Abstract
The initial steps of double-stranded break (DSB) repair by homologous recombination mediated by the 5′–3′ exonuclease/annealing protein pairs, RecE/RecT and Redα/Redβ, were analyzed. Recombination was RecA-independent and required the expression of both components of an orthologous pair, even when the need for exonuclease activity was removed by use of preresected substrates. The required orthologous function correlated with a specific protein–protein interaction, and recombination was favored by overexpression of the annealing protein with respect to the exonuclease. The need for both components of an orthologous pair was observed regardless of whether recombination proceeded via a single-strand annealing or a putative strand invasion mechanism. The DSB repair reactions studied here are reminiscent of the RecBCD/RecA reaction and suggest a general mechanism that is likely to be relevant to other systems, including RAD52 mediated recombination.
Keywords: Specific interaction, DSB repair, homologous recombination, RecE/RecT, Redα/Redβ
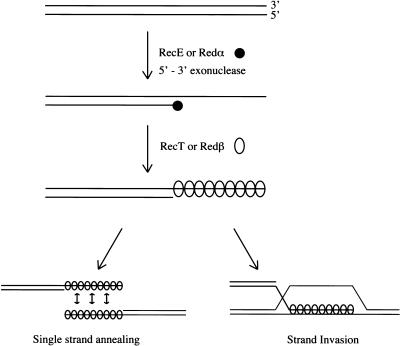
Homologous recombination is of importance to a variety of cellular processes, including the maintenance of genomic integrity, proper segregation of chromosomes in meiosis, and the rescue of stalled replication forks (for review, see Stahl 1996; Kogoma 1996; Cox et al. 2000). Homologous recombination provides a means for repair of DNA double-stranded breaks (DSBs), which can arise during DNA replication as well as after damage by external factors such as irradiation (Edelmann and Kucherlapati 1996; Liang et al. 1998; Richardson et al. 1998; Haber 1999). If DSBs are not repaired, derangements including severe chromosomal defects, carcinogenesis, and cell death can occur (for review, see Kanaar et al. 1998; Pfeiffer 1998). To date studies of DSB repair by homologous recombination have focused on pathways initiated by the prokaryotic protein RecA or its eukaryotic homolog RAD51. However, alternative homologous recombination pathways, which function independently of RecA or RAD51, have been described. For example, inactivation of the RecA/RecBCD pathway in Escherichia coli recBC mutants can be suppressed by sbcA or sbcBC mutations (Barbour et al. 1970; Kushner et al. 1971). In sbcA strains, a cryptic Rac prophage operon is activated to express RecE and RecT (Clark 1974; Willis et al. 1985; Kolodner et al. 1994). RecE is a 5′–3′ exonuclease (Kushner et al. 1971; Joseph and Kolodner 1983) and RecT is a ssDNA-binding protein that promotes ssDNA annealing, strand transfer, and strand invasion in vitro (Clark et al. 1993; Hall et al. 1993; Hall and Kolodner 1994; Noirot and Kolodner 1998). λ Phage contains a similar system to mediate homologous recombination independently of RecA. Here also a phage operon encodes a 5′–3′ exonuclease (Redα; Carter and Radding 1971; Kovall and Matthews 1997) and a ssDNA-binding protein with annealing and strand exchange activity (Redβ; Kmiec and Holloman 1981; Muniyappa and Radding 1986; Karakousis et al. 1998; Li et al. 1998). A variety of studies have concluded that RecE/RecT and Redα/Redβ are functionally equivalent. In particular, recE/recT can substitute for the redα/redβ genes in λ recombination (for review, see Hall and Kolodner 1994; Kolodner et al. 1994). Two models that are not exclusive of each other have been developed to explain DSB repair initiated by the RecE/RecT and Redα/Redβ pathways (Fig. 1; Kobayashi 1992; Stahl et al. 1997). In both models, the 5′–3′ exonuclease, RecE or Redα, resects a DSB to expose a 3′-ended single-stranded region that is then bound by the annealing protein, RecT or Redβ. In the annealing model, the protein–ssDNA filament anneals to a complementary single-stranded region that has arisen from either a similarly prepared DSB or from a DSB produced by DNA replication. In the strand invasion model, the protein–ssDNA filament establishes a D-loop in an unbroken DNA region.
Figure 1.
Current model for double-stranded-break repair initiated by RecE/RecT and Redα/Redβ. For clarity, only one linear end is shown. First, RecE or Redα degrades the DNA in a 5′–3′ direction, starting from the DSB, thereby creating a 3′ ssDNA overhang. Then, RecT or Redβ binds to the ssDNA, forming a recombinogenic proteonucleic filament which is used in recombination, either by single strand annealing or by strand invasion.
Recently we performed a functional screen for a DSB repair reaction that would allow convenient engineering of BACs and other intact circular targets in E. coli. Candidate E. coli hosts were coelectroporated with an intact circular plasmid and a PCR product synthesized to include short flanking regions of homology to the plasmid. Only sbcA hosts gave workable rates of homologous recombination. Subsequently we showed that plasmids expressing the recE/recT or redα/redβ genes convey efficient homologous recombination to recBC hosts (Zhang et al. 1998, Muyrers et al. 1999). For convenience, we refer to use of these protein pairs in a linear plus circular DSB repair reaction as ET recombination.
We explore parameters involved in DSB repair by RecE/RecT and Redα/Redβ by examining different steps of recombination initiation in annealing and ET recombination assays. Our results demonstrate the importance of a specific interaction between the two components of an orthologous pair, which by analogy to other recombination systems suggests a conserved theme for DSB repair.
Results
Both components of either the RecE/RecT or the Redα/Redβ system are required
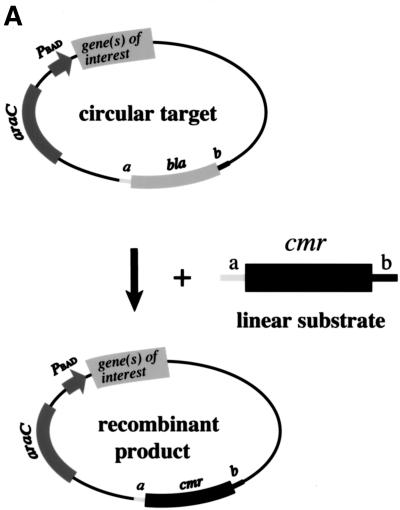
To examine recombination initiation by RecE/RecT and Redα/Redβ, the ET recombination assay illustrated in Figure 2A was used. PCR-generated linear molecules carrying the chloramphenicol resistance gene cmr flanked by homology regions of 50 nucleotides were introduced into host strains containing plasmids from which different combinations of RecE, RecT, Redα, and/or Redβ were expressed. The homology regions directed recombination into the expression plasmid itself, and recombination was scored as the acquisition of chloramphenicol resistance. As shown previously (Zhang et al. 1998), virtually all colonies that acquired antibiotic resistance carried the intended homologously recombined product as determined by DNA restriction analysis (data not shown). Because the first 587 amino acids of full-length RecE are dispensible for exonuclease activity and recombination (Chu et al. 1989), RecE used here started from amino acid 588.
Figure 2.
