Abstract
We have used a purified recombinant chromatin assembly system, including ACF (Acf-1 + ISWI) and NAP-1, to examine the role of histone acetylation in ATP-dependent chromatin remodeling. The binding of a transcriptional activator (Gal4–VP16) to chromatin assembled using this recombinant assembly system dramatically enhances the acetylation of nucleosomal core histones by the histone acetyltransferase p300. This effect requires both the presence of Gal4-binding sites in the template and the VP16-activation domain. Order-of-addition experiments indicate that prior activator-meditated, ATP-dependent chromatin remodeling by ACF is required for the acetylation of nucleosomal histones by p300. Thus, chromatin remodeling, which requires a transcriptional activator, ACF and ATP, is an early step in the transcriptional process that regulates subsequent core histone acetylation. Glycerol gradient sedimentation and immunoprecipitation assays demonstrate that the acetylation of histones by p300 facilitates the transfer of H2A–H2B from nucleosomes to NAP-1. The results from these biochemical experiments suggest that (1) transcriptional activators (e.g., Gal4–VP16) and chromatin remodeling complexes (e.g., ACF) induce chromatin remodeling in the absence of histone acetylation; (2) transcriptional activators recruit histone acetyltransferases (e.g., p300) to promoters after chromatin remodeling has occurred; and (3) histone acetylation is important for a step subsequent to chromatin remodeling and results in the transfer of histone H2A–H2B dimers from nucleosomes to a histone chaperone such as NAP-1. Our results indicate a precise role for histone acetylation, namely to alter the structure of nucleosomes (e.g., facilitate the loss of H2A–H2B dimers) that have been remodeled previously by the action of ATP-dependent chromatin remodeling complexes. Thus, transcription from chromatin templates is ordered and sequential, with precise timing and roles for ATP-dependent chromatin remodeling, subsequent histone acetylation, and alterations in nucleosome structure.
Keywords: Chromatin remodeling, histone transfer, acetylation, p300, ACF
The packaging of DNA into chromatin has important consequences for the regulation of genes transcribed by RNA polymerase II (pol II). Nucleosomes act as general repressors of basal transcription, inhibiting transcriptional initiation and elongation by RNA pol II. The activation of transcriptionally repressed templates requires the actions of sequence-specific DNA-binding transcriptional activators, chromatin remodeling complexes, and histone acetyltransferases (HATs). As genes are transcribed by RNA pol II, the nucleosomal arrays positioned over promoters and transcribed regions are altered (“remodeled”) to counteract their repressive effect (for review, see van Holde et al. 1992; Kornberg and Lorsh 1992; Lewin 1994; Felsenfeld 1996; Kingston et al. 1996; Kingston and Narlikar 1999). Local changes in chromatin structure involve the disruption or alteration of nucleosomal arrays in the vicinity of the promoter and can be mediated by large multipolypeptide chromatin remodeling complexes (for review, see Tsukiyama and Wu 1997; Kadonaga 1998; Varga-Weisz and Becker 1998; Kingston and Narlikar 1999). Chromatin remodeling complexes use ATP to alter nucleosome positioning and structure. All of the known chromatin remodeling complexes contain ATPase subunits that define three distinct families of complexes: SWI2/SNF2-like (e.g., SWI/SNF and RSC), ISWI-like (e.g., ACF, NURF, CHRAC, and ASF) and Mi-2-like (e.g., NURD) (for review, see Kingston and Narlikar 1999).
The SWI/SNF family of remodeling complexes catalyzes two distinct reactions (1) nucleosome remodeling without the loss of nucleosomal components (i.e., histones) and (2) nucleosome transfer (Owen-Hughes and Workman 1996; Lorch et al. 1998, 1999; Schnitzler et al. 1998). SWI/SNF and RSC have been shown to catalyze the stable alteration of nucleosome structures with mononucleosomal substrates (Lorch et al. 1998; Schnitzler et al. 1998). These remodeled nucleosomes retain all four core histones and the DNA, and in many cases can revert back slowly to the original state in the absence of continued remodeling activity. SWI/SNF has also been shown to stimulate the transfer of histones from nucleosomes to nucleoplasmin in the presence of a transcriptional activator protein (Walter et al. 1995; Owen-Hughes and Workman 1996), whereas RSC has been shown to stimulate the transfer of histones from chromatin to acceptor DNA (Lorch et al. 1999).
The ISWI family of remodeling complexes (e.g., ACF, NURF, CHRAC, and ASF) exhibit a variety of assayable ATP-dependent activities related to nucleosome remodeling, including (1) the ordering of disordered nucleosomal arrays (Varga-Weisz et al. 1995; Ito et al. 1997; Tsukiyama et al. 1999), (2) the remodeling of local chromatin structure in the presence of a DNA-binding transcriptional activator protein (Tsukiyama et al. 1994; Tsukiyama and Wu 1995; Ito et al. 1997), (3) the induction of restriction endonuclease sensitivity on nucleosomal arrays (Varga-Weisz et al. 1995, 1997), (4) the stimulation of nucleosome sliding (Hamiche et al. 1999; Langst et al. 1999), and (5) the assembly of free histones into nucleosomes (Ito et al. 1997, 1999). The particular set of activities exhibited differs with each complex in the ISWI family (for review, see Kingston and Narlikar 1999).
