Abstract
Myotonic dystrophy type 2 (DM2) is a dominantly inherited autosomal disease with multi-systemic clinical features and it is caused by expansion of a CCTG tetranucleotide repeat in the first intron of the zinc finger protein 9 (ZNF9) gene in 3q21.The expanded-CCUG-containing transcripts are retained in the cell nucleus and accumulate in the form of focal aggregates which specifically sequester the muscleblind-like 1 (MBNL1) protein, a RNA binding factor involved in the regulation of alternative splicing. The structural organization and composition of the foci are still incompletely known. In this study, the nuclear foci occurring in cultured myoblasts from DM2 patients were characterised at fluorescence and transmission electron microscopy by using a panel of antibodies recognizing transcription and processing factors of pre-mRNAs. MBNL1 proved to co-locate in the nuclear foci with snRNPs and hnRNPs, whereas no co-location was observed with RNA polymerase II, the non-RNP splicing factor SC35, the cleavage factor CStF and the PML protein. At electron microscopy the MBNL1-containing nuclear foci appeared as roundish domains showing a rather homogeneous structure and proved to contain snRNPs and hnRNPs. The sequestration of splicing factors involved in early phases of pre-mRNA processing supports the hypothesis of a general alteration in the maturation of several mRNAs, which could lead to the multiple pathological dysfunctions observed in dystrophic patients.
Key words: myotonic dystrophy type 2 – DM2, myoblast, immunocytochemistry, fluorescence microscopy, electron microscopy, RNA processing.
Myotonic dystrophy type 2 (DM2) is a dominantly inherited autosomal disease with multi-systemic clinical features and it is caused by expansion of a CCTG tetranucleotide repeat in the first intron of the zinc finger protein 9 (ZNF9) gene in 3q21 (Liquori et al., 2001; Ranum and Day, 2004). The expanded-CCUG-containing transcripts are retained in the cell nucleus and accumulate in the form of focal aggregates (Liquori et al., 2001).These aggregates are a biomolecular feature of DMs and have been evidenced in several adult tissues and cell cultures by in situ hybridization (Taneja et al., 1995; Liquori et al., 2001; Mankodi et al., 2003, 2005; Jiang et al., 2004; Schoser et al., 2004; Wheeler et al., 2007; Cardani et al., 2004, 2009). The nuclear foci of mutant mRNA specifically sequester muscleblind-like (MBNL) proteins, i.e. essential splicing factors (Pascual et al., 2006), leading to nuclear depletion and a loss of function of these regulators (Mankodi et al., 2001). MBNL proteins are therefore good markers for the DM2-specific RNA-containing nuclear aggregates (Mankodi et al., 2001; Fardaei et al., 2002; Cardani et al., 2006, 2009). Sequestration of RNA binding proteins is thought to alter the splicing of a large set of pre-mRNAs, including transcripts of the insulin receptor, ClC-1 chloride channel, and other genes relevant to the clinical manifestations (Mankodi et al., 2002; Savkur et al., 2004). Margolis et al. (2006) demonstrated that DM2 foci contain the expanded CCUG tract with no other portions of the ZNF9 transcript. This repetitive RNA persists in the nucleus even after other parts of the intron have been degraded because it has a stable secondary structure or because it is tightly associated with binding proteins (Dere et al., 2004; Margolis et al., 2006; Ranum and Cooper, 2006).
The molecular composition of nuclear foci is far from being fully elucidated, but it is likely to hypothesize that other proteins involved in RNA processing may be sequestered therein, with a possible rebound on the overall nuclear function.
The effective role of nuclear foci in DM disease process is still debated. Recently Junghans (2009) has published his viewpoint on this item in which he discusses if foci may reflect the disease or if may be causative to DM pathogenesis. Studies on molecular composition and mechanisms of formation of nuclear foci may better explain the real role of these aggregates in the disease process.