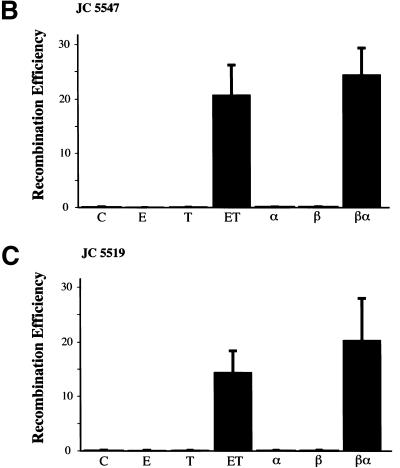
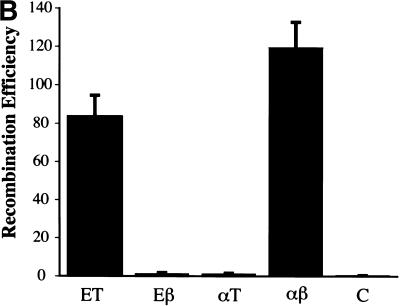
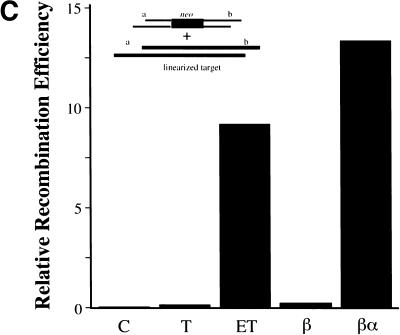
Coexpression of RecE and RecT, or Redα and Redβ, is required for recombination, and RecA does not contribute. (A) Assay used to determine how efficiently different expression constructs performed ET recombination. The gene(s) for the protein(s) of interest were cloned behind the l-arabinose inducible BAD promoter of pBAD24 (Guzman et al. 1995). After l-arabinose induction, electrocompetent cells carrying these constructs were prepared and transformed with a PCR-generated linear substrate consisting of the chloramphenicol resistance gene cmr flanked by homology regions (denoted a and b) at each end. These homology regions were stretches of 50 nucleotides that were also present on the circular target, directly flanking the ampicillin resistance gene bla. Substitution of bla by cmr via recombination through the homology regions resulted in the expression of cmr from the bla promoter, giving rise to chloramphenicol resistant colonies. (B) Recombination in JC5547 (recA−, recBC−; Willetts and Clark 1969) mediated by RecE (E), RecT (T), RecE/RecT (ET), Redα (α), Redβ (β), Redβ/Redα (βα), or no exogenous protein (C). The average of four independent experiments is shown. Error bars represent experimental variation. (C) Same as B except in JC5519 (recA+, recBC−; Willetts and Clark 1969).
First, we tested whether both components of the RecE/RecT or the Redα/Redβ system are required. Homologous recombination did not occur in the absence of any exogenous protein or in the presence of any one component (Fig. 2B). Recombination proceeded efficiently only when both RecE and RecT, or Redα and Redβ, were coexpressed. As controls for the function of the single gene plasmids used, the plasmid expressing only RecE restored recombination in JC8691, a recE− derivative of JC8679 (sbcA, recA+; Clark 1974) and the plasmid expressing only RecT restored recombination in JC8679–ΔrecT, in which recT had been deleted using ET recombination (data not shown). Also, the single gene plasmids were used as parents for the functional constructs described below.
RecA cannot substitute for RecT or Redβ
Although RecA contributes to several examples of RecE/RecT or Redα/Redβ mediated recombination (e.g., Stahl et al. 1978; Muniyappa and Radding 1986; Mythili and Muniyappa 1993; Kolodner et al. 1994; Poteete et al. 1999), it does not contribute to other cases (e.g., Fishel et al. 1981; Silberstein et al. 1990; Nussbaum et al. 1992; Takahashi et al. 1993; Kusano et al. 1994). To test whether RecA contributed to recombination in our assay, or could substitute for any component, parallel experiments were performed in a recA+ host, JC5519 (recA+, recBC−; Willetts and Clark 1969). Very similar results were obtained in the presence and absence of RecA (Figure 2, cf. B and C). Thus, in this assay, RecA cannot substitute for RecT or Redβ, nor affect recombination efficiency.
Recombination requires the orthologous partner
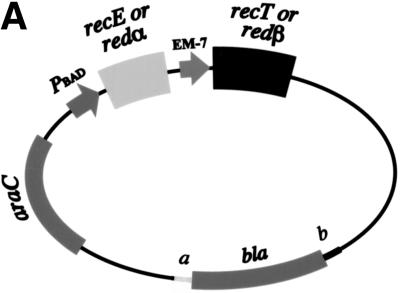
In the current model, RecE or Redα exonucleases initiate recombination from a DSB by generating a 3′-ended ssDNA region. Next, RecT or Redβ binds the ssDNA to create a recombinogenic protein–ssDNA filament (Fig. 1; Kobayashi 1992; Kolodner et al. 1994). The model implies that either exonuclease should be able to cooperate with either annealing protein to initiate recombination. Alternatively, the exonuclease should be dispensible when the linear substrates are preresected to contain ends with 3′ ssDNA overhangs. We tested these predictions, first using constructs from which the orthologous or heterologous pairs were expressed. For practical reasons, the expression strategy was altered. In Figure 2, the protein pairs were expressed from their native operon configurations cloned under the inducible pBAD promoter. In the experiment of Figure 3, the genes were separated so that the exonucleases were placed under the pBAD promoter and the annealing proteins were placed under the strong constitutive EM-7 promoter (Fig. 3A).
Figure 3.
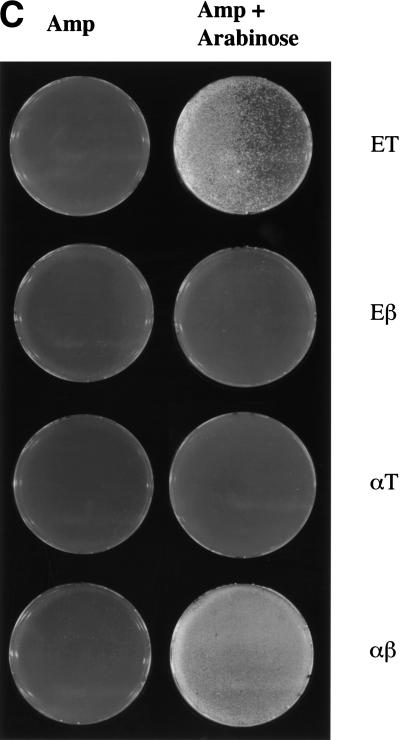
Recombination requires orthologous partners. (A) Diagram of the constructs used. The exonuclease was cloned behind the l-arabinose inducible BAD promoter of pBAD24, whereas the annealing protein was positioned behind the strong, constitutive EM-7 promoter. (B) Recombination efficiencies of JC5547 host cells containing plasmids that expressed pairs of RecE, RecT, Redα, and Redβ as indicated, or no exogenous protein (C), after induction with l-arabinose for 40 min to 2 hr. The average data of four independent experiments is shown. (C) Enhanced survival after UV-irradiation requires the expression of both components of either the RecE/RecT or Redα/Redβ systems. JC5519 cells containing plasmids that expressed the indicated pairs of proteins were plated onto ampicillin only or ampicillin plus l-arabinose plates, UV irradiated, and grown overnight.
Recombinants were obtained when both components of an orthologous protein pair were coexpressed. Neither heterologous pair, however, delivered significant recombination (Fig. 3B). Similarly, repair of UV damage in the JC5519 host required an orthologous pair (Fig. 3C). As expected, recovery from UV damage mediated by RecE/RecT or Redα/Redβ did not occur in the recA− derivative JC5547. As a control for the function of the individual genes present in the two heterologous combinations, the genes for the annealing proteins were cross-cloned into the plasmid containing the orthologous exonuclease to restore recombination proficient orthologous plasmids fully (data not shown).
