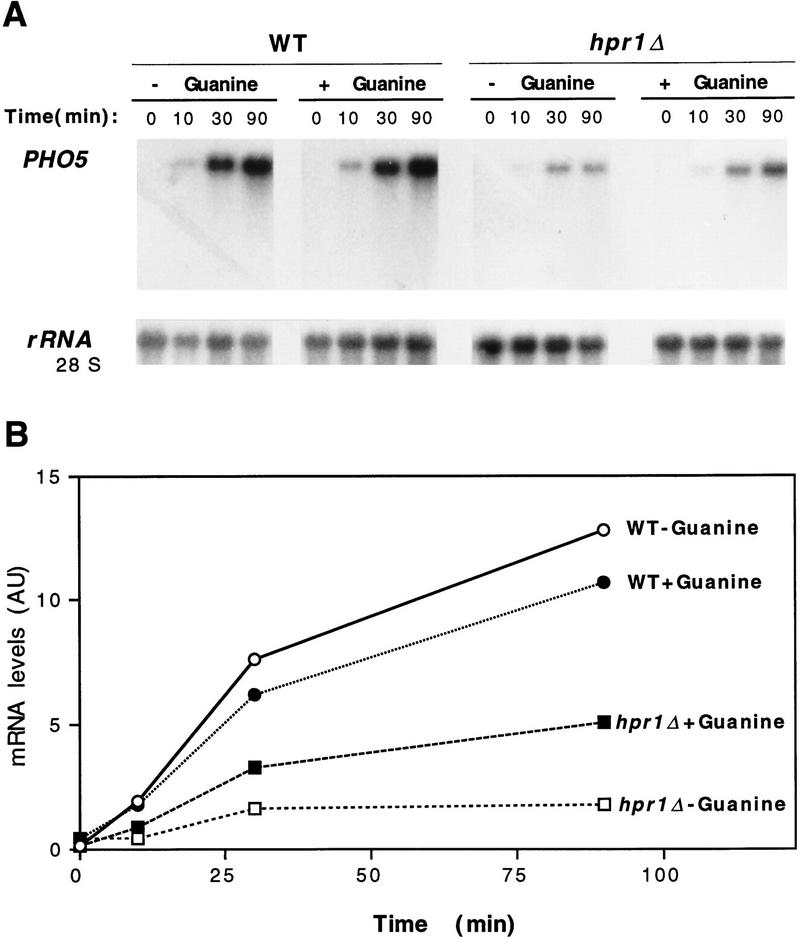
Because transcriptional elongation block, hyper-recombination, and plasmid loss were only associated with the bacterial lacZ sequence in this study, it was important to investigate the role of Hpr1p in transcription and genome instability of yeast DNA sequences. Despite HPR1 not being essential for viability, we considered the possibility that Hpr1p had a general role on transcription elongation of yeast genes, as it is the case of PPR2 encoding elongation factor TFIIS (for review, see Kane 1994). Because it was shown previously that 6-azauracil (6AU) produces a depletion of UTP and GTP responsible for a reduction of transcriptional elongation efficiency, and that the growth of mutants in PPR2 was sensitive to 6AU (Archambault et al. 1992; Exinger and Lacroute 1992), we decided to determine whether the transcription and genetic instability phenotypes conferred by hpr1Δ were also observed in yeast genes in the presence of 6AU. We tried three different concentrations of 6AU (100, 300, and 1000 μg/ml). Both wild-type and hpr1Δ cells, in contrast to transcriptional elongation TFIIS− mutants, were able to grow on SC media containing the three different concentrations, although hpr1Δ growth was more severely affected than wild-type growth (data not shown). When we analyzed transcription of the yeast PHO5 sequence under the GAL1 promoter, however, the kinetics of accumulation of PHO5 mRNA was severely affected in hpr1Δ in the presence of 6AU (Fig. 7). Thus, the level of activation of PHO5 mRNA at 90′ of induction in wild-type cells was sevenfold higher than in hpr1Δ cells. PHO5 transcript levels increased up to sixfold at 90′ of induction in wild-type cells, no increase was observed in hpr1Δ cells in the presence of 6AU. The ACT1 transcript levels were also twofold lower in hpr1Δ versus wild type in the presence of 6AU, indicating that 6AU specifically impairs transcription in hpr1Δ cells (data not shown). To confirm that the observed effect of 6AU on transcription in hpr1Δ cells was caused by impairment of transcription elongation and not by side-effects of 6AU, we determined whether guanine (100 μg/ml) could partially revert the effect of 6AU, as shown previously for mutants of TFIIS and not for 6AU-sensitive mutants unaffected in transcriptional elongation (Archambault et al. 1992). Figure 7 shows that accumulation of PHO5 mRNA was reestablished by guanine in the presence of 6AU.
Figure 7.