In eukaryotic cells, transcription and splicing take place inside nuclear RNA-containing structures recognized, at electron microscopy, as perichromatin fibrils (PF), perichromatin granules (PG), and interchromatin granules (IG) (Spector, 1996; Fakan, 2004). PF are the morphological equivalent of hnRNA transcription and co-transcriptional splicing, while IG represent a storage site for small nuclear RNPs (snRNPs) and non-snRNP splicing factors, and PG are involved in the storage and the nucleus-to-cytoplasm transport of mRNA (recently reviewed in Biggiogera et al., 2007). PF and PG locate at the periphery of condensed chromatin, and IG in the so-called interchromatin space (for a review, see Puvion and Puvion-Dutilleul, 1996): this is a necessary prerequisite for the maturation of nuclear RNAs, so that whenever transcription and/or splicing are altered, the organization, composition, and intranuclear location of RNP-containing structures are affected too (Biggiogera et al., 2004, 2007). It has been shown that transcriptional arrest (irrespective of its physiological or induced origin: Biggiogera et al., 2004) results in the formation of heterogeneous ectopic RNP-derived structures (HERDS) which contain two or more RNP components that normally do not co-locate (Biggiogera et al., 1998; Biggiogera and Pellicciari, 2000). The accumulation within HERDS of RNA and protein factors involved in the processing of nuclear RNAs actually blocks the maturation and export of mRNAs.
The aim of this study was to investigate by means of fluorescence and immunoelectron microscopy the molecular composition of DM2 nuclear foci, in the attempt to elucidate whether other protein besides MBNL might be present therein. Using cultured myoblasts from DM2 patients we have also described, for the first time, the ultrastructural features of the RNA-containing nuclear foci.
Materials and Methods
Sample collection and cell culture
The biopsies used in this study were taken, under sterile conditions, from the biceps brachii of four DM2 patients, after informed consent.The histological diagnosis was performed on serial sections processed for routine histological or histochemical staining, based on the clinical criteria set by the International Consortium for Myotonic Dystrophies (Moxley et al., 2002). The biopsies from donors were trimmed of blood vessels, fat and connective tissues, and rinsed in phosphate-buffered saline (PBS); satellite cells were isolated as reported in Cardani et al. (2009) and placed in HAM's F10 medium (Sigma-Aldrich, Buchs, Switzerland) supplemented with 15% fetal bovine serum (Gibco, Invitrogen, Italy), 0.5 mg/mL albumin from bovine serum (BSA, Sigma - Aldrich), 0.5 mg/mL fetuin, 0.39 µg/mL dexamethasone, 10 ng/ml epidermal growth factor, 0.05 mg/mL insulin, 3 mg/mL glucose, 100 U/mL penicillin and 100 µg/ml streptomycin (Sigma - Aldrich).The myoblasts obtained by this procedure were propagated in plastic flasks at 37°C in a humidified 95% air/5% CO2 atmosphere. For transmission electron microscopy, the myoblasts were fixed in the flasks and then collected by scraping, whereas for light microscopy they were previously planted on glass coverslips to be processed for fluorescence in situ hybridization (FISH) and immunocytochemistry (see below).
Fluorescence microscopy
For fluorescence in situ hybridization (FISH) and immunocytochemistry, myoblasts were fixed with 2% formaldehyde in PBS for 30 min at 4°C.
For FISH, a Texas red labelled (CAGG)5 probe (IDT, Coralville, IA, USA) was used as previously reported by Cardani et al. (2004, 2009). Briefly, fixed myoblasts were permeabilized with 2% acetone in PBS (pre-chilled at −20°C) for 5 min. After washing in PBS, sections were incubated in 30% formamide and 2×SSC for 10 min at room temperature (r.t.), and then hybridized with the probe (1 ng/µL) for 2 h at 37°C in 30% formamide, 2×SSC, 0.02% BSA, 67 ng/µL yeast tRNA, 2 mM vanadyl ribonucleoside complex. Cells were washed in 30% formamide and 2×SSC at 45°C for 30 min, washed five times in PBS for 3 min at r.t., and pre-incubated with normal goat serum (DAKO, Glostrup, Denmark) at a dilution 1:20 in PBS containing 2% BSA for 20 min at r.t.. Immunolabelling for MBNL1 protein was performed using a rabbit polyclonal anti-MBNL1 (kind gift of Prof. C. A. Thornton, University of Rochester, NY, USA) at a dilution of 1:1000 in PBS containing 2% BSA, for 2 hr at r.t., then revealed with an Alexa 488-labelled goat anti-rabbit antibody (Molecular Probes, Invitrogen, Italy) diluted 1:200 in PBS containing 2% BSA for 1 h at r.t.. After incubation, the cells were stained for DNA with 165 nM 4.6 diamidino-2-phenylindole (DAPI) in PBS at r.t. for 30 min, and finally mounted with ProLong (Molecular Probes, Invitrogen).