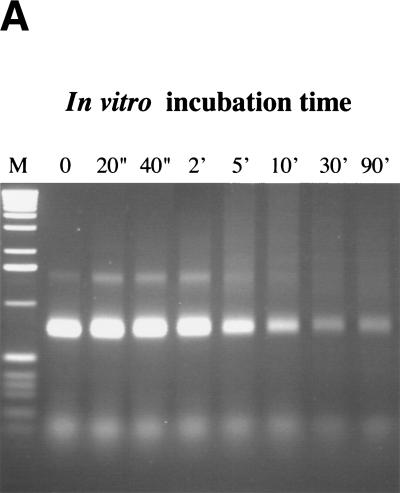
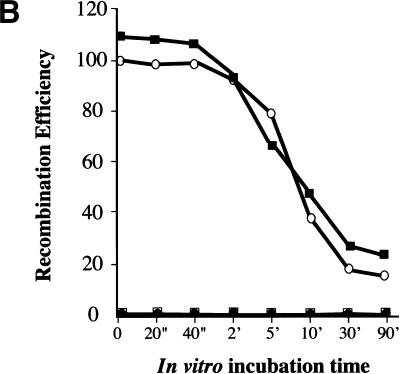
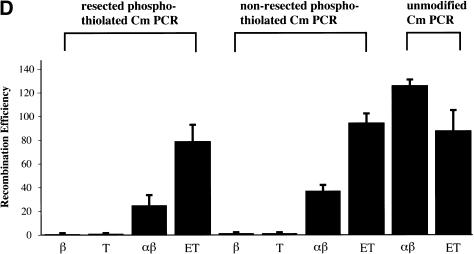
In the second approach, we tested whether reliance on the orthologous 5′–3′ exonuclease could be relieved by use of preresected DNA fragments. Preresected fragments were prepared either by incubation with the nonprocessive T7 gene 6 exonuclease or by use of RecE and linear substrates synthesized to include a triplet of phosphothiolated nucleotides between the homology region and cmr. Using T7 gene 6, a range of incubation temperatures, incubation times, and enzyme concentrations were applied to vary the length of the 3′ ssDNA overhang. An example is shown in Figure 4A. The resected substrates were introduced into cells expressing components of the RecE/RecT or the Redα/Redβ systems. Regardless of preresection, recombinants were only formed in the presence of the orthologous pair, and efficiencies were reduced in approximate concordance with complete exonuclease digestion (Fig. 4B).
Figure 4.
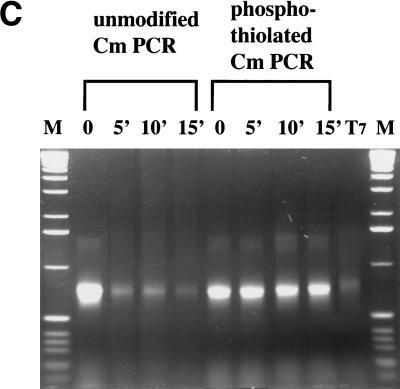
DNA substrates containing in vitro-generated 3′ ssDNA overhangs still require RecE or Redα for recombination. (A) T7 gene 6, a nonprocessive 5′–3′ exonuclease, was used to produce linear substrates (identical to the ones shown in Fig. 2A) containing 3′ ssDNA overhangs of variable length. Aliquots removed during a time course of exonuclease degradation (10 U/μg; 30°C) are shown. M, 1-kb DNA marker (GIBCO BRL). (B) Recombination efficiencies in JC5519 expressing the indicated proteins using the substrates shown in A in the assay of Figure 2A. (○) ET; (●) β; (□ T; (█) αβ. (C) Degradation time courses of unmodified and phosphothiolated substrates after incubation with RecE at 37°C for the indicated times show that the presence of a phosphothiolated triplet inhibited RecE. T7, phosphothiolated substrate incubated with approximately 10 U/μg T7 gene 6 exonuclease; M, 1-kb DNA marker. (D) Recombination obtained using the phosphothiolated substrate after 10 min of RecE digestion in JC5519 expressing the indicated proteins. Results were equivalent using the substrates obtained after five or 15 min of RecE digestion (data not shown). Plasmids expressing RecE/RecT and Redα/Redβ used in B and C are the same as those used in Figure 3.
Precisely defined preresected linear substrates were generated using PCR products containing triplets of phosphothiolated nucleotides between the 50-nucleotide homology regions present on each end, and the cmr gene. As shown in Figure 4C, phosphothiolated triplets blocked RecE exonuclease activity in vitro. Recombination with RecE preresected, phosphotiolated substrates also required an orthologous pair and again, expression of the heterologous pairs or any annealing protein alone did not support significant recombination (Fig. 4D; data not shown). Although phosphothiolated substrates inhibit RecE in vitro, equivalent efficiencies were observed for RecE/RecT recombination in vivo regardless of whether the substrate was unmodified, phosphothiolated, or preresected (Fig. 4D). This indicates that any significant contribution by a 3′–5′ exonuclease can be discounted.However, recombination by Redα/Redβ is affected by the presence of phosphothiolated nucleotides in the substrates.Further work is required to understand this observation.
RecE and RecT protein–protein interaction
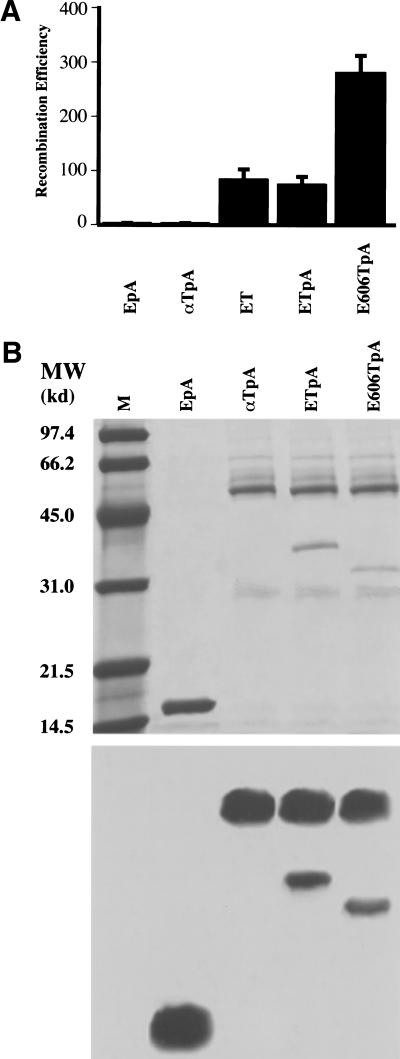
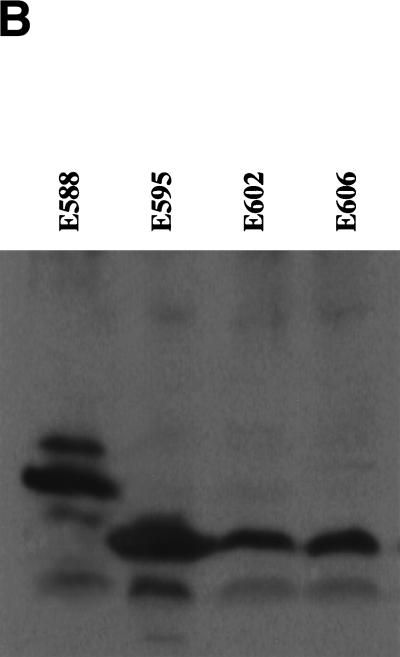
Thus, besides their 5′–3′exonuclease activity, RecE and Redα contribute another essential activity that limits recombination to circumstances in which specific interaction with the orthologous annealing protein is possible. Specific physical interaction between Redα and Redβ has been inferred from observations that these proteins copurify through several steps (Kmiec and Holloman 1981; Muniyappa and Radding 1986). To test whether RecE and RecT interact physically, we expressed RecT as a protein A fusion protein. Positioning protein A at the carboxyl terminus of RecT did not impair recombination (Fig. 5A) or recovery from UV irradiation in the assay of Figure 3C (data not shown). Lysates from cells which expressed the plasmids used in Figure 5A were passed over IgG Sepharose columns. RecE bound to RecT, as did RecE606 (a further amino-terminal truncation of RecE, see below), whereas Redα did not (Fig. 5B). RecE did not interact with protein A alone, demonstrating specificity for RecT. RecE identity was confirmed by Western analysis (Fig. 5B). The physical interaction between RecE and RecT was resistant to DNaseI digestion (data not shown). Thus, RecT can discriminate between RecE and Redα by means of physical interaction.
