Abstract
Genomic and antigenic characterization of Aguacate virus, a tentative species of the genus Phlebovirus, and three other unclassified viruses, Armero virus, Durania virus and Ixcanal virus, demonstrate a close relationship to one another. They are distinct from the other nine recognized species within the genus Phlebovirus. We propose to designate them as a new (tenth) serogroup or species (Aguacate virus) within the genus. The four viruses were all isolated from phlebotomine sandflies (Lutzomyia sp.) collected in Central and South America. Aguacate virus appears to be a natural reassortant and serves as one more example of the high frequency of reassortment in this genus.
Introduction
The family Bunyaviridae currently comprises five genera that are differentiated by antigenic and molecular characteristics: Hantavirus, Nairovirus, Orthobunyavirus, Phlebovirus and Tospovirus (Nichol et al., 2005) (http://www.ictvonline.org/virusTaxonomy.asp?version=2008). Viruses in the genus Phlebovirus are further subdivided into two groups: the sandfly fever and the Uukuniemi groups, primarily on the basis of the presence of the non-structural ORF in the medium (M) segment (Nichol et al., 2005).
Because of the lack of biochemical characterization of most of the named phleboviruses, the International Committee on Taxonomy of Viruses (ICTV) defines species within the genus Phlebovirus by their serological relationships (Nichol et al., 2005). Cross-complement fixation (CF) and plaque-reduction neutralization tests (PRNT) have generally been used for classification of the genus (Tesh et al., 1976, 1982; Travassos da Rosa et al., 1983). Using these criteria, 37 viruses have been assigned to nine Phlebovirus species. Sixteen other named viruses that show little serological relationship to the nine recognized groups are classified as tentative species in the genus.
Here we describe the genetic and antigenic characterization of one of those tentative phleboviruses, Aguacate virus (AGUV) and of three other unclassified viruses: Armero virus (ARMV), Durania virus (DURV) and Ixcanal virus (IXCV). These four viruses are phylogenetically and antigenically related to each other and we propose that they comprise a new (tenth) species of the genus Phlebovirus, tentatively named Aguacate virus. The four viruses were all isolated from sandflies collected in Central or South America in the 1970s and 1980s. Although they are antigenically related to some other members of the phlebotomus fever serogroup by CF test, they are distinct from these other members and from each other by PRNT (Tesh et al., 1974, 1982, 1989; Travassos da Rosa et al., 1983). The present communication is the second report in our effort to develop a more precise classification system for the phleboviruses by sequencing most of the named viruses in the genus in order to clarify their phylogenetic relationships (Palacios et al., 2011).
Results
Serological analysis
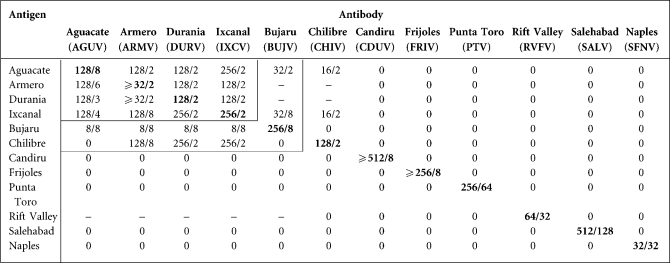
Results obtained in CF tests with AGUV, ARMV, DURV, IXCV and one representative from each of the eight recognized species in the sandfly fever group of phleboviruses are summarized in Table 1. All antisera were reactive with their homologous antigens. Antisera to AGUV, ARMV, DURV and IXCV were markedly cross-reactive with each other in a pattern consistent with a serocomplex comprising these four viruses. Chilibre virus (CHIV) antigen was strongly reactive with ARMV, DURV and IXCV antibodies, and Bujaru virus (BUJV) antigen was slightly reactive to AGUV, ARMV, DURV and IXCV antibodies. Likewise, CHIV and BUJV antibodies reacted slightly with AGUV and IXCV antigens, but the titres were significantly lower than with the homologous antibodies.
Table 1. Results of complement-fixation tests with representative phleboviruses.
Homologous antibody/antigen titres are shown in boldface type. CF titres are expressed as the highest antibody/highest antigen dilution. 0 = <8/Φ.
Genomic characterization
AGUV, ARMV, DURV and IXCV all have the genomic organization characteristic of phleboviruses: three RNA segments that include a large (L) segment encoding the RNA polymerase, a medium segment encoding a polyprotein that includes the non-structural protein (NSm) and both glycoproteins (Gn and Gc), as well as a small (S) segment encoding the nucleocapsid (N) protein (NP) and, in ambisense orientation, a non-structural protein (NSs) (GenBank accession nos HM566137–9, HM566140–2, HM566155–7, HM566161–3, HQ661805–7).