To get information on the intranuclear location of the RNA-containing foci and their composition, a panel of antibodies directed against MBNL1 or transcription, splicing or cleavage factors were used in dual-immunolabelling experiments (Table 1); secondary antibodies labelled with different fluorochromes were used. Briefly, 2% formaldehyde-fixed myoblasts were post-fixed with cold 70% ethanol, and incubated with the primary and the appropriate secondary antibodies (for 2 h and 1 h respectively, at r.t.); immunolabelled myoblasts were stained for DNA with 0.1 µg/mL of Hoechst 33258 in PBS (10 min at r.t.), and finally mounted in a drop of Mowiol (Calbiochem, Milan, Italy).
Table 1. Features of the antibodies used for immunofluorescence analyses.
| Antibody | Origin | Antigen | Dilution | Secondary marker | Reference - Source |
|---|---|---|---|---|---|
| Anti-C8V | chicken polyclonal | hnRNP core proteins | 1:1000 | Alexa 594 labelled Goat anti-chicken IgG | Jones et al., 1980 |
| Anti-Sm | mouse monoclonal | Sm antigen (U1, U2, U4/6, U5 snRNP) | 1:100 | Alexa 594 labelled Goat anti-mouse IgG | Abcam (Cambridge, UK) |
| Anti-SC35 | mouse monoclonal | SC-35 non snRNP splicing factor | 1:200 | Alexa 594 labelled Goat anti-mouse IgG | Sigma Chem Co. (St. Louis MO, USA) |
| Anti-CStF | chicken polyclonal | Cleavage stimulation factor | 1:2000 | Alexa 594 labelled Goat anti-chicken IgG | Veraldi et al., 2001 |
| Anti-RNA polymerase II (clone H14) | mouse monoclonal | Phosphorylated (activated) form of RNA polymerase II | 1:100 | Alexa 594 labelled Goat anti-mouse IgM | Research Diagnostics Inc. (Concorde MA, USA) |
| Anti-PML | mouse monoclonal | Human PML protein | 1:200 | Alexa 594 labelled Goat anti-mouse IgG | Santa Cruz Biotechnology (Santa Cruz CA, USA) |
As negative controls, some slides were processed as described above, but omitting the incubation with the primary antibodies.
An Olympus BX51 microscope equipped with a 100W mercury lamp was used under the following conditions: 330–385 nm excitation filter (excf), 400-nm dichroic mirror (dm), and 420-nm barrier filter (bf), for Hoechst 33258 or DAPI; 450–480 nm excf, 500-nm dm, and 535 nm±20 nm bandpass filter, for the fluorescence of Alexa 488; 540 nm excf, 580 nm dm, and 620 nm bf for Alexa 594 or Texas red. Images were recorded with an Olympus MagniFire camera system, and processed with the Olympus CellF software (Olympus Italia Srl, Segrate, Italy).
Transmission electron microscopy
For conventional morphology, cells were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M Sörensen phosphate buffer at 4°C for 1 h, washed, post-fixed with 1% OsO4 at 4°C for 30 min, dehydrated with acetone and embedded in Epon. For immunoelectron microscopy, cells were fixed with a mixture of 4% paraformaldehyde and 0.2% glutaraldehyde in 0.1 M Sörensen phosphate buffer at 4°C for 1 h, washed, treated with NH4Cl 0.5 M in PBS, dehydrated with ethanol and embedded in Unicryl resin.
Ultrathin sections from Epon-embedded samples were conventionally stained with uranyl acetate and lead citrate.
Ultrathin sections from Unicryl-embedded samples were placed on nickel grids and processed for immunocytochemistry with the same anti-MBNL1 antibody used for fluorescence microscopy. Sections were floated on normal goat serum (NGS) diluted 1:100 in PBS and incubated overnight at 4°C with the primary antibody diluted 1:50 in a solution containing 0.1% BSA and 0.05% Tween 20 in PBS. After rinsing, sections were floated in NGS and then reacted for 20 min with the specific gold-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA) diluted 1:10 in PBS. Finally, the sections were rinsed and air dried. As controls, some grids were treated with the incubation mixture without the primary antibody and then processed as described above.
Moreover, in order to confirm the apparent co-location of MBNL1 with some splicing factors as observed by immunofluorescence microscopy (see the section Results), myoblast samples were also dual-immunolabelled by combining the anti-MBNL1 antibody with antibodies directed against either the Sm complex of U1, U2, U5 and U4/U6 snRNPs (Abcam, Cambridge, MA) or the heterogeneous nuclear RNP (hnRNP) core protein (Jones et al., 1980), according to the procedure described above.