Figure 5.
Protein–protein interaction between RecE and RecT. (A) Recombination efficiencies in JC5547, using the assay of Figure 2A and plasmids that express the indicated proteins. EpA, pBAD RecE, and EM-7 protein A; αTpA, pBAD Redα, and EM-7 RecTpA; ET, pBAD RecE, and EM-7 RecT; ETpA, pBAD RecE, and EM-7 RecTpA; E606TpA, pBAD RecE606 (see Fig. 6), and EM-7 RecTpA. Data represent the average of three independent experiments. (B) SDS-PAGE analysis (top) of the eluates after IgG sepharose chromatography of lysates from cells that coexpressed the indicated proteins. Loading amounts were normalized to similar RecTpA amounts and bands were visualized with Coomassie blue staining. In the EpA lane, only protein A was detected. M, low range protein markers (Bio-Rad). (Bottom) Immunoblots of these eluates using RecE antibodies. In addition to RecE, protein A and RecTpA were detected because of the strong interaction between protein A and the constant domain of IgG. Similar results were obtained using JC5519 or NS3145 (recA−, Genome Systems, Inc.) as the host strain for protein expression (data not shown).
Recombination is favored by overexpression of RecT
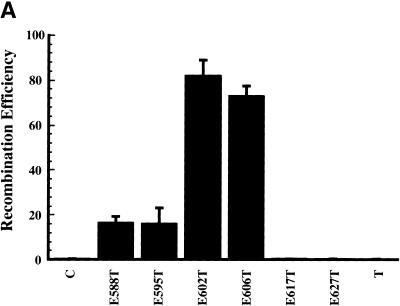
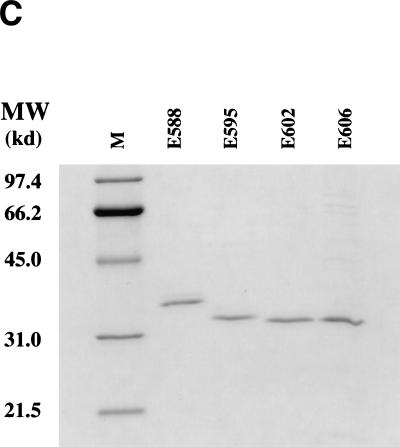
A mutation-sensitive boundary for RecE recombination around amino acid 588 has been defined (Chu et al. 1989). We made a series of fine amino-terminal deletions starting at this point and tested them for activity in recombination. Whereas a deletion of seven amino acids had no effect, recombination efficiency was improved by removal of another seven or 11 amino acids. Larger deletions abolished recombination (Fig. 6A). The increased recombination activity of the RecE602 and Rec606 proteins correlated inversely with a decrease in their expression levels (Fig. 6B). The four active RecE proteins were purified, equalized in concentration (Fig. 6C), and tested for exonuclease activity on linearized dsDNA. The two proteins that showed enhanced activity in recombination (RecE602 and RecE606) showed reduced exonuclease activity at low magnesium concentrations (Fig. 6D). At high magnesium concentrations (10 mm), the differences between exonuclease activities were less, but still observable (data not shown). Thus, reduced exonuclease activity, achieved both by lowered protein expression and by reduced double-stranded exonuclease activity, correlated with improved recombination efficiency.
Figure 6.
Overexpression of RecT relative to RecE enhances recombination efficiency. (A) Recombination efficiencies in JC5547 of amino-terminal RecE deletions, using the assay of Figure 2A, are shown. In these constructs, RecT was coexpressed from the l-arabinose inducible promoter with the following RecE proteins: E588T, RecE588 (the same construct as ET in Fig. 2); E595T, truncated RecE in which seven amino acids were deleted from the amino terminus of RecE588; E602T, deletion of 14 amino acids; E606T, deletion of 18 amino acids; E617T, deletion of 29 amino acids; E627T, deletion of 39 amino acids. T, only RecT expressed; C, no exogenous proteins expressed. (B) Western analysis of the expression level of the RecE proteins after equal induction times. (C) Coomassie stained protein gel of purified RecE proteins after purification and equalization of concentration. (D) Analysis of dsDNA exonuclease activity of purified RecE proteins. Linearized dsDNA was incubated for the times indicated with equal amounts of the indicated RecE proteins, deproteinized, and analyzed on an agarose gel containing ethidium bromide. T, parallel incubation with purified RecT; C, parallel incubation with no added protein; M, 1-kb DNA marker. (E) Recombination efficiencies as a function of RecT expression. The recombination efficiencies are shown using the indicated sbcA host strains carrying a plasmid from which RecT is expressed from the l-arabinose inducible BAD promoter, after 1 hr induction with l-arabinose at the indicated concentrations. (Gray box) JC 8679 ΔrecT; (black box) JC 9604.
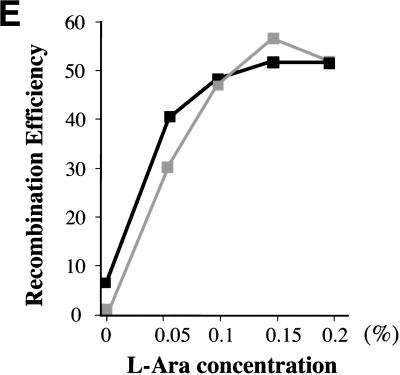
A simple explanation of this improved recombination efficiency is that the lowered level of RecE exonuclease activity alters the ratio of RecE to RecT in a way that favors recombination. This proposition was tested by overexpressing RecT in JC8679–ΔRecT (sbcA, recA+, recT−) and in JC9604 (sbcA, recA−; Clark 1974) from the single gene plasmid used in Figures 2, 4, and 6A. The recombination efficiency increased according to increased expression levels of RecT by arabinose induction (Fig. 6E).
The favorability of a high RecT to RecE expression ratio is in agreement with an observation made during construction of the plasmids pBADETγ and pBADαβγ (Zhang et al. 1998; Muyrers et al. 1999). Initially RecE/RecT or Redα/Redβ were placed under the BAD promoter as operons. Severing the operons to place RecT or Redβ under the strong, constitutive EM-7 promoter improved the recombination efficiency significantly. This effect can be seen by comparing recombination efficiencies presented in Figures 2 and 3. Conversely, placement of RecE or Redα under the EM-7 promoter and RecT or Redβ under the BAD promoter decreased recombination (data not shown). This is consistent with the observation that recombination is favored by relative overexpression of the annealing protein. In contrast, although expression of the exonuclease is required for recombination, its relative overexpression with respect to its annealing protein partner disfavors recombination.
