Abstract
The primary cilium is emerging as a crucial regulator of signaling pathways central to vertebrate development and human disease. We identified atrioventricular canal 1 (avc1), a mouse mutation that caused VACTERL association with hydrocephalus, or VACTERL-H. We showed that avc1 is a hypomorphic mutation of intraflagellar transport protein 172 (Ift172), required for ciliogenesis and Hedgehog (Hh) signaling. Phenotypically, avc1 caused VACTERL-H but not abnormalities in left–right (L–R) axis formation. Avc1 resulted in structural cilia defects, including truncated cilia in vivo and in vitro. We observed a dose-dependent requirement for Ift172 in ciliogenesis using an allelic series generated with Ift172avc1 and Ift172wim, an Ift172 null allele: cilia were present on 42% of avc1 mouse embryonic fibroblast (MEF) and 28% of avc1/wim MEFs, in contrast to >90% of wild-type MEFs. Furthermore, quantitative cilium length analysis identified two specific cilium populations in mutant MEFS: a normal population with normal IFT and a truncated population, 50% of normal length, with disrupted IFT. Cells from wild-type embryos had predominantly full-length cilia, avc1 embryos, with Hh signaling abnormalities but not L–R abnormalities, had cilia equally divided between full-length and truncated, and avc1/wim embryos, with both Hh signaling and L–R abnormalities, were primarily truncated. Truncated Ift172 mutant cilia showed defects of the distal ciliary axoneme, including disrupted IFT88 localization and Hh-dependent Gli2 localization. We propose a model in which mutation of Ift172 results in a specific class of abnormal cilia, causing disrupted Hh signaling while maintaining L–R axis determination, and resulting in the VACTERL-H phenotype.
INTRODUCTION
VACTERL association is a non-random association of congenital anomalies with no known etiology. First described as VATER association by Quan and Smith in 1973, the initial patients were noted to have vertebral anomalies (V), anal atresia (A), tracheoesophageal fistulas (TE) and renal (R) and limb (L) anomalies. The high incidence of congenital heart defects in patients with VATER association was subsequently described, resulting in the inclusive acronym VACTERL (1–3).
The etiology of VACTERL association has not been established. Currently, patients can be diagnosed with VACTERL association if they have two or more of the relevant anomalies (4). In the series reported by Khoury et al. (5), >90% of patients diagnosed with VACTERL association had three or fewer anomalies, and <1% of patients had all six anomalies. There have been few reported instances of familial recurrence of typical VACTERL, and therefore monogenic causation has been deemed unlikely. Interestingly, Mendelian inheritance of a distinct subgroup of VACTERL association, VACTERL with hydrocephalus, or VACTERL-H, has been reported (6). Patients with VACTERL-H meet the diagnostic criteria for VACTERL, and in addition have hydrocephalus. Both autosomal recessive and X-linked recessive inheritance of VACTERL-H have been described ((7,8) reviewed in 9). These inheritance patterns have led some authors to argue that VACTERL-H should be classified as a syndrome (as opposed to an association) despite the unknown genetic etiology, given the evidence for monogenic inheritance (10).
Mice with disrupted Hedgehog (Hh) signaling display phenotypic overlap with VACTERL patients. In particular, Gli2 and Gli3 mutant mice have vertebral anomalies (11); Shh mutants develop truncated limbs (12); Gli3 mutants exhibit polydactyly (13); Shh, Gli2 and Gli3 mutants all show various foregut and hindgut anomalies (14–17); and Shh and Gli2;Gli3 double-mutants show renal anomalies (18–20). These findings have led to speculation that alterations of the Hh signaling pathway may contribute to the pathogenesis of human VACTERL (4). Recently, a human patient with typical VACTERL association was found to have a mutation of HOXD13 (21), a downstream target of Shh, providing possible indirect evidence linking human VACTERL with the Hh signaling pathway. However, no models of VACTERL-H have identified a single-gene etiology to date.
A growing body of work has demonstrated the crucial role played by primary cilia in vertebrate developmental signaling pathways. Multiple studies have begun to elucidate the complex role played by cilia and intraflagellar transport in Hh signaling. Huangfu et al. (22) first made the connection between cilia and Hh signaling, demonstrating that wimple (wim), a null allele of intraflagellar transport protein 172 (Ift172), results in absent cilia and loss of Hh signaling. Subsequently, it has been established that multiple Hh signaling molecules, including Ptch1, Smo, Sufu and the Gli family of transcription factors, are all enriched in cilia (23–27). Intraflagellar transport appears to play an active role in Hh signaling, and IFT proteins are required for both Gli activator and Gli repressor function (25,28,29). Therefore, cilia mutations cause phenotypes consistent with loss of Hh signaling in some developmental contexts and gain of Hh signaling in others. The precise ciliary structural requirements for Hh signaling remain elusive.
Although cilia function has been implicated in both Hh signaling and in left/right axis determination, mechanisms distinguishing these two crucial cilia functions have not been established. Situs abnormalities are a common feature of previously described ciliary mutations. Mutations in ciliogenesis genes that completely eliminate cilia impair both functions, making investigation of the separate requirement for intraciliary molecular signaling from the mechanical role of the cilia in this process unclear. The primary ciliary dyskinesia phenotype, with situs anomalies but no other apparent Hh-related defects, suggests that immotile cilia are unable to create the necessary nodal flow required for L–R determination, but can otherwise carry out signal transduction normally. Conversely, the Oak Ridge polycystic kidney (ORPK) mouse, homozygous for partial loss-of-function of Ift88, demonstrated intact left–right (L–R) axis determination but evidence of abnormal Hh signaling (30), suggesting that it is possible to disrupt Hh signaling while maintaining the ciliary functions needed for situs determination.
Here we report a mouse model for VACTERL-H identified in a forward genetic screen for congenital heart disease (CHD) in mice (31). The recessive N-ethyl,N-nitrosurea (ENU)-induced mutation, avc1 (atrioventricular canal 1), caused the VACTERL-H phenotype including vertebral anomalies, anal atresia, cardiac defects, tracheoesophageal anomalies, renal dysplasia, limb anomalies and hydrocephalous. We identified an avc1-specific polymorphism in Ift172, an intraflagellar transport gene previously implicated in the Hh signaling pathway (22). Avc1 is a hypomorphic allele of Ift172, causing abnormal splicing and decay of the majority of Ift172 transcript. Heart defects in avc1 mutant mice result from the specific loss of an Hh-responsive lineage that generates the atrial septum of the heart, providing the first molecular link between a cilia mutation and CHD. Although avc1 caused significant truncation of nodal cilia, no left/right axis abnormalities were observed in mutant embryos. Using Ift172avc1 and Ift172wim, we generated an Ift172 allelic series to investigate the relationship between Ift172 dosage and cilia architecture, IFT and Hh signaling. We observed that dose reduction of Ift172 caused anomalies of nodal cilia in vivo, primary cilia in vitro and a disruption of Hh signaling in a dose-dependent manner. Truncated mutant cilia displayed both abnormal localization of IFT88 and Gli2 to the distal ciliary tip, suggesting a defect of the distal ciliary axoneme. Our findings implicated structural cilia defects in the pathogenesis of VACTERL and suggested that cilia genes, and in particular IFT genes, are likely candidates for disease-causing mutations in VACTERL-H.