To reduce chromatin contrast and selectively reveal nuclear RNP constituents, the sections were treated according to the EDTA method (Bernhard, 1969), and then observed in a Philips Morgagni TEM operating at 80kV and equipped with a Megaview II camera for digital image acquisition.
Results
Fluorescence microscopy
In DM2 myoblasts, MBNL1 proved to be present in discrete dots (which correspond to the expanded-CCUG-containing foci: Figure 1) as well as in a weakly fluoresceing diffused form throughout the nucleoplasm (consistent with the intranuclear location of splicing factors). Dual immunolabelling experiments demonstrated that MBNL1 does not co-locate in the foci with the non-snRNP splicing factor SC35 (Figure 2) and the PML protein (Figure 3); similar results were obtained also for RNA polymerase II and the cleavage factor CStF (not shown).
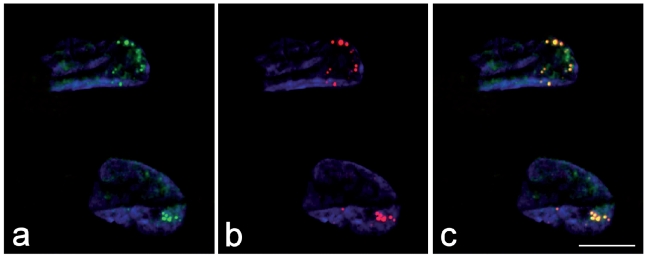
Figure 1.
In the myoblast nuclei, the immunolabelling for MBN1 (green fluorescence in a) and the signal of in situ hybridization for the mutant mRNA containing CCUG repeats (red fluorescence in b) co-locate (yellow fluorescence in the merged image, c). DNA was counterstained with DAPI (blue fluorescence). Bar: 10 µm.
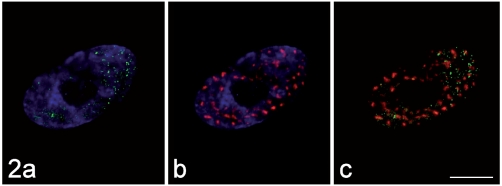
Figure 2.
The immunolabelling for MBNL1 (green fluorescence in a) and that for the non-snRNP splicing factor SC35 (red fluorescence in b) do not co-locate (merged image, c). DNA was counterstained with Hoechst 33258 (blue fluorescence in a, b). Bar: 10 µm.
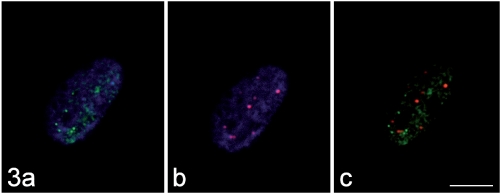
Figure 3.
The immunolabelling for MBNL1 (green fluorescence in a) and that for the PML protein (red fluorescence in b) do not co-locate (merged image, c). DNA was counterstained with Hoechst 33258 (blue fluorescence in a, b). Bar: 10 µm.
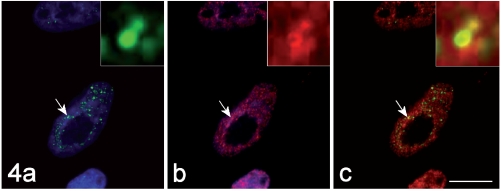
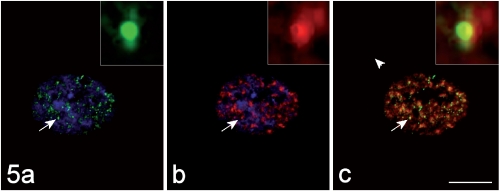
On the contrary, the hnRNP core protein (Figure 4) and the snRNP Sm antigen (Figure 5) were observed in some of the MBNL1-positive foci. Interestingly enough, the immunolabelling for these RNPs appeared as a ring-shaped fluorescence pattern around the brightly fluoresceing MBNL1-contaning core (insets of Figure 4 and 5). In addition, some co-location for the hnRNP core protein and for the Sm antigen was also observed to occur also in the nucleoplasm (Figure 4 and 5).
Figure 4.
The immunolabelling for MBNL1 (green fluorescence in a) and that for the hnRNP core protein (red fluorescence in b) co-locate (yellow fluorescence in the merged image, c). DNA was counterstained with Hoechst 33258 (blue fluorescence in a, b). Bar: 10 µm. In the insets, the co-labelled focus indicated by the arrow is shown at a higher magnification.
