Abstract
We have investigated the role of the RNA Polymerase II (Pol II) carboxy-terminal domain (CTD) in mRNA 5′ capping. Transcripts made in vivo by Pol II with a truncated CTD had a lower proportion of capped 5′ ends than those made by Pol II with a full-length CTD. In addition, the enzymes responsible for cap synthesis, RNA guanylyltransferase, and RNA (guanine-7)-methyltransferase bound directly to the phosphorylated, but not to the nonphosphorylated, form of the CTD in vitro. These results suggest that: (1) Pol II-specific capping of nascent transcripts in vivo is enhanced by recruitment of the capping enzymes to the CTD and (2) capping is co-ordinated with CTD phosphorylation.
Keywords: RNA Pol II, CTD, mRNA cap, RNA guanylyltransferase, RNA 7-methyltransferase
Transcripts made by RNA Polymerase II (Pol II) are distinguished from Pol I and Pol III transcripts by a m7G(5′)ppp(5′)N1 cap (Shatkin 1976) that is added cotranscriptionally (Salditt et al. 1980; Coppola et al. 1983). The cap appears to earmark an RNA molecule for processes that are specific to Pol II transcripts. It enhances splicing, 3′ processing, transport, and translation of mRNA (Konarska et al. 1984; Edery and Sonenberg 1985; Shatkin 1985; Gilmartin et al. 1988; Hamm and Mattaj 1990; Izaurralde et al. 1994; Colot et al. 1996; Cooke and Alwine 1996). The cap also stabilizes mRNA (Furuichi et al. 1977; Green et al. 1983); moreover, decapping is important for triggering degradation (Beelman and Parker 1995).
The capping reaction is catalyzed by three enzymes: (1) RNA triphosphatase, which removes the terminal phosphate; (2) RNA guanylyltransferase, which transfers GMP from GTP to the diphosphate end of RNA to form the GpppN cap; and (3) RNA (guanine-7)-methyltransferase, which adds a methyl group to the N7 position of the cap guanine (for review, see Shuman 1995). In multicellular organisms, a single bifunctional polypeptide functions as phosphatase and guanylyltransferase (Yagi et al. 1984; Takagi et al. 1997; Wang et al. 1997). In budding yeast, discrete triphosphatase and guanylyltransferase subunits copurify as a stable complex distinct from the monomeric methyltransferase (Itoh et al. 1987; Mao et al. 1995).
It is not known how capping enzymes are targeted to the 5′ ends of transcripts made by Pol II and not to transcripts made by other polymerases. One possible explanation for the Pol II specificity of capping is that some unique feature of the polymerase directs the capping enzymes to their substrate. A precedent for this model is provided by the vaccinia capping enzyme, which binds directly to its cognate viral RNA polymerase (Hagler and Shuman 1992). This interaction facilitates the capping of the 5′ end of the nascent mRNA chains as soon as it is extruded from the RNA-binding pocket of the polymerase.
Whereas all three cellular RNA polymerases share extensive homology, the large subunit of Pol II has a unique carboxy-terminal domain (CTD) composed of a tandemly repeated heptad motif whose consensus sequence is absolutely conserved between yeast and mammals (Allison et al. 1985; Corden et al. 1985). The CTD is required for the stimulation of transcription in response to certain enhancers and activators (Allison and Ingles 1989; Scafe et al. 1990; Gerber et al. 1995). It undergoes a cycle of extensive phosphorylation and dephosphorylation that accompanies the transcription cycle. Several kinases can phosphorylate the CTD, including cdk7, a component of the basal transcription factor TFIIH (Roy et al. 1994). Hyperphosphorylation is associated with the transition of the transcription complex to productive elongation (Dahmus 1996). CTD phosphorylation and capping have been elegantly studied in vivo on the Drosophila hsp70 gene. The polymerase pauses at promoter–proximal positions between +25 and +35. Resumption of elongation is correlated with CTD phosphorylation (O’Brien et al. 1994). Capping of hsp70 transcripts coincides closely with polymerase pausing (Rasmussen and Lis 1993).
The CTD binds to specific protein components of the mRNA splicing and cleavage/polyadenylation machinery (Mortillaro et al. 1996; Yuryev et al. 1996) and is required for splicing and polyadenylation in vivo (McCracken et al. 1997; Steinmetz 1997). These observations suggest that the CTD may act as a scaffold for a large “mRNA factory” complex that integrates RNA processing with transcription (Greenleaf 1993; McCracken et al. 1997). Previous experiments have not addressed whether capping is also integrated with transcription by a mechanism involving the CTD. Because the 5′ cap stimulates splicing and polyadenylation (Edery and Sonenberg 1985; Izaurralde et al. 1994; Cooke and Alwine 1996), it remains possible that a capping defect could contribute to reduced processing of transcripts made by Pol II with a truncated CTD. In this paper, we show that CTD truncation does, in fact, reduce mRNA capping in vivo, although this effect cannot fully account for the splicing and polyadenylation defects of transcripts made by the truncated Pol II. Our finding that RNA guanylyltransferase and RNA (guanine-7)-methyltransferase bind directly to the phosphorylated form of the CTD suggests a plausible mechanism for how the CTD targets capping to Pol II transcripts.
Results
GST–eIF4E binding assay for capped transcripts
To analyze 5′ capping of Pol II transcripts in vivo, we used a “pull-down” assay based on the selective binding of m7G-capped RNAs to the translation initiation factor eIF4E (Sonenberg et al. 1978; Edery et al. 1995; Fresco and Buratowski 1996). The specificity of the assay is demonstrated in Figure 1A. A mix of 3′-end labeled m7G-capped rabbit globin mRNA and an uncapped internally-labeled T7 transcript was fractionated by binding to GST–eIF4E, which was immobilized on glutathione beads. Capped globin mRNA was bound nearly quantitatively to the beads and recovered in the pellet fraction (P), whereas most of the uncapped T7 transcript was recovered in the supernatant (S) (Fig. 1A, lanes 6,7). Neither RNA was retained on the beads by GST alone (Fig. 1, lane 5). Similar results were obtained when the labeled globin and T7 transcripts were mixed with 10 μg of total cellular RNA (data not shown). To test the sensitivity of the assay, total RNA from cells transfected with a pGal5 HIV2 CAT reporter gene (see Fig. 1C) was diluted 10- and 100-fold with RNA from untransfected cells, then fractionated by GST–eIF4E binding (Fig. 1B). Reporter gene transcripts were detected by RNase protection with an antisense probe that spanned the start site (see Fig. 2C). The ratio of HIV2 CAT RNA in the pellet and supernatant fractions was not affected significantly by dilution with total cellular RNA (Fig. 1B). This experiment therefore shows that the partitioning of a particular RNA species by GST–eIF4E binding is unaffected by its relative abundance.
Figure 1.
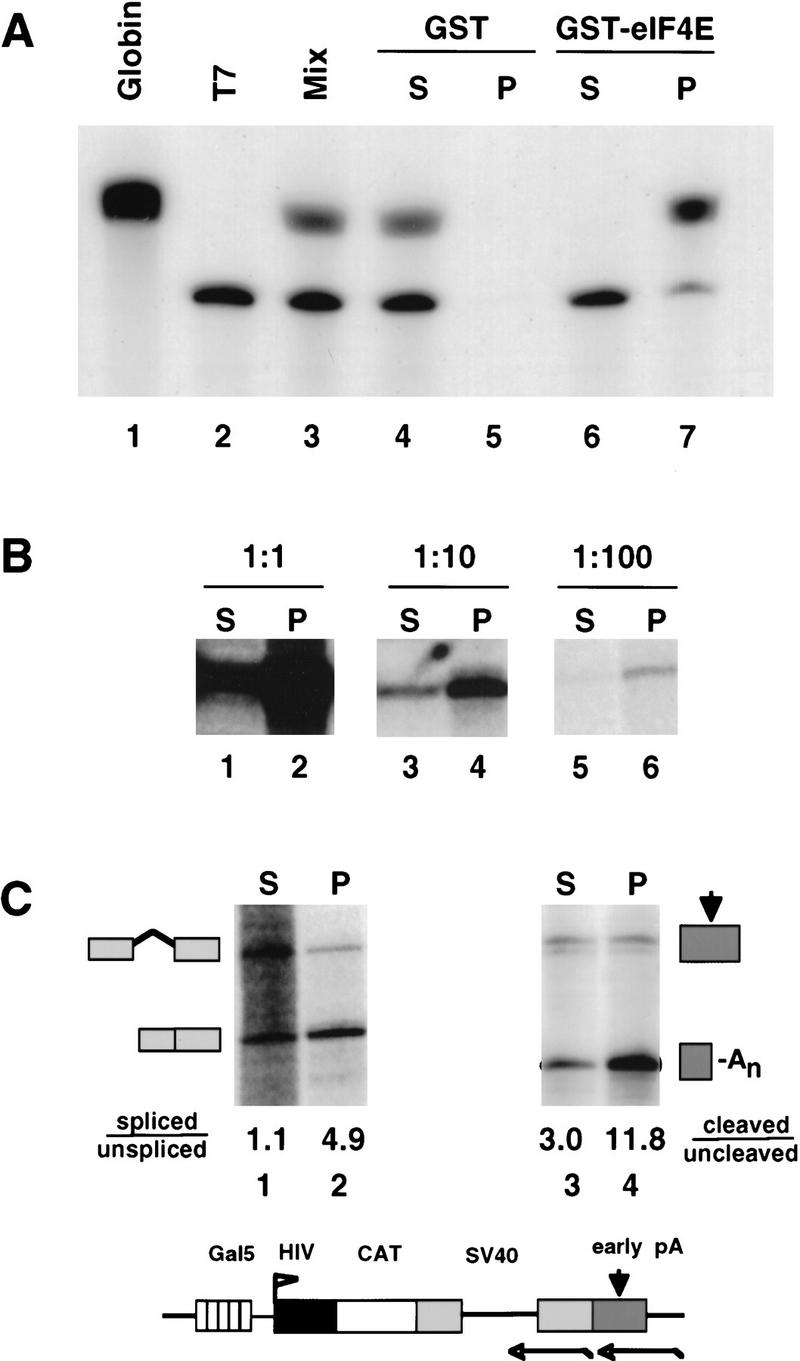
GST–eIF4E pull-down assay of capped RNAs. (A) A mixture (lane 3) of 3H-labeled rabbit globin mRNA (GIBCO-BRL) (capped, lane 1) and a synthetic T7 transcript (uncapped, lane 2) were fractionated using GST (lanes 4,5) or GST–eIF4E (lanes 6,7). Supernatant (S) and pellet (P) fractions were analyzed by electrophoresis and fluorography. (B) RNA from α-amanitin-treated 293 cells transfected with HA–WT α-amanitin-resistant Pol II, pGal5 HIV2 CAT reporter, and Gal4–VP16 activator was serially diluted with RNA from untransfected cells, then fractionated by GST–eIF4E pull-down. A total of 5 μg of RNA was used for each pull-down. Supernatant (S) and pellet (P) fractions were analyzed by RNase protection with a probe spanning the start site (Fig. 2C). The 160-base protection product corresponding to correct HIV2 5′ ends is shown. (C) RNA in the GST–eIF4E supernatant (S) fraction is spliced and polyadenylated less well than RNA in the pellet (P) fraction. GST–eIF4E fractionated RNA from α-amanitin-treated 293 cells expressing HA–WT Pol II and pGal5 HIV2 CAT activated by Gal4–VP16 was analyzed for splicing of the SV40 t intron (lanes 1,2) and cleavage at the SV40 early poly(A) site (lanes 3,4) using antisense RNA probes as indicated in the map. Processed/unprocessed ratios are given after correction for the 32P-labeled U content of the RNase protection products. A map and RNase protection strategy for the pGal5 HIV2 CAT reporter gene is shown.
