Abstract
Chromosome rearrangements are a significant cause of intellectual disability and birth defects. Subtelomeric rearrangements, including deletions, duplications and translocations of chromosome ends, were first discovered over 40 years ago and are now recognized as being responsible for several genetic syndromes. Unlike the deletions and duplications that cause some genomic disorders, subtelomeric rearrangements do not typically have recurrent breakpoints and involve many different chromosome ends. To capture the molecular mechanisms responsible for this heterogeneous class of chromosome abnormality, we coupled high-resolution array CGH with breakpoint junction sequencing of a diverse collection of subtelomeric rearrangements. We analyzed 102 breakpoints corresponding to 78 rearrangements involving 28 chromosome ends. Sequencing 21 breakpoint junctions revealed signatures of non-homologous end-joining, non-allelic homologous recombination between interspersed repeats and DNA replication processes. Thus, subtelomeric rearrangements arise from diverse mutational mechanisms. In addition, we find hotspots of subtelomeric breakage at the end of chromosomes 9q and 22q; these sites may correspond to genomic regions that are particularly susceptible to double-strand breaks. Finally, fine-mapping the smallest subtelomeric rearrangements has narrowed the critical regions for some chromosomal disorders.
INTRODUCTION
Subtelomeric rearrangements at chromosome ends were among the first copy number variations (CNVs) recognized in the human genome (1–5). Unlike CNVs that represent normal variation, subtelomeric rearrangements contribute significantly to intellectual disability, autism and birth defects. The incidence of subtelomeric rearrangements was first recognized when subtelomeric fluorescence in situ hybridization (FISH) testing became a standard cytogenetic test (6,7), and since then subtelomeric rearrangements have been found on every chromosome end (8–10). The detection of subtelomeric rearrangements was greatly improved with the advent of array CGH testing, and clinical array studies have shown that 30–38% of pathogenic chromosome abnormalities involve chromosome ends (10–12). Recent microarray analysis of 15 749 developmentally disabled individuals in the International Standards for Cytogenomic Arrays (ISCA) data set revealed that 26.5 and 16.3% of clinically relevant CNVs lie within the terminal 10 Mb and terminal 5 Mb of chromosomes ends, respectively (13). Subtelomeric rearrangement studies have also paved the way for the discovery of critical regions and genes responsible for specific phenotypes (14,15).
Despite the clinical relevance and incidence of subtelomeric rearrangements, the genomic factors involved in subtelomeric breakage and repair have yet to be investigated comprehensively. However, several groups have analyzed the rearrangement breakpoints in particular chromosome ends, most notably 1p, 9q and 22q. Sequencing breakpoint junctions has revealed that subtelomeric breakpoints do not typically reside at the same site and that breakpoint junctions do not usually bear signatures of homologous recombination (16–21).
To comprehensively evaluate the mutational mechanisms that generate subtelomeric rearrangements, we have taken a large-scale approach to fine-map and sequence breakpoint junctions from a diverse collection of chromosome abnormalities. As is true for the breakpoints of CNVs that represent normal variation (22–24), we find evidence of non-homologous end-joining (NHEJ), non-allelic homologous recombination (NAHR) and DNA replication mechanisms.
RESULTS
To ascertain a diverse collection of subtelomeric rearrangements, we analyzed DNA samples from multiple clinical cytogenetics labs, the Chromosome 18 Clinical Research Center, and a family from the Unique Rare Chromosome Support Group. We accepted samples from individuals with a previous diagnosis of a pathogenic subtelomeric rearrangement that was detected by either subtelomeric FISH or array CGH. Subtelomeric abnormalities of known etiology, for example, the recurrent 3q29 deletion (25) and the recurrent translocation between chromosomes 4p16 and 8p23 (26), were excluded from this study.
To refine subtelomeric breakpoints detected by diagnostic FISH or array CGH, we designed custom oligonucleotide arrays to target chromosome ends with a mean probe spacing of one oligonucleotide per 240 basepairs (bp). We focused on genomic gains and losses involving the terminal 5 Megabases (Mb) of chromosome ends, but also included some larger rearrangements to capture translocation junctions where the breakpoint of one chromosome end was proximal of the terminal 5 Mb. The size of genomic imbalances ranged from 54 kilobases (kb) to 25 Mb, with a median of 2.2 Mb (Fig. 1 and Supplementary Material, Table S1). We analyzed 51 terminal deletions, 11 unbalanced translocations, 10 interstitial deletions, four interstitial duplications and two terminal duplications, for a total of 78 subtelomeric rearrangements (Supplementary Material, Fig. S1). Ten unbalanced translocations involve two chromosome ends; therefore, there are 88 genomic gains and losses corresponding to 78 rearrangements. Since these chromosome abnormalities were ascertained retrospectively from multiple labs, they are not an unbiased collection; however, the large number of rearrangements in our study gave us confidence that diverse mutational mechanisms involved in subtelomeric rearrangements would be captured.
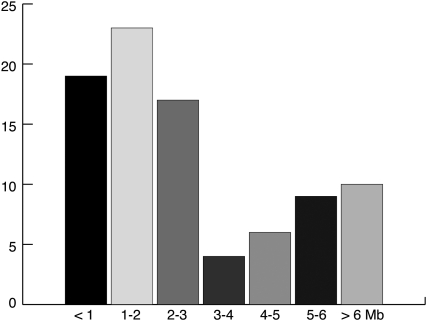
Figure 1.
Sizes of subtelomeric rearrangements. Genomic gains and losses are binned by rearrangement size in Mb (X-axis). The Y-axis indicates the number of rearrangements per size range. The sizes and genomic locations of rearrangements are listed in Supplementary Material, Table S1.
Inheritance and parent of origin
When available, we studied parents via FISH to determine the inheritance of subtelomeric rearrangements. Of the 28 rearrangements where family trios were analyzed, 22 were de novo, three were inherited in an unbalanced form from a mother with a balanced translocation and three were inherited (Supplementary Material, Table S1). In one of the three inherited imbalances, the mother with the same rearrangement was also affected; however, in the other two trios, parents were not cognitively evaluated. Based on these family studies, we conclude that the majority of subtelomeric rearrangements in our study represent large, de novo genomic changes that are most likely pathogenic and responsible for the referring diagnosis.
Determining the parent of origin for de novo subtelomeric rearrangements can shed light on the timing and mechanisms of rearrangement formation. Because most subtelomeric rearrangements are not mosaic in blood, which is the predominant tissue type sampled in clinical cytogenetics testing, we assume that they occur in pre-meiotic cells, during meiosis, or in early embryogenesis. In addition, maternal and paternal parent-of-origin biases have been found for subtelomeric rearrangements involving chromosomes 1p and 18q, respectively (27,28). For six de novo subtelomeric rearrangements in this study, we investigated the parent of origin using Affymetrix Genome-Wide SNP 6.0 chips. We found that rearrangements from LM219, U215 and EGL097 were paternally derived, and rearrangements from EGL205, EGL209 and EGL225 were maternally derived (Supplementary Material, Table S1). Six of the 18q terminal deletions in our study had already been analyzed for parent of origin using microsatellite markers (28). Among this group, five out of six de novo deletions occurred on the paternal allele (Supplementary Material, Table S1).