Figure 5.
The immunolabelling for MBNL1 (green fluorescence in a) and that for the snRNP Sm antigen (red fluorescence in b) co-locate (yellow fluorescence in the merged image, c). DNA was counterstained with Hoechst 33258 (blue fluorescence in a, b). Bar: 10 µm. In the insets, the co-labelled focus indicated by the arrow is shown at a higher magnification.
Fluorescent labelling was negligible in the negative controls (not shown).
Transmission electron microscopy
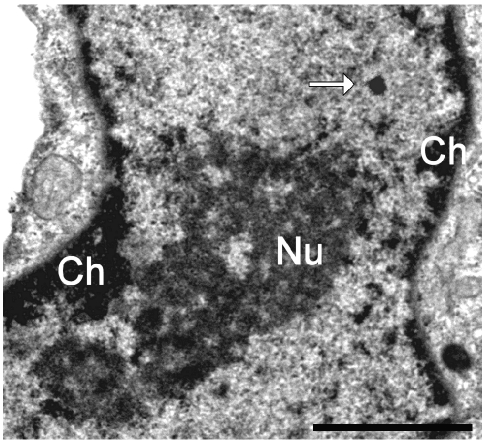
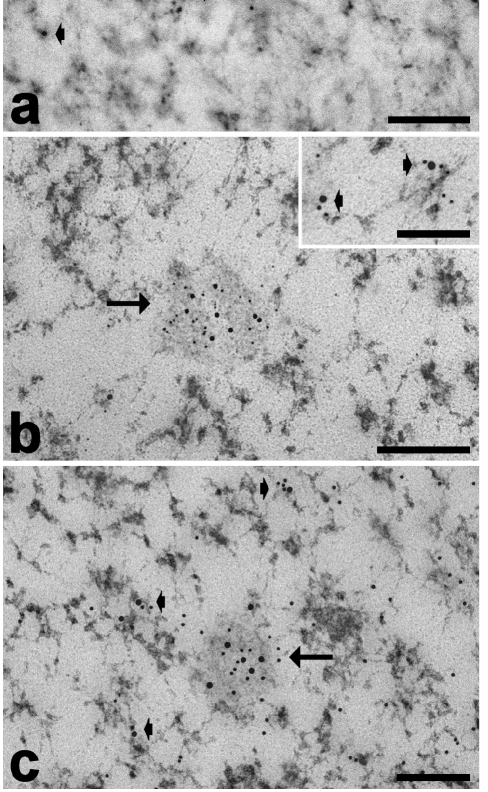
At electron microscopy, myoblast nuclei showed small condensed chromatin clumps (mainly at the nuclear periphery), PG and abundant PF preferentially located at the hedge of condensed chromatin, and small clusters of IG in the interchromatin space; the nucleoli were large with a few fibrillar centres surrounded by the dense fibrillar component and a prominent granular component (Figure 6). In addition to the usual structural constituents, roundish nuclear domains (100–200 nm in diameter) were observed in about 20% of the sectioned nuclei: in Epon-embedded samples they appeared as roundish homogeneously strong electron-dense structures (Figure 4), while in the Unicryl-embedded and EDTA-treated samples they appeared as less electron-dense irregularly sized domains surrounded by fine fibrils spreading out at their periphery (Figure 7). They always occurred in the interchromatin space. The gold-immunolabelling for MBNL1 specifically occurred on PF and was found to accumulate in the nuclear domains described above (Figure 7a). Consistent with the evidence in fluorescence microscopy, MBNL1 was observed to co-locate in the foci with hnRNPs (Figure 7b) and with the snRNP Sm antigen (Figure 7c). On the thin sections, however, the gold grains labelling these RNPs were distributed all over the foci and not at the periphery only.
Figure 6.
Myoblast nucleus, glutaraldehyde fixation, OsO4 postfixation, Epon embedding. A strongly electron-dense roundish nuclear domain (arrow) occurs in the nucleoplasm. Ch: condensed chromatin; Nu: nucleolus. Bar: 1 µm.
Figure 7.
Myoblast nuclei, paraformaldehyde fixation, Unicryl-embedding, EDTA staining. (a) Immunolabelling with anti-MBNL1 antibody: the signal accumulates in the RNP nuclear domain (arrows) and is also associated with PF (arrowheads). (b) Dual immunolabelling with anti-MBNL1 (12 nm) and anti-snRNP (6 nm) antibodies; (c) dual immunolabelling with anti-MBNL1 (18 nm) and anti-hnRNP (12 nm) antibodies: in both cases the probes co-locate in the RNP nuclear domain (arrow) and on PF (arrowheads). Bars: 250 nm (a–c); 100 nm (insets).