Figure 2.
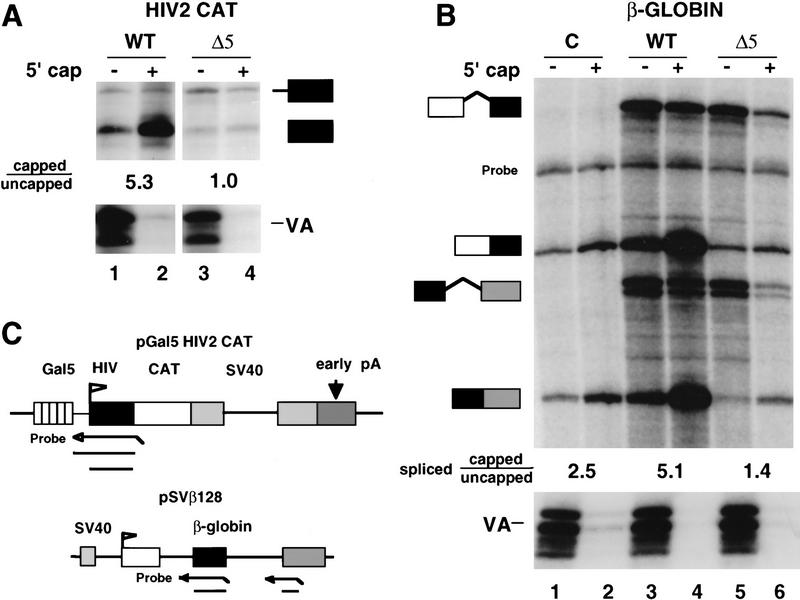
CTD truncation reduces the ratio of capped/uncapped 5′ ends. (A) Analysis of WT and Δ5 transcripts of pGal5 HIV2 CAT by RNase protection as diagrammed. RNA (20 μg) from α-amanitin-treated cells was separated into capped (+) and uncapped (−) fractions and analyzed by RNase protection with probes for the the HIV2 5′ end and VA1. Capped/uncapped ratios for correctly initiated RNAs (lower band) are marked. (B) Analysis of human β-globin RNA transcribed by wild-type (WT) and CTD-truncated (Δ5) α-amanitin-resistant Pol II. The pSVβ128 reporter and Pol II expression vectors or CMV–neo control (C) were transfected transiently into 293 cells. GST–eIF4E fractionated RNA (20 μg) was analyzed as in A by RNase protection with probes for VA control, β-globin intron 1, and intron 2 as diagrammed in C. A band corresponding to incompletely digested intron 2 probe is marked. The capped/uncapped ratios shown are average values for the two “spliced” RNase protected bands. The spliced/unspliced ratios for intron 1 after correcting for 32P-labeled U content are 1.6, 11.5, 0.4, and 2.1 for lanes 3–6, respectively. The spliced/unspliced ratios for intron 2 are 0.7, 5.8, 0.2, and 1.2 for lanes 3–6. (C) Maps and RNase protection strategies for the pGal5 HIV2 CAT and pSVβ128 β-globin reporter genes.
To validate the GST–eIF4E fractionation procedure, we took advantage of the fact that the 5′ cap enhances splicing in vivo (Izzauralde et al. 1994; Fresco and Buratowski 1996; Schwer and Shuman 1996). A genuine uncapped fraction of cellular RNA is therefore expected to contain transcripts that are spliced less well than those in a capped RNA fraction. To test this prediction, RNA from 293 cells transfected with the pGal5 HIV2 CAT reporter was fractionated by GST–eIF4E pull-down and analyzed for splicing of the SV40 t intron (Fig. 1C, see diagram and lanes 1,2). The spliced/unspliced ratio was 4.9 for transcripts bound by GST–eIF4E in the pellet (P), compared with only 1.1 for unbound transcripts in the supernatant (S). This result indicates that GST–eIF4E binding resolves two different populations of RNA—a bound fraction that is predominantly spliced and an unbound fraction that is not spliced as effectively. The difference in the extent of splicing of transcripts in the GST–eIF4E pellet and supernatant strongly suggests that these fractions are enriched in bona fide capped and uncapped RNAs, respectively.
We also observed that transcripts in the GST–eIF4E pellet fraction were cleaved at the poly(A) site more efficiently than those in the supernatant. For HIV2 CAT transcripts with an SV40 early poly(A) site, the cleaved/uncleaved ratio in the GST–eIF4E pellet was 13.8 compared with only 3.0 in the supernatant (Fig. 1C, lanes 3,4). A similar result was observed for CAT transcripts with an SV40 late poly(A) site (see Fig. 3, lanes 1,2). These observations are consistent with in vitro studies showing that the cap enhances 3′ processing (Gilmartin et al. 1988; Cooke and Alwine 1996). These experiments strongly imply that RNAs bound by GST–eIF4E have functional caps, whereas unbound RNAs lack cap structures capable of supporting efficient splicing and 3′ processing.
Figure 3.
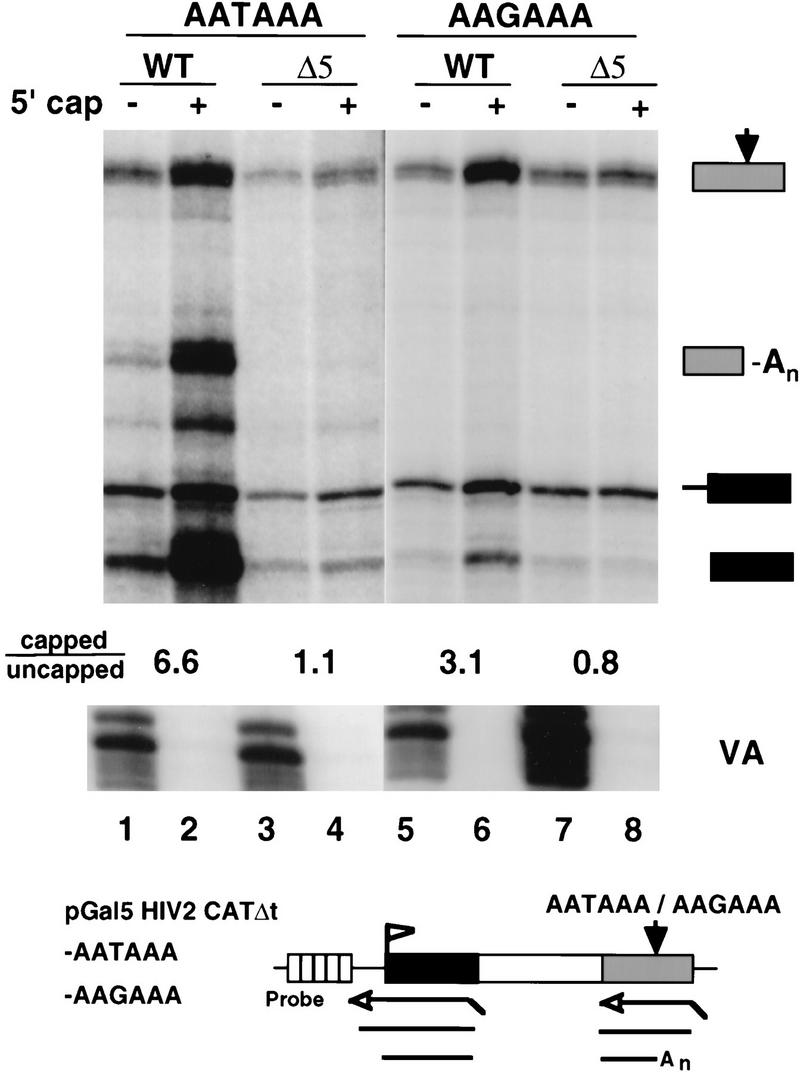
CTD truncation reduces capping of unspliced and unpolyadenylated RNAs. RNase protection of intronless CAT transcripts from pGal5 HIV2 CATΔt–AATAAA and pGal5 HIV2 CATΔt–AAGAAA transcribed by WT or Δ5 Pol II. Total RNA (6 μg) from transfected 293 cells was fractionated by GST–eIF4E binding and analyzed with 5′ and 3′ antisense probes shown in the diagram. The capped/uncapped ratios are shown for correctly initiated transcripts (marked with a solid box). The VA control is indicated. At the poly(A) site, the ratios of cleaved/uncleaved transcripts after correcting for 32P-labeled U content of the RNase protection products are 0.7, 2.1, <0.3, and <0.3 for lanes 1–4, respectively.