Subtelomeric breakpoint junctions
Using array CGH, we fine-mapped 78 subtelomeric rearrangements that represent 88 genomic gains and losses corresponding to 102 breakpoints. The median gap between oligonucleotide probes at breakpoints is 213 bp (Supplementary Material, Table S1). Thus, our high-resolution array CGH experiments narrow the region around chromosome breakpoints to a few hundred bp. However, to truly capture the mode of DNA repair, it is essential to sequence breakpoint junctions. To this end, we have cloned and sequenced 21 breakpoint junctions via a PCR-based strategy.
We inspected breakpoint junction sequences to infer the mutational mechanism responsible for each rearrangement (29). Rearrangements with significant sequence homology (hundreds of bp to hundreds of kb) between the edges of the breakpoint are indicative of NAHR (30). However, breakpoints that lack long stretches of homology may have a few nucleotides of microhomology at breakpoint junctions. Such junctions may be the product of NHEJ (31,32), microhomology-mediated end joining (MMEJ) (33) or DNA replication processes (34–37). Below, we describe breakpoint junctions in the context of the type of subtelomeric rearrangement: terminal deletions, interstitial deletions and unbalanced translocations.
Terminal deletions
We amplified and sequenced 14 out of 51 (27%) of terminal deletion junctions. Beginning with breakpoints determined by array CGH, we designed one primer complementary to the proximal (non-deleted) chromosomal region and used one primer complementary to the telomere repeat sequence (38). After PCR, we cloned and sequenced the breakpoint junctions to capture the post-repair junction sequence.
There are between 2 and 6 nucleotides of microhomology at 10 out of 14 (71%) of terminal deletion junctions (Fig. 2 and Supplementary Material, Fig. S2). End-joining of double-strand breaks in a subtelomere and a telomere repeat sequence would give rise to these types of terminal deletion junctions. Another possibility is that terminal deletions are repaired via de novo synthesis of a new telomere repeat at the site of a double-strand break, a model put forth by others (16,21,38,39). ‘Chromosome healing’ occurs in ciliates when telomerase adds telomeric repeats to non-telomeric double-strand breaks (40). Human telomerase can synthesize telomeres from non-telomeric sites in vitro (41), but this process has not been demonstrated in human cells (42).
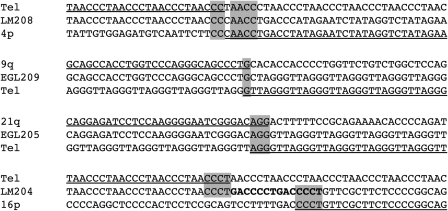
Figure 2.
Subtelomeric breakpoint junctions. Junction sequences are labeled by subject name, and the adjacent regions were derived from the reference genome on the UCSC browser (http://genome.ucsc.edu/) using the March 2006 build (NCBI36/hg18). Regions of the reference genome that align with the junction sequence are underlined. Microhomology at breakpoint junctions is highlighted in grey. We define microhomology as bp in common between adjacent sides of a breakpoint junction, allowing a 1 bp gap on either side of the junction. Bold nucleotides indicate a local duplication at the breakpoint junction of LM204.
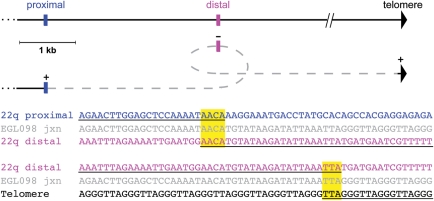
We also identified two terminal deletion junctions that are suggestive of DNA replication processes. The terminal deletion junction derived from LM204 has a 7 bp tandem duplication at the breakpoint, which is typical of sites of serial replication slippage (35) (Fig. 2). Sequencing the junction of EGL098's 22q terminal deletion revealed a 16 bp sequence that did not align to the 22q breakpoint region identified by array CGH or the telomere repeat at the other end of the junction. This sequence corresponds to a region that is 3 kb distal of the 22q breakpoint, but that lies in an inverted orientation relative to the proximal breakpoint on 22q and the telomere repeat (Fig. 3). This type of complex junction has been described in other rearrangement breakpoints and fits the fork stalling and template switching (FoSTeS) model (43–45).
Figure 3.
The breakpoint junction of a 22q terminal deletion suggests a FoSTeS event. Above, the location and orientation of the three segments that make up EGL098's breakpoint junction are shown. The horizontal black line indicates the reference genome showing the proximal side of the breakpoint (blue), the region ∼3 kb distal (pink) and the telomere repeat (black arrowhead). The grey dashed line connects the three segments in the orientation present in the junction sequence. The origin of the telomere sequence at the end of the junction is unknown; however, it is shown relative to the 22q telomere. Below, the junction sequence from EGL098 (grey) is aligned to the reference sequence for the three segments, colored as above. Regions of the reference genome that align with the junction sequence are underlined. Microhomology is highlighted in yellow.
Interstitial deletions
We fine-mapped the breakpoints of 10 interstitial deletions and four interstitial duplications by array CGH and sequenced the breakpoint junctions of three interstitial deletions (Supplementary Material, Table S1). We attempted to amplify three interstitial duplication junctions using a strategy designed to capture breakpoints of tandem or inverted duplications (46), but we were unsuccessful in sequencing any of the duplication junctions.
Sequencing the interstitial deletion junction of EGL094 revealed three nucleotides of microhomology, consistent with NHEJ between two double-strand breaks in the short arm of chromosome 4 (Supplementary Material, Fig. S2). Alternatively, the 54-kb deletion in EGL094 could be the product of a FoSTeS event involving a template switch from one side of the deletion to the other. Interstitial deletions in EGL049 and SCH3 are the product of homologous recombination between Alu elements, and sequencing across the breakpoint junctions of these NAHR events revealed the hybrid repeat transitioning from one element into another (Supplementary Material, Fig. S3). In both rearrangements, the recombining elements are in direct orientation and are in the same repeat class, as would be expected for a true NAHR event. As represented in the reference genome, the distal and proximal AluSp elements that flank EGL049's 17p deletion are 293 and 370 bp, respectively, and 87% identical. The proximal and distal AluYs that flank SCH3's 9q deletion are 302 and 307 bp, respectively, and 88% identical.