RESULTS
avc1: an ENU-derived mouse mutant with phenotypic features of VACTERL-H
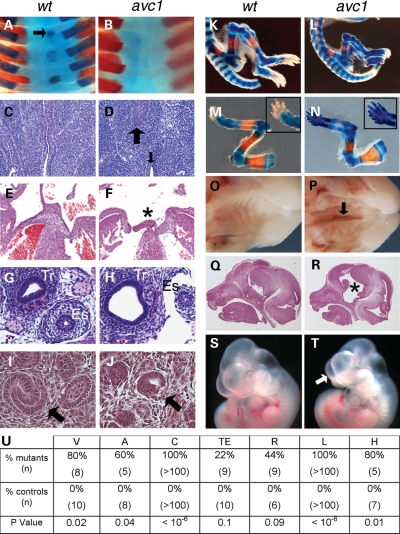
We performed a forward genetic screen for ENU-induced mutations that caused perinatal lethality and CHD in the mouse (31). avc1 caused heritable syndromic atrioventricular septal defects (AVSDs) as part of VACTERL-H association (Fig. 1). avc1 mutant newborns demonstrated hydrocephalus with highly penetrant vertebral, anal, cardiac, limb and palate abnormalities (Fig. 1U). Anomalies of the cervical vertebrae, including absent spinal processes or abnormally formed transverse processes, were present in 8/10 avc1 mutant mice in contrast to 0/10 wild-type littermate controls (P = 0.02; Fig. 1A and B). Mutants had seven cervical vertebrae, as is standard, but each cervical vertebra lacked the vertebral body (Fig. 1B). Thoracic, lumbar and sacral vertebrae appeared normal. Anal atresia, evidenced by discontinuity between the anal squamous epithelium and rectal columnar epithelium, was present in three of five avc1 embryos, whereas all the eight wild-type embryos showed a clear, intact transition between anal and rectal epithelium (P = 0.04; Fig. 1C and D). Cardiac anomalies, atrioventricular septal defects with common atrium, were present in 100% of avc1 mutants analyzed (n > 100; P < 10−6; Fig. 1E and F). Cardiac defects were not observed in any littermate control animals. Bone staining demonstrated anomalies of the long bones of the forelimbs and hindlimbs in mutant embryos (Fig. 1K–N). Mutant embryos demonstrated shortening of the humerus, radius, ulna, femur, tibia and fibula. In addition, all mutant mice demonstrated preaxial polydactyly of both the forelimbs and hindlimbs (n = 119) (Fig. 1N). All newborn animals examined demonstrated abnormal facies, with shortening of the muzzle. Clefts of the secondary palate were similarly noted in 20/20 newborn mutants (P < 0.01) (Fig. 1P). Hydrocephalus was present in four of five mutant animals, but none of seven litter mate wild-type controls (P = 0.01; Fig. 1Q and R).
Figure 1.
Avc1 mutant mice have VACTERL association with hydrocephalus. (A and B) Bone stain demonstrated normal cervical vertebral anatomy in newborn control animal (A), whereas mutant littermate exhibited the absence of vertebral bodies (B). Bone is stained red and cartilage blue. (C and D) Sagital section through an E14.5 control embryo demonstrated anorectal epithelial continuity with normal anatomy (C). Corresponding section through mutant littermate showed anal atresia with discontinuity between the squamous epithelium of the anus (small arrow, D) and the columnar epithelium of the distal rectum (large arrow, D). (E and F) Transverse section through the heart of the E14.5 control embryo showed intact atrial and ventricular septation (E). Corresponding section through the mutant littermate heart showed an atrioventricular septal defect (asterisk) with the common atrium. (G and H) Transverse section of the E14.5 control embryo demonstrated normal foregut anatomy (G). Mutant littermate (H) displayed a hypoplastic esophagus (Es) with an enlarged and abnormal trachea (Tr). (I and J) Transverse section of the E14.5 control embryo showed normal renal histology (I). Corresponding section through the mutant littermate (J) demonstrated dysplasia with hypoplastic glomeruli (arrow). (K–N) Bone stain of the newborn control animal showed normal bones of forelimb and hindlimb (K and M). Mutants exhibited shortening of the long bones of both fore- and hindlimbs (L and N) and preaxial polydactyly in the forelimb (inset, N). (O and P) Gross specimen of newborn wild-type palate (O). Mutants exhibited a cleft of the secondary palate (arrow, P). (Q and R) Sagital section through the head of the newborn avc1 mutant showed ventriculomegaly compared with the littermate control animal of normal ventricular size (Q). (S and T) Gross external morphology of E12.5 avc1 embryo demonstrated abnormal facies with a shortened muzzle (arrow, T) compared with the littermate control of normal size. (U) Table of prevalence of VACTERL-H features in avc1 mutants and littermate controls.
The remaining features of the VACTERL-H phenotype, tracheo-esophageal and renal abnormalities, were present with variable frequency. Esophageal and tracheal anomalies were observed in 2/9 mutant embryos at E14.5 (esophageal stenosis with enlarged, cystic trachea in one case, and esophageal atresia in the other), whereas 0/10 wild-type embryos analyzed showed foregut anomalies (P = 0.1; Fig. 1G and H). Renal dysplasia with hypoplastic glomeruli was observed in four of nine avc1 mutant embryos compared with none of six wild-type (P = 0.09, Fig. 1I and J). No eye abnormalities were observed by gross or histologic analysis of avc1 mutant embryos (n = 5).
Interestingly, no cardiac looping anomalies were noted in any of the mutant embryos. All mutant embryos analyzed (n > 100) showed levocardia with normal, d-looping of the ventricles, as did all wild-type control embryos. Similarly, all mutant embryos demonstrated situs solitus of the abdominal organs. Thus, no situs anomalies were observed in avc1 mutant embryos.
avc1 is an allele of Ift172
A candidate gene approach was used to identify the avc1 mutation. Recombinant chromosomes were identified in affected animals from an intercross between known carriers that located the avc1 mutation between markers D5MIT334 and D5MIT419. The interval containing avc1 corresponded to 0.9 megabases (Mb) on the physical map containing 33 genes. Bioinformatic investigation of the 33 candidate genes revealed no known role for any in heart development. However, one gene in the interval, Ift172, encoded an intraflagellar transport protein required for cilia biogenesis and implicated in the Hh signaling pathway (22).
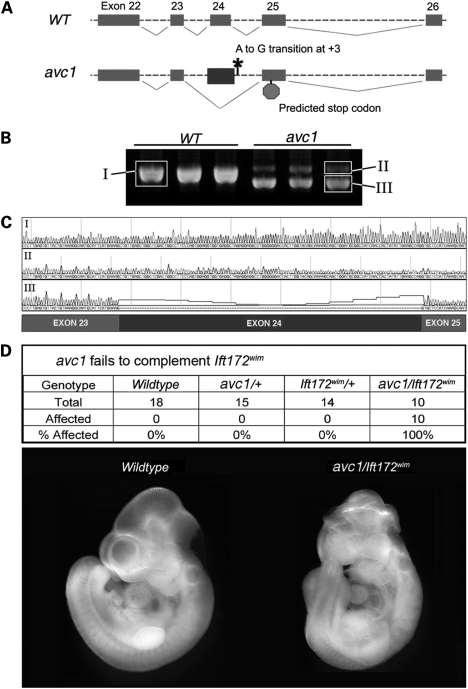
Sequencing of all 48 exons of the Ift172 gene, including adjacent intronic sequence, revealed an A-to-G transition in the splice donor site downstream of exon 24 (Fig. 2A). This polymorphism, located at position +3 in the 3′ intron of exon 24, altered a nucleotide conserved from humans to opossum. We hypothesized that the splice-site sequence alteration may affect the splicing of Ift172 transcripts. We evaluated products of Ift172 transcription by reverse transcriptase polymerase chain reaction (RT-PCR) in wild-type and avc1 mutant embryos, using primers designed to amplify Ift172 transcriptional products generated between exons 22 and 26, spanning the exons adjacent to the sequence polymorphism. Semi-quantitative RT-PCR from wild-type whole-embryonic tissue at E10.5 revealed a single product, whose sequence corresponded to wild-type Ift172 cDNA (Fig. 2B and C). RT-PCR from avc1 mutant whole-embryonic tissue at E10.5 revealed two distinct products. The sequence of the minority species, of higher molecular weight, corresponded to wild-type Ift172 cDNA. However, the sequence of the majority species, of lower molecular weight, revealed exon 24 skipping (Fig. 2B and C). Theoretical translation of the mis-spliced message encountered a frame shift and premature termination in exon 25 (Fig. 2A).