Transcription analysis of a yeast ORF in the presence of 6AU. Northern analysis (A) and kinetics of induction (B) of PHO5 mRNAs driven from the GAL1 promoter in the presence of 6AU with and without guanine are shown. The URA3+ wild-type and hpr1Δ strains transformed with pSCh202 were obtained from overnight cultures in glycerol–lactate synthetic-complete media lacking tryptophan and uracil, and diluted in identical fresh media to an OD600 of 0.5 with 100 μg/ml of 6AU with and without 100 μg/ml of guanine. Galactose was added after 2 hr, and samples were taken for Northern analysis after different times, as specified. The mRNA values are given with respect to rRNA levels (B) (see Materials and Methods). Other details as in Fig. 3.
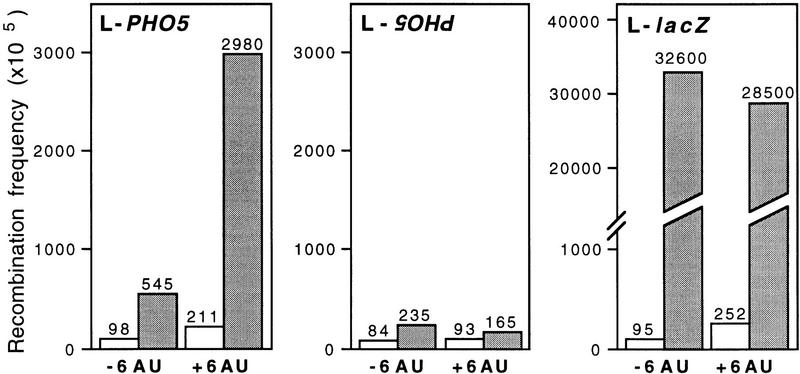
In addition, in the L–PHO5 construct containing the PHO5 ORF between the leu2 repeats in the orientation that is transcribed, recombination was enhanced 14-fold over the wild-type levels in the presence of 6AU, clearly above the increase observed without 6AU (5.5-fold) (Fig. 8). In the L–PHO5 construct with PHO5 in the orientation that is not transcribed, the presence of 6-AU had no effect on recombination (Fig. 8). This result clearly indicates that recombination is increased only when transcription is elongated through PHO5. Indeed, this increase in recombination correlated with a slight decrease of transcript levels in hpr1Δ cells in the presence of 6AU (data not shown). A similar effect was observed with the LA construct, containing the ADE2 gene between the leu2 repeats (data not shown), suggesting that the effect of 6AU on hpr1Δ cells may be associated with elongation through yeast DNA sequences. No increase of recombination in the L–lacZ construct was observed by the the addition of 6AU (Fig. 8), suggesting that transcription-induced recombination in this construct reaches its maximum levels without 6AU.
Figure 8.
Recombination in the presence of 6AU. Recombination analysis of the direct repeat constructs L–lacZ and L–PHO5 carrying the PHO5 ORF in both possible orientations between the leu2 repeats (see Fig. 6). Recombination frequencies were determined in the URA3+ wild-type (open bar) and hpr1Δ (shaded bar) strains transformed with the appropriate plasmids, grown in media with or without 100 μg/ml of 6AU.
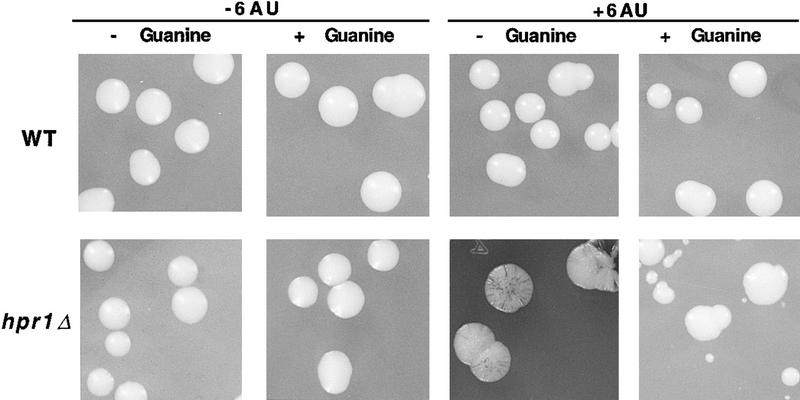
Finally, the use of centromeric plasmid pRS314-LA permitted us to determine directly by visual inspection the effect of hpr1Δ on genomic instability in the presence of 6AU. Figure 9 shows that the ADE2 gene was extremely unstable in hpr1Δ cells treated with 6AU, as shown by its red-sectoring phenotype caused by the Ade− segregants, in contrast to untreated hpr1Δ cells or wild-type cells, regardless of being treated with 6AU. The recombination frequency leading to Ade− recombinants in pRS314-LA is too low (<5 × 10−3) to be detected by this sectoring assay. Therefore, the observed red-sectoring phenotype is mostly caused by plasmid loss, as confirmed by genetic analysis. As expected, the phenotype of increased plasmid loss observed in hpr1Δ cells in the presence of 6AU was reverted by guanine (Fig. 9), consistent with being caused by transcription-elongation blockage.