Unbalanced translocations
We fine-mapped the breakpoints of 11 unbalanced translocations and sequenced four breakpoint junctions. There is little or no microhomology at the translocation junctions in 18q–71c, LM218 and EGL102, consistent with NHEJ between double-strand breaks in two different chromosomes (Supplementary Material, Fig. S2). 18q–71c's rearrangement was originally identified as a terminal deletion by array CGH; however, breakpoint sequencing revealed a cryptic translocation between 18q and a segmental duplication that maps to the end of chromosome 4p. Since segmental duplications at the ends of chromosomes are extremely polymorphic (47), we cannot be certain that the translocated segment was derived from 4p and not another chromosome end.
Sequencing across the unbalanced translocation junction in LM221 revealed a hybrid LINE at the breakpoint, consistent with interchromosomal NAHR (Supplementary Material, Fig. S3). The translocation was mediated by homologous recombination between L1PA2 elements in direct orientation on chromosomes 6 and 16. The L1PA2s on 6p and 16q in the reference genome are 5767 and 6003 bp, respectively, and 96% identical.
Subtelomeric breakpoint hotspots
Analysis of subtelomeric breakpoint junctions reveals mechanisms of DNA repair; however, the factors involved in the initial double-strand break or DNA replication error are also critically important for understanding the mechanisms of rearrangement formation. Previous studies using FISH or BAC-based array CGH strategies suggested that some subtelomeric breakpoints are relatively close to one another, pointing to a potential recurrent breakage site (48). However, these approaches resolved breakpoints to tens or hundreds of kilobase regions, at best.
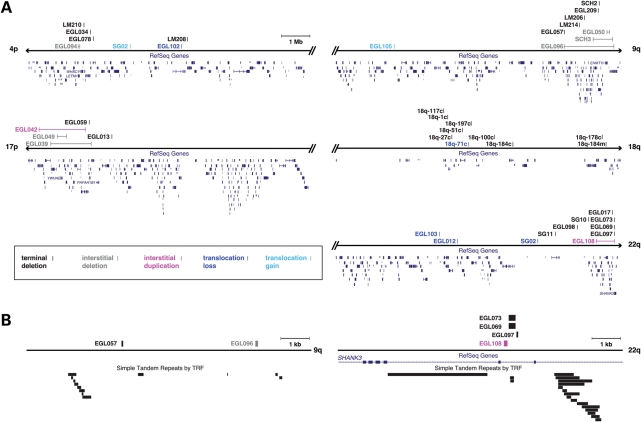
Higher-resolution array CGH and sequencing of subtelomeric breakpoints allowed us to distinguish truly recurrent breakpoints from ‘nearby’ breakpoints. We focused on five chromosome ends where we had at least eight breakpoints to sample a representative number of breakpoints per subtelomere. As shown in Figure 4, the spectrum of subtelomeric breakpoints in 4p, 9q, 17p, 18q and 22q differ among chromosome ends. Breakpoints in 18q are distributed throughout the end of the chromosome, consistent with previous studies (28). Breakpoints in 9q appear to cluster, including breakpoints from EGL057 and EGL096 that are only 4.6 kb apart (Fig. 4 and Supplementary Material, Table S1). These breakpoints are not identical, but may cluster due to a local genomic architecture that is susceptible to rearrangement. This 9q hotspot lies ∼2 Mb from the end of the chromosome and does not overlap with other 9q breakpoint clusters reported in the literature (21,48).
Figure 4.
Breakpoints in 4p, 9q, 17p, 18q and 22q. (A) The terminal 10 Mb of the chromosome are shown per chromosome end. Arrowheads indicate the start (p) and end (q) of the sequence for each chromosome in the human genome assembly, as represented in the hg18 (March 2006, NCBI36) build (http://genome.ucsc.edu/). Double slash marks indicate a break in the chromosome depiction at the edges of the terminal 10 Mb; a scale bar is shown at the top of the figure. Vertical lines indicate the location of breakpoints as listed in Supplementary Material, Table S1; paired interstitial deletion and duplication breakpoints are connected by a horizontal line. Breakpoints are labeled to the left of the vertical line with subject names as in Supplementary Material, Table S1. Breakpoints of terminal deletions, interstitial deletions, terminal duplications, interstitial duplications and translocations are color coded as shown. Breakpoints of unbalanced translocations that correspond to genomic losses (dark blue) and gains (light blue) are colored differently to distinguish monosomic and trisomic segments, respectively. (B) Zoomed in views of the 9q and 22q hotspots. Ten kilobase regions of 9q and 22q surrounding hotspots are shown. Breakpoints and genes are color-coded as in (A). Tandem repeats as detected by the Tandem Repeats Finder (TRF) (77) are shown as black rectangles.
Remarkably, four of the 12 breakpoints in 22q lie within 320 bp in chromosome band 22q13.33. Breakpoints from three terminal deletions (EGL069, EGL073 and EGL097) and one interstitial duplication (EGL108) lie between exons 8 and 9 of the SHANK3 gene, a breakpoint also reported before in the studies of 22q terminal deletions (19,49). These data suggest that this site is a hotspot for genomic rearrangement, most often manifested as a terminal deletion that causes the 22q13 monosomy syndrome (OMIM 606232). Though the interstitial duplication present in EGL108 has a distal breakpoint within the same hotspot region, without determining the orientation of the duplication, we cannot know how this rearrangement will impact the organization of the SHANK3 gene.
DISCUSSION
Subtelomeric rearrangements have important implications for human disease and evolution. Genomic studies of subtelomeres have revealed that human chromosome ends are subject to elevated rates of meiotic recombination, sister chromatid exchange and DNA transfer (47,50–52). For over 40 years, subtelomeric rearrangements have been recognized as a significant cause of intellectual disability. Previous studies showed that, unlike some chromosome abnormalities that cause genomic disorders, subtelomeric rearrangements do not typically have recurrent breakpoints (8–10). This led some to propose that subtelomeric rearrangements are the product of ‘random chromosome breakage’ (53).
Here we show that subtelomeric rearrangements arise via distinct mutational mechanisms. We sequenced 21 breakpoint junctions and found that 18 junctions (86%) have little or no homology and that three junctions (14%) have hundreds of bp of sequence homology between recombining segments, typical of NAHR. The frequency of these classes of events is similar to what is seen in CNVs from control populations, where NAHR accounts for 9 (22), 10–15 (23) and 22% (24) of sequenced breakpoint junctions.