Annealing vs. strand invasion mechanisms
The initiation of recombination by RecE/RecT or Redα/Redβ is completed by establishing a hybrid DNA region by either a single-strand annealing or a strand invasion mechanism. To examine this aspect of DSB repair, the linear plus circular ET assay described was compared with a linear plus linear variation. For single-strand annealing, two linear molecules sharing terminal homology regions are required. Because strand invasion relies on the invasion of an intact duplex, only one linear molecule is required (see Fig. 1). Thus, we reasoned that the ET format may represent a strand invasion pathway, whereas the linear plus linear assay represents an annealing pathway. To compare the two, cells expressing RecE/RecT or Redα/Redβ were cotransformed with a PCR-generated linear substrate and either an intact circular target, or a target that was prelinearized between the homology regions by restriction digestion. A series of PCR-generated substrates that contained ends of variable homology length to the target were tested. Two differences between the annealing and the ET assays emerged. For both RecE/RecT and Redα/Redβ, the absolute recombination efficiency was higher at any tested length of homology region in the annealing than in the ET assay. Furthermore, two linear molecules were more efficiently recombined with increasing homology region length (Fig. 7A). In contrast, recombination between a circular and a linear target became more efficient only until a homology length of 100 for RecE/RecT, or 120 for Redα/Redβ, after which it remained the same (Fig. 7B). Similar results were obtained in JC5547 or JC5519, demonstrating RecA independence, and as in Figure 2, one component alone did not promote recombination (data not shown). Use of a different RecE/RecT or Redα/Redβ expression configuration (using the EM-7 promoter for RecT or Redβ), delivered the same qualitative results. Thus, the initiation of recombination by either RecE/RecT or Redα/Redβ in annealing and putative strand invasion assays can be qualitatively and quantitatively distinguished.
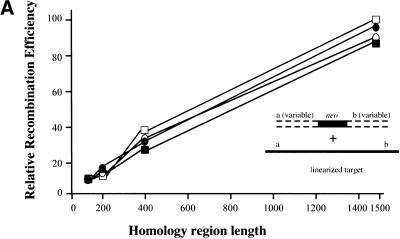
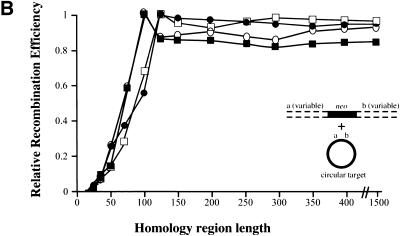
Figure 7.
Different RecE/RecT and Redα/Redβ recombination mechanisms revealed by different substrates. (A) Relative recombination efficiencies achieved using RecE/RecT (ET) or Redα/Redβ (βα) expression constructs in either JC5547 or JC5519 as indicated, as a function of homology region length, are shown. Linear molecules, which were generated to contain homology regions of variable length at their ends, were allowed to recombine with a prelinearized target generated by restriction digestion. Recombination efficiencies from one experiment are shown relative to the maximum recombination efficiency of B, which was set to 1. Recombination efficiencies are thus presented as relative and are not directly comparable to those shown in previous figures. The experiment was repeated twice, producing similar results. (█) 5547ET; (○) 5519ET; (●) 5547βα; (□) 5519βα. (B) Same as A, except that the target was left intact. The maximum absolute recombination efficiency found was arbitrarily set to 1, and all other recombination efficiencies were related to this maximum. Recombination efficiencies from one experiment are shown. The experiment was repeated five times, producing similar results. The experiments shown in A and B were done in parallel with the same batches of competent cells and PCR fragments. Colony numbers observed in these experiments were adjusted to relative recombination efficiencies by relating to the maximums obtained in B, which were set to 1. (█) 5547ET; (○) 5519ET; (●) 5547βα; (□) 5519βα. (C) Recombination efficiencies in the presence of the indicated expression constructs in JC5519 using two linear molecules, both of which were preresected in vitro using T7 gene six exonuclease. Data from one representative experiment is shown, in which pools of preresected molecules (which shared 400 nucleotide homology regions and were generated using six different incubation conditions with T7 gene 6 exonuclease) were recombined. C, no exogenous protein expressed; β, expression of Redβ only; βα, coexpression of Redβ and Redα; T, expression of RecT only; ET, coexpression of RecE and RecT. Recombination efficiencies are shown relative to the maximum of B.
A further test of the dependence of RecT on RecE, or Redβ on Redα, is shown in Figure 7C. Both linear substrates of Figure 7A were treated with T7 gene 6 exonuclease to generate 3′ ssDNA overhangs of various lengths. These preresected substrates were pooled and coelectroporated into strains expressing components of the RecE/RecT and the Redα/Redβ systems. Although some recombination occured in the presence of an annealing protein alone, the copresence of the orthologous exonuclease increased the efficiency ≥20-fold. Like recombination between a linear and a circular molecule, recombination between two linear molecules thus depends on the presence of both components of RecE/RecT or Redα/Redβ.
Discussion
The initial steps of DSB repair mediated by two protein pairs, RecE/RecT and Redα/Redβ, were examined. Recombination mediated by these pairs is mechanistically and functionally equivalent. Both pairs are a combination of a 5′–3′ exonuclease with a ssDNA-binding protein that has annealing activity, and recE/recT can functionally substitute for the redα/redβ genes in several recombination assays (for review, see Kolodner et al. 1994; Hall and Kolodner 1994). Furthermore, both pairs delivered very similar efficiencies of recombination when compared side-by-side in the same assay (Figs. 2–4,7). However, we show that the existing two-step model of initiation, in which the only contribution of the exonuclease involves the resection of a DSB to expose a single-stranded region for binding by the annealing protein (Fig. 1), is not sufficient. Recombination required expression of both the exonuclease and annealing proteins even when preresected DSBs were used to replace the need for exonuclease activity or when RecA was used to replace the annealing protein. Furthermore, recombination required the expression of both members of an orthologous pair and was not recovered with expression of the heterologous pairs.
The simplest explanation of the additional orthologous function lies with a specific protein–protein interaction. The existence of a specific protein–protein interaction between Redα and Redβ has been inferred from their coelution through several biochemical purification steps (Kmiec and Holloman 1981; Muniyappa and Radding 1986). We observe specific protein–protein interaction between RecE and RecT, but not between Redα and RecT (Fig. 5B). Thus, the evidence for specific protein–protein interactions correlates with the required orthologous function. Several observations presented argue against a simple one-to-one molar relationship between the exonuclease and annealing protein. These include: (1) empirical work showing that recombination was enhanced by plasmid-based expression configurations producing an excess of the annealing protein over the exonuclease, and diminished by the alternative configuration (Figs. 2,3, and data not shown); (2) our finding that fine deletions at the amino terminus of RecE enhanced recombination. When examined in detail, it was found that these deletions decreased both RecE expression levels and exonuclease activity (Fig. 6A–D); (3) overexpression of RecT in sbcA strains enhanced recombination (Fig. 6E). This combination of an exonuclease with a relatively overexpressed annealing protein and a protein–protein interaction is reminiscent of the relationship between the RecBCD exonuclease and RecA proteins. After encountering a chi site, RecBCD resects in the 5′–3′ direction and serves as a loading factor that assists RecA binding to the emerging 3′-ended strand, thereby allowing efficient competition with SSB, the high affinity single-stranded binding protein, which both competes with and contributes to RecA filament formation (Anderson and Kowalczykowski 1997, 1998; Eggleston and West 1997). By analogy, RecE and Redα could promote loading of their corresponding annealing proteins, allowing the formation of a recombinogenic proteonucleic filament in the presence of SSB. In the simplest case conceivable, one exonuclease complex is bound to the dsDNA end, while the protruding ssDNA becomes coated with many molecules of the annealing protein, thereby explaining our observations that recombination is favored by higher levels of annealing protein than exonuclease.