Figure 2.
Aberrant splicing of Ift172 in Avc1 mutant embryos. (A) Sequencing of Ift172 in Avc1 mutants demonstrated an A-to-G transition at position +3 of exon 24 splice donor. Mis-splicing and exon 24 skipping resulted in theoretical transcription encountering a stop site in exon 25. (B) RT-PCR spanning Ift172 exons 22 to 26 from three wild-type (left lanes) and three Avc1 mutant (right lanes) E10.5 embryos. Multiple splice products were obtained from Avc1 but not wild-type embryos. (C) Sequence tracings from gel-purified RT-PCR products shown in (B). RT product from wild-type embryos (I) and minority product from Avc1 embryos (II) demonstrated wild-type cDNA sequence. The majority product from the Avc1 embryos (III) demonstrated exon 24 skipping. (D) Avc1 failed to complement Ift172wim. Embryos with either the avc1 or Ift172wim alleles appeared normal. Embryos carrying both alleles were all affected.
The results of RT-PCR from avc1 mutant embryos suggested that the sequence alteration identified in Ift172 had functional consequences on Ift172 splicing. However, this perturbation of Ift172 had yet to be directly linked to the phenotype observed in avc1 mutant embryos. Because the avc1 allele was generated by ENU mutagenesis, it remained feasible that an undiscovered mutation affecting another gene in the physical interval containing avc1 contributed to the mutant phenotype. We sought to directly test whether avc1 and Ift172 were allelic. We performed a complementation test between avc1 and Ift172wim, an Ift172 null allele that causes mid-gestation embryonic lethality and loss of Hh signaling (22). The embryos from an intercross between Ift172wim/+ and avc1/+ mice were evaluated. Mice carrying both the Ift172wim and avc1 alleles demonstrated cardiac, limb and craniofacial abnormalities, whereas none of the heterozygous carriers of either mutant allele demonstrated phenotypic abnormalities (Fig. 2D). These findings demonstrated that Ift172wim and avc1 failed to complement and provided further evidence that the identified mutation of the Ift172 gene is responsible for the phenotype observed in avc1 mutant mice. As Ift172avc1 mice maintained some wild-type transcript and die at birth whereas Ift172wim mice die at E10.5, we conclude that avc1 is a hypomorphic allele. No homozygous avc1 mice were observed at weaning from 200 consecutive litters.
Whole-mount in situ hybridization against Ift172 in control embryos showed widespread expression of Ift172 in tissues affected by avc1 and known to be Hh-responsive (Fig. 3A–C, left column). Ift172avc1 mutant embryos demonstrated greatly reduced but detectable expression (Fig. 3A–C, right column). These observations suggested that the majority product of mis-spliced mRNA in the mutant embryos underwent nonsense-mediated decay. To directly test this hypothesis, we performed western blotting with an antibody against the N-terminus of IFT172 from E10.5 embryonic extracts. avc1 mutant embryos expressed 18% of the amount of IFT172 compared with littermate wild-type controls after normalization (Fig. 3D). Heterozygous avc1 mutant embryos expressed 63% of the amount of IFT172 compared with littermate wild-type controls after normalization (Fig. 3D). These results provide additional evidence that the mis-spliced product of Ift172 RNA undergoes nonsense-mediated decay in avc1 mutant embryos, and that avc1 causes a dose decrease in IFT172 protein levels.
Figure 3.
Reduced expression of Ift172 in avc1 mutant embryos. (A–C) Whole-mount in situ hybridization against Ift172 at E8 (A), E9 (B) and E10 (C) showed widespread expression of Ift172 in tissues with known requirements for Hh-signaling in control embryos (left column) with minimal expression in the heart. Ift172 mRNA was barely detectable in mutant littermates at each time point, with faint staining primarily along the neural tube (A–C, right column). (D) IFT172 protein was quantitatively determined by western blot analysis using a rabbit antibody raised against the N-terminus of IFT172. Lanes represent whole-embryo extracts from wild-type (lane 1), avc1/+ heterozygote (lane 2) or avc1/avc1 mutant (lane 3) embryos at E10.5, with γ-tubulin used as a loading control. The filled arrowhead indicates wild-type IFT172 protein and the two asterisks indicate major non-specific bands at ∼65 and 105 kDa. IFT172 band intensities normalized to γ-tubulin: wild-type = 1.00 (±0.14 SD), avc1/+ = 0.6297 and avc1/avc1 = 0.1826.
Ift172 mutant embryos have defects in Hh signaling
Given the known role of Ift172 in Hh signaling and the suspected link between defective Hh signaling and VACTERL syndrome, we sought to evaluate the impact of reduced Ift172 expression on Hh signaling in Ift172avc1 mutant mice. We assessed the integrity of the Hh signaling pathway in Ift172avc1 mutants in two ways, by whole-mount analysis of an Hh reporter and biochemically. First, we qualitatively evaluated the expression pattern of Patched1 (Ptch1) and Gli1. The expression of Ptch1, the primary Hh membrane receptor, and Gli1, a member of the Gli transcription factor family responsible for transducing the Hh signal, are Hh dependent. We compared the expression of Ptch1 using a previously reported Ptch1-lacZ Hh-reporter mouse line (32) and Gli1 by in situ hybridization in Ift172avc1 mutants and wild-type littermate controls at E10.5 (Fig. 4). In wild-type embryos, Ptch1 and Gli1 expression were confined to regions with known Hh signaling mirroring the expression of Ift172 (Fig. 4A). In comparison, Ift172avc1 mutant littermates demonstrated a global reduction in the expression of Ptch1 and Gli1 (Fig. 4B). Consistent with previous analysis describing an extra-cardiac population of Hh-receiving progenitor cells that subsequently migrate into the heart to form the atrial septum (33), there was minimal evidence of Hh signaling within the heart itself (Fig. 4A and B, lower panels).
Figure 4.
Hh signaling defects in avc1 mutants. (A and B) Whole-mount analysis of Hh-signaling-responsive genes Ptch1 and Gi1 in E10 embryos. Ptch-lacZ expression (upper panels) recapitulated known Hh-signaling pattern in wild-type (A) embryos, and with qualitatively reduced level of expression in avc1 mutant littermates (B). Gli1 expression is demonstrated in a similar pattern by in situ hybridization and was qualitatively reduced in avc1 mutant embryos (middle and lower panels). The arrow denotes Gli1 expression in the second heart field, behind the heart. (C) Full-length (Gli3–190) and processed (Gli3–83) forms of Gli3 were detected by western blot in wild-type, Ift172avc1, Ift172avc1/wim and Ift172wim embryos. The average processed Gli3–83 to full-length Gli3-190 protein ratios were 1.33, 0.39, 0.17 and 0.01, respectively. (D and E) Hh-receiving atrial septum progenitors were marked in R26RGli1CreERT2 embryos by the administration of tamoxifen at E7.5 and 8.5 and visualized at E13.0. In wild-type embryos, the Hh-receiving lineage filled the atrial septum (D). In avc1 mutant embryos, the Hh-receiving lineage was absent and a large atrioventricular septal defect was observed (E).