Background values were negligible in all immunolabelling experiments (not shown).
Discussion
Combining fluorescence and immunoelectron microscopy on DM2 cultured myoblasts, we obtained information on the molecular composition of the typical nuclear foci were the expanded-CCUG-containing transcripts accumulate; in addition these foci were recognized as distinct nuclear domains at the ultrastructural level.
Experiments of simultaneous in situ hybridization and immunolabelling confirmed that in cultured myoblasts the mutant RNA-containing nuclear foci sequester the splicing factor MBNL1 (Cardani et al., 2009), thus indicating that the molecular features observed in DM2 adult tissues are reproduced in in vitro muscle cells (Mankodi et al., 2003; Cardani et al., 2004; Schoser et al., 2004).
The RNA-containing nuclear foci observed at fluorescence microscopy can be also recognized at electron microscopy as roundish electron-dense domains which do contain RNPs (as demonstrated by the EDTA regressive staining), and proved to be immunopositive for MBNL1. These nuclear domains are always surrounded by PF but never occur in proximity of IG clusters, consistent with previous observations by immunofluorescence techniques (Holt et al., 2007). They morphologically resemble the so-called amorphous bodies, i.e., RNP-containing nuclear aggregates which accumulate in several Vertebrate tissues during hibernation, and are storage sites for many protein factors involved in pre-mRNA processing (e.g., Malatesta et al., 1994, 1999, 2006). The diffuse immunolabelling for MBNL1 observed in myoblast nuclei at fluorescence microscopy was found to correspond to the ultrastructural labelling of PF, where transcription and early splicing of pre-mRNA take place (Fakan, 2004); this distribution is in agreement with the functional role of MBNL1 as an RNA-binding protein involved in the regulation of alternative splicing (Pascual et al., 2006). Fluorescence dual-immunolabelling experiments demonstrated that RNA polymerase II, the non-snRNP splicing factor SC35, the cleavage factor CStF, and the PML protein do not co-locate with MBNL1 in the foci, whereas a co-occurrence herein was observed for the hnRNP core protein and the snRNP Sm antigen. This co-location was confirmed by immunoelectron microscopy not only on the RNP-containing nuclear domains but also on PF; this latter finding corresponds to the diffuse co-location observed at light microscopy. Interestingly, in electron micrographs the labelling for hnRNPs and snRNPs was distributed over the whole nuclear domains, whereas at fluorescence microscopy a bright ring-shaped signal for hnRNPs or snRNPs was found to surround the MBNL1-rich core of the foci. To explain this inconsistence, one should consider that at electron microscopy the antibodies can interact with their antigens even inside the foci, since the immunolabelling is performed on sections; on the contrary, whole cells are used for the immunofluorescent labelling, so that the penetration of the probe could be hindered by the apparently dense structure of the RNP-containing foci. The intense fluorescence for hnRNPs and snRNPs around the foci could also be explained by the high content in these two splicing factors on PF which surround the MBNL1-containing core. The presence of hnRNPs and snRNPs indicates that the DM2-specific nuclear foci share similarities in their molecular composition with both the amorphous bodies and the HERDS, i.e., the heterogeneous aggregates containing at least two nuclear RNA-associated protein factors which usually do not co-locate. Amorphous bodies form during hibernation, when the transcription and splicing rate is drastically reduced, and disassemble upon arousal, releasing the pre-mRNA processing factors accumulated therein, in order to rapidly resume the nuclear activities (Malatesta et al., 2001). HERDS were observed in different cell systems, whenever RNA transcription is altered, and have been suggested as a possible marker of transcriptional arrest (Biggiogera and Pellicciari, 2000): under normal conditions, a balance exists between the amount of newly synthesized pre-mRNA and the proteins involved in their processing but, if the amount of newly formed hnRNA decreases, RNA-associated proteins become exceedingly predominant and accumulate in HERDS. The present results suggest that an exceeding amount of RNA (i.e., the expanded-CCUG-containing transcripts retained in the nucleus of DM2 cells) can determine the sequestration in the foci not only of MBNL1 but also of other pre-mRNA processing factors.