NAHR events give rise to reciprocal deletions and duplications, as well as translocations (26,54,55). However, the majority of NAHR-mediated chromosome rearrangements described in the literature are the product of recombination between large segmental duplications also known as low-copy repeats (56,57). NAHR events between smaller interspersed repeats, such as LINEs and Alus, are overlooked by most array-based studies and are only captured via sequencing (24,58,59). Though the deletions and translocation detected in SCH3, EGL049 and LM221 were all originally identified by clinical diagnostic array CGH, the NAHR substrates responsible for the rearrangements were only detected via high-resolution array CGH and sequencing. Thus, NAHR events in clinically recognized subtelomeric rearrangements are likely underestimated. Furthermore, sequence variation in interspersed repeats may affect the propensity of two elements to recombine. The recombining repeats in SCH3, EGL049 and LM221 were 88, 87 and 96% identical, respectively, as represented in the human genome assembly. However, we have not sequenced the parental repeats that recombined in these individuals, and it is possible that gene conversion between repeats may make some more homologous than represented in the reference genome (60,61).
Like other studies of pathogenic CNV breakpoints (62), the majority of breakpoint junctions in our study lack significant sequence homology and may arise via NHEJ, MMEJ, DNA replication errors, or de novo telomere synthesis. NHEJ and MMEJ can be distinguished in experimental systems by disrupting proteins in either repair pathway and monitoring the amount of microhomology at breakpoint junctions (31–33), but we find that the amount of microhomology at subtelomeric breakpoint junctions does not fall into discrete categories that would distinguish NHEJ from MMEJ. We presume that translocations with little sequence homology at the edges of the junction are the product of and end-joining process between two double-strand breaks. However, terminal deletions may be the product of end-joining or de novo telomere synthesis (16,38,39).
Errors in DNA replication can also give rise to chromosome rearrangements, and some replication events reveal characteristic breakpoint junctions (17,34–37,62). Serial replication slippage generates short deletions and duplications at breakpoints, (35) like the breakpoint junction in LM204 (Fig. 2). Analyses of non-recurrent breakpoint junctions suggest that FoSTeS may be responsible for breakpoints with insertions of short duplicated and/or inverted segments (43–45,63). Of the 21 breakpoint junctions we sequenced, only the 22q breakpoint from EGL098 had an inverted insertion consistent with FoSTeS. This 16 bp insertion did not affect our PCR-based junction sequencing; however, other larger insertions would escape detection by this method.
Sequencing breakpoint junctions reveals the mechanism of DNA repair, points to the site of chromosome breakage and determines rearrangement organization, which could impact the regulation of nearby genes. Though sequencing breakpoint junctions was a high priority for this study, we were only able to sequence 21 out of 78 (27%) junctions (Supplementary Material, Table S1). We attempted to amplify 58 breakpoints as described in Methods, and captured breakpoint junctions in 21 rearrangements, giving us an overall success rate of 21 out of 58 (36%). In some cases, we exhausted the subject DNA stock before we could optimize PCR conditions, but in others we tried multiple PCR conditions and primer pairs. These recalcitrant breakpoints likely constitute more complex breakpoint junctions than predicted by high-resolution array CGH. Small local deletions and duplications are not uncommon at breakpoint junctions (18,20,21,62) and we suspect that many of the missing junctions in our study fall into this category.
We were able to identify one such cryptic rearrangement by amplifying the translocation junction in 18q–71c, which appeared to be a terminal deletion by other methods. However, we were unable to sequence the junctions of any terminal or interstitial duplications. Other groups have had similar problems in sequencing duplication junctions (22,23,43), which leads us to suspect that many are more complicated than perfect tandem or inverted duplications. Capturing complex junctions will require an unbiased approach that does not depend on inferring the correct rearrangement structure.
There are likely many factors involved in initiating the double-strand breaks that give rise to subtelomeric rearrangements. Large data sets of sequenced breakpoints coupled with functional assays of chromosome fragility will help us determine the roles of DNA sequence and chromatin in double-strand breaks and DNA replication errors. As described in Vissers et al. (62), other pathogenic CNV breakpoints are enriched in sequence motifs that may stimulate CNV formation. Given the enrichment of tandem repeats at chromosome ends (47,64) and subtelomeric hotspots on 9q and 22q that coincide with tandem repeats (Fig. 4), we suspect that some subtelomeric rearrangements are initiated by breakage in particularly fragile DNA sequences. Tandem repeats have been found at other subtelomeric breakpoints (18,21), and are overrepresented in normal CNV breakpoint data sets (22,63). Like fragile sites in the human genome (65), variation in repeat track length or sequence composition could affect the risk for chromosome breakage at subtelomeric hotspots.
Furthermore, the concentration of particular repetitive sequences at chromosome ends could explain differences in the density and location of breakpoints among subtelomeres (Fig. 4). Chromosome ends enriched in ‘fragile motifs’ would be expected to be more susceptible to new rearrangements. This has been suggested for breakpoints in 1p and 9q that are particularly GC-rich and concentrated in specific classes of repetitive sequences predicted to form secondary structures (20,21,66). Of course, embryonic lethal subtelomeric rearrangements will also contribute to the ascertainment of rearrangements of chromosome ends. Subtelomeric rearrangements in gene-poor chromosome ends (e.g. 18q) are on average larger than rearrangements from gene-rich chromosome ends (e.g. 9q) (Fig. 4 and Supplementary Material, Table S1), likely reflecting the lethality of large genomic gains and losses that encompass many genes.
Determining the parental origin of chromosome rearrangements can also yield mechanistic insights into how rearrangements form and at what time in development they occur (67,68). Analysis of six trios in our study revealed an equal number of maternally and paternally derived subtelomeric rearrangements. These data are too limited to assess parental bias and include only two sequenced junctions. Thus, we cannot determine if particular classes of subtelomeric rearrangements have a parental bias. In a large cohort of 18q terminal deletions, Heard et al. (28) found a significant paternal bias as 71 out of 81 deletions were paternal in origin. In contrast, analysis of the parent of origin of 40 de novo 1p terminal deletions revealed that 24 and 16 deletions occurred on the maternal and paternal chromosomes, respectively (27). In their analysis of 9q subtelomeric rearrangements, Yatsenko et al. (21) found that 11 and six rearrangements occurred on the paternal and maternal alleles, respectively. It is possible that subtelomeric rearrangements involving particular loci or chromosome ends have a parental bias; however, as a group, subtelomeric rearrangements do not appear to have a significant parent-of-origin effect.