We next quantitatively assessed Hh signaling in Ift172avc1 embryos, examining the proteolytic processing of the Gli3 transcription factor. In mammals, Gli3 exists in two forms, a full-length protein that acts as a transcriptional activator (Gli3-190) and a processed form that acts as a transcriptional repressor (Gli3-83). Processing of Gli3 from the full-length activator form to the truncated repressor form is regulated by Hh signaling. Using western blotting and densitometry to quantify protein levels in tissue extracted from mutant and wild-type embryos, we found that there appeared to be a direct relationship between Ift172 dosage and Gli3 processing (Fig. 4C). In wild-type embryos, the ratio of processed to unprocessed Gli3 was 1.33. The ratios of processed to unprocessed Gli3 in avc1 mutants, avc1/wim compound heterozygote mutants and wim mutants were 0.39, 0.17 and 0.01, respectively (Fig. 4). This represented a 70.7% reduction of Gli3 processing in Ift172avc1 mutant embryos, an 87.2% reduction in Ift172avc1/wim embryos and a 99.2% reduction in Ift172wim embryos. Thus, the Ift172avc1 allele disrupted the Hh signaling, but to a lesser extent than the Ift172wim allele. These results supported the conclusion that Ift172avc1 is a hypomorphic allele of Ift172 and causes significant but incomplete loss of embryonic Hh signaling.
The intersection between the requirement for cilia and Hh signaling in heart development had not been demonstrated. We asked whether a decrement in Hh signaling could account for the atrioventricular septal defects observed in avc1 mutant embryos. Hh-receiving atrial septum progenitor cells were marked by Gli1 expression in Gli1:CreERT2;R26R embryos (33) and their contribution to the atrial septum was evaluated in wild-type and avc1 mutant embryos using genetic inducible fate mapping. Hh-receiving atrial septum progenitors were marked in R26RGli1-CreERT2 embryos by the administration of tamoxifen at E7.5 and E8.5 (33). The Hh-receiving lineage generated the atrial septum by E13.0 in control embryos (Fig. 4D). In contrast, the Hh-dependent atrial septum lineage was absent from avc1 mutant hearts at E13.0 (Fig. 4E). Thus, Ift172avc1 caused a structural cardiovascular phenotype, an atrioventricular septal defect, via deficient Hh signaling.
Ift172 mutant embryos display truncated cilia
To evaluate the effects of reduced Ift172 expression on cilia structure in vivo, we used scanning electron microscopy to analyze the ventral node of Ift172avc1 mutant, Ift172avc1/wim mutant and wild-type littermate embryos at E7.5 (Fig. 5). We observed a dose-dependent requirement for Ift172 in the production of full-length nodal cilia. The mean nodal cilia length was 3.56 ± 0.05 µm in wild-type E7.5 embryos (Fig. 5A, D, G and H), 2.26 ± 0.08 µm in Ift172avc1 embryos (Fig. 5B, E, G and H) and 0.81 ± 0.02 µm in Ift172avc1/wim embryos (Fig. 5C and F–H). Reduced Ift172 dosage therefore impairs ciliogenesis in vivo at the ventral node.
Figure 5.
Truncated nodal cilia in Ift172 mutant mutants. (A–H) Scanning electron microscopy was performed on the ventral node of E7.5 wild-type (A and D), Ift172avc1 (B and E) and Ift172avc1/wim (C and F) mutant littermate embryos. In contrast to the length of wild-type nodal cilia (3.56 ± 0.05 µm), Ift172avc1/avc1 nodal cilia were foreshortened (2.26 ± 0.08 µm, P < 0.001), and Ift172avc1/wim nodal cilia were severely truncated (0.81 ± 0.02 µm, P < 0.0001) (G and H). Four wild-type, four avc1/avc1 and five avc1/wim embryos were analyzed.
To better characterize the mechanistic basis for the quantitative decrement in Hh signaling observed in Ift172avc1 mutants, we evaluated the primary cilia of mutant mouse embryonic fibroblasts (MEFs). We predicted that fibroblast primary cilia would be abnormal in the avc1 mutant setting and tested this hypothesis by isolating and comparing fibroblasts from wild-type, Ift172avc1 and Ift172avc1/wim embryos. Analysis of the Ift172 allelic series revealed a dose-dependent requirement for Ift172 in the synthesis of primary cilia. Using acetylated tubulin immunohistochemistry, cilia were identified on 90.8% of wild-type cells (Fig. 6A, D and G), 41.5% of Ift172avc1 mutant cells (Fig. 6B, E and G) and 28.4% of Ift172avc1/wim cells (Fig. 6C, F and G).
Figure 6.
Abnormal number and morphology of primary cilia in Ift172 mutant fibroblasts. (A–G) Fibroblasts were isolated from Ift172avc1 and Ift172avc1/wim mutant and wild-type littermate embryos. After serum-starving to promote cilia growth, primary cilia were visualized in the cultured cells with acetylated tubulin (red). Arrows identify primary cilia. Nuclei were stained with DAPI (blue). Nearly all wild-type fibroblasts were ciliated (90.8%) (A, D and G), although significantly fewer Ift172avc1 (41.5%) (B, E and G) and Ift172avc1/wim (28.4%) (C, F and G) mutant fibroblasts were ciliated (P-values 8.12 × 10−173 and 4.06 ×10−239, respectively). (H) Ift172avc1 and Ift172avc1/wim mutant cilia were significantly shorter on average than wild-type cilia (2.18 and 1.62 versus 2.62 µm, P-values of 1.20 × 10−6 and 1.71 × 10−30, respectively). (I) Histogram analysis of primary cilia length demonstrates two distinct cilia populations, a full-length population and a truncated population that are approximately half normal length. Wild-type cilia were mostly full-length, Ift172avc1 cilia were divided evenly between the two populations and Ift172avc1/wim cilia appeared mostly truncated.
The documented deficiency in Hh signaling appeared greater in avc1 mutants than could be explained by the decreased percentage of ciliated cells (avc1 mutants showed an ∼70% reduction in Gli3 processing, but only an ∼54% reduction in the number of ciliated fibroblasts). We hypothesized that the cilia present on mutant cells may be abnormal and unable to support normal Hh signaling. We proceeded to structurally and functionally evaluate the cilia present on avc1 and avc1/wim mutant MEFs. Quantitative analysis of primary cilia length revealed that there was a direct relationship between Ift172 dosage and cilia length. Cilia on fibroblasts derived from wild-type embryos were significantly longer than the cilia on cells derived from Ift172avc1 embryos which were in turn significantly longer than cilia on cells from Ift172avc1/wim compound heterozygote embryos (Fig. 6H).
Closer analysis of cilia length suggested that there were two distinct populations of cilia present, full length and truncated to approximately half length, rather than a normal distribution of cilia length within each genotype. Histogram analysis of the length of cilia on Ift172avc1/avc1 fibroblasts demonstrated two peaks, rather than a normal distribution of lengths. Ift172avc1 cilia were approximately equally divided between two populations, a ‘full length' population, with a peak centered at ∼2.5 µm, and a ‘truncated' population, with a peak centered at ∼1.5 µm (Fig. 6I). Analysis of wild-type fibroblast cilia length revealed a greater population of ‘full length' cilia, with a much smaller population of ‘truncated' cilia, although analysis of Ift172avc1/wim fibroblasts revealed a large ‘truncated' population, with a much smaller population of the ‘full length' cilia (Fig. 6I). Cilia of both types were present in all genotypes, and the range of cilia length was nearly identical between the three genotypes (Fig. 6I). This observation suggested that the observed difference in mean cilia length was due to varying the relative proportions between the normal and truncated cilium populations, as opposed to a generalized reduction in cilium length in mutant fibroblasts.