Notably, electron microscopy (EM) analysis of RecT-DNA fibers showed that addition of RecE resulted in formation of filamentous nucleoprotein structures at the DNA ends (Thresher et al. 1995). These structures could reflect RecE-promoted loading of RecT onto DNA or a structural contribution by RecE to the recombinogenic complex. In either case, as our data demonstrate that both components of these protein pairs are required for recombination, it is likely that the RecE/RecT filamentous structures have more relevance than the repetitive fibers or spherical particles seen in EM with RecT alone. By analogy, we predict that EM analysis of Redα/Redβ proteonucleic complexes will reveal relevantly different structures compared to the repetitive Redβ fibers and rings described previously (Passy et al. 1999).
After establishment of RecE/RecT or Redα/Redβ protein–ssDNA filaments, the next step in homologous recombination is either annealing to a complementary single-stranded region or strand invasion of an intact duplex (see Fig. 1). Previous work with these proteins demonstrates that they are efficient annealing proteins, but their ability to perform strand invasion is controversial. Using DSB assays, evidence suggestive of strand invasion, in the presence or absence of RecA, has been presented (Kobayashi and Takahashi 1988; Takahashi and Kobayashi 1990; Nussbaum et al. 1992; Poteete et al. 1999). When the issue of annealing versus strand invasion by Redα/Redβ was explicitly addressed, it was concluded that the vast majority of recombination events was attributable to annealing (Stahl et al. 1997). However, those investigators noted that a small proportion of recombination events could have arisen by another mechanism. Biochemical evidence for strand invasion by RecT in vitro has been presented previously (Noirot and Kolodner 1998). However, those in vitro studies may not have settled the issue to the satisfaction of all, as RecT mediated strand invasion was observed using supercoiled templates in the absence of magnesium and SSB. In a recent review (Kusminov 1999), the perspective was presented that none of this evidence proves that RecE/RecT or Redα/Redβ, in the absence of RecA, can utilize a strand invasion mechanism.
To the debate, we add the observation that ET recombination efficiencies plateau with increasing homology region length (Fig. 7B). Beyond a homology region length of 100 (RecE/RecT) or 120 (Redα/Redβ) nucleotides, no significant increase or decrease in the number of recombinants was observed. In contrast, when two linear substrates were used in parallel annealing pathway experiments, recombination efficiencies increased linearly with increasing length of homology (Fig. 7A). Thus, there is a significant operational difference between linear plus linear (annealing) and linear plus circular (ET) recombination. Even at the most efficient homology region length for ET recombination (100 or 120 nucleotides), the annealing pathway provides about 10-fold more recombinants. At longer lengths, absolute numbers of the annealing pathway recombinants increase to >80-fold those obtained with ET recombination. Because most former studies of annealing versus strand invasion mechanisms have used long homology regions, annealing pathway events would be expected to dominate, with potential strand invasion events occurring at very low frequencies. In contrast to former investigations, ET recombination presents an example of a recombination assay in which the annealing pathway is not readily available and thus cannot obscure other mechanisms.
In a strand invasion mechanism, the plateau observed beyond 100–120 nucleotides with ET recombination may represent a functional limit of homology searching attributable to a limiting aspect of either the RecE/RecT or Redα/Redβ recombinogenic complexes or the triple stranded intermediates. If RecE/RecT and Redα/Redβ can only use annealing mechanisms in the absence of RecA, then the second DSB probably arises from the replication fork (Kuzminov 1999). In this case, the plateau could reflect a limiting aspect of replication-associated DSB exposure before processing by other mechanisms. However, any explanation needs to incorporate our consistent observation that RecE/RecT efficiencies plateau at 100 nucleotides of homology whereas Redα/Redβ efficiencies plateau at 120 nucleotides. This difference is consistent across several different conditions of RecE/RecT or Redα/Redβ expression, including low to high expression of both protein pairs or varied exonuclease to annealing protein ratios (data not shown).
Regardless of whether annealing or putative strand invasion pathways are used, the expression of both components of the RecE/RecT or Redα/Redβ systems is required for efficient recombination. This is true even for recombination between two preresected linear molecules (Fig. 7C) and indicates that, in agreement with the loading model analogy described above, functional cooperation between orthologous proteins is required at the step of proteonucleic filament formation.
Specific cooperation between an exonuclease and its partner annealing protein may be a central theme in DSB repair by homologous recombination. Besides its importance in RecBCD/RecA mediated proteonucleic filament formation, it is likely to underly recombination by several potential analogs of the RecE/RecT and the Redα/Redβ systems in different organisms that have been suggested (Kolodner et al. 1994; Thresher et al. 1995). Also, a recent analysis of the Redβ–ssDNA fiber by electron microscopy led to the proposition that RAD52, RecT, and Redβ are similar (Passy et al. 1999). Although we, and others, have been unable to identify any protein sequence similarities between any of RecT, Redβ, and any RAD52, the proposition is provocative. Like RecT and Redβ, RAD52 binds to ssDNA and promotes annealing in vitro (Mortensen et al. 1996). Furthermore, RAD52 performs in vivo recombination with or without RAD51, the eukaryotic homolog of RecA (for review, see Haber 1999), as do RecT or Redβ with or without RecA. Although RAD52 mediates homologous recombination primarily by an annealing mechanism, genetic evidence suggests the possibility of strand invasion at a low frequency, in the absence of RAD51 (Ivanov et al. 1996; Baertsch et al. 2000). Given these apparent functional analogies, and our finding that RecT and Redβ require their partner exonuclease for recombination, it will be interesting to determine whether a partner 5′–3′ exonuclease exists for RAD52, because a 5′–3′ exonuclease/RAD52 pair may have relevant different properties from RAD52 alone.
Materials and methods
Plasmids and oligonucleotides
All plasmids were based on pBAD24 (Guzman et al. 1995) and were constructed by conventional methods or by ET recombination (Zhang et al. 1998). Similar expression levels of the various proteins expressed from these plasmids were confirmed by SDS-PAGE followed by Coomassie staining and/or Western blotting. Sequences of the oligonucleotides used to generate the linear substrate of Figure 2A are: 5′-TGAGACAATAACCCTGATAAATGCTTCAATAATATTGAAAAAGGAAGAGTATGGAGAAAAAAATCACTGGATATACCACCG-3′ (homology region a is underlined); 5′-TACAGGGCGCGTAAATCAATCTAAAGTATATATGAGTAAACTTGGTCTGACAGTTACG-CCCCGCCCTGCCACTCATCGCA-3′ (homology region b is underlined). Oligonucleotides were synthesized by the EMBL oligonucleotide service. Details of plasmids and oligonucleotides used are available upon request.