In addition to the actions of the ATP-dependent chromatin remodeling complexes described above, transcription by RNA pol II is enhanced by factors with intrinsic HAT activity (for review, see Roth and Allis 1996; Grunstein 1997; Wade and Wolffe 1997). The discovery that increased levels of histone acetylation are often associated with transcriptionally active genes (e.g., Hebbes et al. 1988) first suggested a link between histone acetylation and transcriptional activation. Because acetylation neutralizes the positively charged lysine residues of the amino-terminal tails of histones and decreases their affinity for DNA, it has been speculated that this might increase the mobility of nucleosomes on DNA or facilitate structural changes in the nucleosomes (for review, see Roth and Allis 1996; Grunstein 1997; Wade and Wolffe 1997). Many proteins with HAT activity, including Gcn5p and p300/CBP, function as transcriptional coactivators. For example, studies have shown that the HAT activity of Gcn5p is critical for the transcriptional activation of target genes in vivo (Kuo et al. 1998; Wang et al. 1998). Likewise, the HAT activity of p300 was found to be critical for p300 coactivator activity with a variety of activators on chromatin templates in vitro (Kraus et al. 1999). The HAT proteins and complexes are targeted to promoters by sequence-specific DNA-binding transcriptional activators. Thus, transcriptional regulation by activator proteins involves two distinct classes of chromatin modifying complexes, HATs and ATP-dependent chromatin remodeling complexes.
One important question relating to the transcription of genes assembled into chromatin is how the activities of these two classes of chromatin modifying complexes are coordinated during the transcription process. Initially, it was thought that histone acetylation by HATs helps to facilitate nucleosome remodeling mediated by ATP-dependent chromatin remodeling complexes. However, recent in vivo chromatin immunoprecipitation experiments in yeast suggest that chromatin remodeling may precede histone acetylation (Cosma et al. 1999; Krebs et al. 1999). We present the results of biochemical studies that support these in vivo studies. In addition, we demonstrate that histone acetylation is important for a step subsequent to chromatin remodeling and results in the transfer of histone H2A–H2B dimers from nucleosomes to the histone chaperone NAP-1. Our results indicate a precise role for histone acetylation, namely to alter the structure of nucleosomes (e.g., facilitate the loss of H2A–H2B dimers) that have been remodeled previously by the action of ATP-dependent chromatin remodeling complexes.
Results and Discussion
Transcription factor binding facilitates the acetylation of nucleosomal core histones by p300
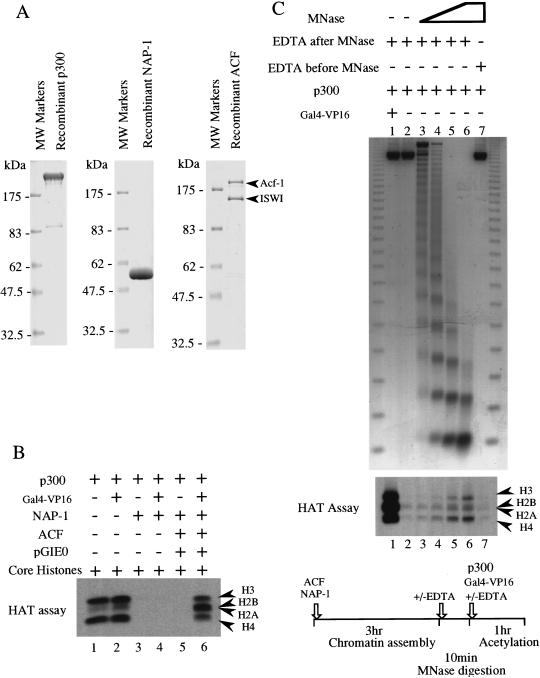
In these studies, we have examined the role of histone acetylation in nucleosome remodeling by ACF, an ATP-utilizing chromatin assembly and remodeling factor. Recently, it has been shown that purified recombinant Drosophila ACF (dAcf-1 and dISWI) can assemble regularly spaced nucleosomes in conjunction with purified native Drosophila NAP-1, a core histone chaperone (Ito et al. 1999). For our current studies, we have used a completely recombinant purified chromatin assembly system (dAcf-1, dISWI, and dNAP-1; Fig. 1A) to assemble regularly spaced nucleosomes on plasmid templates. To examine the effects of histone acetylation, we used the HAT p300, which we expressed in insect cells using a baculovirus expression vector and purified to near homogeneity (Fig. 1A; Kraus and Kadonaga 1998). Note that ACF itself does not possess significant HAT activity (data not shown), so the only HAT activity in our assays is from p300. As a model transcriptional activator protein, we used Gal4–VP16, a hybrid protein that consists of the DNA-binding domain of the yeast transcriptional activator Gal4 fused to the transcriptional activation domain of the herpes virus protein VP16. The DNA template (pGIE0) contains five Gal4 binding sites upstream of the adenovirus E4 promoter.