Ift172 mutant cilia demonstrate abnormal IFT88 and Gli2 localization
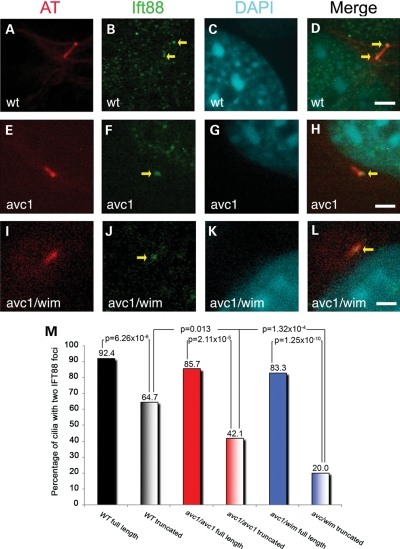
We predicted that Ift172 mutant cilia might have impaired intraflagellar transport, resulting in aberrant cilia structure and Hh signaling. To test this hypothesis, we performed immunocytochemistry with an antibody against IFT88, an essential component of IFT complex B, on wild-type, Ift172avc1 and Ift172avc1/wim cultured fibroblasts. It had been previously reported that IFT88 concentrates at two sites within the cilium, one at the cilium base and one at the distal tip (25). Interestingly, full-length cilia (>2 µm in length) demonstrated normal IFT88 distribution, with foci of staining at the distal and proximal ends of the axoneme, regardless of genotype (Fig. 7). We found that 92.4% of full-length wild-type cilia (Fig. 7A–D and M), 85.7% of full-length Ift172avc1 cilia (Fig. 7E–H and M) and 83.3% of full-length Ift172avc1/wim cilia (Fig. 7I–M) showed normal IFT88 distribution and there was no significant difference between the distribution of IFT88 in full-length cilia of all genotypes (wild-type versus Ift172avc1, P = 0.13; wild-type versus Ift172avc1/wim, P = 0.074). In contrast, the majority of truncated cilia showed only a single focus of IFT88 staining: only 42.1% of truncated Ift172avc1 cilia and 20.0% of truncated Ift172avc1/wim cilia showed the normal IFT88 distribution (wild-type versus avc1: P = 5.54 × 10−11 and wild-type versus avc1/wim: P = 1.53 × 10−25, respectively). Even truncated cilia on wild-type MEFs showed the normal distribution of IFT88 only 64.7% of the time, significantly less often than full-length wild-type cilia (P = 6.26 × 10−6). These results identified two distinct populations of primary cilia in cultured MEFs, full length and truncated, and documented abnormal IFT88 localization in the truncated population. A reduction in Ift172 dosage therefore established a truncated class of cilia with abnormal IFT.
Figure 7.
Abnormal Ift88 localization in Ift172 mutant primary cilia. (A–M) Immunofluorescence was performed on serum-starved cultured fibroblasts from wild-type (A–D), Ift172avc1 (E–H) and Ift172avc1/wim (I–L) embryos. Cilia were stained with acetylated tubulin (red) and IFT88 was stained with a specific anti-IFT88 antibody (green). Yellow arrows identify foci of Ift88 fluorescence. Size standard equals 3 µm. Cilia of each genotype were analyzed in two groups, full length (>2 µm) and truncated (<2 µm). The majority of full-length cilia of all genotypes displayed the normal distribution of IFT88, with one focus at the distal tip and one focus at the proximal tip (92.4% of wild-type cilia, 85.7% of Ift172avc1 cilia and 83.3% of Ift172avc1/wim cilia) (M). There was not a significant difference in IFT88 distribution between any of these groups (wild-type versus Ift172avc1 P = 0.13; wild-type versus Ift172avc1/wim P = 0.074; Ift172avc1 versus Ift172avc1/wim P = 0.49) (M). Truncated mutant cilia revealed a different distribution (M); the majority of Ift172avc1 truncated cilia (57.9%) and the majority of Ift172avc1/wim truncated cilia (80.0%) had a single focus of IFT88 staining. IFT88 distribution was significantly different between truncated cilia and full-length cilia of all genotypes. IFT88 distribution was also significantly different between truncated wild-type cilia and either truncated Ift172avc1 cilia (P = 0.028) or truncated Ift172avc1/wim cilia (P= 1.65 × 10−6).
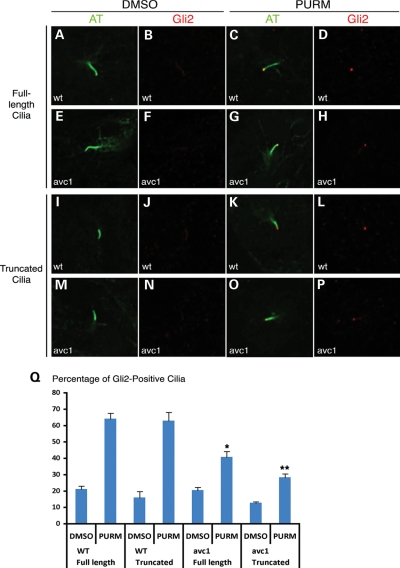
We hypothesized that the observed IFT defect in Ift172 mutant cilia may impair the localization of Hh signaling components to the distal ciliary axoneme, providing a possible molecular explanation for the observed Hh signaling defects. We directly evaluated endogenous Gli2 localization by immunohistochemistry in wild-type and avc1 mutant MEFs to test this hypothesis. Gli2 undergoes localization to the distal tip of the ciliary axoneme by an Hh-dependent mechanism (34,35). We found that after the activation of Hh signaling with purmorphamine (PURM) (36), >60% of wild-type control MEFs contained concentrated Gli2 at the distal tip of the ciliary axoneme (Fig. 8A–D and I–L). There was no difference in the percentage of Gli2-positive wild-type full-length cilia (64 ± 3.5%) versus truncated cilia (65 ± 5.2%; P = 1). However, Gli2 localized to significantly fewer cilia in Ift172avc1 mutant MEFs after PURM treatment (Fig. 8Q). Compared with wild-type cilia, Gli2 localized to significantly fewer full-length mutant cilia (41 ± 3.4%; P = 0.003) and truncated mutant cilia (28.5 ± 2.2%; P = 0.001) (Fig. 8E–H and M–Q). These results indicate that avc1 impairs Hh-dependent Gli2 localization to the distal ciliary axoneme, a possible mechanism for the Hh signaling decrement observed in avc1 mutant embryos.
Figure 8.
Abnormal Gli2 localization in truncated avc1 mutant cilia. (A–P) Immunofluorescence was performed on serum-starved fibroblasts isolated from Ift172avc1 mutant embryos and their wild-type littermates. Fibroblasts were incubated with PURM to activate the Hh signaling pathway. Cilia were then stained with acetylated tubulin (green) and Gli2 with a specific anti-Gli2 antibody (red). Full-length and truncated cilia were analyzed separately. (Q) Approximately 64% of full-length or truncated wild-type cilia demonstrated Gli2 localization to the distal tip of the axoneme, whereas only 41% of full-length avc1 mutant cilia (*P = 0.003) and 28% of truncated avc1 mutant cilia (**P = 0.001) demonstrated similar localization.
DISCUSSION
We report that VACTERL-H may be added to the list of confirmed ciliopathies in mice, demonstrating that avc1, a novel hypomorphic allele of Ift172, results in VACTERL-H. We document abnormalities in cilia structure, intraflagellar transport and Hh signaling that provide mechanistic insight into how avc1 causes the VACTERL-H phenotype. Our results provided empirical evidence for the previously suspected but unsubstantiated link between cilia, Hh signaling and VACTERL-H. We demonstrated for the first time the cellular mechanism causing a congenital heart defect in a cilia mutant: loss of an Hh-dependent lineage that generates the atrial septum, resulting in AVSDs with common atrium. Furthermore, by documenting the presence of a specific class of truncated cilia with disrupted IFT and Hh signaling in avc1 mutants, we add a novel piece to the complex understanding of how cilia mutations impact the Hh signal transduction pathway and L–R determination.