Recombination efficiency assay
JC5547 (recA−, recBC−; Willetts and Clark 1969) was transformed by the pBAD24-based expression plasmid carrying the gene(s) of interest. Electrocompetent cells were prepared as described previously (Zhang et al. 1998); further experimental details are available (Muyrers et al. 2000; see http://www.embl-heidelberg.de/ExternalInfo/stewart/index.html). Cells were electroporated with 0.3 μg PCR fragment consisting of a 50 nucleotides 5′ homology region (denoted a in Fig. 2A) ending at the ATG of bla, followed by the chloramphenicol resistance gene (cmr) and the 50 nucleotides 3′ homology region (b in Fig. 2A) starting immediately after the stop codon of bla. Recombination replaced bla with cmr, thereby cmr was expressed from the bla promoter, allowing colonies to grow on LB plates that contained 50 μg/ml chloramphenicol. Colony numbers observed in a given experiment were adjusted to values shown as recombination efficiency by adjusting for batch-to-batch variations in transformation efficiency. Thus, recombination efficiency was calculated by dividing the observed number of chloramphenicol resistant recombinants by the transformation efficiency of the competent cell preparation, defined as the amount of colonies obtained by transforming a standard amount (0.5 ng) of pBR322 plasmid. Correct recombinants were routinely confirmed by DNA restriction analysis.
UV protection assay
Similar numbers of JC5519 host cells containing the appropriate expression plasmid were plated on LB plates containing 100 μg/ml ampicillin, in the presence or absence of 0.1% arabinose. Plates were preincubated for 2 hr at 37°C and then exposed to UV light at 312 nm for 25 sec by inverting them on a transilluminator (UVT-20M, Herolab). Cell survival was determined after overnight incubation at 37°C.
In vitro generation of 3′ ssDNA overhangs
T7 gene 6 exonuclease digestion was done in the buffer provided by the supplier (Amersham, Inc.), using ∼1.5 μg linear substrate per reaction. The amounts of enzyme used varied between four and 200 units, incubation temperatures varied between 4°C and 37°C, and incubation times varied between 15 sec and 90 min. In total, 12 different combinations of incubation temperature and enzyme concentration were tested using a range of incubation times. Substrates carrying a phosphothiolated triplet at the border of the homology regions and cmr were PCR amplified using oligonucleotides carrying this modification. Approximately 1.5 μg substrate was digested with 0.5 μg RecE in reaction buffer composed of 10 mm Tris-HCl (pH 7.9), 10 mm MgCl2, 50 mm NaCl, 1 mm DTT. All in vitro digestions were terminated by extraction with a mixture of phenol/chloroform/isoamylalcohol (Amresco) after which the resected substrates were precipitated, washed, and dissolved in ddH2O. An aliquot was checked on an agarose gel for DNA degradation, and ∼0.3 μg was used for recombination efficiency assays.
RecE and RecT protein A interaction assay
Host cells carrying the plasmids of interest were grown to OD600 0.1 and induced with l-arabinose for 3 hr. Cells were then pelleted and resuspended in 4 ml protein A buffer (25 mm NaCl, 20 mm Tris at pH 7.4, 0.1% NP40, 5 mm MgCl2) containing 20% glycerol and stored at −80°C. Cells were lysed by incubation at 4°C for 45 min with 0.2 mg/ml lysozyme, protein inhibitor cocktail at the recommended concentration (Roche) and 10 mm spermidine (Sigma). Clear lysates, obtained by ultracentrifugation, were stored at −80°C. Aliquots were passed over a Poly-Prep chromatography column (BioRad Laboratories) containing IgG Sepharose 6 fast flow (Amersham Pharmacia Biotech), washed with 25 ml protein A buffer, and eluates were collected as recommended by the manufacturer. These were then lyophilized and analyzed by SDS-PAGE and Western immunoblotting using a routinely generated rabbit anti-serum to purified RecE. The protein–protein interaction between RecE and RecT was stable up to ≤200 mm NaCl.
In vitro RecE exonuclease assay
Amino-terminal truncations of RecE were purified as described (Luisi-DeLuca et al. 1988) from the NS3145 host strain (Genome Systems, Inc.) carrying RecE expression plasmids. Following purification to >90% as determined by Coomassie-stained protein gels, the concentrations of the proteins were equalized and confirmed by Western analysis using the rabbit-derived anti-RecE serum. Approximately 0.3 μg EcoRV-linearized pSVpaZ11 (Buchholz et al. 1996) was used as the substrate for the exonuclease assay. Exonuclease assays were performed at 37°C in a buffer composed of 25 mm NaCl, 20 mm Tris at pH 7.5, 0.3 mm MgCl2 (or 10 mm MgCl2), 1 μg/ml BSA, and 0.5 mm DTT, using ∼10 ng of the RecE mutant to be tested.
Recombination mechanism assays
Supercoiled pSVpaZ11 (Buchholz et al. 1996) was used as the circular target. To obtain the linear target, pSVpaZ11 was linearized by EcoRV digestion at its unique recognition site in pSVpaZ11, approximately in the middle of lacZ. PCR-generated linear DNA substrates containing various lengths of homology to pSVpaZ11 were generated as follows: First, the neomycin resistance gene neo, which gives resistance to kanamycin in prokaryotes, was introduced at the EcoRV site on pSVpaZ11 by ET recombination. The resulting plasmid, pSVneo, was then used as the template for PCR amplification of the neo gene using PCR-primer pairs at various distances from the neo promoter at the 5′ end, and from the stop codon of neo at the 3′ end. These distances were thus equal to the length of the identical sequence (the homology region) shared between the neo-containing linear PCR fragments and pSVpaZ11. Recombination efficiencies were determined and normalized as described above, using resistance to kanamycin for selection.
Acknowledgments
We thank Inhua Muyrers-Chen, Michelle Meredyth, and Vladimir Benes for critical reading of the manuscript. This work was supported in part by grants from the Volkswagen Foundation, Program on Conditional Mutagenesis, and the NIH, National Institute for Aging. Y.Z. was a recipient of an EMBO fellowship. F.B. is a fellow of The Leukemia & Lymphoma Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL stewart@embl-heidelberg.de; FAX 49-6221-387518.