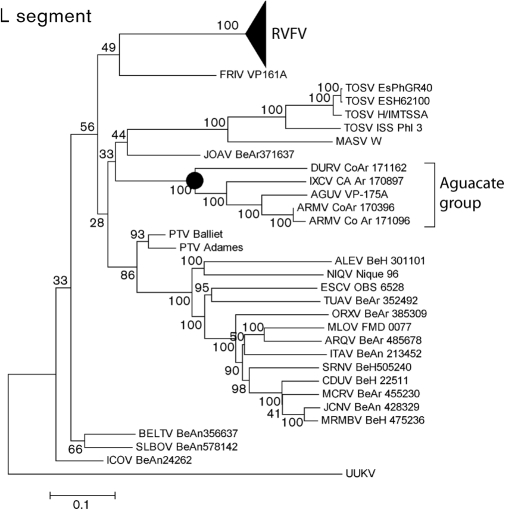
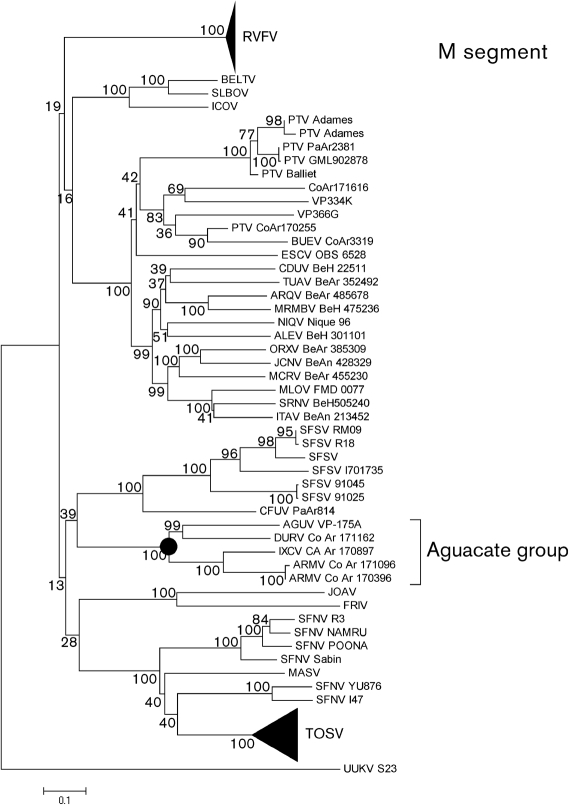
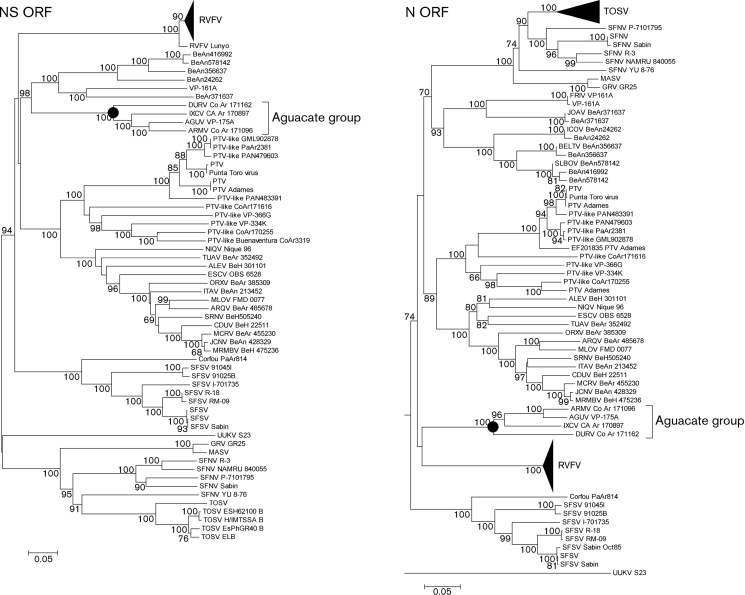
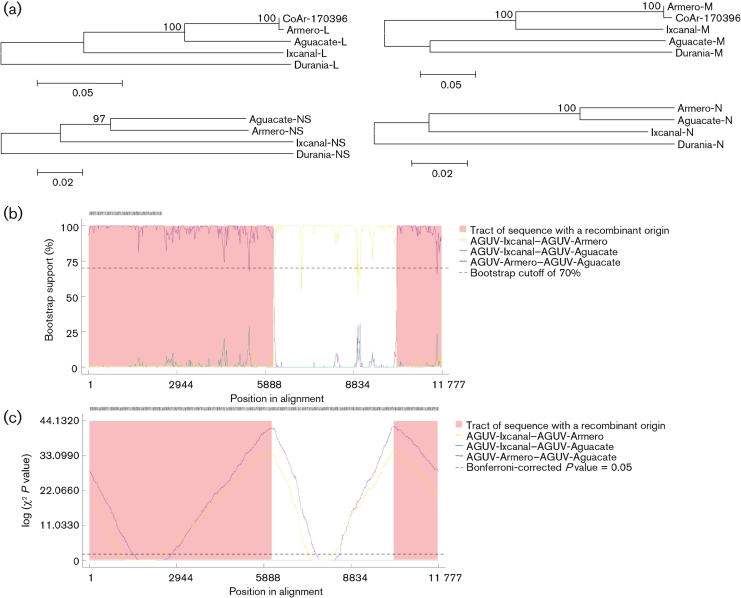
In the phylogenetic analysis, major nodes that represented viruses belonging to the same species or antigenic complex were clearly distinct and confirmed previously reported topologies (Charrel et al., 2009; Collao et al., 2009). The four viruses reported here clustered together, defining an Aguacate virus antigenic-complex node (Figs 1, 2 and 3). Partial sequencing data for BUJV and CHIV demonstrate that the viruses reported here do not belong to the BUJV antigenic complex (data not shown). Similar topology was observed in phylogenetic trees at the nucleotide level (data not shown). Branching inconsistencies observed inside this node suggested the possibility of reassortment. Therefore, we searched for evidence of reassortment using concatenated full genomes. These analyses indicate that AGUV is a reassortant virus that has obtained its M segment from an as-yet unknown virus, probably of the same antigenic complex (Fig. 4).