The complete range of the VACTERL-H phenotypes is observed in Ift172avc1 mutant embryos with variable penetrance. Although several null alleles of IFT protein genes result in the absence of cilia, hypomorphic ciliary mutations allowing functional analysis of mutant cilia are less well described. One previously described hypomorphic allele of an IFT complex B protein, the ORPK allele of IFT88 (reviewed in 30), allows survival to adulthood and a milder subset of phenotypes than those caused by avc1. Like avc1 mutants, ORPK mice have also been shown to have truncated cilia. The ‘gasping' (gsp) phenotype reported by Ermakov et al. (37) demonstrates some similarities to VACTERL and to avc1, with abnormalities of foregut and hindgut, as well as polydactyly. The gsp1, gsp3 and gsp6 lines were all reported to have truncated nodal cilia, although the responsible gene has not been identified. Identification of avc1 as a loss-of-function mutation in an IFT complex B gene provides the first identification of a monogenic cause for VACTERL-H association, which may now be considered a syndrome.
The avc1 allele of Ift172 decouples the role of the cilium in L–R patterning from its role in Hh signaling: L–R axis determination was preserved, whereas Hh signaling was significantly disrupted. The generation of L–R asymmetry is at least partially dependent on nodal flow established by rotating nodal monocilia (26), and L–R patterning abnormalities are common in cilia mutants (see reference 38 for partial list). For example, Ift172wim mutants demonstrated absent nodal cilia and randomized L–R patterning [e.g. randomized heart looping (22)]. In contrast, Ift172avc1 mutants demonstrated foreshortened nodal cilia but normal L–R patterning (e.g. normal heart looping in all cases). Our observations suggested that the proximal end of the cilium, maintained in avc1 mutants, may be sufficient to support normal rotation of nodal cilia and establishment of nodal flow, although insufficient to support normal Hh signal transduction. Although further studies are required to evaluate this hypothesis more definitively, this model could explain the decoupling of L–R axis formation from Hh signaling defects observed in avc1 mutant mice.
Our results suggest that decreased Ift172 dosage affects cilia synthesis in two distinct ways: (i) cells are less likely to produce cilia at all in the setting of decreased Ift172 dosage and (ii) the cilia that are formed are more likely to be truncated with abnormalities in IFT and Hh signaling. Fibroblasts with no Ift172 make no cilia, and carry out no Hh signaling transduction (22). Our analysis of the Ift172 allelic series utilizing our avc1 allele with the wim null allele of Ift172 suggests a dose-dependent requirement of Ift172 for the synthesis of functional primary cilia. Furthermore, we report that a hypomorphic allele of Ift172 leads to disrupted IFT in the setting of a specific class of foreshortened cilia. In avc1 and avc1/wim mutant MEFs, we observed a distinct class of cilia that are approximately half normal length (Fig. 6) with associated molecular abnormalities of the distal axoneme, e.g. loss of IFT88 and Gli2 localization (Figs 7 and 8, respectively). Thus, as the Ift172 dosage was decreased, both the number of ciliated fibroblasts and the percentage of full-length, fully functional cilia decreased. The relative proportions of cells with full-length, fully functional cilia, cells with truncated, dysfunctional cilia and cells with absent cilia will likely determine the degree to which the Hh signal transduction pathway is disrupted by any given cilia mutation.
These observations are consistent with a model of the mammalian cilia as a composite structure composed of distinct functional units, with the distal tip specifically required for cilium-based signaling. Work in Caenorhabditis elegans has demonstrated that differences in the IFT molecular components determine the distinction between axonemal core and distal tip, and mutants lacking the distal singlet tips demonstrate signaling defects (reviewed in 39). A similar model governing mammalian cilia-based signaling would predict that cells with abnormalities of the distal ciliary axoneme would demonstrate defective signal transduction, as we observed in Ift172 mutant MEFs. The observed quantitative loss of Hh signaling and defective or absent distal axoneme in Ift172 mutant cilia support a model requiring the distal ciliary axoneme for Hh signal transduction. A specific defect of the distal axoneme of Ift172 mutant cilia in mice is consistent with the required role of IFT172 in the transition of intraflagellar transport at the cilia tip of Chlamydomonas (40). However, this model does not explain our additional observation that Gli2 localization is impaired in normal-length avc1 mutant cilia (Fig. 8Q). This observation implies that IFT172 may also have a role in Gli2 trafficking and Hh signaling independent of its role in ciliogenesis. More work will need to be done to further elucidate the role of IFT172 on the trafficking of Hh signaling molecules within the ciliary axoneme.
The list of confirmed ciliopathies includes Bardet–Biedl, nephronophthisis, Senior–Loken syndrome, Alstrom syndrome, Meckel syndrome, Joubert syndrome, oral–facial–digital syndrome type I, Jeune asphyxiating thoracic dystrophy, Ellis–van Creveld syndrome, Leber congenital amaurosis, Kartagener syndrome and polycystic kidney disease [reviewed in (41)]. Here we provide evidence that avc1, a hypomorphic mutation of Ift172, caused a ciliopathy with VACTERL-H-like features in mice. By providing a direct link between a single IFT gene mutation and VACTERL-H phenotypes, this work illustrates the diversity of phenotypes that can result from IFT gene mutations. IFT gene mutations have been significantly under-represented in the human ciliopathy literature. Establishing a Mendelian etiology for VACTERL-H in mice, we suggest that IFT genes, particularly IFT complex B genes including Ift172, be considered candidate genes for human VACTERL-H.
MATERIALS AND METHODS
Mouse strains
Avc1 was identified in a screen for recessive ENU-induced mutations that caused perinatal lethality and structural heart defects. Identification and characterization of wim has been previously described (22). The Gli1CreERT2 line was obtained from the Joyner Laboratory (Sloan-Kettering Institute, NY, USA). Activation of CreERT2 was accomplished by oral gavage with 2 mg of tamoxifen per dose in corn oil to pregnant dams, demonstrated to achieve optimal activation of CreERT2 without toxicity (33).
Patched1-LacZ (B6;129-Ptch1tm1Mps/J) and R26R (B6;129S4-Gt(ROSA)26Sortm1Sor/J) mice were obtained from The Jackson Laboratory, and genotyping was performed as described (www.jax.org). All mouse experiments were performed in a mixed B6/129/SvEv background. All experiments involving mice were carried out according to a protocol reviewed and approved by the Institutional Animal Care and Use Committee of The University of Chicago, in compliance with the USA Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Genetic mapping and molecular identification of avc1
SNPs that were previously identified as polymorphic between inbred strains of mouse, including C57BL/6J, FVB/NJ and C3H/FeJ (42) were genotyped on the Illumina platform (42) using DNA from affected mice. Additional polymorphic markers from publically available databases were utilized for high-resolution mapping. Avc1 was mapped to a 0.9 Mb interval on chromosome 5 between D5Mit229 and D5Mit420 in a mapping cross with more than 1000 opportunities for recombination. The only sequence alteration detected in the avc1-containing molecular interval was the A-to-G transition at position +3 in the Ift172 exon 24 splice donor site.
Embryo dissection and X-Gal staining
Embryos were dissected from maternal tissue samples and tail samples were taken for genotyping. Mice or dissected organs were then fixed for 1 h in 4% paraformaldehyde and stained with X-gal staining solution [5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6, 2 mm MgCl2, 0.02% NP-40, 0.01% deoxycholate, 0.1% X-Gal in PBS] when appropriate or fixed in 10% neutral buffered formalin if no β-galactosidase-producing allele was present. To facilitate staining, embryos older than E10.5 were dissected open prior to staining. Mice were embedded in paraffin and sectioned at 5 µm. If X-gal stain was present, slides were counter-stained with 50% eosin for 1 s. If no X-gal stain was present, the slides were stained with hematoxylin and eosin.