CTD truncation reduces the fraction of capped HIV2 CAT transcripts
The capping of transcripts made in vivo by RNA Pol II with the full-length CTD or a truncated CTD was compared using the GST–eIF4E binding assay (Fig. 2A). In this experiment, we transfected 293 cells with expression vectors for α-amanitin-resistant Pol II large subunits with the full-length CTD (HA–WT) or only five of the 52 heptad repeats (HA–Δ5) (Gerber et al. 1995). Treatment of the transfected cells with α-amanitin inhibits the endogenous Pol II and permits analysis of RNAs made by the drug-resistant Pol II. In addition to the HA–WT or HA–Δ5 vectors, the cells were cotransfected with the pGal5 HIV2 CAT reporter construct and expression vector for the chimeric Gal4–VP16 transactivator. The adenovirus VA1 gene, which directs the synthesis of uncapped Pol III transcripts, was cotransfected as a control. α-Amanitin was added 12 hr after transfection, and RNA was harvested after 60–65 hours, when most mRNAs in the steady-state population have been made by Pol II that has incorporated the drug-resistant subunit (McCracken et al. 1997). CTD truncation inhibits transcription in response to activators such as Gal4–VP16, therefore Δ5 transcripts are always less abundant than WT transcripts in transfected cells (Gerber et al. 1995). Because RNA abundance does not affect the capped/uncapped ratio determined by GST–eIF4E fractionation (Fig. 1B), we have not attempted to control further for the lower level of transcripts made by Δ5 Pol II.
RNA from α-amanitin treated cells was separated into capped (+) and uncapped (−) fractions and analyzed by RNase protection (Fig. 2A). Uncapped VA RNAs were recovered in the supernatant fractions, as expected. HIV CAT reporter transcripts were analyzed with an antisense probe that spanned the HIV2 TAR sequence and transcriptional start site, as diagrammed in Figure 2C. This probe yields a 160-base protection product (marked with a solid box in Fig. 2A), which corresponds to correctly initiated transcripts. Most HIV2 CAT transcripts made by RNA polymerase containing the full-length CTD (wild type) were bound by GST–eIF4E, showing that they were capped (Fig. 2A, lanes 1,2). Short HIV TAR trancripts 120–150 bases long (data not shown) were capped to an equivalent extent as longer transcripts. The salient observation was that a high proportion of correctly initiated HIV2 CAT transcripts produced by Pol II with a truncated CTD (Δ5) did not contain a m7G cap structure (Fig. 2A, lanes 3,4). The ratio of HIV2 CAT RNA in the capped (pellet) fraction to that in the uncapped (supernatant) fraction was 5.3 for WT but only 1.0 for Δ5 transcripts. Note that because uncapped RNA is less stable than capped RNA, the steady-state level of uncapped transcripts is a minimum estimate of the amount synthesized. This difference in capping of WT and Δ5 transcripts was also observed after a second round of GST–eIF4E fractionation (data not shown). Transcription by CTD-truncated polymerase results in a relative increase in the amount of a second, longer RNAse protection product (Fig. 2A, solid box with thick line extension) that includes transcripts that read around the plasmid as a result of defective polyadenylation and termination (McCracken et al. 1997). Capping of this subset of transcripts was also reduced by CTD truncation. The effect of Δ5 Pol II on RNA capping is not attributable to a change in the level of guanylyltransferase, which was equivalent in α-amanitin-treated WT and Δ5 expressing cells (see Materials and Methods; data not shown). In summary, these results show that truncation of the CTD reduces the proportion of HIV2 CAT transcripts with capped 5′ ends.
CTD truncation reduces the fraction of capped globin transcripts
We examined the effect of CTD truncation on capping of a different transfected reporter gene, human β-globin (Fig. 2B). The pSVβ128 globin reporter (see Fig. 2C) was transfected transiently into 293 cells with HA–WT or HA–Δ5 expression vectors for α-amanitin-resistant Pol II large subunit or with CMV–neo as a control. Total RNA from transfected cells was fractionated into capped and uncapped populations and analyzed by RNase protection with probes spanning the splice acceptor sites for β-globin introns 1 and 2 (as diagrammed in Fig. 2C). The ratio of capped/uncapped β-globin mRNAs was reduced significantly when the gene was transcribed by Pol II with the truncated CTD. The capped/uncapped ratios for transcripts spliced at the first and second introns were 4.4 and 5.7, respectively, (average 5.1) for WT Pol II (Fig. 2B, lanes 3,4) but only 1.5 and 1.3 (average 1.4) for Δ5 (Fig. 2B, lanes 5, 6). CTD truncation also reduced the ratio of capped/uncapped transcripts for unspliced precursors. The average capped/uncapped ratio for the two RNAse protection products corresponding to unspliced precursors in Figure 2B was 0.65 for wild-type and 0.27 for Δ5 (Fig. 2B, lanes 3–6).
The CMV–neo transfected cells serve as a control for the background of β-globin transcripts made by endogenous Pol II in the presence of α-amanitin (Fig. 2B, lanes 1, 2). The transcription rate in cells expressing WT and Δ5 α-amanitin-resistant Pol II is significantly higher than in the CMV–neo control cells as shown by the accumulation of unspliced precursors (Fig. 2B, cf. lanes 1 and 2 with 3–6) and by direct measurement using the nuclear runon assay (data not shown). Moreover, the CMV–neo control overestimates the level of background transcription in HA–WT- and HA–Δ5-expressing cells because the excess of α-amanitin-resistant Pol II competes with endogenous Pol II (McCracken et al. 1997). Background β-globin transcripts made in the control cells were capped at an intermediate level (capped/uncapped ratio of 2.5, Fig. 2B, lanes 1,2). The most likely reason for this is that the low rate of transcription in these cells combined with the long half-life of β-globin mRNA results in accumulation of decapped degradation intermediates. In summary, the experiments in Figure 2 show that when either the β-globin or HIV2 CAT gene was transcribed by Pol II with a truncated CTD, there was a marked decrease in the fraction of transcripts that had a 5′ cap.
Is the CTD effect on capping direct or indirect?
CTD truncation could affect the fraction of capped transcripts, either directly or indirectly. RNAs made by Pol II with a truncated CTD are not spliced or cleaved and polyadenylated as efficiently as those made by wild-type Pol II (McCracken et al. 1997). In addition, the unprocessed transcripts made by Δ5 Pol II were almost entirely nuclear, whereas the processed RNAs made by WT Pol II were predominantly cytoplasmic (data not shown). Failure to splice, polyadenylate, or transport to the cytoplasm could indirectly reduce the fraction of capped 5′ ends if unprocessed precursors were targeted for rapid decapping in the nucleus. To address this issue, we examined transcripts of a modified HIV2 CAT gene lacking the SV40 t intron and containing the SV40 late poly(A) site (pGal5 HIV2 CATΔt–AATAAA). This reporter was transfected into 293 cells with expression vectors for Gal4–VP16 and α-amanitin-resistant Pol II subunits. RNase protection with an antisense probe did not show any evidence of cryptic splicing within the entire CAT sequence with either WT or Δ5 Pol II (data not shown). RNA was fractionated into capped and uncapped populations and analyzed by RNase protection with probes spanning the initiation and poly(A) sites (Fig. 3, lanes 1–4). This experiment showed that capped/uncapped ratio for correctly initiated unspliced transcripts was 6.6 for WT (Fig. 3, lanes 1,2) and 1.1 for Δ5 (Fig. 3, lanes 3,4). We conclude that failure to splice can not explain the low percentage of Δ5 transcripts with a 5′ cap.
To test the possibility that failure to process the 3′ end could indirectly reduce the fraction of capped RNAs, we examined transcripts from an isogenic, intronless CAT gene with a mutant poly(A) site (pGal5 HIV2 CATΔt–AAGAAA). Regardless of whether or not the polymerase had a full-length CTD, transcripts of this gene were almost entirely nuclear and were not cleaved or terminated efficiently resulting in a relative accumulation of transcripts that read around the plasmid (solid box with thick line extension in Fig. 3 lanes 5–8). The capped/uncapped ratio for correctly initiated WT transcripts (solid box in Fig. 3) from pGal5 HIV2 CAT Δt–AAGAAA was 3.1 compared with 0.8 for Δ5 (Fig. 3, cf. lanes 5 and 6 with 7 and 8). These results show that the effect of CTD truncation on capping is also observed with an unspliced and unpolyadenylated RNA species.
In summary, these observations are not consistent with the idea that defective splicing or 3′ end processing is responsible for the reduced proportion of capped transcripts observed when the Pol II CTD was truncated. Instead they suggest that the CTD directly enhances capping or inhibits decapping.