Figure 9.
Plasmid instability in the presence of 6AU. Yeast colonies of URA3+ wild-type and hpr1Δ strains transformed with centromeric plasmid pRS314-LA growing on SC − Trp with or without 100 μg/ml of 6AU and with or without 100 μg/ml of guanine.
These results indicate that when transcription elongation is impaired, yeast DNA sequences, and not only bacterial sequencies such as lacZ, become genetically unstable in hpr1Δ cells.
Discussion
The main conclusion of this work is that HPR1, originally identified by the hyper-recombination phenotype conferred by hpr1 mutations, participates in transcriptional elongation. We show that the block of transcriptional elongation produced in hpr1Δ cells is responsible for the high frequency of deletions between repeats and the high frequency of plasmid-loss. Our work not only shows a role for Hpr1p in transcriptional elongation but also indicates that transcriptional elongation blocks may be an important source of genome instability and provides a molecular mechanism to explain transcription-induced recombination.
Hpr1p functions in transcriptional elongation
Our experiments indicate that Hpr1p stimulates transcriptional elongation through lacZ in vivo. Because previous reports suggesting that Hpr1p is required for transcription activation are based on the analysis of expression of a lacZ reporter fused to different yeast regulated promoters (Fan and Klein 1994; Zhu et al. 1995), we should reinterpret those results as being caused by the incapacity of hpr1Δ cells to elongate transcription through lacZ. This is independent of whether the promoters used are constitutive or regulated (Figs. 1 and 2) and whether the lacZ sequence is proximal or distal to the promoter from which it is transcribed (Figs. 3, 5, and 6). Because no effect on transcription was observed when the yeast PHO5 ORF was used as reporter (Figs. 1 and 2), we conclude that Hpr1p is not involved in transcriptional activation. Our results solve the paradox of why activation of endogenous yeast genes is not affected by hpr1Δ (Fan and Klein 1994; Zhu et al. 1995). When minor effects are detected, as is the case of SUC2 (Zhu et al. 1995), they are likely to be caused by a reduced efficiency in elongation. Certainly, our study raises serious concerns about using lacZ to study transcription in yeast and invalidates previous conclusions suggesting a role for Hpr1p in transcriptional activation of promoters (Zhu et al. 1995).
The strong effect of hpr1Δ on transcriptional elongation through lacZ is not observed in yeast genes such as PHO5 at 30°C (Fig. 3), even though a weak effect is detected at 37°C (S. Chávez and A. Aguilera, unpubl.). The effect of hpr1Δ on elongation of yeast genes may be masked by other redundant functions. Indeed, the observation that in the presence of the transcriptional elongation inhibitor 6-azauracil, PHO5 mRNA accumulation is strongly impaired in hpr1Δ mutants, a phenotype partially reverted by guanine, suggests that indeed Hpr1p has a general role in transcriptional elongation of yeast genes. Because hpr1Δ are viable in 6-azauracil, the role of Hpr1p in transcriptional elongation must be different from that of TFIIS, a factor required for the relief of an arrested RNA Pol II (for review, see Reines 1994). The function of Hpr1p cannot be related to TFIIS, because TFIIS is inhibited by Sarkosyl (Reines 1992), used in our run-on experiments. If that were the case, no difference between wild type and hpr1Δ should have been observed in the pattern of the pausing of RNA Pol II at lacZ (Fig. 4). Instead, Hpr1p might be required for preventing arrest of RNA pol II, as it has been proposed for TFIIF, elongin, or ELL (eleven-nineteen lysine-rich leukemia) (Price et al. 1989; Aso et al. 1995).
We do not know the nature of the signal on which RNA Pol II is blocked in the absence of Hpr1p. DNA bends or a particular chromatin structure of the bacterial lacZ sequence (for review, see Kane 1994; Bentley 1995) may determine the need for Hpr1p for its transcription, in particular at the upstream 170 nucleotides where the major hpr1-dependent transcriptional block maps (Fig. 4). In this sense, it is noteworthy that sin1(spt2) mutations, affected in a HMG1-like gene, suppress the transcriptional phenotype of hpr1Δ (Zhu et al. 1995) and that SIN1-2 and the imbalance of histones H2A/H2B or H3/H4 produced synthetic lethality in hpr1Δ (Fan and Klein 1994; Zhu et al. 1995). In addition, promoter-proximal pausing during transcriptional elongation in the human hsp70 gene depends on nucleosome templates in vitro (Brown et al. 1996). Other types of signals cannot be dismissed, however, such as those depending on RNA stems (Reeder and Hawley 1996). In any case, the transcriptional block caused by hpr1Δ at lacZ is different from RNA Pol II transcriptional pausing in other eukaryotic genes, because the latter is controlled by transactivators as an integral part of the initiation step (Yankulov et al. 1994), whereas the block at lacZ is independent of (1) its position relative to the promoter, whether proximal or distal (Figs. 5 and 6), and (2) the type of promoter to which it is fused, whether regulated or constitutive (Figs. 1 and 2). In this sense, it is particularly relevant the observation that the lacZ coding sequence abolishes production of full-length transcript when fused to the 3′ untranslated region (3′ UTR) of PHO5 (Fig. 5).