Fig. 1.
Phylogenetic analysis of the available phlebovirus L ORF sequences. Neighbour-joining analysis at the amino acid level was performed because of the high variability of the underlying nucleotide sequences. The statistical significance of the tree topology was evaluated by bootstrap resampling of the sequences 1000 times. GenBank accession numbers used in this tree: RVFV (DQ375395, DQ375396, DQ375397, DQ375398, DQ375399, DQ375427, DQ375428, DQ375426, DQ375434, DQ375433, DQ375431, DQ375432, DQ375430, DQ375429, DQ375416, DQ375417, DQ375415, DQ375418, DQ375423, DQ375422, DQ375424, DQ375421, DQ375420, DQ375419, DQ375402, DQ375401, DQ375400, DQ375425, DQ375413, DQ375414, DQ375412, DQ375403, DQ37540, DQ375411, DQ375409, DQ375410, DQ375408, DQ375407, DQ375405, DQ375406, X56464), FRIV (EF076027), TOSV (FJ153280, FJ153279, FJ153281, X68414), MASV (EU725771), JOAV (EF076026), DURV (HM566155), IXCV (HM566161), AGUV (HM566137), ARMV (HQ661805, HM566140), PTV (DQ363409, DQ363408), ALEV (HM119401), NIQV (HM119425), ESCV (HM119410), TUAV (HM119431), ORXV (HM119434), MLOV (HM119413), ARQV (HM119404), ITAV (HM119416), SRNV (HM119428), CDUV (HM119407), MCRV (HM119419), JCNV (HM466934), MRMBV (HM119422), BELTV (EF076023), ICOV (EF076024), SLBOV (EF076025) and Uukuniemi virus (UUKV) (D10759).
Fig. 2.
Phylogenetic analysis of the available phlebovirus M ORF sequences. Neighbour-joining analysis at the amino acid level was performed because of the high variability of the underlying nucleotide sequences. The statistical significance of the tree topology was evaluated by bootstrap resampling of the sequences 1000 times. GenBank accession numbers used in this tree: RVFV (M11157, DQ380199, DQ380202, DQ380201, DQ380200, DQ380207, DQ380206, M25276, DQ380208, DQ380204, DQ380203, DQ380205, DQ380210, DQ380209, DQ380221, DQ380198, DQ380197, DQ380196, DQ380214, DQ380211, DQ380213, DQ380212, DQ380219, DQ380218, DQ380220, DQ380217, DQ380215, DQ380216, DQ380195, DQ380222, DQ380187, DQ380186, DQ380183, DQ380185, DQ380184, DQ380194, DQ380193, DQ380192, DQ380191, DQ380190, DQ380189, DQ380188), BELTV (EF076018), SLBOV (EF076020), ICOV (EF076019), PTV (DQ363407, AY129745, AY129751, AY129747, M11156, AY129746, AY129752, AY129748, AY129750), BUEV (AY129749), ESCV (HM119411), CDUV (HM119408), TUAV (HM119432), ARQV (HM119405), MRMBV (HM119423), NIQV (HM119426), ALEV (HM119402), ORXV (HM119435), JCNV (HM466935), MCRV (HM119420), MLOV (HM119414), SRNV (HM119429), ITAV (HM119417), SFSV (AY129741, AY129742, U30500, AY129743, AY129740, AY129739), CFUV (AY129744), AGUV (HM566138), DURV (HM566156), IXCV (HM566162), ARMV (HM566141), ARMV (HQ661806), JOAV (EF076021), FRIV (EF076022), SFNV (AY129736, AY129738, AY129734, AY129733, AY129735, AY129732), MASV (EU725772), UUKV (M17417) and TOSV (AY129737, FJ153284, FJ153283, FJ153282, DQ479891, DQ479903, DQ479906, DQ479890, DQ479914, DQ479912, DQ479898, DQ479910, DQ479907, DQ479909, DQ479916, DQ479913, DQ479915, DQ479892, DQ479893, DQ479900, DQ479902, DQ479899, DQ479908, X89628, DQ479911, DQ479904, DQ479896, DQ479895, DQ479905, DQ479894, DQ479897 DQ479901).