Association of the mammalian capping enzyme with the phosphorylated CTD
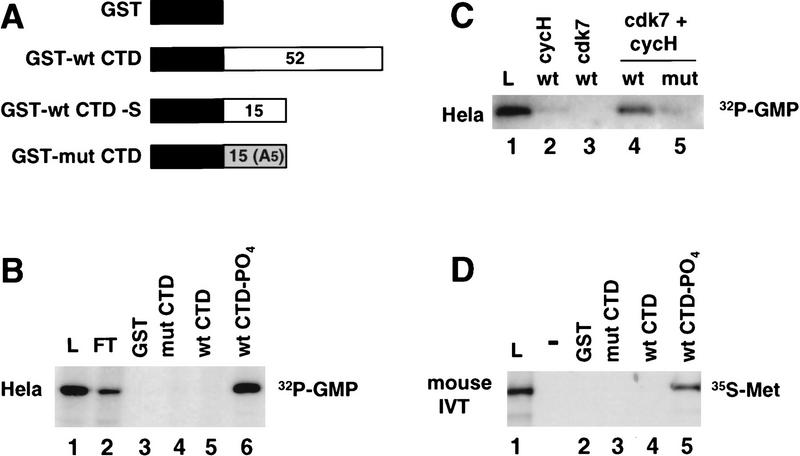
The functional evidence in Figures 2 and 3 shows that the CTD is important for producing transcripts with a high proportion of capped 5′ ends. To investigate whether the CTD interacts physically with the enzymes responsible for cap synthesis, we performed CTD affinity chromatography of HeLa extract. For this experiment, four affinity resins were prepared containing immobilized GST, GST–mutant CTD (mut), GST–wild-type CTD (wt), and phosphorylated GST–wild-type CTD (wt-PO4) (McCracken et al. 1997) (Fig. 4A). The wild-type CTD fusion protein contained the full-length murine CTD (52 repeats; consensus YSPTSPS). In some experiments, we also used a short form of the wild-type CTD fusion containing only the amino-terminal 15 heptad repeats (GST–wt CTD–S). The mutant CTD fusion protein contained 15 repeats of the consensus heptad sequence with a Ser to Ala substitution at position 5 (West and Corden 1995). In Figure 4, B and D, the GST–wt CTD resin containing 52 heptad repeats was phosphorylated using HeLa nuclear extract as a source of kinase activity; the resin was then washed extensively in a high-salt buffer to remove cellular proteins. The phosphorylation reaction resulted in addition of approximately three phosphates per molecule of GST–CTD on average. This level of phosphorylation is sufficient to change its conformation resulting in retarded migration in an SDS gel (data not shown) (Zhang and Corden 1991). The kinase reaction also conferred strong reactivity with the H14 monoclonal antibody against the phosphorylated CTD (data not shown) (Bregman et al. 1995). HeLa nuclear extract was chromatographed sequentially on these four columns in the order shown in Figure 4B and then eluted with high salt.
Figure 4.
Mammalian guanylyltransferase binds specifically to the phosphorylated CTD. (A) GST–CTD fusion proteins used for affinity chromatography. The number of heptad repeats in each is shown. (B) HeLa guanylyltransferase binds to the phosphorylated CTD. Nuclear extract was chromatographed sequentially on GST, GST–mut CTD, GST–wt CTD, and GST–wt CTD–PO4 that had been phosphorylated in HeLa nuclear extract and extensively washed in high-salt buffer (see Materials and Methods). GST–mut CTD was used at a higher ligand concentration to compensate for its shorter length. Load (L), flowthrough (FT), and high-salt eluates were assayed for the 68-kD guanylyltransferase by [32P]GMP labeling. 0.033% of L and FT fractions were analyzed and 0.4% of the eluates. (C) GST–wt CTD and GST–mut CTD were phosphorylated in reactions with baculoviral cyclin H, cdk7, or both cdk7 and cycH. HeLa extract was bound to these resins and the load (2%) and high-salt eluates (5%) as assayed for guanylyltransferase. (D) Specific binding of in vitro-translated mouse guanylyltransferase to the phosphorylated CTD. [35S]Met-labeled in vitro translation product of amino acids 1–590 of mouse guanylyltransferase homolog (see Fig. 5) was bound to GST–CTD resins. Load (L; 5% of total) and high-salt eluates (30%) from the resins were electrophoresed on an SDS gel and autoradiographed.
Guanylyltransferase activity in the eluates was assayed by the transfer of 32P-labeled GMP from [α32P]GTP to the capping enzyme to form a 68-kD covalent enzyme–GMP adduct (Shuman 1982). Formation of the 68-kD enzyme–guanylate complex required a divalent cation, either Mg2+ or Mn2+ (Shuman 1982) (data not shown). We observed specific binding of the 68-kD HeLa guanylyltransferase to the phosphorylated wild-type type CTD, but not to the nonphosphorylated wild-type or mutant CTD columns (Fig. 4B, lanes 4–6). An additional control experiment showed that the guanylyltransferase did not bind to a CTD–affinity column that had been reacted with the crude HeLa CTD–kinase preparation in the presence of ATP, but in the absence of magnesium (data not shown). In summary, these results show a phosphorylation-dependent interaction between human guanylyltransferase and the Pol II CTD.
To investigate further the specificity of guanylyltransferase binding, we phosphorylated the CTD with recombinant cdk7–cycH kinase. This kinase subunit of TFIIH specifically phosphorylates Ser-5 of the heptad repeat (Roy et al. 1994; Gebara et al. 1997). The GST–wt CTD was phosphorylated with baculovirus-expressed cdk7 and cycH alone and in combination. (The third subunit of the kinase, MAT1, could not be used because of a high level of contaminating kinase activity in the preparation.) Specific phosphorylation (0.5 mole PO4/mole GST–wt CTD) occurred only when both cycH and cdk7 were combined. Less than 10% of this level of phosphorylation occurred when cdk7–cyclin H was used to phosphorylate the GST–mut CTD with Ala at position 5. When HeLa extract was chromatographed on these resins, guanylyltransferase bound only to the wild-type CTD column that had been reacted with the heterodimeric kinase (Fig. 4C, lane 4). This result shows that phosphorylation of Ser-5 of the heptad repeat is sufficient to permit guanylyltransferase binding. Reaction of GST–wtCTD with recombinant human cdc2/cyclin B (New England Biolabs), which phosphorylates both Ser-2 and Ser-5 (Gebara et al. 1997), also permitted binding of guanylyltransferase (data not shown).
Recombinant mouse capping enzyme binds the phosphorylated CTD
The only guanylyltransferase sequence isolated previously from a higher eukaryote is that from the nematode Caenorhabditis elegans (Takagi et al. 1997; Wang et al. 1997). On the basis of sequence homology, we isolated a complete cDNA of a homologous mouse gene that was used to test for binding of the recombinant protein to the CTD. Comparison of the predicted amino acid sequence of the mouse protein with capping enzymes identified previously strongly suggests that it is a bifunctional protein composed of an amino-terminal RNA triphosphatase domain and a carboxy-terminal RNA guanylyltransferase domain. It has 43% amino acid identity with the C. elegans protein over its entire length (Fig. 5). The predicted molecular mass of the 597-residue protein is 68,683 daltons, in good agreement with the observed size of the human enzyme–GMP intermediate. The mouse protein contains the six defining motifs of the covalent nucleotidyl transferase superfamily (red boxes in Fig. 5) (Shuman and Schwer 1995) and has conserved all 16 residues identified by mutagenesis as essential for Ceg1 function (Wang et al. 1997) (asterisks in Fig. 5). Moreover, all 13-amino-acid side chains of the Chlorella virus capping enzyme that contact GTP in the cocrystal (Hakansson et al. 1997) (arrowheads in Fig. 5) are conserved in the mouse protein. The amino-terminal portion of the mouse capping enzyme contains the (I/V)HC × AG × GR(S/T)G signature motif (green box in Fig. 5) of the dual specificity Ser/Tyr protein phosphatase family (Denu et al. 1996) that includes the conserved cysteine at the catalytic site. The recombinant mouse protein when expressed as a GST fusion in Escherichia coli has guanylyltransferase activity (data not shown). Furthermore the mouse gene complements the yeast ceg1 mutation (data not shown).
Figure 5.
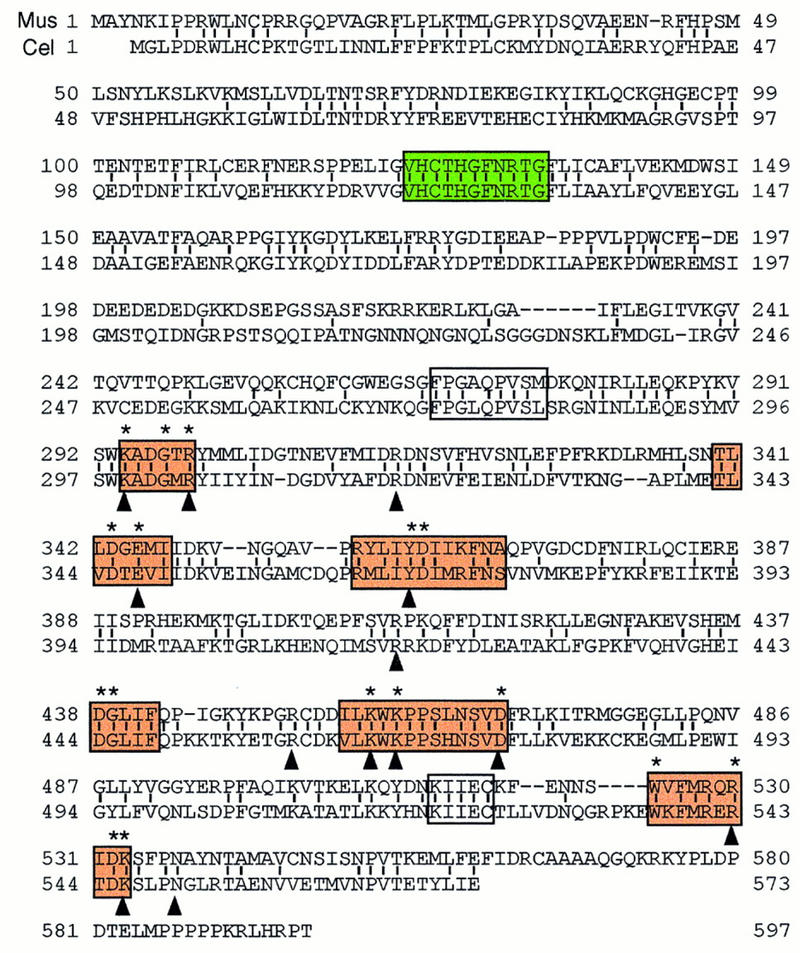
Sequence alignment of the mouse and C. elegans capping enzymes. The predicted mouse sequence was derived from two overlapping cDNA clones. The C. elegans sequence is from Takagi et al. (1997) and Wang et al. (1997). Amino acid identity, denoted by vertical lines, is 43%. Amino acids defined by alanine-scanning mutagenesis as essential for the activity of the S. cerevisiae capping enzyme Ceg1 are denoted by asterisks above the aligned sequences. The six nucleotidyl transferase superfamily motifs (I, III, IIIa, IV, V, and VI) are shown in bold black boxes. Motifs P (FPG × QPVS ×) and Vc (KIIEC) are in fine black boxes. The phosphatase motif (VHCTHGFNRTG) is in a gray box. Residues in proximity to the GTP moiety in the Chlorella virus capping enzyme–GTP cocrystal are indicated by arrowheads (Hakansson et al. 1997).