Finally, our results open the possibility that the synthetic lethality of hpr1 top1 mutants (Aguilera and Klein 1990) could be related to the role of Hpr1p on transcription elongation, because topoisomerase I is also needed in RNA Pol II transcription (Schultz et al. 1992). In this sense, it is important to note that mutations in different TOP genes also confer hyper-recombination of different DNA repeats in yeast (Christman et al. 1988; Wallis et al. 1989). An in vitro-reconstituted system would be required to assess whether the transcriptional blocks caused by hpr1Δ is caused by nucleosomes, DNA sequence, or an RNA secondary structure. In any case, our results show clearly that in vivo transcriptional elongation is blocked at lacZ in a promoter-independent manner and impaired at DNA sequences such as PHO5 in the presence of 6AU. We find no evidence for a role of Hpr1p on transcription activation of promoters.
Transcriptional elongation blocks lead to genome instability
Our work shows that hyper-recombination and plasmid instability in hpr1Δ cells is produced in association with a transcriptional elongation block. This explains our previous observation that hyper-recombination was dependent on transcription progression through particular DNA regions that include the bacterial tet and amp sequences (Prado et al. 1997). A strong increase in deletions between repeats and plasmid loss is observed in hpr1Δ cells only when transcription is elongated through the lacZ sequence (Fig. 6) or progresses through yeast DNA sequences (Fig. 7) in the presence of the transcriptional elongation inhibitor 6AU. In this sense, it is particularly relevant that in the absence of 6AU in hpr1Δ cells, a weak but significant increase in recombination was observed in the L–PHO5 construct (sixfold) when transcription elongated through PHO5, but no effect was observed when PHO5 was not transcribed. This strengthens our conclusion that Hpr1p participates in transcription elongation through yeast DNA sequences. Our results provide evidence that blockage of transcription elongation leads to genomic instability of DNA repeats and offers an alternative model to explain transcription-induced recombination to those proposed previously (Blackwell et al. 1986; Voelkel-Meiman et al. 1987; Stewart and Roeder 1989; Thomas and Rothstein 1989; Nickoloff 1992). Recombination in hpr1Δ is different from previously reported cases of transcription-induced recombination, in which an increase in recombination is associated with an increase in transcript levels.
To explain how a transcriptional block can induce both a deletion between repeats and the loss of a plasmid or chromosome, we propose that a stalled transcriptional elongation complex may induce DNA breaks or may cause an arrest of the replication fork, that would be either repaired or bypassed, respectively, by recombinational repair (see Fig. 10). Concerning the possibility that genomic instability could arise as a consequence of the arrest of the replication fork after colliding with the blocked RNA Pol II (Fig. 10), it has been shown recently that DNA replication forks transiently arrest at highly transcribed DNA regions in yeast (Deshpande et al. 1996), and it is known that mutations in DNA Pol I and Pol III lead to an increase in chromosome loss and recombination (Hartwell and Smith 1985; Aguilera and Klein 1988). If the replication fork collided with the blocked RNA Pol II, either the 3′ end of the nascent DNA could invade the other DNA-repeat region beyond the block, generating a deletion by one-ended invasion, or a cut could occur in the template leading to a double-strand break (Fig. 10e) that would be repaired by a deletion event through either single-strand annealing (Lin et al. 1984) or one-ended invasion (Prado and Aguilera 1995). The latter possibility would fit with the recent observation that replication arrests cause double-strand breaks in Escherichia coli (Michel et al. 1997), and could explain the high frequency of recombination induced by converging replication and transcription machineries (Vilette et al. 1992). Finally, if DNA breaks induced by transcriptional blocks were not repaired, they would cause plasmid or chromosome loss (Fig. 9; Santos-Rosa and Aguilera 1994).
Figure 10.