Figure 1.
The binding of Gal4–VP16 to chromatin templates assembled with recombinant ACF and NAP-1 facilitates the acetylation of nucleosomal core histones by p300. (A) SDS-polyacrylamide gel analysis of purified recombinant p300, NAP-1, and ACF. Affinity-tagged proteins were expressed in Sf9 cells by infection with recombinant baculoviruses. The proteins were purified by affinity or traditional chromatography as described in Materials and Methods. (B) The binding of Gal4–VP16 facilitates the acetylation of nucleosomal core histones by p300. In lanes 1–4, free core histones were incubated with p300 and 3H-acetyl CoA in the presence or absence of the factors indicated. In lanes 5 and 6, chromatin assembled with ACF and NAP-1 was incubated with p300 and 3H-acetyl CoA in the presence or absence of the factors indicated. The samples were analyzed on 12% polyacrylamide gels and the acetylated core histones were detected by fluorography. (C) Digestion of chromatin with micrococcal nuclease (MNase) increases the acetylation of nucleosomal core histones by p300. Chromatin templates assembled with ACF and NAP-1 were incubated with increasing amounts of MNase for 10 min prior to the addition of EDTA to stop the digestion reactions. The extent of MNase digestion was determined by deproteinizing aliquots of the chromatin and analyzing the DNA fragments on ethidium bromide-stained agarose gels (top). Gal4–VP16 and p300 plus 3H-acetyl CoA were added to aliquots of digested chromatin as indicated and incubated for 1 hr (see schematic, bottom). The samples were analyzed on 12% polyacrylamide gels and the acetylated core histones were detected by fluorography (middle).
First, we investigated the acetylation of core histones in regularly spaced nucleosomal arrays assembled with ACF and NAP-1. p300 acetylated H2A, H2B, H3, and H4 (core histones) as free histones both in the presence or absence of Gal4–VP16 (Fig. 1B, lanes 1,2). In the presence of NAP-1, which binds to all four core histones in vitro (Bulger et al. 1995; Ito et al. 1996), p300 failed to acetylate free core histones in the presence or absence of Gal4–VP16 (Fig. 1B, lanes 3,4). We speculate that the NAP-1 inhibited the access of p300 to the amino-terminal tails of the core histones. p300 also failed to acetylate core histones assembled into a regularly spaced nucleosomal array in the absence of Gal4–VP16 (Fig. 1B, lane 5). Interestingly, however, p300 did acetylate the nucleosomal core histones in the presence of Gal4–VP16 (Fig. 1B, lane 6). Consistent with previous studies (Bannister and Kouzarides 1996; Ogryzko et al. 1996; Kraus et al. 1999), we observed a preference of p300 for the acetylation of H3–H4 relative to H2A–H2B as free histones but not as nucleosomal histones (Fig. 1B, cf. lanes 1 and 2 with lane 6).
In contrast to the results shown in Figure 1B, previous studies have shown that p300 can acetylate core histones assembled into nucleosomes, even in the absence of a DNA-binding transcriptional activator (Ogryzko et al. 1996; Schiltz et al. 1999). However, those studies used nucleosomes digested with micrococcal nuclease as the substrate for p300, whereas we used undigested polynucleosomal arrays. In another study, the acetylation of histones in undigested polynucleosomal arrays by p300 was found to occur with ∼2% of the efficiency of free core histones (Kraus et al. 1999). To determine whether the extent of digestion with micrococcal nuclease could affect the ability of p300 to acetylate nucleosomal histones, we performed the experiment shown in Figure 1C. As expected, the acetylation of core histones in chromatin assembled with ACF and NAP-1 was dependent on the extent of digestion with micrococcal nuclease (Fig. 1C). These results suggest that the trimming of DNA wrapped around the histone octamer by micrococcal nuclease increases the accessibility of histone tails to HATs such as p300.
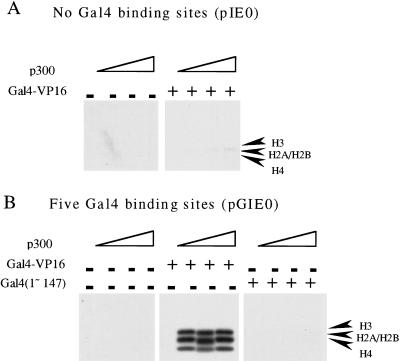
The results in Figure 1B indicate that the binding of Gal4–VP16 to nucleosomal templates helps to increase the accessibility of nucleosomal histone tails to the p300 HAT. To clarify the contribution of DNA binding by Gal-VP16 and the role of the VP16 activation domain in this process, we performed the experiments shown in Figure 2. When we used a template lacking Gal4 binding sites (pIE0), p300 was unable to acetylate nucleosomal core histones, even in the presence of Gal4–VP16 (Fig. 2A). However, in control experiments, acetylation was observed in the presence of Gal4–VP16 using a template containing five Gal4 binding sites (Fig. 2B). Next, we examined the ability of p300 to acetylate nucleosomal core histones in the presence of a Gal4 derivative lacking the VP16 activation domain [Gal4(1–147)]. As shown in Figure 2B, the VP16 activation domain was required for the acetylation of nucleosomal core histones by p300. Thus, the binding of a sequence-specific factor to nucleosomal DNA and the presence of an activation domain are both required for the acetylation of nucleosomal core histones by p300. These results suggest that the VP16 activation domain of Gal4–VP16 is able to target p300 to nucleosomes via the Gal4 DNA-binding component. Our observations are consistent with previous experiments showing that (1) acidic activation domains can recruit HATs to promoters (Utley et al. 1998; Ikeda et al. 1999); (2) p300 functions as a coactivator of Gal4–VP16-mediated transcription (Kraus et al. 1999); and (3) the HAT activity of p300 is required for full enhancement of Gal4–VP16-mediated transcription (Kraus et al. 1999).