Some of the pathological features of DM are thought to be caused by the mis-regulated alternative splicing of RNA due, at least in part, to the mis-localization of MBNL1 (review in Osborne and Thornton, 2006). The sequestration of other splicing factors involved in early phases of pre-mRNA processing demonstrated by the present study strengthens the idea of a general alteration in the maturation of several mRNAs, which could lead to the multiple pathological dysfunctions observed in dystrophic patients.The occurrence of hnRNPs and snRNPs in some (but not all the) RNA-containing foci suggests that a possible limiting factor for their sequestration could be the amount of CCUG repeats and/or MBNL1 accumulated therein; taking into account that the severity of the DM2 phenotype relates to the amplification of tetraplet repeats, this opens interesting perspectives for future studies.
Acknowledgments
We are particularly grateful to Prof. C. A. Thornton for providing us with the anti-MBNL1 antibody, and to Prof. T.E. Martin for the anti-hnRNP core protein antibody. Thanks are due to Mrs P. Veneroni for the excellent technical assistance. F.P. is a PhD student in receipt of a fellowship from the Dottorato di Ricerca in Biologia cellulare (University of Pavia). Supported by the University of Pavia (Fondo di Ateneo per la Ricerca, 2008).
References
- Bernhard W. A new staining procedure for electron microscopic cytology. J Ultrastruct Res. 1969;27:250–65. doi: 10.1016/s0022-5320(69)80016-x. [DOI] [PubMed] [Google Scholar]
- Biggiogera M, Bottone MG, Scovassi AI, Soldani C, Vecchio L, Pellicciari C. Rearrangement of nuclear ribonucleoprotein (RNP)-containing structures during apoptosis and transcriptional arrest. Biol Cell. 2004;96:603–15. doi: 10.1016/j.biolcel.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Biggiogera M, Cisterna B, Spedito A, Vecchio L, Malatesta M. Perichromatin fibrils as early markers of transcriptional alterations. Differentiation. 2007;76:57–65. doi: 10.1111/j.1432-0436.2007.00211.x. [DOI] [PubMed] [Google Scholar]
- Biggiogera M, Bottone M.G, Pellicciari C. Nuclear RNA is extruded from apoptotic cells. J Histochem Cytochem. 1998;46:999–1006. doi: 10.1177/002215549804600903. [DOI] [PubMed] [Google Scholar]
- Biggiogera M, Pellicciari C. Heterogeneous Ectopic RNP-Derived Structures (HERDS) are markers of transcriptional arrest. FASEB J. 2000;14:828–34. doi: 10.1096/fasebj.14.5.828. [DOI] [PubMed] [Google Scholar]
- Cardani R, Baldassa S, Botta A, Rinaldi F, Novelli G, Mancinelli E, et al. Ribonuclear inclusions and MBNL1 nuclear sequestration do not affect myoblast differentiation but alter gene splicing in myotonic dystrophy type 2. Neuromuscul Disord. 2009;19:335–43. doi: 10.1016/j.nmd.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Cardani R, Mancinelli E, Rotondo G, Sansone V, Meola G. Muscleblind-like protein 1 nuclear sequestration is a molecular pathology marker of DM1 and DM2. Eur J Histochem. 2006;50:177–82. [PubMed] [Google Scholar]
- Cardani R, Mancinelli E, Sansone V, Rotondo G, Meola G. Biomolecular identification of (CCTG)n mutation in myotonic dystrophy type 2 (DM2) by FISH on muscle biopsy. Eur J Histochem. 2004;48:437–42. doi: 10.4081/918. [DOI] [PubMed] [Google Scholar]
- Dere R, Napierala M, Ranum LPW, Wells RD. Hairpin structure-forming property of the (CCTG-CAGG) tetranucletide repeats contributes to the genetic instability associated with myotonic dystrophy type 2. J Biol Chem. 2004;279:41715–26. doi: 10.1074/jbc.M406415200. [DOI] [PubMed] [Google Scholar]
- Fakan S. Ultrastructural cytochemical analyses of nuclear functional architecture. Eur J Histochem. 2004;48:5–14. [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, et al. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Gen. 2002;11:805–14. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- Holt I, Mittal S, Furling D, Butler-Browne GS, Brook JD, Morris GE. Defective mRNA in myotonic dystrophy accumulates at the periphery of nuclear splicing speckles. Genes Cells. 2007;12:1035–48. doi: 10.1111/j.1365-2443.2007.01112.x. [DOI] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Gen. 2004;13:3079–88. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Jones RE, Okamura CS, Martin TE. Immunofluorescent localization of the proteins of nuclear ribonucleoprotein complexes. J Cell Biol. 1980;86:235–43. doi: 10.1083/jcb.86.