We tested whether the mouse capping enzyme bound to the Pol II CTD. The protein was translated in vitro and labeled with [35S]Met and then tested for binding to GST–CTD affinity resins. In the experiment shown in Figure 4D the in vitro translation product corresponding to amino acids 1–590 bound specifically to the phosphorylated wild-type CTD but not nonphosphorylated or mutant CTD resins. The recombinant mouse guanylyltranferase therefore behaves identically to the native HeLa enzyme with respect to its specificity for binding to the Pol II CTD.
Binding of yeast capping and methylating enzymes to phosphorylated CTD
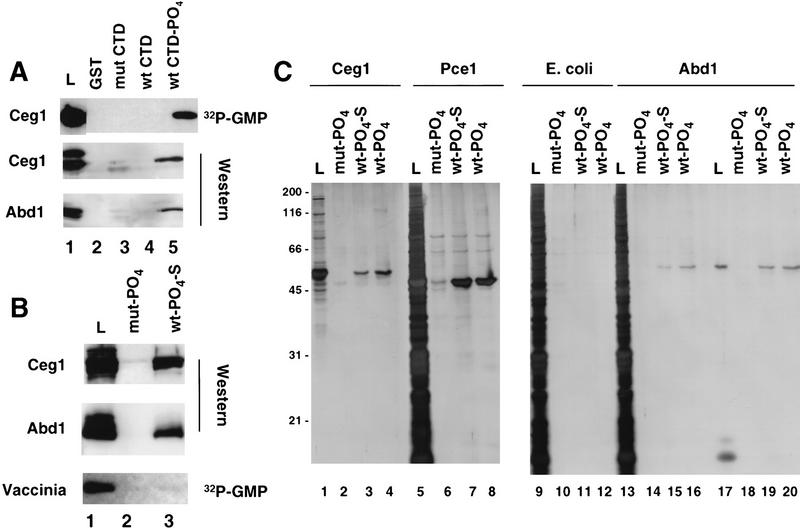
The cap guanylyltransferase and methyltransferase enzymes of Saccharomyces cerevisiae are encoded by the CEG1 and ABD1 genes, respectively (Shibagaki et al. 1992; Mao et al. 1995). To test whether the association of the capping apparatus with the CTD is conserved in budding yeast, we asked whether the Ceg1 and Abd1 proteins bind to GST–CTD affinity chromatography resins. Note that the CTD heptad consensus sequence is conserved completely between yeast and mammals. A partially purified yeast protein fraction was chromatographed on CTD affinity resins and the high-salt eluates were analyzed for the presence of capping enzymes. Specific binding of the 51-kD Ceg1 protein to the phosphorylated CTD column was detected both by label transfer of [32P]GMP and by Western blotting with anti-Ceg1 antibodies (Fig. 6A, lane 5). The 50-kD methyltransferase Abd1 also bound specifically to the phosphorylated CTD column, as demonstrated by Western blotting of column eluates with anti-Abd1 antibodies (Fig. 6A, lane 5). No significant binding of either Ceg1 or Abd1 to mutant CTD or nonphosphorylated wild-type CTD was detected. Ceg1- and Abd1-binding was not mediated by RNA, as it was unaffected by treating the extract with RNase A and T1. In summary, these data show that there is an evolutionarily conserved interaction between capping enzymes and the phosphorylated CTD.
Figure 6.
Yeast capping enzymes bind directly to the phosphorylated CTD. (A) Yeast guanylyltransferase, Ceg-1, methyltransferase, and Abd-1 bind to the phosphorylated CTD. Yeast extract partially purified on BioRex-70 was incubated with the GST–CTD resins, and the load (0.5%) and high-salt eluates (2%) were assayed by [32P]GMP labeling and Western blotting with antisera against Ceg1 and Abd1. Fivefold more protein was used for the Western blots. Several nonspecific bands detected by the antibodies in the load fraction were not bound to the CTD resins. (B) GST–CTD mutant (mut, lane 2) and wild-type (wt; 15 heptad repeats, lane 3) were phosphorylated with HeLa extract, washed extensively (see Materials and Methods), and used as ligands for affinity chromatography with bacterial fractions containing recombinant Ceg1, Abd1, or purified vaccinia capping enzyme. In lanes 1–6 the load (L, 0.74% of total, lane 1) and high salteluates (1.34%, lanes 2,3) were analyzed by Western blotting with anti-Ceg1 and Abd1 antisera. Vaccinia virus capping enzyme (4.7 units loaded, BRL) was assayed by [α32P]GTP labeling of the load (6% of total) and high-salt eluates (5%). (C) Silver-stained SDS gels of bacterially expressed yeast capping enzymes chromatographed on phosphorylated GST–CTD resins. Load (L; 10% of total) and high-salt eluates (20%) of recombinant S. cerevisiae and S. pombe guanylyltransferases (Ceg1 and Pce1) and S. cerevisiae methyltransferase (Abd1) were analyzed. As a negative control, total E. coli extract from a nonexpressing strain was used (lanes 9–12). Abd1 was chromatographed as a highly purified fraction (lanes 17–20) or after mixing with E. coli extract (lanes 13–16).
Direct binding of capping enzymes to the phosphorylated CTD
To test whether association of the capping enzymes with the phosphorylated CTD results from a direct protein–protein contact, bacterially expressed recombinant Ceg1 and Abd1 were chromatographed on phosphorylated CTD resins. The mouse guanylyltransferase was not tested because we have not succeeded in producing it in sufficient quantities in soluble form in bacteria. In this experiment, we used a short, wild-type mouse CTD fusion (GST–wt CTD–S) containing only the amino-terminal 15 heptad repeats; therefore, the wild-type and mutant CTD fusion proteins were of equal length (Fig. 4A). The extent of phosphate incorporation into the short, wild-type CTD during reaction within HeLa nuclear extract was 3.0 mole/mole versus .04 mole/mole for the mut CTD. Recombinant Ceg1 and Abd1 both bound to the phosphorylated short wild-type CTD–S as detected by Western blotting; however, neither protein bound to the mutant CTD column (Fig. 6B). As a specificity control, vaccinia virus capping enzyme was chromatographed on these resins and found not to bind to the mutant or wild-type Pol II CTD (Fig. 6B). Vaccinia capping enzyme does bind specifically to vaccinia virus RNA polymerase (Hagler and Shuman 1992). Although the large subunit of vaccinia polymerase is homologous to that of Pol II, the viral enzyme has no CTD (Broyles and Moss 1986).
The binding specificity of recombinant yeast capping enzymes to the CTD was also assessed by analyzing the polypeptide compositions of the high-salt eluates from the affinity chromatography resins. Soluble bacterial lysates containing S. cerevisiae guanylyltransferase (Ceg1) or Schizosaccharomyces pombe guanylyltransferase (Pce1) (Shuman et al. 1994) were incubated with phosphorylated mutant and wild-type CTD resins (both 15 and 52 heptad repeats) and the eluates were analyzed by silver staining of SDS gels (Fig. 6C). Highly purified recombinant Abd1 was chromatographed directly and after mixing with crude extract from nonexpressing bacteria (Fig. 6C). Whereas there was little or no binding of bacterial polypeptides to any of the CTD resins (Fig. 6C, lanes 9–12), recombinant 51-kD Ceg1 (Fig. 6C, lanes 1–4), 47-kD Pce1 (Fig. 6C, lanes 5–8), and 50-kD Abd1 polypeptides (Fig. 6C, lanes 13–20) all specifically bound to the wild-type and not the mutant CTD–PO4 resins. The identity of the bound proteins was confirmed by Western blotting and [32P]GMP labeling (data not shown). Note that highly purified Abd1 was effectively separated from a low-molecular-weight contaminant by binding to the phosphorylated wild-type CTD (Fig. 6C, lanes 17 vs. 19 and 20). In summary, the effective one-step purification of capping enzymes in these chromatography experiments demonstrates a highly specific interaction of guanylyltransferase and (guanine-7)-methyltransferase with the phosphorylated CTD.