Figure 2.
The acetylation of nucleosomal core histones by p300 requires Gal4-binding sites in the template and the VP16 activation domain. Plasmid DNA templates were assembled into chromatin by ACF and NAP-1. After a 4 hr incubation to allow for the completion of chromatin assembly, Gal4–VP16 or Gal4(1-147) (i.e., the Gal4 DNA-binding domain alone), as well as increasing amounts of purified recombinant p300, were added as indicated in the presence 3H-acetyl CoA. The samples were analyzed on 12% polyacrylamide gels and the acetylated core histones were detected by fluorography. (A) Experiments using a plasmid (pIE0) lacking Gal4 binding sites. (B) Experiments using a plasmid (pGIE0) containing five Gal4 binding sites.
Prior activator-mediated, ATP-dependent chromatin remodeling is required for the efficient acetylation of nucleosomal core histones by p300
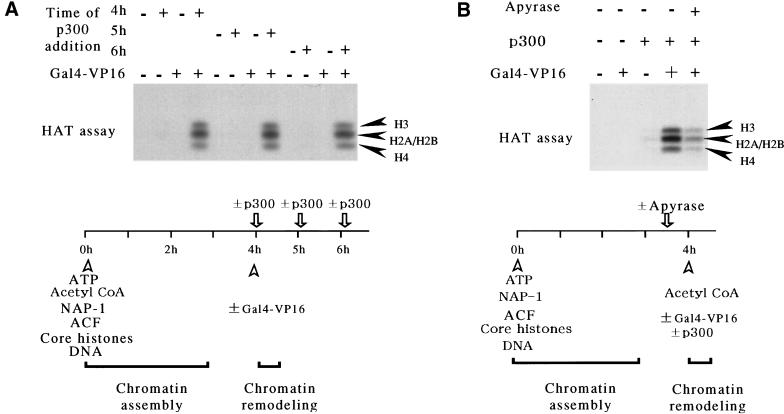
To explore the temporal relationship between activator-mediated chromatin remodeling and core histone acetylation, we added p300 at different time points after the completion of chromatin assembly (Fig. 3A). When p300 was added 1, 2, or 3 hr post-chromatin assembly (at t = 4, 5, or 6 hr in our experimental scheme; see Fig. 3A, bottom) in the presence of acetyl CoA but in the absence of Gal4–VP16, it was unable to acetylate the core histones in the assembly reaction. However, when added post-assembly in the presence of Gal4–VP16, p300 was able to acetylate all four core histones. Note that with this purified chromatin assembly system (ACF + NAP-1), the addition of Gal4–VP16 (e.g., at the 4, 5, and 6 hr time points shown in Fig. 3A) can mediate promoter-proximal chromatin remodeling (Ito et al. 1997; data not shown).
Figure 3.
Prior activator-mediated, ATP-dependent chromatin remodeling is required for the efficient acetylation of nucleosomal core histones by p300. (A) Activator-dependent acetylation of nucleosomal histones by p300. The pGIE0 plasmid was assembled into chromatin by ACF and NAP-1 in the presence 3H-acetyl CoA. After a 4 hr incubation to allow for the completion of chromatin assembly, Gal4–VP16 was added to one set of samples as indicated. At different time points relative to the start of chromatin assembly, purified recombinant p300 was added as indicated, followed by a 30 min incubation (see schematic, bottom). The samples were analyzed on 12% polyacrylamide gels and the acetylated core histones were detected by fluorography (top). (B) ATP-dependent chromatin remodeling is required for the efficient acetylation of nucleosomal core histones by p300. The pGIE0 plasmid was assembled into chromatin by ACF and NAP-1. After a 3.5 hr incubation to allow for the completion of chromatin assembly, apyrase was added as indicated. Subsequently, 3H-acetyl CoA was added, as well as p300 and Gal4–VP16 as indicated (see schematic, bottom). The samples were analyzed on 12% polyacrylamide gels and the acetylated core histones were detected by fluorography (top).