Most subtelomeric rearrangements involve many genes that could be responsible for an abnormal phenotype. Nevertheless, smaller rearrangements can refine critical regions involved in developmental disabilities and birth defects. Although we lack the detailed phenotypic information for the subjects in our study that would allow specific genotype–phenotype correlations, it is worth mentioning some of the smaller rearrangements that may narrow the genes involved in the referring diagnosis (Fig. 4). EGL039 and EGL049 have overlapping interstitial deletions of 17p13.3 that include the YWHAE gene, but not PAFAH1B1. Mutations in PAFAH1B1, also known as LIS1, cause lissencephaly (69) (OMIM 601545). EGL039 was referred with an indication of mild intellectual disability and short stature; EGL049 was referred for developmental delay. Parental analysis revealed that both deletions are de novo (Supplementary Material, Table S1), which is typical of pathogenic chromosome rearrangements. In addition, recent studies have reported similar deletions of 17p13.3 in children with mild intellectual disability and moderate-to-severe growth restriction (70,71). Thus, our data support the conclusion that deletions of this part of 17p13.3 cause a less severe phenotype than larger deletions that also include PAFAH1B1. We also identified an interstitial duplication of an overlapping region of 17p13.3; however, no phenotypic information was available for subject EGL042, who carries the duplication. Duplications of this region have reported in children with intellectual disability, macrosomia and dysmorphic facial features (72).
The smallest chromosome imbalance in our study is a 54-kb deletion of 4p16.3 that includes only one gene, LETM1 (Fig. 4). LETM1 lies within the Wolf–Hirschhorn syndrome (WHS) critical region 2 (73) (OMIM 194190), but subject EGL094 exhibited no facial features of WHS; rather, she was referred for testing at 1 year of age, presenting with microtia, renal agenesis, Duane anomaly and a congenital heart defect. These data suggest that loss of LETM1 is not responsible for the characteristic facial features in WHS and that other candidate genes in the critical region may be involved.
Overall, we have shown that subtelomeric rearrangements are a heterogeneous class of chromosome abnormalities caused by diverse mutational mechanisms. Most breakpoint junctions do not have significant sequence homology between recombining segments, consistent with end-joining, DNA replication errors, or de novo telomere synthesis. NAHR between interspersed repeats mediates interstitial deletions and a translocation, though we found no recurrent NAHR-mediated events in this group of subtelomeric rearrangements. We suspect that the shorter stretch of sequence homology when compared with larger segmental duplications makes Alu–Alu and L1–L1 recombination less frequent. We also identified one subtelomeric rearrangement with sequenced junctions that are consistent with the FoSTeS model of replicative DNA repair. Ultimately, as with other CNVs in the human genome, there is no single mechanism to account for subtelomeric rearrangement formation.
MATERIALS AND METHODS
Human subjects
Samples from subjects with subtelomeric rearrangements were ascertained from Emory Genetics Laboratory (EGL), Signature Genomic Laboratories (SG), Seattle Children's Hospital (SCH), the Chromosome 18 Clinical Research Center (18q-) (28,74), the Unique Rare Chromosome Support Group (U) and the Ledbetter and Martin laboratories (LM) (75,76) (Supplementary Material, Table S1). Male (GM10851) and female (GM15510) reference cell lines were obtained from Coriell Cell Repositories. This study was approved by the Emory University Institutional Review Board. Samples were de-identified or obtained with informed consent per the study protocol at Emory and/or collaborating institutions.
Subjects were referred for diagnostic testing for a number of indications, typically intellectual disabilities, autism and birth defects. Detailed phenotypic information for subjects was not available. Subtelomeric rearrangements were originally analyzed in clinical diagnostic laboratories with different array comparative genomic hybridization (CGH) platforms or subtelomeric FISH assays. Array CGH results were confirmed by FISH in diagnostic laboratories using standard FISH methodologies, except in the case of four rearrangements: interstitial deletions from EGL050 and EGL094 and interstitial duplications from EGL072 and EGL108 were too small to analyze via FISH. FISH confirmed the imbalance detected by array CGH (eliminating false positives), determined the rearrangement structure, and guided our breakpoint sequencing strategy. We confirmed all subtelomeric rearrangements ascertained from clinical diagnostic labs in our targeted array CGH experiments. Thus, there were no false positives in our subtelomeric rearrangement dataset.
Targeted array CGH
Using a 244K platform from Agilent Technologies, we designed four custom arrays to cover the terminal 5 Mb of 41 chromosome ends (excluding acrocentric p arms and the Y chromosome), providing a mean probe spacing of one oligonucleotide per 240 bp. Oligonucleotides were designed using Agilent's eArray program (https://earray.chem.agilent.com/earray/). To minimize non-unique oligonucleotides that would not be informative in array CGH, we performed an HD probe search to prefer existing ‘catalog probes’ and we used the most stringent ‘similarity score filter’ designed to select probes that hybridize to only one genomic location. The unique identifiers (AMADIDs) for the array designs are 021634, 021635, 021636 and 021637 for chromosomes 1–5, 6–10, 11–17 and 18-X, respectively. For breakpoints outside of the terminal 5 Mb, we designed other targeted arrays (AMADIDs 08181, 23978, 27686, 29464 and 97831). All array designs are available upon request.
We extracted genomic DNA from most cell lines and peripheral blood samples using the Gentra Puregene DNA Extraction Kit (Qiagen, Valencia, CA). SG DNA samples were prepared from blood using the Qiagen M48 Biorobot for automated DNA extraction with standard conditions (Qiagen, Valencia, CA). Subject DNA was co-hybridized with reference DNA from either GM10851 or GM15510. Arrays were scanned using a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA) or the Agilent high-resolution C scanner (Agilent Technologies, Santa Clara, CA), and signal intensities were evaluated using Feature Extraction Version 9.5.1.1 software (Agilent Technologies, Santa Clara, CA). We used DNA Analytics Version 4.0 software (Agilent Technologies, Santa Clara, CA) to analyze the array data and call breakpoints (Supplementary Material, Table S1 and Fig. S1).
Breakpoint junction PCR
Starting with breakpoints identified by array CGH, we designed PCR primers to amplify putative breakpoint junctions. For terminal deletions, we designed a primer complementary to the intact (non-deleted) side of the junction and paired this primer with one of two telomere primers, 5′-CCCTAACCCTAACCCTAACCCTAACCCTAA-3′ or 5′-TATGGATCCCTAACCCTGACCCTAACCC-3′ (38). For unbalanced translocations, we designed PCR primers to amplify the junction from the derivative chromosome to the translocated segment. We amplified interstitial deletion junctions by designing primers complementary to the edges of the deletion. We performed PCR using TaKaRa Ex Taq polymerase (Clontech Laboratories, Inc., Madison, WI) with 1× PCR buffer, 0.2 mm dNTP, 8 pmol of each primer and 50–100 ng of DNA template. PCR conditions for amplifying terminal deletions were: 94°C for 1 min; 10 cycles at 94°C for 30 s, 65°C for 1 min (decreasing 1°C per cycle), 72°C for 3 min; 20 cycles at 94°C for 30 s, 59°C for 1 min, 72°C for 3 min; and the final extension at 72°C for 5 min. Conditions for other PCRs were: 94°C for 1 min, 30 cycles at 94°C for 30 s, 56°C for 30 s and 72°C for 1 min/kb of expected product, followed by the final extension at 72°C for 5 min. We purified PCR products from agarose gels using the QIAquick gel extraction kit (Qiagen, Valencia, CA), and then cloned them into a TOPO-TA vector following the manufacturer's protocol (Invitrogen, Carlsbad, CA). We transformed the ligated construct into SURE 2 Supercompetent Cells (Agilent Technologies, Cedar Creek, TX) following the manufacturer's protocol. We propagated plasmids in recombination-deficient SURE 2 Escherichia coli to prevent rearrangement of the cloned insert.