Discussion
The CTD is required for efficient targeting of the cap to Pol II transcripts
The 5′ cap is a unique modification of cellular transcripts made by Pol II. RNAs made by Pol I, Pol III, or viral polymerases are not capped in vivo. It is not known how capping is specifically directed to the 5′ ends of Pol II transcripts. This specificity is clearly not dictated by the transcripts themselves, insofar as guanylyltransferases are capable of capping any RNA substrate (even homopolymeric RNAs) containing a 5′ diphosphate terminus. The interaction of vaccinia virus capping enzyme with the viral RNA polymerase provided precedent for the suggestion that cellular capping enzymes would interact with some component of the Pol II transcription complex (Hagler and Shuman 1992). Our results now show that the CTD, a unique feature of Pol II is important for efficient capping in vivo and is directly recognized by the cellular capping enzymes.
To assess the role of the CTD in mRNA capping, we compared transcripts made by full-length wild-type (WT) and CTD-truncated (Δ5) α-amanitin-resistant Pol II (Gerber et al. 1995). The ratio of capped/uncapped transcripts decreased significantly when reporter genes were transcribed by the truncated Pol II relative to wild type (Fig. 2). Because the effect on capping was also observed for transcripts from a gene lacking introns and a functional poly(A) site (Fig. 3), it cannot be explained as a secondary result of reduced splicing and 3′ processing. The observation that capping enzymes bind directly to the phosphorylated CTD argues that the CTD functions to enhance cap formation and to target capping to nascent Pol II RNAs. We noted that up to 50% of the transcripts were capped when transcribed by Pol II with only 5 heptad repeats in the CTD. The capping enzymes may bind with lower affinity to the truncated CTD or may be recruited to the transcription elongation complex via independent lower-affinity interactions with other components of the Pol II complex. Our results do not eliminate the possibility that the CTD, in addition to recruiting capping enzymes to the ternary complex, could also inhibit cap removal, for example, by excluding access of decapping enzymes to the nascent RNA.
The capping defect cannot account for poor splicing and 3′ processing of transcripts made by CTD-truncated Pol II
Our results are consistent with previous studies showing the cap is important for subsequent RNA processing events (Gilmartin et al. 1988; Izaurralde et al. 1994; Colot et al. 1996; Cooke and Alwine 1996; Fresco and Buratowski 1996; Schwer and Shuman 1996). The uncapped RNA fraction in vivo was enriched for unspliced β-globin (Fig. 2B) and Gal5 HIV2–CAT transcripts (Fig. 1C). Uncapped RNA was also enriched for precursors that were not cleaved at the poly(A) site (Figs. 1C and 3).
Does failure to put a 5′ cap on Δ5 transcripts account for the defects in splicing and polyadenylation of these transcripts? We reasoned that if reduced capping causes the splicing and 3′ processing defects of Δ5 transcripts, then those RNAs made by Δ5 that do receive a cap should be processed normally. If, on the other hand, CTD truncation also inhibits splicing and 3′ processing by other mechanisms, then processing defects should occur even among those Δ5 RNAs that do have a cap. Cleavage at the SV40 late poly(A) site was measured in the capped fraction of WT and Δ5 transcripts from the pGal5 HIV2 CATΔt–AATAAA reporter gene (Fig. 3). The cleaved/uncleaved ratio for capped WT transcripts (Fig. 3, lane 2) was 2.1 compared with less than 0.3 for the capped fraction of Δ5 transcripts (Fig. 3, lane 4). Reduced capping therefore cannot completely explain why Δ5 transcripts are cleaved less well than WT transcripts at the poly(A) site.
The extent to which the capping defect can account for reduced splicing was examined for human β-globin transcripts (Fig. 2B). Splicing of both β-globin introns was inhibited by CTD truncation as we observed previously for other introns (McCracken et al. 1997). Moreover, with WT Pol II, the ratio of spliced/unspliced RNA for β-globin introns 1 and 2 was five- to sevenfold higher in the capped fraction than in the uncapped fraction (see legend to Fig. 2B and cf. lanes 3 and 4). Most importantly, capped Δ5 transcripts were spliced significantly less well than capped WT transcripts. The spliced/unspliced ratios for introns 1 and 2 were 11.5 and 5.8, respectively, for capped WT transcripts (Fig. 2B, lane 4) but only 2.1 and 1.2 for Δ5 transcripts (Fig. 2B, lane 6). In other experiments we also observed reduced splicing of the SV40 t intron among capped Δ5 transcripts relative to capped WT transcripts (data not shown). In summary, these observations therefore indicate that failure to put on a 5′ cap cannot completely account for the reduced splicing or 3′ processing of Δ5 transcripts. The results do not exclude the possibility that capping of Δ5 transcripts occurs too late to support efficient splicing and 3′ processing. Conversely, the results suggest that the effects of the CTD on capping, splicing and polyadenylation are at least partially independent of one another. This idea is supported by the fact that capping, splicing, and 3′ processing factors have all been found to bind the CTD either directly or indirectly.
Capping enzymes bind exclusively to the phosphorylated CTD
Guanylyltransferase in extracts from HeLa and S. cerevisiae cells as well as the recombinant mouse, S. cerevisiae, and S. pombe enzymes all bound to the CTD in a manner that was highly dependent on its phosphorylation. Native and recombinant S. cerevisiae methyltransferase Abd1 also bound specifically to the phosphorylated CTD. In contrast, in the same chromatography experiment, the polyadenylation factors CstF and CPSF bound equally well to the wild-type CTD whether or not it was phosphorylated (McCracken et al. 1997). The interactions of polyadenylation factors and capping enzymes with the CTD therefore clearly have distinct properties. The phosphorylated amino-terminal 15 heptad repeats of the CTD were sufficient for binding of both capping enzymes and mutation of Ser-5 to Ala prevented binding. Recombinant cdk7–cycH, which phosphorylates Ser-5, was sufficient to confer binding of human guanylyltransferase. Furthermore, recombinant guanylyltransferase and methyltransferase were the only major silver-stained bands retained when crude bacterial extracts were chromatographed on phosphorylated CTD resins. Phosphorylation causes a global conformational change in the CTD that could be recognized by the capping enzymes (Zhang and Corden 1991). Alternatively, they could recognize specific sites of phosphorylation. Our results show that extensive CTD phosphorylation may be not required for recognition by capping enzymes. When GST–wt CTD was phosphorylated by recombinant cdk7–cycH, an average incorporation of only 0.5 mole PO4/mole was sufficient to confer some binding (Fig. 4C).
Independent and direct binding of guanylyltransferase and methyltransferase to the phosphorylated CTD may have an important function in coordinating cap synthesis. S. cerevisiae guanylyltransferase copurifies with an 80-kD polypeptide that contains the RNA triphosphatase component of the capping apparatus (Itoh et al. 1987). This enzyme converts the 5′ triphosphate end of the primary transcript to a diphosphate that can then be capped by the guanylyltransferase Ceg1. The triphosphatase could, therefore, be recruited to Pol II by virtue of the association between the CTD and Ceg 1. In C. elegans, the phosphatase and guanylyltransferase activities reside within a single bifunctional polypeptide (Takagi et al. 1997; Wang et al. 1997). The sequence of the mouse capping enzyme homolog (Fig. 5) strongly suggests that the mammalian enzymes are also bifunctional. The yeast and worm cap methyltransferase are not physically associated with their respective guanylyltransferases (Mao et al. 1995; Wang and Shuman 1997). The direct and independent binding of guanylyltransferase and methyltransferase to the phosphorylated CTD provides a plausible solution to the problem of how all capping activities are recruited simultaneously to the nascent 5′ end of the mRNA. Recruitment of both polypeptides to the phosphorylated CTD may assist the concerted action of these two enzymes, which is required to drive the equilibrium of the capping reaction in the forward direction by preventing reversal of the guanylation step (Shuman 1995).
Implications for regulation and coordination of capping with transcription
The specificity of capping enzyme binding to the phosphorylated CTD has implications for how capping is integrated with transcription. Phosphorylation of the CTD is an important step in the regulation of transcription after initiation has occurred. CTD phosphorylation is correlated with the transition of paused Pol II complexes into an actively elongating state (O’Brien et al. 1994). Our results suggest that CTD phosphorylation contributes to a high efficiency of capping. Capping could, therefore, be closely coordinated with conversion of a promoter-proximal arrested ternary complex into an actively elongating complex. A CTD phosphorylation requirement for capping could ensure that abortive transcripts do not get capped and, therefore, do not compete for cap-binding proteins.
Capping is usually regarded as a constitutive step in mRNA biosynthesis, although the question of whether capping is regulated is difficult to study because of the instability of uncapped transcripts. In one case where it was studied, a difference in the extent of capping in vivo between nascent hsp70 and hsp27 transcripts was reported (Rasmussen and Lis 1993). Our observations suggest that factors such as transactivators could potentially affect capping by modulating CTD phosphorylation (O’Brien et al. 1994; Parada and Roeder 1996). The results reported here also raise the possibility that phosphorylation of the CTD acts as a switch with the dual function of converting the polymerase to a processive form and enhancing cap formation, which in turn stimulates subsequent splicing and 3′ processing.