Our results suggest that the accessibility of histone tails can modulate the extent of histone acetylation by p300. Presumably, accessibility of the histone tails could be modulated by ATP-dependent chromatin remodeling factors (e.g., ACF), as well as by experimental manipulation using micrococcal nuclease (as shown in Fig. 1C). To explore further the requirement for prior activator-mediated, ATP-dependent chromatin remodeling in the acetylation of nucleosomal histones by p300, we inhibited the ATP-dependent remodeling activity of ACF by the addition of apyrase (Fig. 3B). Apyrase depletes ATP by catalyzing the hydrolysis of the pyrophosphate bonds in ATP. Previous chromatin remodeling experiments showed that the depletion of ATP by apyrase can inhibit promoter-proximal disruption of a periodic nucleosomal array by Gal4–VP16 without affecting the binding of Gal4–VP16 to the template (Pazin et al. 1994). As shown in Figure 3B, the addition of apyrase reduced substantially the extent of nucleosomal histone acetylation by p300. These data indicate that activator-mediated, ATP-dependent chromatin remodeling is essential for subsequent nucleosomal core histone acetylation by p300.
The relationship between ATP-dependent chromatin remodeling and core histone acetylation is an important question (for review, see Kingston and Narlikar 1999). Collectively, our data are consistent with the model that (1) activators and chromatin remodeling factors induce chromatin remodeling in the absence of histone acetylation; (2) HATs such as p300 are recruited to the promoter by DNA-bound activators; and (3) histone acetylation is important for a step subsequent to chromatin remodeling. Our results are a good biochemical counterpart to the recent in vivo chromatin immunoprecipitation experiments of Cosma et al. (1999) and Krebs et al. (1999). In these studies, analysis of the Saccharomyces cerevisiae HO promoter showed that the association of the SWI/SNF complex with the promoter is required for subsequent recruitment of histone acetylases (Gcn5p/SAGA complex) and increased acetylation of nucleosomal histones at the promoter (Cosma et al. 1999; Krebs et al. 1999). Together, these studies raise the question of the role of histone acetylation in transcriptional regulation if transcription factor binding and chromatin remodeling occur independently of histone acetylation.
p300 facilitates the transfer of H2A–H2B from nucleosomes to NAP-1 in the presence of acetyl CoA
The term chromatin remodeling is used somewhat loosely and perhaps imprecisely (for review, see Kadonaga 1998). Chromatin remodeling is defined typically as a change in nucleosome structure as assessed by one of several different assays, usually involving digestion with nucleases. The assays include (1) the loss of regularly repeating DNA ladders generated by micrococcal nuclease digestion and (2) the loss of a characteristic 10-bp repeat of DNase I digestion in a rotationally positioned nucleosome. However, these chromatin remodeling assays are somewhat indirect and cannot discriminate nucleosome transfer from nucleosome sliding or local changes in nucleosome structure. Thus, we used a different approach to determine the fate of histones during ACF-mediated chromatin remodeling.
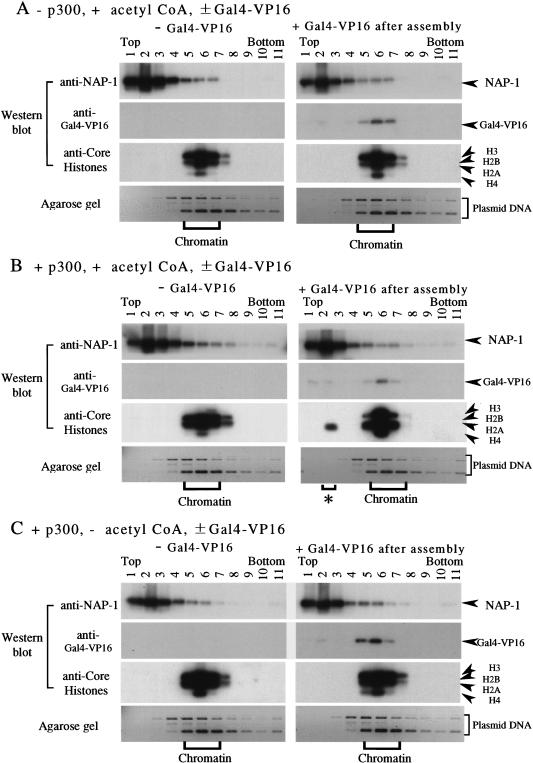
We assembled regularly spaced nucleosomal arrays with ACF and NAP-1 using the pGIE0 plasmid. After chromatin assembly, Gal4–VP16, p300, and acetyl CoA were added in various combinations. After incubation for 1 hr, the reactions were subjected to glycerol density gradient sedimentation. The presence of NAP-1, Gal4–VP16, and core histones in the different fractions were monitored by Western blotting, whereas the presence of the plasmid DNA was monitored by agarose gel electrophoresis and ethidium bromide staining. As shown in Figure 4, the chromatin templates sedimented near the middle of the gradient (peak in fraction 6), whereas NAP-1 and free Gal4–VP16 sedimented near the top of the gradient (peak in fraction 2). We used this assay to monitor histone transfer during chromatin remodeling in the presence or absence of p300.