Fig. 3.
Phylogenetic analysis of the available phlebovirus N and NS ORF sequences. Neighbour-joining analysis at the amino acid level was performed because of the high variability of the underlying nucleotide sequences. The statistical significance of the tree topology was evaluated by bootstrap resampling of the sequences 1000 times. GenBank accession numbers used in this tree: TOSV (EF120631, AY705941, FJ153285, EF120631, AY705940, AY705935, EF120628, AY705938, EF120632, AY705936, AY705943, AY705942, EF201833, FJ153286, EF120630, AY705934, EF120629, AY705933, AY705939, AY705937, AY766034), SFNV (EF201830, AY705944, EF201829, EF201832, EF201828, EF201831), MASV (EU725773), GRV (GU135608), FRIV (EF076017, EF201819), JOAV (EF076016, EF201820), ICOV (EF076014, EF201818), BELTV (EF076013, EF201817), SLBOV (EF076015, EF201816, EF201815), PTV (K02736, EF201834, DQ363406, EF201844, EF201843, EF201841, EF201837, EF201835, EF201836, EF201838, EF201842, EF201840), ALEV (HM119403), NIQV (HM119427), ESCV (HM119412), TUAV (HM119433), ORXV (HM119436), ARQV (HM119406), MLOV (HM119415), SRNV (HM119430), ITAV (HM119418), CDUV (HM119409), MCRV (HM119421), JCNV (HM466936), MRMBV (HM119424), ARMV (HM566142), AGUV (HM566139), IXCV (HM566163), DURV (HM566157), CFUV (EF201821), SFSV (EF201824, EF201823, EF201827, EF201826, EF201825, AJ811547, J04418, EF201822), UUKV (M33551) and RVFV (DQ380177, DQ380178, DQ380179, DQ380180, DQ380181, DQ380173, DQ380158, DQ380176, DQ380174, EF530207, DQ380175, DQ380159, AF134532, DQ380157, DQ380156, DQ380155, DQ924959, DQ380171, DQ380169, DQ380170, DQ380165, DQ380166, DQ380167, DQ380168, DQ380161, DQ380160, DQ380164, AF134531, DQ380182, DQ380162, DQ380163, DQ380172, DQ380144, DQ380143, DQ380150, DQ380149, DQ380148, DQ380147, DQ380153, DQ380145, DQ380146, AF134535, AF134533, DQ380152, AF134534, DQ380151, X53771, AF134530, X53771, DQ380154).
Fig. 4.
Evidence supporting AGUV as a reassortant. (a) Phylogenetic, (b) bootscan and (c) chimera analysis all demonstrate AGUV as being a reassortant.
ORFs
RNA-dependent RNA polymerase (L).
The four members of the Aguacate virus antigenic complex show high conservation with previously conserved functional domains described in phlebovirus sequences (Supplementary Figs S1 and S2, available in JGV Online) (Aquino et al., 2003; Palacios et al., 2011; Poch et al., 1989; Xiong & Eickbush, 1990).
Polyprotein (M).
The polyprotein is cotranslationally cleaved into three protein products: NSm, Gn and Gc. The topology of the polyprotein of the Aguacate virus antigenic complex is similar to the predicted topology for TOSV, PTV, RVFV and CDUV (Gerrard & Nichol, 2002; Grò et al., 1997; Ihara et al., 1985; Matsuoka et al., 1996; Valentini et al., 2008). Signal sequences, transmembrane domains and predicted cleavage sites for the protein products are conserved (Supplementary Fig. S3, available in JGV Online).
NP.
Similarly, all the functional domains described in the NP are conserved (Gerrard & Nichol, 2002; Le May et al., 2005; Palacios et al., 2011) (Supplementary Fig. S4, available in JGV Online).
Discussion
Despite the clinical importance of phleboviruses as pathogens of both humans and livestock, we have only limited insight into their phylogenetic diversity. Until now, speciation of phleboviruses has been largely driven by antigenic studies. However, it is no longer feasible to test newly isolated phleboviruses by serology against all other known members of the genus because of their abundance and diversity, the frequency of recombination events (Collao et al., 2010; Palacios et al., 2011) and the fact that some of these viruses do not produce readable plaques in cell culture or produce illness in newborn mice. Thus, high-throughput sequencing offers another option for their characterization and classification. We are presently sequencing the genomes of the known phleboviruses to determine their taxonomic relationships; this is one of several planned publications on our findings.