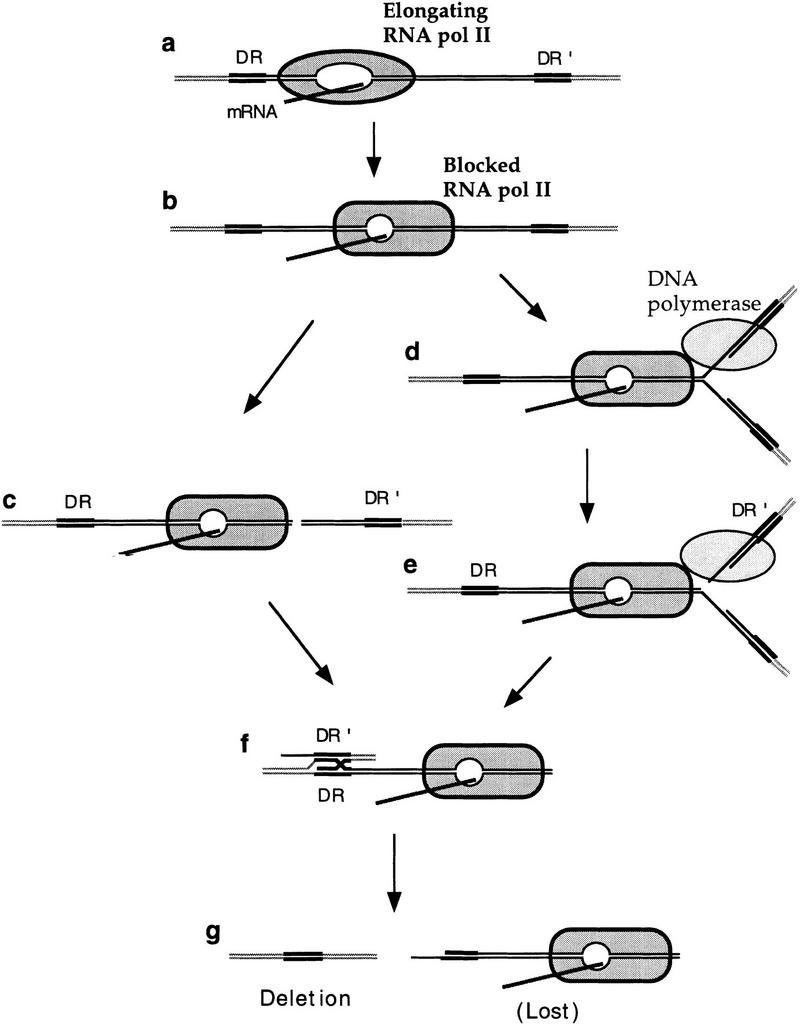
Alternative models to explain induction of genomic instability by a transcriptional elongation block. An elongating RNA Pol II (a) would be blocked at particular DNA sequences in the absence of Hpr1p in a region located between direct repeats DR and DR′ (b). The RNA Pol II–DNA complex may facilitate DNA breaks, whether or not mediated by a nuclease (c) or may impede progression of the replication fork (d). The collapsed replication fork could eventually facilitate the break of the template strand leading to a double strand break (e), although this step might not be necessary. From either step (c–e), and presumably after exonuclease digestion of one DNA strand, strand pairing between DR′ and DR (f) facilitated by either single-strand annealing, one-ended invasion, or DNA polymerase strand slippage would cause a deletion event and the loss of the intervening region (lost) (g), explaining the hyper-recombination phenotype of hpr1Δ. Otherwise the DNA molecule, either a plasmid or a chromosome, would be lost, explaining the high levels of chromosome and plasmid loss of hpr1Δ cells.
Although we do not have evidence that the transcription machinery itself could participate in the recruitment of recombination proteins, similar to what is believed to occur in transcription-coupled nucleotide excision repair (for review, see Basthia et al. 1996; Hoeijmakers et al. 1996), this is a possibility that cannot be dismissed. Thus, some transcription factors present in an elongating RNA Pol II might serve to recruit the recombination machinery that would repair the DNA breaks occurring as a consequence of a transcriptional block. In this sense, it is noteworthy that the identification of Rad51p in the human RNA Pol II holoenzyme (Maldonado et al. 1996) and the finding that Srb2p and Hrs1p, two RNA Pol II general transcription factors, are required for hpr1Δ-induced recombination (Piruat and Aguilera 1996; Piruat et al. 1997).
In summary, regardless of the mechanism leading to deletions in association to transcriptional elongation blocks, our work shows that failures in transcriptional elongation may be an important source of genome instability, as assessed by recombination between repeats and plasmid instability.