Breakpoint sequence analysis
We purified plasmid DNA (Qiagen Miniprep kit, Valencia, CA) and submitted plasmids for sequencing (Beckman Coulter Genomics, Danvers, MA). DNA sequences were analyzed by comparing reads to the human genome reference assembly (NCBI36/hg18) using the BLAT tool on the UCSC Genome Browser (http://genome.ucsc.edu/). All junction sequences are listed in Supplementary Material, Figs S2 and S3.
Parent-of-origin studies
Parent of origin was determined for six rearrangements by analyzing proband and parental DNA using the Affymetrix Human Genome-Wide SNP Array 6.0 at Emory University. For other family trios, parental DNA was not available. Genotyping was performed with the Birdseed algorithm, as implemented in Affymetrix Power Tools software. For genomic losses, parent of origin was determined by inferring the origin of the missing allele in the proband. For genomic gains, we analyzed informative SNPs in SNP cluster graphs to determine the origin of the additional allele.
WEB RESOURCES
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
UCSC Genome Browser, http://genome.ucsc.edu/
Tandem Repeats Finder (TRF), http://tandem.bu.edu/trf/trf.html
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported by a grant from the NIH (MH092902 to M.K.R.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank participating families, referring clinicians and the Unique Rare Chromosome Disorder Support Group for their contributions to this project. We thank Erin B. Kaminsky for sharing unpublished data and for insightful discussions. Ian Goldlust, Heather Mason-Suares, Brooke Weckselblatt, Tara Wood and Becca Cozad performed array CGH and sequenced breakpoint junctions. We thank Cheryl Strauss for editorial assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lejeune J., Lafourcade J., Berger R., Vialatte J., Boeswillwald M., Seringe P., Turpin R. 3 Cases of Partial Deletion of the Short Arm of a 5 Chromosome. C. R. Hebd. Seances. Acad. Sci. 1963;257:3098–3102. [PubMed] [Google Scholar]
- 2.Hirschhorn K., Cooper H.L., Firschein I.L. Deletion of short arms of chromosome 4–5 in a child with defects of midline fusion. Humangenetik. 1965;1:479–482. doi: 10.1007/BF00279124. [DOI] [PubMed] [Google Scholar]
- 3.Wolf U., Reinwein H., Porsch R., Schroter R., Baitsch H. Deficiency on the short arms of a chromosome No. 4. Humangenetik. 1965;1:397–413. [PubMed] [Google Scholar]
- 4.Lamb J., Wilkie A.O., Harris P.C., Buckle V.J., Lindenbaum R.H., Barton N.J., Reeders S.T., Weatherall D.J., Higgs D.R. Detection of breakpoints in submicroscopic chromosomal translocation, illustrating an important mechanism for genetic disease. Lancet. 1989;2:819–824. doi: 10.1016/s0140-6736(89)92995-4. [DOI] [PubMed] [Google Scholar]
- 5.Wilkie A.O., Lamb J., Harris P.C., Finney R.D., Higgs D.R. A truncated human chromosome 16 associated with alpha thalassaemia is stabilized by addition of telomeric repeat (TTAGGG)n. Nature. 1990;346:868–871. doi: 10.1038/346868a0. [DOI] [PubMed] [Google Scholar]
- 6.National Institutes of Health and Institute of Molecular Medicine Collaboration. A complete set of human telomeric probes and their clinical application. Nat. Genet. 1996;14:86–89. doi: 10.1038/ng0996-86. [DOI] [PubMed] [Google Scholar]
- 7.Knight S.J., Lese C.M., Precht K.S., Kuc J., Ning Y., Lucas S., Regan R., Brenan M., Nicod A., Lawrie N.M., et al. An optimized set of human telomere clones for studying telomere integrity and architecture. Am. J. Hum. Genet. 2000;67:320–332. doi: 10.1086/302998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravnan J.B., Tepperberg J.H., Papenhausen P., Lamb A.N., Hedrick J., Eash D., Ledbetter D.H., Martin C.L. Subtelomere FISH analysis of 11 688 cases: an evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. J. Med. Genet. 2006;43:478–489. doi: 10.1136/jmg.2005.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballif B.C., Sulpizio S.G., Lloyd R.M., Minier S.L., Theisen A., Bejjani B.A., Shaffer L.G. The clinical utility of enhanced subtelomeric coverage in array CGH. Am. J. Med. Genet. A. 2007;143:1850–1857. doi: 10.1002/ajmg.a.31842. [DOI] [PubMed] [Google Scholar]
- 10.Shao L., Shaw C.A., Lu X.Y., Sahoo T., Bacino C.A., Lalani S.R., Stankiewicz P., Yatsenko S.A., Li Y., Neill S., et al. Identification of chromosome abnormalities in subtelomeric regions by microarray analysis: a study of 5380 cases. Am. J. Med. Genet. A. 2008;146A:2242–2251. doi: 10.1002/ajmg.a.32399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaffer L.G., Kashork C.D., Saleki R., Rorem E., Sundin K., Ballif B.C., Bejjani B.A. Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J. Pediatr. 2006;149:98–102. doi: 10.1016/j.jpeds.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Baldwin E.L., Lee J.Y., Blake D.M., Bunke B.P., Alexander C.R., Kogan A.L., Ledbetter D.H., Martin C.L. Enhanced detection of clinically relevant genomic imbalances using a targeted plus whole genome oligonucleotide microarray. Genet. Med. 2008;10:415–429. doi: 10.1097/GIM.0b013e318177015c. [DOI] [PubMed] [Google Scholar]
- 13.Kaminsky E.B., Kaul V., Paschall J., Church D.M., Bunke B., Kunig D., Moreno-De-Luca D., Moreno-De-Luca A., Mulle J.G., Warren S.T., et al. An evidence-based approach to establish the functional and clinical significance of CNVs in intellectual and developmental disabilities. Genet. Med. 2011 doi: 10.1097/GIM.0b013e31822c79f9. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleefstra T., Brunner H.G., Amiel J., Oudakker A.R., Nillesen W.M., Magee A., Genevieve D., Cormier-Daire V., van Esch H., Fryns J.P., et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am. J. Hum. Genet. 2006;79:370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durand C.M., Betancur C., Boeckers T.M., Bockmann J., Chaste P., Fauchereau F., Nygren G., Rastam M., Gillberg I.C., Anckarsater H., et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ballif B.C., Yu W., Shaw C.A., Kashork C.D., Shaffer L.G. Monosomy 1p36 breakpoint junctions suggest pre-meiotic breakage-fusion-bridge cycles are involved in generating terminal deletions. Hum. Mol. Genet. 2003;12:2153–2165. doi: 10.1093/hmg/ddg231. [DOI] [PubMed] [Google Scholar]