Antibody production
Chemically synthesized peptides corresponding to KLRRDY YQWLMDTQQEER were sent to Pocono Rabbit Farm and Laboratory, Inc., for IFT172 antibody production in rabbits and antibody purification. The synthesized peptide corresponds to amino acids 738–755 of IFT172 encoded in exon 22 (NCBI m37 mouse assembly) and shared by both wild-type and avc1 proteins.
Western blotting
Western blots were performed in triplicate. Mouse embryos were harvested in PBS at 4°C, an aliquot taken for genotyping and the remainder transferred into 0.5 ml of RIPA lysis buffer maintained at 4°C (Roche CompleteTM protease inhibitor, 0.5 m Tris–HCl, pH 7.4, 1.5 m NaCl, 2.5% deoxycholic acid, 10% NP-40, 10 mm EDTA). Tissues were lysed with rotor–stator-type homogenizer for 30 s. Lysates were rotated for 15 min and centrifuged for 15 min at 12 000g. The resulting supernatants were removed, assayed for protein concentration, normalized by the addition of RIPA buffer and diluted into Laemmli sample buffer (Bio-Rad, 161-0737EDU). The resulting samples were run into 8% Tris–glycine gels for SDS–PAGE and transferred to nitrocellulose blots for immunodetection. Following block (5% milk/TBST for 30 min) and wash (TBST 3×), primary antibodies (a gift of B. Wang) were added at 1:100 in 2.5% BSA/TBST and incubated overnight at 4°C. Alkaline phosphatase-conjugated secondary antibody was utilized and blots were detected with an Amersham ECL-Plus detection kit (RPN2124).
Bone staining
Bone and cartilage staining was performed on E18.5 embryos as described by McLeod (43).
Primary MEFs
MEFs were isolated from Ift172avc1/avc1 embryos at E13.5 and Ift172avc1/wim embryos at E12.5. Culture conditions were as described by Ocbina and Anderson (28).
Immunofluorescence
Cells were grown to confluence on eight well-chambered slides (Lab-Tek II). Fixation and immunofluorescence staining were performed as described by Ocbina and Anderson (28), with the following minor modifications: blocking solution was composed of PBS with 10% v/v normal donkey serum and 0.1% v/v Triton X-100. All antibodies were diluted in 1:1 blocking solution and PBS. The following primary antibodies were used: mouse anti-acetylated-alpha-tubulin (Sigma-Aldrich, T7451) at a dilution of 1:1000; rabbit anti-Ift88 (a gift from B. Yoder, University of Alabama, Birmingham, AL, USA) at a dilution of 1:500 and rabbit anti-Gli2 (a gift from Paotien Chuang) at a dilution of 1:400. Alexa488, Alexa568, goat anti-mouse AlexaFluor 488 and goat anti-rabbit AlexaFluor 594-conjugated secondary antibodies (Invitrogen) were each used at a dilution of 1:500.
Electron microscopy
E8.0 embryos were dissected in PBS at room temperature and immediately fixed in 2.5% glutaraldehyde and 2% PFA in 0.1 m sodium cacodylate buffer, pH 7.4 (Electron Microscopy Sciences), for at least 1 h at room temperature. Embryos were stored in a fixative for an additional 24 h at 4°C before processing as described in Huangfu and Anderson (44). Scanning electron micrograph images were taken on a Zeiss SUPRA 25 FESEM.
In situ hybridization
In situ hybridization was performed using digoxigenin-labeled probes. Protocol was as described in (45), with the following changes: embryos were washed six times in maelic acid buffer for 1 h (MAB; 0.1 m maelic acid, pH 7.5, 0.15 m NaCl, 0.1% Tween-20 and 0.002 m levamisole), followed by a 16 h overnight wash at room temperature. Color reactions were allowed to develop overnight, and images were taken prior to storage in 80% glycerol. The in situ probe for Ift172 message spanned the 30th to 40th exons and for Gli1 as described (33).
FUNDING
This work was funded by grants from the NIH [R01 HL092153 (I.P.M.)], the March of Dimes (I.P.M.), the AHA (I.P.M.) and the Schweppe Foundation (I.P.M.).
ACKNOWLEDGEMENTS
The authors thank Bradley Yoder and the University of Alabama at Birmingham Hepato/Renal Fibrocystic Diseases Core Center (UAB HRFDCC DK074038) for the IFT88 antibody.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Temtamy S.A., Miller J.D. Extending the scope of the VATER association: definition of the VATER syndrome. J. Pediatr. 1974;85:345–349. doi: 10.1016/s0022-3476(74)80113-7. doi:10.1016/S0022-3476(74)80113-7. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman R.L. Birth defects and oral contraceptives. Lancet. 1973;1:1396. doi: 10.1016/s0140-6736(73)91731-5. doi:10.1016/S0140-6736(73)91731-5. [DOI] [PubMed] [Google Scholar]
- 3.Nora A.H., Nora J.J. A syndrome of multiple congenital anomalies associated with teratogenic exposure. Arch. Environ. Health. 1975;30:17–21. doi: 10.1080/00039896.1975.10666626. [DOI] [PubMed] [Google Scholar]
- 4.Kim J.H., Kim P.C.W., Hui C-c. The VACTERL association: lessons from the Sonic hedgehog pathway. Clin. Genet. 2001;59:306–315. doi: 10.1034/j.1399-0004.2001.590503.x. doi:10.1034/j.1399-0004.2001.590503.x. [DOI] [PubMed] [Google Scholar]
- 5.Khoury M.J., Cordero J.F., Greenberg F., James L.M., Erickson J.D. A population study of the VACTERL association: evidence for its etiologic heterogeneity. Pediatrics. 1983;71:815–820. [PubMed] [Google Scholar]
- 6.Corsello G., Giuffre L. VACTERL with hydrocephalus: a further case with probable autosomal recessive inheritance. Am. J. Med. Genet. 1994;49:137–138. doi: 10.1002/ajmg.1320490133. [DOI] [PubMed] [Google Scholar]
- 7.Herman T.E., Siegel M.J. VACTERL-H syndrome. J. Perinat. 2002;22:496–498. doi: 10.1038/sj.jp.7210765. [DOI] [PubMed] [Google Scholar]
- 8.Lomas F.E., Dahlstrom J.E., Ford J.H. VACTERL with hydrocephalus: family with X-linked VACTERL-H. Am. J. Med. Genet. 1998;76:74–78. doi: 10.1002/(sici)1096-8628(19980226)76:1<74::aid-ajmg14>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 9.Lurie I.W., Ferencz C. VACTERL-hydrocephaly, DK-phocomelia, and cerebro-cardio-radio-reno-rectal community. Am. J. Med. Genet. 1997;70:144–149. doi: 10.1002/(sici)1096-8628(19970516)70:2<144::aid-ajmg8>3.0.co;2-y. doi:10.1002/(SICI)1096-8628(19970516)70:2<144::AID-AJMG8>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 10.Wang H., Hunter A.G., Clifford B., McLaughlin M., Thompson D. VACTERL with hydrocephalus: spontaneous chromosome breakage and rearrangement in a family showing apparent sex-linked recessive inheritance. Am. J. Med. Genet. 1993;47:114–117. doi: 10.1002/ajmg.1320470124. [DOI] [PubMed] [Google Scholar]
- 11.Mo R., Freer A.M., Zinyk D.L., Crackower M.A., Michaud J., Heng H.H., Chik K.W., Shi X.M., Tsui L.C., Cheng S.H., Joyner A.L., Hui C. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- 12.Chiang C., Litingtung Y., Lee E., Young K.E., Corden J.L., Westphal H., Beachy P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. doi:10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 13.Masuya H., Sagai T., Moriwaki K., Shiroishi T. Multigenic control of the localization of the zone of polarizing activity in limb morphogenesis in the mouse. Dev. Biol. 1997;182:42–51. doi: 10.1006/dbio.1996.8457. doi:10.1006/dbio.1996.8457. [DOI] [PubMed] [Google Scholar]
- 14.Motoyama J., Liu J., Mo R., Ding Q., Post M., Hui C.C. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nat. Genet. 1998;20:54–57. doi: 10.1038/1711. doi:10.1038/1711. [DOI] [PubMed] [Google Scholar]