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghans RP. Dystrophya myotonia: why focus on foci? Eur J Hum Gen. 2009;17:543–53. doi: 10.1038/ejhg.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–7. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Cardinali A, Battistelli S, Zancanaro C, Martin TE, Fakan S, et al. Nuclear bodies are usual constituents in tissues of hibernating dormice. Anat Rec. 1999;254:389–95. doi: 10.1002/(SICI)1097-0185(19990301)254:3<389::AID-AR10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Frigato E, Baldelli B, Battistelli S, Foà A, Bertolucci C. Influence of temperature on the liver circadian clock in the ruin lizard Podarcis sicula. Microsc Res Tech. 2007;70:578–84. doi: 10.1002/jemt.20437. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Luchetti F, Marcheggiani F, Fakan S, Gazzanelli G. Disassembly of nuclear bodies during arousal from hibernation: an in vitro study. Chromosoma. 2001;110:471–7. doi: 10.1007/s004120100166. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Zancanaro C, Martin TE, Chan EKL, Amalric F, Lührmann R, et al. Cytochemical and immunocytochemical characterization of nuclear bodies during hibernation. Eur J Cell Biol. 1994;65:82–93. [PubMed] [Google Scholar]
- Mankodi A, Lin XL, Blaxal BC, Swanson MS, Thornton CA. Nuclear RNA foci in the heart in myotonic dystrophy. Circ Res. 2005;97:1152–5. doi: 10.1161/01.RES.0000193598.89753.e3. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Teng-Umnuay P, Krym M, Henderson D, Swanson M, Thornton CA. Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann Neurol. 2003;54:760–8. doi: 10.1002/ana.10763. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–70. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Margolis JM, Schoser BG, Moseley ML, Day JW, Ranum LP. DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum Mol Genet. 2006;15:1808–15. doi: 10.1093/hmg/ddl103. [DOI] [PubMed] [Google Scholar]
- Moxley RT, 3rd, Meola G, Udd B, Ricker K. Report of the 84th ENMC workshop:PROMM (proximal myotonic myopathy) and other myotonic dystrophy-like syndromes: 2nd workshop. 13–15th October 2000, Loosdrecht:The Netherlands. Neuromuscul Disord. 2002;12:306–17. doi: 10.1016/s0960-8966(01)00284-x. [DOI] [PubMed] [Google Scholar]
- Osborne RJ, Thornton CA. RNA-dominant diseases. Hum Mol Genet. 2006;15:R162–9. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- Pascual M, Vicente M, Monferrer L, Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- Puvion E, Puvion-Dutilleul F. Ultrastructure of the nucleus in relation to transcription and splicing: roles of perichromatin fibrils and interchromatin granules. Exp Cell Res. 1996;229:217–25. doi: 10.1006/excr.1996.0363. [DOI] [PubMed] [Google Scholar]
- Ranum LPW, Day JW. Myotonic Dystrophy: RNA Pathogenesis Comes into Focus. Am Jf Hum Genetics. 2004;74:793–804. doi: 10.1086/383590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–77. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA, Dalton JC, Moseley ML, Ranum LP, et al. Insulin receptor splicing alteration in myotonic dystrophy type 2. Am J Hum Genet. 2004;74:1309–13. doi: 10.1086/421528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoser BGH, Ricker K, Schneider-Gold C, Hengstenberg C, Dürre J, Bültmann B, et al. Sudden cardiac death in myotonic dystrophy type 2. Neurology. 2004;63:2402–4. doi: 10.1212/01.wnl.0000147335.10783.e4. [DOI] [PubMed] [Google Scholar]
- Spector DL. Nuclear organization and gene expression. Exp Cell Res. 1996;229:189–97. doi: 10.1006/excr.1996.0358. [DOI] [PubMed] [Google Scholar]
- Taneja KL, McCurrach M, Schalling M, Housmann D, Singer RH. Foci of trinucleotide repeats in nuclei of myotonic dystrophy cells and tissue. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C. HnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol Cell Biol. 2001;21:1228–38. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Krym MC, Thornton CA. Ribonuclear foci at the neuromuscular junction in myotonic dystrophy type 1. Neuromuscul Disord. 2007;17:242–7. doi: 10.1016/j.nmd.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Thornton CA. Myotonic dystrophy: RNA-mediated muscle disease. Curr Opin Neurol. 2007;20:572–6. doi: 10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]