The results of this study demonstrate that AGUV, ARMV, DURV and IXCV are closely related to one another in terms of sequence and serology and that they are more distantly related to members of the other nine recognized species within the genus Phlebovirus. We propose that this warrants their designation as a new (tenth) species (Aguacate virus) within the genus. Interestingly, the prototype Aguacate virus strain (VP-175A) appears to be a natural reassortant among the members of the serological complex. Reassortment among RNA segments of related viruses in the family Bunyaviridae (including phleboviruses) has been reported before (Beaty et al., 1985; Briese et al., 2006, 2007; Collao et al., 2010; Dunn et al., 1994; Nunes et al., 2005; Palacios et al., 2011; Saeed et al., 2001); it is thought to be a mechanism which permits rapid evolution of viruses in this taxon. Although Bird et al. (2008) characterized the diversity of RVFV as being low, reassortment may explain the abundance and diversity observed between different phleboviruses circulating in nature and the difficulty in their precise identification and taxonomic classification.
Methods
Viruses
The phlebovirus strains used in this study were Aguacate (AGUV) strain VP-175A, Armero (ARMV) strains Co Ar 171096 and Co Ar 170396, Durania (DURV) strain Co Ar 171162, Ixcanal (IXCV) strain CA Ar 170897, Bujaru (BUJV) Be An 47693, Candiru (CDUV) Be H 22511, Chilibre (CHIV) strain VP-118D, Frijoles (FRIV) strain VP-161A, Punta Toro (PTV) strain D 4021A, Rift Valley fever (RVFV) strain MP-12, Salehabad (SALV) strain I-81 and sandfly fever Naples (SFNV) Sabin strain. All virus stocks were obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch.
AGUV.
Multiple isolates of AGUV were made from separate pools of male and female sandflies (Lutzomyia sp.) collected in secondary tropical forest at two localities (El Aguacate, central Panama, and Limbo, in the Panama Canal Zone) in Panama between 1969 and 1971 (Tesh et al., 1974). The prototype strain (VP-175A) was isolated in Vero cell culture. Initially, it did not produce illness or death in newborn mice, but it was subsequently adapted by serial intracerebral (ic) blind passage (Tesh et al., 1974).
ARMV.
Strains Co Ar 171096 and Co Ar 170396 were isolated in Vero cells from pools of female sand flies (Lutzomyia sp.) collected in a secondary forest in the municipality of Mariquita, Department of Tolima, Colombia, in 1986 (Tesh et al., 1989). It produced illness in newborn mice approximately 9 days after ic inoculation.
DURV.
Strain Co Ar 171162 was isolated in Vero cells from a pool of female sand flies (Lutzomyia sp.) collected in 1986 from a coffee plantation 8 km from the town of Durania, Norte de Santander Department, Colombia (Tesh et al., 1989). On first passage in infant mice, it produced hind-limb paralysis and death within 13 days of ic inoculation (Tesh et al., 1989).
IXCV.
Strain CA Ar 170897 was isolated in Vero cells from a pool of male sand flies (Lutzomyia sp.) collected in 1982 in the vicinity of two small villages (Aldeas Ixcanal and Puerto Progreso) in the riparian habitat of the Cato river valley in Guatemala. IXCV did not initially produce disease in newborn mice, but it was subsequently adapted by serial passage (Tesh et al., 1989).
Antigens and immune reagents.
Methods used to prepare antigens for the CF tests and immune ascitic fluids have been described previously (Beaty et al., 1989; Travassos da Rosa et al., 1983; Xu et al., 2007). Antigens and antibodies were both prepared in mice.
CF tests.
CF tests were performed by using the microtitre technique (Beaty et al., 1989; Xu et al., 2007), using 2 U of guinea pig complement and overnight incubation of the antigen and antibody at 4 °C. CF titres were recorded as the highest dilutions giving 3+ or 4+ fixation of complement (0–25 % haemolysis).
Genome sequencing.