Figure 4.
p300 facilitates the transfer of H2A–H2B from nucleosomes to NAP-1 in the presence of acetyl CoA. The pGIE0 plasmid was assembled into chromatin by ACF and NAP-1. After a 3 hr incubation, Gal4–VP16 (125 nm), p300 (100 nm), and unlabeled acetyl CoA (30 μm) were added as indicated, followed by incubation with the chromatin templates for 1 hr. The samples were then loaded on a 15%–40% glycerol gradient and sedimented at 60 k rpm for 3 hr at 4°C in a SW60 (Beckman) rotor. Fractions were collected from the gradients, separated on 12% polyacrylamide-SDS gels, and analyzed by Western blotting with antibodies against NAP-1, Gal4–VP16, and core histones using an 125I protein A detection method. The presence of plasmid DNA in the fractions was analyzed by agarose gel electrophoresis followed by ethidium bromide staining. (A) Without p300, with acetyl CoA. (B) With p300 and acetyl CoA. (C) With p300, without acetyl CoA.
In the absence of Gal4–VP16 and p300, the core histones and plasmid DNA were assembled into nucleosomes and sedimented near the middle of the gradient (Fig. 4A, left panel). The addition of Gal4–VP16 in the absence of p300 did not affect the distribution of the core histones in the gradient (i.e., they remained assembled into chromatin) (Fig. 4A, right panel). Thus, the binding of Gal4–VP16 by itself was not sufficient for core histone transfer from the supercoiled plasmid template to the NAP-1 chaperone. However, in the presence of p300, Gal4–VP16 stimulated the transfer of histones H2A–H2B to the peak NAP-1-containing fraction (fraction 2; Fig. 4B, right panel, asterisk). This transfer was dependent on the presence of Gal4–VP16 (cf. with Fig. 4B, left panel) and p300 (cf. with Fig. 4A, right panel). Thus p300, in conjunction with the binding of Gal4–VP16 to the chromatin template, facilitates the dissociation of the histone octamer and the transfer of H2A–H2B from nucleosomes to the NAP-1 fraction. This activity was dependent on the presence of acetyl CoA, the acetyl donor for p300 HAT activity (Fig. 4C).
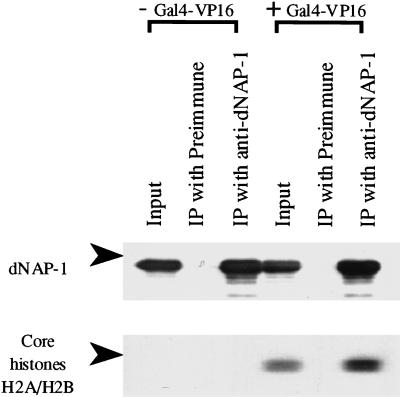
The experiments described above demonstrated cosedimentation of the histones released from the chromatin template in the presence of Gal4–VP16, p300, and acetyl CoA with NAP-1. To demonstrate an interaction between the histones and NAP-1 in those fractions, the material in the peak NAP-1-containing fractions (Fig. 4B, fraction 2) was immunoprecipitated with a NAP-1 antibody and then analyzed by Western blotting for NAP-1 and core histones. As shown in Figure 5, histones H2A–H2B coimmunoprecipitated with the NAP-1 in the presence, but not in the absence, of Gal4–VP16. In contrast, neither anti-p300 nor anti-ACF antibodies coprecipitated core histones from the NAP-1-containing fractions. Thus, the histones transferred from the chromatin templates associate with NAP-1, but not with p300 and ACF. The results in Figures 4 and 5 indicate that the acetylation of nucleosomal core histones by p300 facilitates the transfer of H2A–H2B dimers from nucleosomes to NAP-1.
Figure 5.
Acetylated H2A–H2B interacts with NAP-1. Coimmunoprecipitation of NAP-1 and H2A–H2B acetylated by p300 in the presence of Gal4–VP16. The second fractions from the top of the gradients shown in Figure 4B were analyzed by coimmunoprecipitation with preimmune serum and NAP-1 antiserum. The resulting immunoprecipitates were analyzed by 12% polyacrylamide-SDS gel electrophoresis followed by Western blotting for core histones (125I protein A detection method) and NAP-1 (enhanced chemiluminescent detection method).
Histone acetylation and the fate of nucleosomes during chromatin remodeling
Numerous studies have demonstrated an important role for chromatin remodeling and histone acetylation in activator-mediated transcriptional regulation with chromatin templates. However, the role of histone acetylation in chromatin remodeling has been mostly speculative. As suggested in numerous recent reviews, the acetylation of the amino-terminal tails of the core histones in nucleosomes is thought to facilitate chromatin remodeling by loosening interactions between the histones and DNA, thus leading to increased nucleosome mobility (for review, see Roth and Allis 1996; Grunstein 1997; Wade and Wolffe 1997). However, two lines of evidence suggest that this may not be the case. First, in biochemical studies, ATP-dependent chromatin-remodeling by chromatin remodeling complexes can occur in the absence of histone hyperacetylation (Lorch et al. 1998; Schnitzler et al. 1998; Ito et al. 1999; Tsukiyama et al. 1999). Second, in vivo studies in Saccharomyces cerevisiae suggest that chromatin remodeling precedes histone acetylation at the endogenous HO promoter (Cosma et al. 1999; Krebs et al. 1999). In our experiments, we have uncovered a connection between the fate of nucleosomes during chromatin remodeling and histone acetylation.