Materials and methods
Strains and plasmids
Strains used were the wild-type W303-1A (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) and its isogenic hpr1Δ mutant U678-4C. Isogenic URA3+ derivatives of W303-1A and U678-4C were obtained by transformation with the 0.9-kb PstI–SmaI URA3 fragment. All plasmids used were based on the pRS series of single-copy autonomously replicating vectors. Plasmids p416GAL1–lacZ, p426TEF–lacZ and p416ADH–lacZ contain the lacZ coding region fused to the GAL1, the TEF2, and the ADH1 promoters, respectively (Mumberg et al. 1994, 1995). Plasmid pSCh202, containing the GAL1–PHO5 fusion was constructed by cloning the 1.6-kb EcoRI–HindIII PHO5 coding region from pJDB207–PHO5 (Eco) (B. Meyhack, CIBA-GEIGY, Switzerland) immediately downstream of the GAL1 promoter in p416GAL1 (Mumberg et al. 1994). Plasmids pSCh204 and pSCh205 containing the L–lacZ constructs were made by inserting the 3-kb BamHI lacZ gene from pPZ (Straka and Hörz 1991), in both possible orientations, into the BglII site of pRS314LB, located between the two leu2 direct repeats (Prado and Aguilera 1995). Similarly, pSCh206 and pSC207 carrying the L–PHO5 constructs were made by inserting the 1.5-kb PstI PHO5 gene from pSCh202, in both possible orientations, into the PstI site of pRS314LB, also located between the two leu2 direct repeats. Plasmid p314LADE2 is pRS314LB with the ADE2 gene between the leu2 repeats (Prado and Aguilera 1997). Plasmid p306PHO5lacZ contains the lacZ gene fused to the PHO5 promoter in the integrative pRS306 vector (Piruat et al. 1997). This plasmid was integrated at the URA3 locus on chromosome V by transforming strains W303-1A and U678-4C after linearization with ApaI.
Plasmid pSCh212 contains the GAL1–PHO5–lacZ construct, which consists of the complete lacZ ORF transcriptionally fused to the 3′-end UTR of the PHO5 coding sequence placed under the GAL1 promoter. Construction of this plasmid was achieved by creating a BglII site right after the PHO5 stop codon by directed mutagenesis of plasmid pSCh202 with the 23-mer oligonucleotide TAGTTATACAGATCTATTGTCTC, being CTA the stop codon in the transcribed strand and G and T the 2 bases mutated from an original A each to create the BglII site. Directed mutagenesis was performed with the Bio-Rad kit according to Bio-Rad recommendations. In the newly created BglII site, we inserted a BglII–NheI–NruI–SphI synthetic polylinker and a lacZ 3.0-kb BamHI fragment in the orientation that is naturally transcribed.
Enzymatic assays
For the analysis of GAL1-driven transcription, yeast transformants with the appropriate pRS-derived plasmids were inoculated in selective synthetic medium lacking uracil with either 2% glucose or 2% galactose to an OD600 of 0.1. After 8 hr at 30°C either β-galactosidase or acid phospatase activity was assayed as described (Guarente 1983; Haguenauer-Tsapis and Hinnen 1984) in either permeabilized cells or whole cells, respectively. For the analysis of PHO5-driven transcription yeast cells from an SC + 2% glucose mid-log phase, cultures were resuspended in SC (−Pi) or SC supplemented with 7.5 mm Na2PO4 (+Pi) as described (Piruat et al. 1997)
Northern analysis
Three micrograms of total RNA prepared from exponential cultures in the appropriate selective medium were subject to electrophoresis on formaldehyde-agarose gels, transferred to Hybond-N filters, and UV crosslinked prior to hybridization at 42°C in 50% formamide 5× SSPE, 1× Denhardt’s solution, 1% SDS with the corresponding [32P]dCTP-labeled DNA probes (Prado et al. 1997). The filters were first hybridized with either the lacZ, PHO5, or LEU2 probe, and then rehybridized with the ACT1 probe, with previous removal of the former signals. Quantification of mRNA levels was performed in a PhosphorImager and are given in arbitrary units. All values were normalized with respect to either the amount of ACT1 mRNA or 28S rRNA detected. The rRNA was detected by hybridization with a 32P-oligolabeled 589-bp 28S rRNA internal fragment obtained by PCR by use of the 19-mer oligonucleotides TTGGAGAGGGCAACTTTGG and CAGGATCGGTCGATTGTGC.
Run-on analysis
Run-on analysis was performed according to previously described protocols (Elion and Warner 1986; Osborne and Guarente 1989) with the modifications of C. Birse (pers. comm.). One hundred micrograms of DNA denatured with NaOH from each lacZ fragment were immobilized on Hybond-N+ filters with a pR600 Slot Blot (Hoefer, USA). Two percent glucose or 2% galactose were added to yeast exponential cultures in glycerol-lactate synthetic medium lacking uracil at a OD600 of 0.05. After 5 hr, cells were washed in cold TMN buffer (10 mm Tris-HCl at pH 7.4, 5 mm MgCl2, 10 mm NaCl), resuspended in 0.9 ml H2O and incubated for 20 min with 0.5% N-lauryl-sarcosine. After centrifuging, the supernatant was carefully discarded and the run-on reaction was performed in 150 μl of 20 mm Tris-HCl at pH 7.7, 200 mm KCl, 32 mm MgCl2, 2 mm DTT, 0.5 mm ATP, CTP, GTP, and 120 μCi of [32P]UTP(>3000 Ci/mmole). The reaction was stopped with 1 ml of TMN containing unlabeled UTP. After washing with iced H2O, total RNA was extracted as described (Köhrer and Dombey 1991), resuspended in 40 μl of H2O, and frozen at −20°C. Before hybridization, the RNA was treated with 40 mm NaOH in ice, and neutralized with 40 mm HCl and 100 mm Tris-HCl at pH 7.5. Hybridization with the previously prepared Hybond-N+ filter containing the different lacZ DNA probes was performed as described (Prado et al. 1997) by use of E. coli tRNA at a final concentration of 100 μg/ml. Radiolabeled mRNA bound to each DNA fragment was quantified in a Fujix β-radiation Analyzer.