Viral stocks were extracted using TRIzol LS (Invitrogen). Total RNA extracts were treated with DNase I (DNA free; Ambion). cDNA was generated using a Superscript II system (Invitrogen) employing random hexamers linked to an arbitrary 17-mer primer sequence (Palacios et al., 2007). The resulting cDNA was treated with RNase H and then randomly amplified by PCR with a 9 : 1 mixture of primer corresponding to the 17-mer sequence and the random hexamer-linked 17-mer primer (Palacios et al., 2007). Products >70 bp were selected by column chromatography (MinElute; Qiagen) and ligated to specific adapters for sequencing on a 454 Genome Sequencer FLX (454 Life Sciences) without fragmentation (Cox-Foster et al., 2007; Margulies et al., 2005; Palacios et al., 2008). Software programs accessible through the analysis applications at the GreenePortal website (http://tako.cpmc.columbia.edu/Tools/) were used for removal of primer sequences, redundancy filtering and sequence assembly. Sequence gaps were completed by PCR by using primers based on pyrosequencing data. Amplification products were size-fractionated on 1 % agarose gels, purified (MiniElute; Qiagen) and directly sequenced in both directions with ABI Prism Big Dye Terminator 1.1 Cycle Sequencing kits on ABI Prism 3700 DNA Analyzers (Perkin-Elmer Applied Biosystems). For the termini of each segment, a primer with the 8 nt conserved sequence was used for a specific reverse transcription reaction with additional arbitrary nucleotides on the 5′ end (5′-AAGCAGTGGTATCAACGCAGAGTACACACAAAG-3′; the boldface portion indicates the conserved nucleotides). This primer is designed to bind to the 3′ end of the genomic RNA and the 3′ end of the mRNA. The sequences of the genomes were verified by classical dideoxy sequencing by using primers designed from the draft sequence to create products of 1000 bp with 500 bp overlap.
Phylogenetic analysis
A set of phlebovirus sequences (70 for the L segment, 122 for the M segment, 131 for the N gene and 98 for the NS gene), comprising all sequences available from GenBank (June 2010), was used to determine the phylogenic relationships of AGUV, ARMV, DURV and IXCV. All sequences were aligned using the clustal algorithm (as implemented in mega version 3) at the amino acid level, with additional manual editing to ensure the highest possible quality of alignment. Neighbour-joining analysis at the amino acid level was performed because of the observed high variability of the underlying nucleotide sequences. Nucleotide trees were also produced using neighbour-joining analysis and the Kimura two-paramater model. The statistical significance of tree topology was evaluated by bootstrap resampling of the sequences 1000 times. Phylogenetic analyses were performed using mega software (Kumar et al., 2004).
Detection of recombination events.
Systematic screening for the presence of recombination patterns was achieved by using the nucleotide alignments and the recombination detection program (rdp) (Martin & Rybicki, 2000). The algorithms bootscan (Salminen et al., 1995), MaxChi (Smith, 1992), chimaera (Posada & Crandall, 2001), lard (Holmes, 1998) and phylip Plot (Felsenstein, 1989) were also used for the detection of recombination.
Sequence analysis.
geneious 4.8.3 (Biomatters) was used for sequence assembly and analysis. Topology and targeting predictions were generated by employing SignalP, NetNGlyc, tmhmm (http://www.cbs.dtu.dk/services), TopPred2 (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::toppred), and integrated predictions in Geneious (Bendtsen et al., 2004; Claros & von Heijne, 1994; Kahsay et al., 2005; Käll et al., 2004; Krogh et al., 2001).
Acknowledgements
This work was supported by Google.org, National Institutes of Health award AI57158 (North-east Biodefense Center – Lipkin), and USAID Predict funding source code 07-301-7119-52258 (Center for Infection and Immunity) and the Department of Defense. A. T. R., R. T., and H. G. were supported by NIH contract HHSN27220100004OI/HHSN27200004/DO4.
Footnotes
Present address: School of Medicine and Biomedical Sciences, University of Buffalo, Buffalo, New York, USA.
The GenBank/EMBL/DDBJ accession numbers for the sequences of the three RNA segments making up the genomes of AGUV, ARMV, DURV and IXCV are HM566137–9, HM566140–2, HM566155–7, HM566161–3 and HQ661805–7.
Four supplementary figures are available with the online version of this paper.