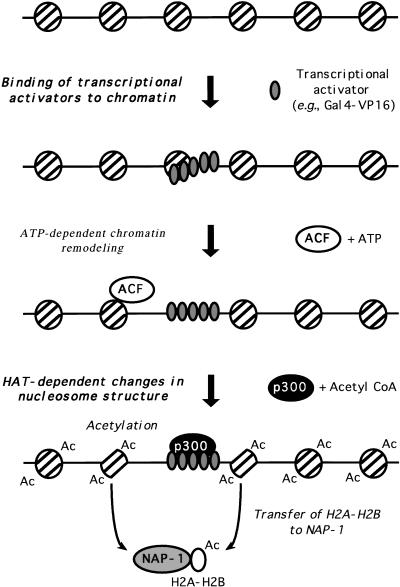
The results from our biochemical experiments suggest the following conclusions: (1) Transcriptional activators (e.g., Gal4–VP16) and chromatin remodeling complexes (e.g., ACF) induce chromatin remodeling in the absence of histone acetylation; (2) transcriptional activation domains target HATs to promoters after chromatin remodeling has occurred; and (3) histone acetylation by p300 is important for a step subsequent to chromatin remodeling and results in the transfer of histone H2A–H2B dimers from nucleosomes to a histone chaperone such as NAP-1 (see Fig. 6). Conclusions 1 and 2 are consistent with the results of recent in vivo cross-linking and immunoprecipitation studies in yeast (Cosma et al. 1999; Krebs et al. 1999) and other biochemical studies (Utley et al. 1998; Ikeda et al. 1999; Wang et al. 2000), but raise a question concerning the role of histone acetylation in chromatin remodeling. If prior activator-mediated chromatin remodeling precedes histone acetylation, what role could histone acetylation have in chromatin remodeling? Conclusion 3, which is a novel conclusion from our experiments, suggests an answer to this question, namely that the role of histone acetylation is to alter the structure of nucleosomes (e.g., facilitate the loss of H2A–H2B dimers) that have been remodeled previously by the action of ATP-dependent chromatin remodeling complexes.
Figure 6.
Model for histone transfer. ATP-dependent chromatin remodeling precedes histone acetylation by p300. Post-remodeling histone acetylation facilitates the transfer of histone H2A–H2B dimers to a histone chaperone.
Our experiments indicate that NAP-1 acts as a histone acceptor after the acetylation of nucleosomal histones by p300. There are two likely causes for this effect: (1) Acetylation might reduce the affinity of H2A–H2B for nucleosomes, with subsequent removal of the H2A–H2B dimers from chromatin by NAP-1; or (2) the acetylation of H2A–H2B might increase the affinity of H2A–H2B dimers for NAP-1. Our experiments do not distinguish between these two possibilities. Furthermore, although we have used NAP-1 as a histone acceptor in our experiments, this role might be filled in the cell by some other histone chaperone (e.g., CAF-1 or nucleoplasmin) or some other cellular factor.
What role might an altered nucleosome structure/composition have in transcriptional activation? The presence of altered (i.e., H2A–H2B-depleted) nucleosomes at a transcriptionally active, chromatin-remodeled promoter may help to maintain an open chromatin structure conducive to multiple rounds of activated transcription. This and other possibilities will need to be examined further in future studies.
We have used a purified recombinant chromatin assembly system to analyze the temporal relationship between chromatin remodeling and histone acetylation. This biochemical approach has allowed us to manipulate carefully various factors (e.g., ACF and p300) and cofactors (e.g., ATP and acetyl CoA) to determine their roles at different times during the process of chromatin remodeling and histone acetylation. Our results have revealed a previously unknown role for the acetylation of histones by p300, namely to facilitate histone transfer from nucleosomes to NAP-1. This experimental system should continue to prove useful for the examination of detailed mechanisms of factor-mediated chromatin remodeling and histone acetylation. For example, the localization of histone acetylation with respect to the enhancer and promoter is an important issue that will need to be clarified in the future.
Materials and methods
Purification of recombinant proteins
Recombinant p300 and ACF were prepared essentially as described previously (Kraus and Kadonaga 1999; Ito et al. 1999). His6-tagged NAP-1 was purified from baculovirus-infected Sf9 cells by Ni-NTA affinity chromatography, followed by three conventional chromatographic steps: Q-Sepharose FF, SP-Sepharose FF, and Q-Sepharose FF (all media were from Pharmacia). Peak fractions of the final Q-Sepharose FF column were pooled and used in the assays. Gal4–VP16 was purified as described previously (Chasman et al. 1989).