References
- Anderson DG, Kowalczykowski SC. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- ————— SSB protein controls RecBCD enzyme nuclease activity during unwinding: A new role for looped intermediates. J Mol Biol. 1998;282:275–285. doi: 10.1006/jmbi.1998.2013. [DOI] [PubMed] [Google Scholar]
- Barbour SD, Nagaishi H, Templin A, Clark AJ. Biochemical and genetic studies of recombination proficiency in Escherichia coli. II. Rec+ revertants caused by indirect suppression of rec− mutations. Proc Natl Acad Sci. 1970;67:128–135. doi: 10.1073/pnas.67.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baertsch S, Kang LE, Symington LS. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol Cell Biol. 2000;20:1194–1205. doi: 10.1128/mcb.20.4.1194-1205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Angrand PO, Stewart AF. A simple assay to determine the functionality of Cre or FLP recombination targets in genomic manipulation constructs. Nucleic Acids Res. 1996;24:3118–3119. doi: 10.1093/nar/24.15.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter DM, Radding CM. The role of exonuclease and beta protein of phage lambda in genetic recombination. II. Substrate specificity and the mode of action of lambda exonuclease. J Biol Chem. 1971;246:2502–2512. [PubMed] [Google Scholar]
- Chu CC, Templin A, Clark AJ. Suppression of a frameshift mutation in the recE gene of Escherichia coli K-12 occurs by gene fusion. J Bacteriol. 1989;171:2101–2109. doi: 10.1128/jb.171.4.2101-2109.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ. Progress toward a metabolic interpretation of genetic recombination of Escherichia coli and bacteriophage lambda. Genetics. 1974;78:259–271. doi: 10.1093/genetics/78.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Sharma V, Brenowitz S, Chu CC, Sandler S, Satin L, Templin A, Berger I, Cohen A. Genetic and molecular analyses of the C-terminal region of the recE gene from the Rac prophage of Escherichia coli K-12 reveal the recT gene. J Bacteriol. 1993;175:7673–7682. doi: 10.1128/jb.175.23.7673-7682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Kucherlapati R. Role of recombination enzymes in mammalian cell survival. Proc Natl Acad Sci. 1996;93:6225–6227. doi: 10.1073/pnas.93.13.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston AK, West SC. Recombination initiation: Easy as A, B, C, D... chi? Curr Biol. 1997;7:R745–R749. doi: 10.1016/s0960-9822(06)00394-0. [DOI] [PubMed] [Google Scholar]
- Fishel RA, James AA, Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981;294:184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. DNA repair. Gatekeepers of recombination. Nature. 1999;398:665–667. doi: 10.1038/19423. [DOI] [PubMed] [Google Scholar]
- Hall SD, Kolodner RD. Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc Natl Acad Sci. 1994;91:3205–3209. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SD, Kane MF, Kolodner RD. Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. J Bacteriol. 1993;175:277–287. doi: 10.1128/jb.175.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph JW, Kolodner R. Exonuclease VIII of Escherichia coli. I. Purification and physical properties. J Biol Chem. 1983;258:10411–10417. [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JH, van Gent DC. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- Karakousis G, Ye N, Li Z, Chiu SK, Reddy G, Radding CM. The beta protein of phage lambda binds preferentially to an intermediate in DNA renaturation. J Mol Biol. 1998;276:721–731. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- Kmiec E, Holloman WK. Beta protein of bacteriophage lambda promotes renaturation of DNA. J Biol Chem. 1981;256:12636–12639. [PubMed] [Google Scholar]
- Kobayashi I. Mechanisms for gene conversion and homologous recombination: The double-strand break repair model and the successive half crossing-over model. Adv Biophys. 1992;28:81–133. doi: 10.1016/0065-227x(92)90023-k. [DOI] [PubMed] [Google Scholar]
- Kobayashi I, Takahashi N. Double-stranded gap repair of DNA by gene conversion in Escherichia coli. Genetics. 1988;119:751–757. doi: 10.1093/genetics/119.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T. Recombination by replication. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- Kolodner R, Hall SD, Luisi-DeLuca C. Homologous pairing proteins encoded by the Escherichia coli recE and recT genes. Mol Microbiol. 1994;11:23–30. doi: 10.1111/j.1365-2958.1994.tb00286.x. [DOI] [PubMed] [Google Scholar]
- Kovall R, Matthews BW. Toroidal structure of lambda-exonuclease. Science. 1997;277:1824–1827. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- Kusano K, Sunohara Y, Takahashi N, Yoshikura H, Kobayashi I. DNA double-strand break repair: Genetic determinants of flanking crossing-over. Proc Natl Acad Sci. 1994;91:1173–1177. doi: 10.1073/pnas.91.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner SR, Nagaishi H, Templin A, Clark AJ. Genetic recombination in Escherichia coli: The role of exonuclease I. Proc Natl Acad Sci. 1971;68:824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Karakousis G, Chiu SK, Reddy G, Radding CM. The beta protein of phage lambda promotes strand exchange. J Mol Biol. 1998;276:733–744. doi: 10.1006/jmbi.1997.1572. [DOI] [PubMed] [Google Scholar]
- Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi-DeLuca C, Clark AJ, Kolodner RD. Analysis of the recE locus of Escherichia coli K-12 by use of polyclonal antibodies to exonuclease VIII. J Bacteriol. 1988;170:5797–5805. doi: 10.1128/jb.170.12.5797-5805.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniyappa K, Radding CM. The homologous recombination system of phage lambda. Pairing activities of beta protein. J Biol Chem. 1986;261:7472–7478. [PubMed] [Google Scholar]
- Muyrers JPP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyrers JPP, Zhang Y, Stewart AF. ET-cloning: Think recombination first. In: Setlow JK, editor. Genetic engineering: Principles and methods. 2000. , (In press) Plenum Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- Mythili E, Muniyappa K. Formation of linear plasmid multimers promoted by the phage lambda Red-system in lon mutants of Escherichia coli. J Gen Microbiol. 1993;139:2387–2397. doi: 10.1099/00221287-139-10-2387. [DOI] [PubMed] [Google Scholar]
- Noirot P, Kolodner RD. DNA strand invasion promoted by Escherichia coli RecT protein. J Biol Chem. 1998;273:12274–12280. doi: 10.1074/jbc.273.20.12274. [DOI] [PubMed] [Google Scholar]
- Nussbaum A, Shalit M, Cohen A. Restriction-stimulated homologous recombination of plasmids by the RecE pathway of Escherichia coli. Genetics. 1992;130:37–49. doi: 10.1093/genetics/130.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passy SI, Yu X, Li Z, Radding CM, Egelman EH. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc Natl Acad Sci. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P. The mutagenic potential of DNA double-strand break repair. Toxicol Lett. 1998;96–97:119–129. doi: 10.1016/s0378-4274(98)00058-7. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Fenton AC, Murphy KC. Roles of RuvC and RecG in phage lambda red-mediated recombination. J Bacteriol. 1999;181:5402–5408. doi: 10.1128/jb.181.17.5402-5408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: Suppression of chromosomal translocations. Genes & Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein Z, Maor S, Berger I, Cohen A. Lambda Red-mediated synthesis of plasmid linear multimers in Escherichia coli K12. Mol Gen Genet. 1990;223:496–507. doi: 10.1007/BF00264459. [DOI] [PubMed] [Google Scholar]
- Stahl F. Meiotic recombination in yeast: Coronation of the double-strand-break repair model. Cell. 1996;87:965–968. doi: 10.1016/s0092-8674(00)81791-2. [DOI] [PubMed] [Google Scholar]
- Stahl FW, Stahl MM, Malone RE. Red-mediated recombination of phage lambda in a recA−recB− host. Mol Gen Genet. 1978;159:207–211. [Google Scholar]
- Stahl MM, Thomason L, Poteete AR, Tarkowski T, Kuzminov A, Stahl FW. Annealing vs. invasion in phage lambda recombination. Genetics. 1997;147:961–977. doi: 10.1093/genetics/147.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Kobayashi I. Evidence for the double-strand break repair model of bacteriophage lambda recombination. Proc Natl Acad Sci. 1990;87:2790–2794. doi: 10.1073/pnas.87.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi NK, Kusano K, Yokochi T, Kitamura Y, Yoshikura H, Kobayashi I. Genetic analysis of double-strand break repair in Escherichia coli. J Bacteriol. 1993;175:5176–5185. doi: 10.1128/jb.175.16.5176-5185.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thresher RJ, Makhov AM, Hall SD, Kolodner R, Griffith JD. Electron microscopic visualization of RecT protein and its complexes with DNA. J Mol Biol. 1995;254:364–371. doi: 10.1006/jmbi.1995.0623. [DOI] [PubMed] [Google Scholar]
- Willetts NS, Clark AJ. Characteristics of some multiply recombination-deficient strains of Escherichia coli. J Bacteriol. 1969;100:231–239. doi: 10.1128/jb.100.1.231-239.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DK, Satin LH, Clark AJ. Mutation-dependent suppression of recB21 recC22 by a region cloned from the Rac prophage of Escherichia coli K-12. J Bacteriol. 1985;162:1166–1172. doi: 10.1128/jb.162.3.1166-1172.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Buchholz F, Muyrers JPP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]