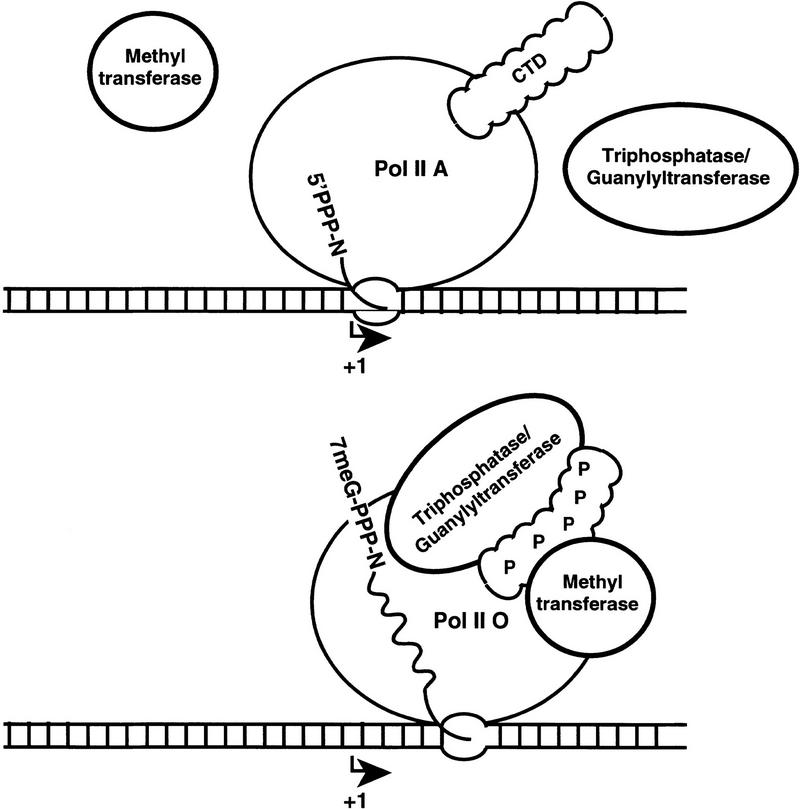
Previously, splicing-related factors and polyadenylation factors were found to associate with the carboxy-terminal domain of Pol II. These interactions may be responsible for the cotranscriptional nature of splicing and 3′ processing (for review, see Steinmetz 1997). The association of capping enzymes with the phosphorylated CTD adds further support to the idea that this domain has a central role in linking mRNA transcription with processing. Although our results do not exclude additional mechanisms, the binding of capping enzymes to a unique and highly conserved domain of Pol II provides an attractive explanation for how the cap is specifically targeted to transcripts made by Pol II (Fig. 7).
Figure 7.
Model for recruitment of capping enzymes to the phosphorylated CTD.
Materials and methods
Plasmids and RNase protection probes
pGal5 HIV2 CAT, pGal5 HIV2 CATΔt–AATAAA, and pGal5 HIV2 CATΔt–AAGAAA, pSPVA, and Gal4–VP16 plasmids and RNase protection probes were described previously (McCracken et al. 1997). HA–WT and HA–Δ5 expression vectors for α-amanitin-resistant Pol II were a gift of J. Corden (Gerber et al. 1995). pSVβ128 is a derivative of pSVHNβΔ128 (Treisman and Maniatis 1985) containing the SV40 enhancer 128 bases upstream of the start site. β-Globin RNase protection probes were generated from PCR-amplified DNA fragments spanning intron 1–exon 2 (positions +178–486) and intron 2–exon 3 (positions +1307–1496) cloned into Bluescript KS−.
Transfections
293 cells were transfected transiently with 5 μg of reporter plasmid, 0.5 or 1 μg (Fig. 2A) of Gal4–VP16 expression plasmid, 0.5 μg of pSPVA, and where indicated, 5 μg (Fig. 2A) or 0.5 μg (Figs. 2B and 3) of Pol II expression vector by calcium phosphate precipitation. α-Amanitin (2.5 μg/ml) was added 12–15 hr after transfection and cells were collected at 60–65 hr post-transfection. RNA was prepared by guanidine isothiocyanate/acid phenol extraction followed by DNase I treatment. Nuclear and cytoplasmic RNA were prepared after lysis in 10 mm HEPES at pH 7.6, 10 mm NaCl, 3 mm CaCl2, and 0.5% NP-40.
Pull-down assay for capped RNAs
GST–eIF4E was expressed using a plasmid kindly provided by Dr. J. Pelletier (McGill University, Montreal, Canada). The protein was prepared as described for protein A–eIF4E (Edery et al. 1995) except that glutathione beads were used. Total cellular RNA up to a maximum of 20 μg in 100 μl of cap-binding buffer [17 mm HEPES at pH7.9, 83 mm NaCl, 0.08 mm EDTA, 0.8 mm dithiothreitol (DTT), 17% glycerol, 0.08% NP-40, 1.1% polyvinyl alcohol, 200 μg/ml of poly(U), and 17 μg/ml (dG:dC): (dG:dC)] was heated for 2 min at 95°C and cooled on ice. DTT (6 mm) was added, followed by RNAguard (36 units, Pharmacia) and 30 μg of GST–eIF4E. After incubation for 1 hr at 4°C, this mixture was added to 25 μl of packed GST beads that had been preblocked in binding buffer. After 1 hr of mixing at 4°C, the beads were collected by centrifugation. The supernatant was pooled with the first wash in 200 μl of wash buffer (20 mm HEPES at pH 7.9, 0.1 m NaCl, 0.1 mm EDTA, 1 mm DTT, 20% glycerol, and 0.1% NP-40). The beads were then washed twice more in 1 ml of wash buffer, and the capped RNA was eluted with 0.5 ml of 0.3 m Na acetate pH 4.5 and 1.0% SDS. Glycogen (1 μg) was added to all samples and carrier Torula RNA was added to the pellet fraction to maintain the same nucleic acid concentration as the supernatant. Samples were extracted with phenol/chloroform, precipitated, and analyzed by RNase protection.
Ligands for affinity chromatography
Full-length wild-type murine GST–CTD (52 repeats) was expressed from pet 21a GST–CTD, a derivative of pGCTD (Peterson et al. 1992). GST–mut CTD (A5)15 was expressed from a plasmid provided by Dr. J. Corden (West and Corden 1995). Short, wild-type murine GST–wt CTD–S (15 repeats) was expressed from pet 21a GST–CTDΔ15 made using a PCR-amplified fragment encoding the amino-terminal 15 heptad repeats of murine CTD. The phosphorylated CTD was prepared by incubating the resin at a final substrate ligand concentration of 1 mg/ml in a reaction with HeLa nuclear extract (1.6 mg/ml) in 20 mm HEPES at pH 7.9, 10 mm MgCl2, 2 mm DTT, 2 mm ATP, 20 mm creatine phosphate, 0.05 mg/ml of creatine phosphokinase, 2 μg/ml of BSA, 20 mm β-glycerophosphate, 2 μm microcystine, 2 mm benzamidine, 0.01 μg/μl of aprotinin, 0.01 μg/μl of pepstatin A, 0.02 μg/μl of leupeptin, and 80 μm PMSF. GST–mut CTD (0.3 mole PO4/mole) and GST–wt CTD–S (3 mole PO4/mole) were phosphorylated in solution under similar conditions and then bound to glutathione agarose at a ligand concentration of 5 mg/ml gel. The glutathione beads were preblocked with E. coli extract then washed in high- and low-salt buffers. Phosphorylation of the GST–long wild-type CTD (0.5 mole PO4/mole) or GST mut CTD (0.04 mole PO4/mole) with baculoviral cycH and cdk7 was performed in solution using 0.08 mg/ml of each subunit. After phosphorylation, the resin was washed exhaustively with 1m NaCl in column buffer (20 mm HEPES at pH7.9, 0.1 mm EDTA, 1 mm DTT, and 20% glycerol), and then with 0.1 m NaCl in the same buffer. Phosphorylation of wild-type but not mut CTD by both HeLa extract and recombinant, cdk7–cycH conferred reactivity with the H14 monoclonal antibody (Bregman et al. 1995).
Affinity chromatography
Large-scale sequential chromatography was described (McCracken et al. 1997). The GST fusion proteins were loaded onto glutathione agarose at 4 mg/ml except for the GST–mut CTD, which was at 8 mg/ml. HeLa nuclear extract (33 mg) (Dignam et al. 1983) was passed sequentially over four 0.2-ml columns at 4°C, washed with 2 ml of column buffer containing 0.1 m KCl, and eluted in 0.5 ml of column buffer containing 0.6 m KCl—0.3% to 0.4% of the total protein bound to each column. During sequential passage over the four columns, the sample was not diluted significantly.
For small-scale batch chromatography of yeast, HeLa, and bacterial proteins, 25–50 μl of the above resins were incubated with 180–400 μl of extract in binding buffer (20 mm HEPES at pH 7.9, 0.1 mm EDTA, 1 mm DTT, 20% glycerol, 0.1 m NaCl, 0.5 μm microcystine, 1 mm β glycerophosphate, and 0.1% NP-40). Reticulocyte lysate in vitro translation reactions were diluted fivefold in binding buffer before incubation with the resins. After binding for 1 hr at 4°C, the supernatant was removed and the beads were washed three to four times with 400–500 μl of binding buffer, then eluted with 40–100 μl of binding buffer plus 0.9 m NaCl.
Yeast extract was a Biorex 70 0.1–0.3 m K acetate fraction (0.37 mg/ml). Recombinant Ceg1 (6 μg per 200 μl binding reaction) was a partially purified hydroxylapatite fraction. Abd1 (2.5 μg per 200 μl binding reaction) was used as a highly purified phosphocellulose fraction (Mao et al. 1995) or mixed with crude bacterial extract from nonexpressing cells. Pce1 (Shuman et al. 1994) was a crude bacterial lysate.
Recombinant yeast capping enzymes
The guanylyltransferases of S. cerevisiae (Ceg1) and S. pombe (Pce1) were expressed in bacteria under the control of an inducible bacteriophage T7 promoter and recovered in soluble bacterial lysates as described (Schwer and Shuman 1994; Shuman et al. 1994). Abd1 was expressed with an amino-terminal His tag and purified from bacterial lysates by Ni-agarose and phosphocellulose column chromatography as described (Wang and Shuman 1997).
Antibodies
Rabbit anti-Ceg1 (Schwer and Shuman 1996) and anti-Abd1 (Wang and Shuman 1997) antibodies have been described.