Chromatin assembly reactions and histone acetyltransferase assays
Chromatin assembly reactions were performed essentially as described by Bulger et al. (1995) and Ito et al. (1996) using supercoiled plasmid DNA. A standard reaction contained plasmid DNA (0.4 μg), purified core histones from Drosophila embryos (0.33 μg), purified recombinant dNAP-1, purified recombinant ACF, ATP (3 mm), and an ATP-regenerating system (30 mm phosphocreatine and 1 μg/ml creatine phosphokinase). Where specifically indicated, 3H Acetyl CoA (1 μm), cold acetyl CoA (10 μm), and p300 were added. For fluorography, part of the reaction was subjected to 12% SDS gel electrophoresis. Gels were Coomassie stained and prepared for fluorography with Enhance (NEN).
Density gradient analysis of chromatin
After chromatin was assembled and remodeled as indicated, reaction mixtures were subjected to 15%–40% glycerol density gradient sedimentation in a SW60 (Beckman) rotor at 60,000 rpm for 3 hr at 4°C. Part of the gradient fractions were digested with proteinase K, deproteinized with phenol-chloroform, and subjected to 1% agarose gel electrophoresis followed by ethidium bromide staining. The remaining portions of the samples were TCA precipitated and analyzed by 12% SDS polyacrylamide gel electrophoresis followed by Western blot analysis or fluorography as indicated. For quantification of core histones, Western blots were performed with anti-core histone antibodies and detected by 125I protein A.
Acknowledgments
We thank Dr. Jim Kadonaga and Dr. Yasuhisa Nogi for reading and commenting on the manuscript. We are grateful to Dr. Carl Wu for the ISWI cDNA. We thank Dr. Kiyoshi Yasui for his discussion throughout the course of this work. We are also grateful to Dr. Keisuke S. Iwamoto for his help with writing the manuscript. This work was supported by grants from the Japanese Society for the Promotion of Science (JSPS), the Research for the Future Program, the Uehara Memorial Foundation, and the Sankyo Foundation of Life Science to Takashi Ito, and a Career Award from the Burroughs Wellcome Fund to W. Lee Kraus.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tito@saitama-med.ac.jp; FAX 81-492-94-9751.
References
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Bulger M, Ito T, Kamakaka RT, Kadonaga JT. Assembly of regularly spaced nucleosome arrays by Drosophila chromatin assembly factor 1 and a 56-kDa histone-binding protein. Proc Natl Acad Sci. 1995;92:11726–11730. doi: 10.1073/pnas.92.25.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman DI, Leatherwood J, Carey M, Ptashne M, Kornberg RD. Activation of yeast polymerase II transcription by herpesvirus VP16 and GAL4 derivatives in vitro. Mol Cell Biol. 1989;9:4746–4749. doi: 10.1128/mcb.9.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Steger DJ, Eberharter A, Workman JL. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol Cell Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Kobayashi R, Kadonaga JT. Drosophila NAP-1 is a core histone chaperone that functions in ATP- facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Ito T, Levenstein M, Fyodorov D, Kutach AK, Kobayashi R, Kadonaga JT. The Acf1 and ISWI subunits of ACF function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes & Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT. Eukaryotic transcription: An interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Bunker CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes & Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Kadonaga JT. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes & Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— . Ligand- and cofactor-regulated transcription with chromatin templates. In: Picard D, editor. Steroid/nuclear receptor superfamily: A practical approach. Oxford/New York: Oxford University Press; 1999. pp. 167–189. [Google Scholar]
- Kraus WL, Manning ET, Kadonaga JT. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs JE, Kuo MH, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes & Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes & Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- Lewin B. Chromatin and gene expression: Constant questions, but changing answers. Cell. 1994;79:397–406. doi: 10.1016/0092-8674(94)90249-6. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Cairns BR, Zhang M, Kornberg RD. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Zhang M, Kornberg RD. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T, Workman JL. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 1996;15:4702–4712. [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Kamakaka RT, Kadonaga JT. ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science. 1994;266:2007–2011. doi: 10.1126/science.7801129. [DOI] [PubMed] [Google Scholar]
- Roth SY, Allis CD. Histone acetylation and chromatin assembly: A single escort, multiple dances? Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- Schnitzler G, Sif S, Kingston RE. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- ————— Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes & Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley RT, Ikeda K, Grant PA, Cote J, Steger DJ, Eberharter A, John S, Workman JL. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- van Holde KE, Lohr DE, Robert C. What happens to nucleosomes during transcription? J Biol Chem. 1992;267:2837–2840. [PubMed] [Google Scholar]
- Varga-Weisz PD, Becker PB. Chromatin-remodeling factors: Machines that regulate? Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/s0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Blank TA, Becker PB. Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J. 1995;14:2209–2216. doi: 10.1002/j.1460-2075.1995.tb07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Wade PA, Wolffe AP. Histone acetyltransferases in control. Curr Biol. 1997;7:R82–R84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- Walter PP, Owen-Hughes TA, Cote J, Workman JL. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol Cell Biol. 1995;15:6178–6187. doi: 10.1128/mcb.15.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu L, Berger SL. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes & Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Grossman SR, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]