References
- Aquino V. H., Moreli M. L., Moraes Figueiredo L. T. (2003). Analysis of oropouche virus L protein amino acid sequence showed the presence of an additional conserved region that could harbour an important role for the polymerase activity. Arch Virol 148, 19–28 10.1007/s00705-002-0913-4 [DOI] [PubMed] [Google Scholar]
- Beaty B. J., Sundin D. R., Chandler L. J., Bishop D. H. (1985). Evolution of bunyaviruses by genome reassortment in dually infected mosquitoes (Aedes triseriatus). Science 230, 548–550 10.1126/science.4048949 [DOI] [PubMed] [Google Scholar]
- Beaty B. J., Calisher C. H., Shope R. E. (1989). Arboviruses. In Diagnostic Procedures for Viral, Rickettsial and Chlamydial Infections, pp. 797–855 Edited by Schmidt N. J., Emmons R. W. Washington, DC: American Public Health Association [Google Scholar]
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340, 783–795 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- Bird B. H., Githinji J. W., Macharia J. M., Kasiiti J. L., Muriithi R. M., Gacheru S. G., Musaa J. O., Towner J. S., Reeder S. A., et al. (2008). Multiple virus lineages sharing recent common ancestry were associated with a large Rift Valley fever outbreak among livestock in Kenya during 2006–2007. J Virol 82, 11152–11166 10.1128/JVI.01519-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Bird B., Kapoor V., Nichol S. T., Lipkin W. I. (2006). Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa. J Virol 80, 5627–5630 10.1128/JVI.02448-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Kapoor V., Lipkin W. I. (2007). Natural M-segment reassortment in Potosi and Main Drain viruses: implications for the evolution of orthobunyaviruses. Arch Virol 152, 2237–2247 10.1007/s00705-007-1069-z [DOI] [PubMed] [Google Scholar]
- Charrel R. N., Moureau G., Temmam S., Izri A., Marty P., Parola P., da Rosa A. T., Tesh R. B., de Lamballerie X. (2009). Massilia virus, a novel Phlebovirus (Bunyaviridae) isolated from sandflies in the Mediterranean. Vector Borne Zoonotic Dis 9, 519–530 10.1089/vbz.2008.0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros M. G., von Heijne G. (1994). TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci 10, 685–686 [DOI] [PubMed] [Google Scholar]
- Collao X., Palacios G., Sanbonmatsu-Gámez S., Pérez-Ruiz M., Negredo A. I., Navarro-Marí J. M., Grandadam M., Aransay A. M., Lipkin W. I., et al. (2009). Genetic diversity of Toscana virus. Emerg Infect Dis 15, 574–577 10.3201/eid1504.081111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collao X., Palacios G., de Ory F., Sanbonmatsu S., Pérez-Ruiz M., Navarro J. M., Molina R., Hutchison S. K., Lipkin W. I., et al. (2010). Granada virus: a natural phlebovirus reassortant of the sandfly fever Naples serocomplex with low seroprevalence in humans. Am J Trop Med Hyg 83, 760–765 10.4269/ajtmh.2010.09-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Foster D. L., Conlan S., Holmes E. C., Palacios G., Evans J. D., Moran N. A., Quan P. L., Briese T., Hornig M., et al. (2007). A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318, 283–287 10.1126/science.1146498 [DOI] [PubMed] [Google Scholar]
- Dunn E. F., Pritlove D. C., Elliott R. M. (1994). The S RNA genome segments of Batai, Cache Valley, Guaroa, Kairi, Lumbo, Main Drain and Northway bunyaviruses: sequence determination and analysis. J Gen Virol 75, 597–608 10.1099/0022-1317-75-3-597 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1989). phylip - phylogeny inference package (version 3.2). Cladistics 5, 164–166 [Google Scholar]
- Gerrard S. R., Nichol S. T. (2002). Characterization of the Golgi retention motif of Rift Valley fever virus GN glycoprotein. J Virol 76, 12200–12210 10.1128/JVI.76.23.12200-12210.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grò M. C., Di Bonito P., Fortini D., Mochi S., Giorgi C. (1997). Completion of molecular characterization of Toscana phlebovirus genome: nucleotide sequence, coding strategy of M genomic segment and its amino acid sequence comparison to other phleboviruses. Virus Res 51, 81–91 10.1016/S0168-1702(97)00076-2 [DOI] [PubMed] [Google Scholar]
- Holmes E. C. (1998). Molecular epidemiology of dengue virus–the time for big science. Trop Med Int Health 3, 855–856 10.1046/j.1365-3156.1998.00332.x [DOI] [PubMed] [Google Scholar]
- Ihara T., Smith J., Dalrymple J. M., Bishop D. H. (1985). Complete sequences of the glycoproteins and M RNA of Punta Toro phlebovirus compared to those of Rift Valley fever virus. Virology 144, 246–259 10.1016/0042-6822(85)90321-6 [DOI] [PubMed] [Google Scholar]
- Kahsay R. Y., Gao G., Liao L. (2005). An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 21, 1853–1858 10.1093/bioinformatics/bti303 [DOI] [PubMed] [Google Scholar]