Guanylyltransferase assays
Guanylyltransferase was assayed as described previously (Shuman 1982) by incubating samples in 50 mm Tris-Cl at pH 7.5, 5 mm MnCl2, 0.5 μm GTP, 2.5 mm DTT, and 0.15 μCi/μl of [α-32P]GTP for 10 min at 37°C. The reaction was stopped by addition of SDS sample buffer, and analyzed by gel electrophoresis and autoradiography. In Figure 4C, the concentration of unlabeled GTP was 25 nm.
Recombinant cdk7 and cyclin H
Sf9 cells were infected (m.o.i. 5.0) with recombinant pBlueBacHis2 Baculovirus (Invitrogen) encoding amino-terminally His tagged human cyclin H or Xenopus Cdk7. Cells were harvested at 72 hr postinfection, swollen in 10 volumes of hypotonic buffer (20 mm HEPES at pH 7.9, 10 mm KCl, 10 mm β-mercaptoethanol, 10 mm β-glycerophosphate, 0.5 mm sodium vanadate, 2 mm NaF, 1 mm PMSF, 2 mm Benzamidine, 1μg/ml of leupeptin, 2 μg /ml of pepstatin, and 1 μg/ml of E64), and lysed with a Dounce homogenizer. Imidazole, NaCl, NP-40, and glycerol were added to final concentrations of 20 mm and 400 mm, 0.2% and 10%, respectively, to make lysis buffer. The cleared lysates were passed through His-bind resin (Novagen), washed with 10 volumes of lysis buffer, then 10 volumes of lysis buffer plus 40 mm imidazole. Cyclin H and Cdk7 were collected in elution buffer (20 mm HEPES at pH 7.9, 500 mm imidazole, 400 mm KCl, 10 mm β-mercaptoethanol, 10 mm β-glycerophosphate, 0.5 mm sodium vanadate, 2.0 mm NaF, and protease inhibitors), dialyzed against the same buffer minus imidazole, and concentrated by ultrafiltration.
Mouse guanylyltransferase homolog
Two overlapping cDNA clones were identified on the basis of homology between the C. elegans guanylyltransferase sequence and expressed sequence tags from two mouse bone marrow cDNA libraries. Both clones were sequenced completely on both strands. In one clone, a 66-base intron flanked by consensus splice sites had been excised, resulting in an open reading frame (ORF) with an in-frame termination codon after residue 480. [35S]Met-labeled protein was expressed by TNT (Promega).
Acknowledgments
We thank J. Pelletier, J. Corden, and S. Suggs for gifts of plasmids; R. Treisman, B. McNeil, P. Atadja, and S. Mason for helpful discussions; and H. Agah, M. Luchico, and S. Pang provided superb technical and secretarial help. This work was supported, in part, by a grant from the Medical Research Council of Canada to D.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
The sequence data described in this paper have been submitted to GenBank under accession no. AF034568.
Footnotes
E-MAIL david.bentley@utoronto.ca; FAX (416) 204-2278.
References
- Allison LA, Ingles CJ. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc Natl Acad Sci. 1989;86:2794–2798. doi: 10.1073/pnas.86.8.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Beelman CA, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles SS, Moss B. Homology between RNA polymerases of poxviruses, prokaryotes, and eukaryotes: Nucleotide sequence and transcriptional analysis of vaccinia virus genes encoding 147-kDa and 22-kDa subunits. Proc Natl Acad Sci. 1986;83:3141–3145. doi: 10.1073/pnas.83.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Stutz F, Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes & Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- Cooke C, Alwine JC. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol Cell Biol. 1996;16:2579–2584. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola JA, Field AS, Luse DS. Promoter-proximal pausing by RNA polymerase II in vitro: Transcripts shorter than 20 nucleotides are not capped. Proc Natl Acad Sci. 1983;80:1251–1255. doi: 10.1073/pnas.80.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL, Cadena DL, Ahearn JM, Jr, Dahmus ME. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Denu JM, Stuckey JA, Saper MA, Dixon JE. Form and function in protein dephosphorylation. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I, Chu LL, Sonenberg N, Pelletier J. An efficient strategy to isolate full-length cDNAs based on an mRNA cap retention procedure (CAPture.) Mol Cell Biol. 1995;15:3363–3371. doi: 10.1128/mcb.15.6.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I, Sonenberg N. Cap-dependent RNA splicing in a HeLa nuclear extract. Proc Natl Acad Sci. 1985;82:7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresco LD, Buratowski S. Conditional mutants of the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA. 1996;2:584–596. [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, LaFiandra A, Shatkin A. 5′-Terminal structures and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- Gebara MM, Sayre MH, Corden JL. Phosphorylation of the carboxy-terminal repeat domain in RNA polymerase II by cyclin-dependent kinases is sufficient to inhibit transcription. J Cell Biochem. 1997;64:390–402. [PubMed] [Google Scholar]
- Gerber HP, Hagmann M, Seipel K, Georgiev O, West MA, Litingtung Y, Schaffner W, Corden JL. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- Gilmartin GM, McDevitt MA, Nevins JR. Multiple factors are required for specific RNA cleavage at a poly(A) addition site. Genes & Dev. 1988;2:578–587. doi: 10.1101/gad.2.5.578. [DOI] [PubMed] [Google Scholar]
- Green MR, Maniatis T, Melton DA. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983;32:681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Greenleaf AL. Positive patches and negative noodles: Linking RNA processing to transcription? Trends Biol Sci. 1993;18:117–119. doi: 10.1016/0968-0004(93)90016-g. [DOI] [PubMed] [Google Scholar]
- Hagler J, Shuman S. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science. 1992;255:983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- Hakansson K, Doherty AJ, Shuman S, Wigley DB. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- Hamm J, Mattaj IW. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- Itoh N, Yamada H, Kaziro Y, Mizumoto K. Messenger RNA guanylyltransferase from Saccharomyces cerevisiae. Large scale purification, subunit functions, and subcellular localization. J Biol Chem. 1987;262:1989–1995. [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Konarska MM, Padgett RA, Sharp PA. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol Cell Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan GH, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-Terminal domain of RNA polymerase II couples messenger RNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei XY, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Parada CA, Roeder RG. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- Peterson SR, Dvir A, Anderson CW, Dynan WS. DNA binding provides a signal for phosphorylation of the RNA polymerase II heptapeptide repeats. Genes & Dev. 1992;6:426–438. doi: 10.1101/gad.6.3.426. [DOI] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on 3 drosophila heat-shock genes. Proc Natl Acad Sci. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Adamczewski JP, Seroz T, Vermeulen W, Tassan JP, Schaeffer L, Nigg EA, Hoeijmakers J, Egly JM. The MO15 cell-cycle kinase is associated with the TFIIH transcription DNA-repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Salditt G-M, Harpold M, Chen K-S, Darnell JE. The addition of 5′ cap structures occurs early in hnRNA synthesis and prematurely terminated molecules are capped. Cell. 1980;19:69–78. doi: 10.1016/0092-8674(80)90389-x. [DOI] [PubMed] [Google Scholar]
- Scafe C, Chao D, Lopes J, Hirsch JP, Henry S, Young RA. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- Schwer B, Shuman S. Mutational analysis of yeast mRNA capping enzyme. Proc Natl Acad Sci. 1994;91:4328–4332. doi: 10.1073/pnas.91.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Conditional inactivation of mRNA capping enzyme affects yeast pre-mRNA splicing in vivo. RNA. 1996;2:574–583. [PMC free article] [PubMed] [Google Scholar]
- Shatkin A. Capping of eukaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- Shatkin AJ. mRNA cap binding proteins: Essential factors for initiating translation. Cell. 1985;40:223–224. doi: 10.1016/0092-8674(85)90132-1. [DOI] [PubMed] [Google Scholar]
- Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. mRNA capping enzyme. Isolation and characterization of the gene encoding mRNA guanylytransferase subunit from Saccharomyces cerevisiae. J Biol Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- Shuman S. RNA capping by HeLa cell RNA guanylyltransferase. Characterization of a covalent protein-guanylate intermediate. J Biol Chem. 1982;257:7237–7245. [PubMed] [Google Scholar]
- ————— Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res & Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- Shuman S, Schwer B. RNA capping enzyme and DNA ligase: A superfamily of covalent nucleotidyl transferases. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- Shuman S, Liu Y, Schwer B. Covalent catalysis in nucleotidyl transfer reactions: Essential motifs in Saccharomyces cerevisiae RNA capping enzyme are conserved in Schizosaccharomyces pombe and viral capping enzymes and among polynucleotide ligases. Proc Natl Acad Sci. 1994;91:12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Morgan M, Merrick W, Shatkin A. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc Natl Acad Sci. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ. Pre-messenger RNA processing and the CTD of RNA Polymerase II– the tail that wags the dog. Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- Takagi T, Moore C, Diehn F, Buratowski S. An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- Treisman R, Maniatis T. Simian virus 40 enhancer increases number of RNA polymerase II molecules on linked DNA. Nature. 1985;315:73–75. doi: 10.1038/315072a0. [DOI] [PubMed] [Google Scholar]
- Wang SP, Shuman S. Structure-function analysis of the mRNA cap methyltransferase of Saccharomyces cerevisiae. J Biol Chem. 1997;272:14683–14689. doi: 10.1074/jbc.272.23.14683. [DOI] [PubMed] [Google Scholar]
- Wang S, Deng L, Ho C, Shuman S. Phylogeny of mRNA capping enzymes. Proc Natl Acad Sci. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Corden J. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Mizumoto K, Kaziro Y. Limited tryptic digestion of messenger RNA capping enzyme from Artemia salina. Isolation of domains for guanylyltransferase and RNA 5′-triphosphatase. J Biol Chem. 1984;259:4695–4698. [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi R, Gentile C, Gebara M, Corden J. The CTD of RNA polymerase II interacts with a novel set of SR-like proteins. Proc Natl Acad Sci. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Corden JL. Phosphorylation causes a conformational change in the carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991;266:2297–2302. [PubMed] [Google Scholar]