- 17.Ballif B.C., Wakui K., Gajecka M., Shaffer L.G. Translocation breakpoint mapping and sequence analysis in three monosomy 1p36 subjects with der(1)t(1;1)(p36;q44) suggest mechanisms for telomere capture in stabilizing de novo terminal rearrangements. Hum. Genet. 2004;114:198–206. doi: 10.1007/s00439-003-1029-y. [DOI] [PubMed] [Google Scholar]
- 18.Gajecka M., Glotzbach C.D., Jarmuz M., Ballif B.C., Shaffer L.G. Identification of cryptic imbalance in phenotypically normal and abnormal translocation carriers. Eur. J. Hum. Genet. 2006;14:1255–1262. doi: 10.1038/sj.ejhg.5201710. [DOI] [PubMed] [Google Scholar]
- 19.Bonaglia M.C., Giorda R., Mani E., Aceti G., Anderlid B.M., Baroncini A., Pramparo T., Zuffardi O. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J. Med. Genet. 2006;43:822–828. doi: 10.1136/jmg.2005.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gajecka M., Gentles A.J., Tsai A., Chitayat D., Mackay K.L., Glotzbach C.D., Lieber M.R., Shaffer L.G. Unexpected complexity at breakpoint junctions in phenotypically normal individuals and mechanisms involved in generating balanced translocations t(1;22)(p36;q13) Genome Res. 2008;18:1733–1742. doi: 10.1101/gr.077453.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yatsenko S.A., Brundage E.K., Roney E.K., Cheung S.W., Chinault A.C., Lupski J.R. Molecular mechanisms for subtelomeric rearrangements associated with the 9q34.3 microdeletion syndrome. Hum. Mol. Genet. 2009;18:1924–1936. doi: 10.1093/hmg/ddp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perry G.H., Ben-Dor A., Tsalenko A., Sampas N., Rodriguez-Revenga L., Tran C.W., Scheffer A., Steinfeld I., Tsang P., Yamada N.A., et al. The fine-scale and complex architecture of human copy-number variation. Am. J. Hum. Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad D.F., Bird C., Blackburne B., Lindsay S., Mamanova L., Lee C., Turner D.J., Hurles M.E. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat. Genet. 2010;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kidd J.M., Graves T., Newman T.L., Fulton R., Hayden H.S., Malig M., Kallicki J., Kaul R., Wilson R.K., Eichler E.E. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willatt L., Cox J., Barber J., Cabanas E.D., Collins A., Donnai D., FitzPatrick D.R., Maher E., Martin H., Parnau J., et al. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am. J. Hum. Genet. 2005;77:154–160. doi: 10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giglio S., Calvari V., Gregato G., Gimelli G., Camanini S., Giorda R., Ragusa A., Guerneri S., Selicorni A., Stumm M., et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am. J. Hum. Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heilstedt H.A., Ballif B.C., Howard L.A., Lewis R.A., Stal S., Kashork C.D., Bacino C.A., Shapira S.K., Shaffer L.G. Physical map of 1p36, placement of breakpoints in monosomy 1p36, and clinical characterization of the syndrome. Am. J. Hum. Genet. 2003;72:1200–1212. doi: 10.1086/375179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heard P.L., Carter E.M., Crandall A.C., Sebold C., Hale D.E., Cody J.D. High resolution genomic analysis of 18q- using oligo-microarray comparative genomic hybridization (aCGH) Am. J. Med. Genet. A. 2009;149A:1431–1437. doi: 10.1002/ajmg.a.32900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hastings P.J., Lupski J.R., Rosenberg S.M., Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lupski J.R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 31.Lieber M.R., Ma Y., Pannicke U., Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 32.Hefferin M.L., Tomkinson A.E. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 33.McVey M., Lee S.E. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosco G., Haber J.E. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics. 1998;150:1037–1047. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J.M., Chuzhanova N., Stenson P.D., Ferec C., Cooper D.N. Intrachromosomal serial replication slippage in trans gives rise to diverse genomic rearrangements involving inversions. Hum. Mutat. 2005;26:362–373. doi: 10.1002/humu.20230. [DOI] [PubMed] [Google Scholar]
- 36.McEachern M.J., Haber J.E. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 37.Smith C.E., Llorente B., Symington L.S. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 38.Flint J., Craddock C.F., Villegas A., Bentley D.P., Williams H.J., Galanello R., Cao A., Wood W.G., Ayyub H., Higgs D.R. Healing of broken human chromosomes by the addition of telomeric repeats. Am. J. Hum. Genet. 1994;55:505–512. [PMC free article] [PubMed] [Google Scholar]
- 39.Lamb J., Harris P.C., Wilkie A.O., Wood W.G., Dauwerse J.G., Higgs D.R. De novo truncation of chromosome 16p and healing with (TTAGGG)n in the alpha-thalassemia/mental retardation syndrome (ATR-16) Am. J. Hum. Genet. 1993;52:668–676. [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G.L., Blackburn E.H. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991;67:823–832. doi: 10.1016/0092-8674(91)90077-c. [DOI] [PubMed] [Google Scholar]
- 41.Morin G.B. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 42.Melek M., Shippen D.E. Chromosome healing: spontaneous and programmed de novo telomere formation by telomerase. Bioessays. 1996;18:301–308. doi: 10.1002/bies.950180408. [DOI] [PubMed] [Google Scholar]
- 43.Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 44.Bauters M., Van Esch H., Friez M.J., Boespflug-Tanguy O., Zenker M., Vianna-Morgante A.M., Rosenberg C., Ignatius J., Raynaud M., Hollanders K., et al. Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res. 2008;18:847–858. doi: 10.1101/gr.075903.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang F., Khajavi M., Connolly A.M., Towne C.F., Batish S.D., Lupski J.R. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arlt M.F., Mulle J.G., Schaibley V.M., Ragland R.L., Durkin S.G., Warren S.T., Glover T.W. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am. J. Hum. Genet. 2009;84:339–350. doi: 10.1016/j.ajhg.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linardopoulou E.V., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeScipio C., Spinner N.B., Kaur M., Yaeger D., Conlin L.K., Ambrosini A., Hu S., Shan S., Krantz I.D., Riethman H. Fine-mapping subtelomeric deletions and duplications by comparative genomic hybridization in 42 individuals. Am. J. Med. Genet. A. 2008;146A:730–739. doi: 10.1002/ajmg.a.32216. [DOI] [PubMed] [Google Scholar]