Frequency of recombination and plasmid stability
The frequency of Leu+ recombinants was calculated as described previously (Prado and Aguilera 1995). Plasmid stability was assayed by determining the median frequency of plasmid-containing cells (Ura+) from six independent colonies isolated from nonselective YEP-rich medium containing either glucose or galactose. It is important to note that plasmid-instability is only observed in nonselective media, and that all other experiments (Figs. 1, 2, 3, 4) were performed in selective media, in which the proportion of plasmid-containing cells was the same in wild-type and hpr1Δ cells (>95%). Transcriptional elongation inhibitor 6-azauracil was used at the concentration of 100 μg/ml.
Acknowledgments
We thank C. Birse for providing the run-on protocols and related technical tips, B. Meyhack and M. Funk for plasmids, J. Jordano, M. Beato, and F. Prado for critical reading of the manuscript, and E. Marsh and W. Reven for style correction. This work was supported by a grant from Direccion General de Ciencia y Tecnologica (DGICYT, Spain) to A.A. S.C. was an M. Curie postdoctoral fellow from the European Community Training and Mobility of Researchers (TMR) program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL aguilo@cica.es; FAX (34) 5-4557104.
References
- Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol Cell Biol. 1990;10:1439–1451. doi: 10.1128/mcb.10.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Faye G, Bentley DL. Distinct activated and non-activated RNA polymerase II complexes in yeast. EMBO J. 1996;15:4654–4664. [PMC free article] [PubMed] [Google Scholar]
- Archambault J, Lacroute F, Rue A, Friesen JD. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): A multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- Basthia PK, Wang Z, Friedberg EC. DNA repair and transcription. Curr Opin Genet Dev. 1996;6:146–150. doi: 10.1016/s0959-437x(96)80043-8. [DOI] [PubMed] [Google Scholar]
- Bentley DL. Regulation of transcriptional elongation by RNA polymerase II. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Moore MW, Yancopoulos GD, Suh H, Lutzker S, Selsing E, Alt FW. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986;324:585–589. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes & Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- Christman MF, Dietrich FS, Fink GR. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988;55:413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- Dröge P. Transcription-driven site-specific DNA recombination in vitro. Proc Natl Acad Sci. 1993;90:2759–2763. doi: 10.1073/pnas.90.7.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dul JL, Drexler H. Transcription stimulates recombination. I. Specialized transduction of Escherichia coli λtrp phages. Virology. 1988;162:466–470. doi: 10.1016/0042-6822(88)90488-6. [DOI] [PubMed] [Google Scholar]
- Eick D, Wedel A, Heumann H. From initiation to elongation: Comparison of transcription by prokaryotic and eukaryotic RNA polymerases. Trends Genet. 1994;10:292–296. doi: 10.1016/0168-9525(90)90013-v. [DOI] [PubMed] [Google Scholar]
- Elion EA, Warner JR. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- Fan H-Y, Klein HL. Characterization of mutations that suppress the temperature-sensitive growth of the hpr1Δ mutant of Saccharomyces cerevisiae. Genetics. 1994;137:1–12. doi: 10.1093/genetics/137.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H-Y, Cheng KK, Klein HL. Mutations in the RNA polymerase II transcription machinery suppress the hyper-recombination mutant hpr1Δ of Saccharomyces cerevisiae. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Schaer P, Munz P, Kohli J. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1991;11:289–298. doi: 10.1128/mcb.11.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Haguenauer-Tsapis R, Hinnen A. A deletion that includes the signal peptidase cleavage sites impairs processing, glycosylation and secretion of cell surface yeast acid phosphatase. Mol Cell Biol. 1984;4:2668–2675. doi: 10.1128/mcb.4.12.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985;110:381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JHJ, Egly J-M, Vermeulen W. TFIIH: A key component in multiple DNA transactions. Curr Opin Genet Dev. 1996;6:26–33. doi: 10.1016/s0959-437x(96)90006-4. [DOI] [PubMed] [Google Scholar]
- Kane CM. Transcript elongation and gene regulation in eukaryotes. In: Conaway RC, Conaway JW, editors. Transcription: Mechanisms and Regulation. New York, NY: Raven Press; 1994. pp. 279–296. [Google Scholar]
- Keil RL, Roeder GS. cis-Acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- Klar AJS, Strathern JN, Hicks JB. A position-effect control for gene transposition: State of the expression of yeast mating-type genes affect their ability to switch. Cell. 1981;25:517–524. doi: 10.1016/0092-8674(81)90070-2. [DOI] [PubMed] [Google Scholar]