- Käll L., Krogh A., Sonnhammer E. L. (2004). A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338, 1027–1036 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5, 150–163 10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Le May N., Gauliard N., Billecocq A., Bouloy M. (2005). The N terminus of Rift Valley fever virus nucleoprotein is essential for dimerization. J Virol 79, 11974–11980 10.1128/JVI.79.18.11974-11980.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y. J., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. R. E., Rybicki E. (2000). RDP: detection of recombination amongst aligned sequences. Bioinformatics 16, 562–563 10.1093/bioinformatics/16.6.562 [DOI] [PubMed] [Google Scholar]
- Matsuoka Y., Chen S. Y., Holland C. E., Compans R. W. (1996). Molecular determinants of Golgi retention in the Punta Toro virus G1 protein. Arch Biochem Biophys 336, 184–189 10.1006/abbi.1996.0547 [DOI] [PubMed] [Google Scholar]
- Nichol S. T., Beaty B. J., Elliott R. M., Goldbach R., Plyusnin A., Schmaljohn C. S., Tesh R. B. (2005). Family Bunyaviridae. In Virus Taxonomy Eighth Report of the International Committee on Taxonomy of Viruses, pp. 695–716 Edited by Fauquet C. M., Mayo M. A., Desselberger J., Ball L. A. San Diego: Elsevier [Google Scholar]
- Nunes M. R., Travassos da Rosa A. P., Weaver S. C., Tesh R. B., Vasconcelos P. F. (2005). Molecular epidemiology of group C viruses (Bunyaviridae, Orthobunyavirus) isolated in the Americas. J Virol 79, 10561–10570 10.1128/JVI.79.16.10561-10570.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Quan P. L., Jabado O. J., Conlan S., Hirschberg D. L., Liu Y., Zhai J., Renwick N., Hui J., et al. (2007). Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis 13, 73–81 10.3201/eid1301.060837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Druce J., Du L., Tran T., Birch C., Briese T., Conlan S., Quan P. L., Hui J., et al. (2008). A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med 358, 991–998 10.1056/NEJMoa073785 [DOI] [PubMed] [Google Scholar]
- Palacios G., Tesh R., Travassos da Rosa A., Savji N., Sze W., Jain K., Serge R., Guzman H., Guevara C., et al. (2011). Characterization of the Candiru antigenic complex (Bunyaviridae: Phlebovirus), a highly diverse and reassorting group of viruses affecting humans in tropical America. J Virol..</year> 10.1128/JVI.02275-10 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. (1989). Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8, 3867–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K. A. (2001). Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A 98, 13757–13762 10.1073/pnas.241370698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M. F., Wang H., Suderman M., Beasley D. W., Travassos da Rosa A., Li L., Shope R. E., Tesh R. B., Barrett A. D. (2001). Jatobal virus is a reassortant containing the small RNA of Oropouche virus. Virus Res 77, 25–30 10.1016/S0168-1702(01)00262-3 [DOI] [PubMed] [Google Scholar]
- Salminen M. O., Carr J. K., Burke D. S., McCutchan F. E. (1995). Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses 11, 1423–1425 10.1089/aid.1995.11.1423 [DOI] [PubMed] [Google Scholar]
- Smith J. M. (1992). Analyzing the mosaic structure of genes. J Mol Evol 34, 126–129 10.1007/BF00182389 [DOI] [PubMed] [Google Scholar]
- Tesh R. B., Chaniotis B. N., Peralta P. H., Johnson K. M. (1974). Ecology of viruses isolated from Panamanian phlebotomine sandflies. Am J Trop Med Hyg 23, 258–269 [DOI] [PubMed] [Google Scholar]
- Tesh R. B., Saidi S., Gajdamovic S. J., Rodhain F., Vesenjak-Hirjan J. (1976). Serological studies on the epidemiology of sandfly fever in the Old World. Bull World Health Organ 54, 663–674 [PMC free article] [PubMed] [Google Scholar]
- Tesh R. B., Peters C. J., Meegan J. M. (1982). Studies on the antigenic relationship among phleboviruses. Am J Trop Med Hyg 31, 149–155 [DOI] [PubMed] [Google Scholar]
- Tesh R. B., Boshell J., Young D. G., Morales A., Ferra de Carrasquilla C., Corredor A., Modi G. B., Travassos da Rosa A. P., McLean R. G., et al. (1989). Characterization of five new phleboviruses recently isolated from sand flies in tropical America. Am J Trop Med Hyg 40, 529–533 [PubMed] [Google Scholar]
- Travassos da Rosa A. P., Tesh R. B., Pinheiro F. P., Travassos da Rosa J. F., Peterson N. E. (1983). Characterization of eight new phlebotomus fever serogroup arboviruses (Bunyaviridae: Phlebovirus) from the Amazon region of Brazil. Am J Trop Med Hyg 32, 1164–1171 [DOI] [PubMed] [Google Scholar]
- Valentini M., Valassina M., Savellini G. G., Cusi M. G. (2008). Nucleotide variability of Toscana virus M segment in strains isolated from clinical cases. Virus Res 135, 187–190 10.1016/j.virusres.2008.01.016 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. (1990). Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9, 3353–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Liu D., Nunes M. R., Da Rosa A. P., Tesh R. B., Xiao S. Y. (2007). Antigenic and genetic relationships among Rift Valley fever virus and other selected members of the genus Phlebovirus (Bunyaviridae). Am J Trop Med Hyg 76, 1194–1200 [PubMed] [Google Scholar]