- 15.Litingtung Y., Lei L., Westphal H., Chiang C. Sonic hedgehog is essential to foregut development. Nat. Genet. 1998;20:58–61. doi: 10.1038/1717. doi:10.1038/1717. [DOI] [PubMed] [Google Scholar]
- 16.Ramalho-Santos M., Melton D.A., McMahon A.P. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 2000;127:2763–2772. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]
- 17.Mo R., Kim J.H., Zhang J., Chiang C., Hui C.C., Kim P.C. Anorectal malformations caused by defects in sonic hedgehog signaling. Am. J. Pathol. 2001;159:765–774. doi: 10.1016/S0002-9440(10)61747-6. doi:10.1016/S0002-9440(10)61747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J., Carroll T.J., McMahon A.P. Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development. 2002;129:5301–5312. doi: 10.1242/dev.129.22.5301. [DOI] [PubMed] [Google Scholar]
- 19.Bose J., Grotewold L, Ruther U. Pallister–Hall syndrome phenotype in mice mutant for Gli3. Hum. Mol. Genet. 2002;11:1129–1135. doi: 10.1093/hmg/11.9.1129. doi:10.1093/hmg/11.9.1129. [DOI] [PubMed] [Google Scholar]
- 20.Hu M.C., Mo R., Bhella S., Wilson C.W., Chuang P.T., Hui C.C., Rosenblum N.D. Gli3-dependent transcriptional repression of Gli1, Gli2 and kidney patterning genes disrupts renal morphogenesis. Development. 2005;133:569–578. doi: 10.1242/dev.02220. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Barcelo M.M., Wong K.K., Lui V.C., Yuan Z.W., So M.T., Ngan E.S., Miao X.P., Chung P.H., Khong P.L., Tam P.K. Identification of a HOXD13 mutation in a VACTERL patient. Am. J. Med. Genet. A. 2008;146A:3181–3185. doi: 10.1002/ajmg.a.32426. doi:10.1002/ajmg.a.32426. [DOI] [PubMed] [Google Scholar]
- 22.Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., Anderson K.V. Hedgehog signaling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. doi:10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 23.Rohatgi R., Milenkovic L., Scott M.P. Patched1 regulates Hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. doi:10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 24.Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., Reiter J.F. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. doi:10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 25.Haycraft C.J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E.J., Yoder B.K. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein Polaris for processing and function. PLoS. 2005;1:480–488. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;99:829–837. doi: 10.1016/s0092-8674(00)81705-5. doi:10.1016/S0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Zhou Z., Walsh C.T., McMahon A.P. Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc. Natl Acad. Sci. USA. 2009;106:2623–2628. doi: 10.1073/pnas.0812110106. doi:10.1073/pnas.0812110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ocbina P.J.R., Anderson K.V. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: analysis in mouse embryonic fibroblasts. Dev. Dyn. 2008;237:2030–2038. doi: 10.1002/dvdy.21551. doi:10.1002/dvdy.21551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu A., Wang B., Niswander L.A. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 30.Lehman J.M., Michaud E.J., Schoeb T.R., Aydin-Son Y., Miller M., Yoder B.K. The Oak Ridge polycystic kidney mouse: modeling ciliopathies of mice and men. Dev. Dyn. 2008;237:1960–1971. doi: 10.1002/dvdy.21515. doi:10.1002/dvdy.21515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamp A., Peterson M.A., Svenson K.L., Bjork B.C., Hentges K.E., Rajapaksha T.W., Moran J., Justice M.J., Seidman J.G., Seidman C.E., Moskowitz I.P., Beier D.R. Genome-wide identification of mouse congenital heart disease loci. Hum. Mol. Genet. 2010;19:3105–3113. doi: 10.1093/hmg/ddq211. doi:10.1093/hmg/ddq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodrich L.V., Milenkovic L., Higgins K.M., Scott M.P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. doi:10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 33.Hoffmann A.D., Peterson M.A., Friedland-Little J.M., Anderson S.A., Moskowitz I.P. Sonic hedgehog is required in pulmonary endoderm for atrial septation. Development. 2009;136:1761–1770. doi: 10.1242/dev.034157. doi:10.1242/dev.034157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen M.H., Wilson C.W., Li Y.J., Law K.K., Lu C.S., Gacayan R., Zhang X., Hui C.C., Chuang P.T. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. doi:10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J., Kato M., Beachy P.A. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc. Natl Acad. Sci. USA. 2009;106:21666–21671. doi: 10.1073/pnas.0912180106. doi:10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinha S., Chen J.K. Purmorphamine activates the Hedgehog pathway by targeting Smoothened. Nat. Chem. Biol. 2006;2:29–30. doi: 10.1038/nchembio753. [DOI] [PubMed] [Google Scholar]
- 37.Ermakov A., Stevens J.L., Whitehill K., Robson J.E., Pieles G., Brooker D., Goggolidou P., Powles-Glover N., Hacker T., Young S.R., et al. Mouse mutagenesis identifies novel roles for left-right patterning genes in pulmonary, craniofacial, ocular, and limb development. Dev. Dyn. 2009;238:581–594. doi: 10.1002/dvdy.21874. doi:10.1002/dvdy.21874. [DOI] [PubMed] [Google Scholar]
- 38.Davenport J.R., Yoder B.K. An incredible decade for the primary cilium: a look at a once-forgotten organelle. Am. J. Physiol. Renal Physiol. 2005;289:1159–1169. doi: 10.1152/ajprenal.00118.2005. doi:10.1152/ajprenal.00118.2005. [DOI] [PubMed] [Google Scholar]
- 39.Scholey J.M., Anderson K.V. Intraflagellar transport and cilium-based signaling. Cell. 2006;125:439–442. doi: 10.1016/j.cell.2006.04.013. doi:10.1016/j.cell.2006.04.013 [Review] [DOI] [PubMed] [Google Scholar]
- 40.Pedersen L.B., Miller M.S., Geimer S., Leitch J.M., Rosenbaum J.L., Cole D.G. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr. Biol. 2005;15:262–266. doi: 10.1016/j.cub.2005.01.037. doi:10.1016/j.cub.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 41.Baker K., Beales P.L. Making sense of cilia in disease: the human ciliopathies. Am. J. Med. Genet. C Semin. Med. Genet. 2009;151C:281–295. doi: 10.1002/ajmg.c.30231. [DOI] [PubMed] [Google Scholar]
- 42.Moran J.L., Bolton A.D., Tran P.V., Brown A., Dwyer N.D., Manning D.K., Bjork B.C., Li C., Montgomery K., Siepka S.M., et al. Utilization of a whole genome SNP panel for efficient genetic mapping in the mouse. Genome. Res. 2006;16:436–440. doi: 10.1101/gr.4563306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLeod M.J. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. doi:10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 44.Huangfu D., Anderson K.V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl Acad. Sci. USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. doi:10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biris K.K., Dunty W.C., Jr., Yamaguchi T.P. Mouse Ripply2 is downstream of Wnt3a and is dynamically expressed during somitogenesis. Dev. Dyn. 2007;236:3167–3172. doi: 10.1002/dvdy.21342. [DOI] [PubMed] [Google Scholar]