- Köhrer K, Dombey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–401. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Kotani H, Kmiec EB. Transcription activates RecA-promoted homologous pairing of nucleosomal DNA. Mol Cell Biol. 1994;14:1949–1955. doi: 10.1128/mcb.14.3.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauster R, Reynaud C-A, Martenson I-L, Peter A, Bucchini D, Jami J, Weill J-C. Promoter, enhancer and silencer elements regulate rearrangement of an immuglobulin transgene. EMBO J. 1993;12:4615–4623. doi: 10.1002/j.1460-2075.1993.tb06150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Skerle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: Role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson CW, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- McCormack WT, Thompson CB. Chicken IgL variable region gene conversions display pseudogene donor preference and 5′ to 3′ polarity. Genes & Dev. 1990;4:548–558. doi: 10.1101/gad.4.4.548. [DOI] [PubMed] [Google Scholar]
- Michel B, Ehrlich SD, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nevo-Caspi Y, Kupiec M. Transcriptional induction of Ty recombination in yeast. Proc Natl Acad Sci. 1994;88:12711–12715. doi: 10.1073/pnas.91.26.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff JA. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol Cell Biol. 1992;12:5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne BI, Guarente L. Mutational analysis of a yeast transcriptional terminator. Proc Natl Acad Sci. 1989;86:4097–4101. doi: 10.1073/pnas.86.11.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piruat JI, Aguilera A. Mutations in the yeast SRB2 general transcription factor suppress hpr1-induced recombination and show defects in DNA repair. Genetics. 1996;143:1533–1542. doi: 10.1093/genetics/143.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piruat, J.I., S. Chávez and A. Aguilera. 1997. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics (in press). [DOI] [PMC free article] [PubMed]
- Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: Different requirements for the RAD1, RAD10 and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Piruat JI, Aguilera A. Recombination between DNA repeats in yeast hpr1Δ cells is linked to transcription elongation. EMBO J. 1997;16:2826–2835. doi: 10.1093/emboj/16.10.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH, Sluder AE, Greenlaf AL. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder TC, Hawley DK. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem. 1992;267:3795–3800. [PMC free article] [PubMed] [Google Scholar]
- ————— . Nascent RNA cleavage by transcription elongation complexes. In: Conaway RC, Conaway JW, editors. Transcription: Mechanisms and regulation. New York, NY: Raven Press; 1994. pp. 263–278. [Google Scholar]
- Ross J. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 1995;12:171–175. doi: 10.1016/0168-9525(96)10016-0. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Aguilera A. Increase in incidence of chromosome instability and non-conservative recombination between repeats in Saccharomyces cerevisiae hpr1Δ strains. Mol & Gen Genet. 1994;245:224–236. doi: 10.1007/BF00283271. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Clever B, Heyer W-D, Aguilera A. The yeast HRS1 gene encodes a polyglutamine-rich nuclear protein required for spontaneous and hpr1-induced deletions between direct repeats. Genetics. 1996;142:705–716. doi: 10.1093/genetics/142.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MC, Brill SJ, Ju QD, Sternglanz R, Reeder RH. Topoisomerases and yeast rRNA transcription: Negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes & Dev. 1992;6:1332–1341. doi: 10.1101/gad.6.7.1332. [DOI] [PubMed] [Google Scholar]
- Stewart SE, Roeder GS. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straka A, Hörz W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 1991;10:361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Johnson BL, Campbel C. The effect of target site transcription on gene targeting in human cells in vitro. Nucleic Acids Res. 1995;23:2784–2790. doi: 10.1093/nar/23.14.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura H, Pandit S, Jigami Y, Sternglanz R. Mutations in GCR3, a gene involved in the expression of glycolytic genes in Saccharomyces, suppress the temperature-sensitive growth of hpr1 mutants. Genetics. 1996;142:1095–1103. doi: 10.1093/genetics/142.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilette D, Uzest M, Ehrlich SD, Michel B. DNA transcription and repressor binding affect deletion formation in Escherichia coli plasmids. EMBO J. 1992;11:3629–3634. doi: 10.1002/j.1460-2075.1992.tb05447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Keil RL, Roeder GS. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Yankulov K, Blau J, Purton T, Roberts S, Bentley DL. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Peterson CL, Christman MF. HPR1 encodes a global positive regulator of transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1698–1708. doi: 10.1128/mcb.15.3.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]