- 49.Wong A.C., Ning Y., Flint J., Clark K., Dumanski J.P., Ledbetter D.H., McDermid H.E. Molecular characterization of a 130-kb terminal microdeletion at 22q in a child with mild mental retardation. Am. J. Hum. Genet. 1997;60:113–120. [PMC free article] [PubMed] [Google Scholar]
- 50.Lynn A., Kashuk C., Petersen M.B., Bailey J.A., Cox D.R., Antonarakis S.E., Chakravarti A. Patterns of meiotic recombination on the long arm of human chromosome 21. Genome Res. 2000;10:1319–1332. doi: 10.1101/gr.138100. [DOI] [PubMed] [Google Scholar]
- 51.Matise T.C., Sachidanandam R., Clark A.G., Kruglyak L., Wijsman E., Kakol J., Buyske S., Chui B., Cohen P., de Toma C., et al. A 3.9-centimorgan-resolution human single-nucleotide polymorphism linkage map and screening set. Am. J. Hum. Genet. 2003;73:271–284. doi: 10.1086/377137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudd M.K., Friedman C., Parghi S.S., Linardopoulou E.V., Hsu L., Trask B.J. Elevated rates of sister chromatid exchange at chromosome ends. PLoS Genet. 2007;3:e32. doi: 10.1371/journal.pgen.0030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ledbetter D.H., Martin C.L. Cryptic telomere imbalance: a 15-year update. Am. J. Med. Genet. C Semin. Med. Genet. 2007;145:327–334. doi: 10.1002/ajmg.c.30149. [DOI] [PubMed] [Google Scholar]
- 54.Shaffer L.G., Lupski J.R. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- 55.Ou Z., Stankiewicz P., Xia Z., Breman A.M., Dawson B., Wiszniewska J., Szafranski P., Cooper M.L., Rao M., Shao L., et al. Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes. Genome Res. 2011;21:33–46. doi: 10.1101/gr.111609.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 57.Rudd M.K., Keene J., Bunke B., Kaminsky E.B., Adam M.P., Mulle J.G., Ledbetter D.H., Martin C.L. Segmental duplications mediate novel, clinically relevant chromosome rearrangements. Hum. Mol. Genet. 2009;18:2957–2962. doi: 10.1093/hmg/ddp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lehrman M.A., Schneider W.J., Sudhof T.C., Brown M.S., Goldstein J.L., Russell D.W. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pousi B., Hautala T., Heikkinen J., Pajunen L., Kivirikko K.I., Myllyla R. Alu-Alu recombination results in a duplication of seven exons in the lysyl hydroxylase gene in a patient with the type VI variant of Ehlers-Danlos syndrome. Am. J. Hum. Genet. 1994;55:899–906. [PMC free article] [PubMed] [Google Scholar]
- 60.Kass D.H., Batzer M.A., Deininger P.L. Gene conversion as a secondary mechanism of short interspersed element (SINE) evolution. Mol. Cell. Biol. 1995;15:19–25. doi: 10.1128/mcb.15.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanco P., Shlumukova M., Sargent C.A., Jobling M.A., Affara N., Hurles M.E. Divergent outcomes of intrachromosomal recombination on the human Y chromosome: male infertility and recurrent polymorphism. J. Med. Genet. 2000;37:752–758. doi: 10.1136/jmg.37.10.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vissers L.E., Bhatt S.S., Janssen I.M., Xia Z., Lalani S.R., Pfundt R., Derwinska K., de Vries B.B., Gilissen C., Hoischen A., et al. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum. Mol. Genet. 2009;18:3579–3593. doi: 10.1093/hmg/ddp306. [DOI] [PubMed] [Google Scholar]
- 63.Conrad D.F., Pinto D., Redon R., Feuk L., Gokcumen O., Zhang Y., Aerts J., Andrews T.D., Barnes C., Campbell P., et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rudd M.K. Encyclopedia of Life Sciences. John Wiley & Sons, Ltd; 2007. Subtelomeres: Evolution in the Human Genome. Kehrer-Sawatzki (ed), [Google Scholar]
- 65.Sutherland G.R. Rare fragile sites. Cytogenet. Genome Res. 2003;100:77–84. doi: 10.1159/000072840. [DOI] [PubMed] [Google Scholar]
- 66.Gajecka M., Pavlicek A., Glotzbach C.D., Ballif B.C., Jarmuz M., Jurka J., Shaffer L.G. Identification of sequence motifs at the breakpoint junctions in three t(1;9)(p36.3;q34) and delineation of mechanisms involved in generating balanced translocations. Hum. Genet. 2006;120:519–526. doi: 10.1007/s00439-006-0222-1. [DOI] [PubMed] [Google Scholar]
- 67.Crow J.F. The origins, patterns and implications of human spontaneous mutation. Nat. Rev. Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 68.Bandyopadhyay R., Heller A., Knox-DuBois C., McCaskill C., Berend S.A., Page S.L., Shaffer L.G. Parental origin and timing of de novo Robertsonian translocation formation. Am. J. Hum. Genet. 2002;71:1456–1462. doi: 10.1086/344662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lo Nigro C., Chong C.S., Smith A.C., Dobyns W.B., Carrozzo R., Ledbetter D.H. Point mutations and an intragenic deletion in LIS1, the lissencephaly causative gene in isolated lissencephaly sequence and Miller-Dieker syndrome. Hum. Mol. Genet. 1997;6:157–164. doi: 10.1093/hmg/6.2.157. [DOI] [PubMed] [Google Scholar]
- 70.Bruno D.L., Anderlid B.M., Lindstrand A., van Ravenswaaij-Arts C., Ganesamoorthy D., Lundin J., Martin C.L., Douglas J., Nowak C., Adam M.P., et al. Further molecular and clinical delineation of co-locating 17p13.3 microdeletions and microduplications that show distinctive phenotypes. J. Med. Genet. 2010;47:299–311. doi: 10.1136/jmg.2009.069906. [DOI] [PubMed] [Google Scholar]
- 71.Schiff M., Delahaye A., Andrieux J., Sanlaville D., Vincent-Delorme C., Aboura A., Benzacken B., Bouquillon S., Elmaleh-Berges M., Labalme A., et al. Further delineation of the 17p13.3 microdeletion involving YWHAE but distal to PAFAH1B1: four additional patients. Eur. J. Med. Genet. 2010;53:303–308. doi: 10.1016/j.ejmg.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 72.Bi W., Sapir T., Shchelochkov O.A., Zhang F., Withers M.A., Hunter J.V., Levy T., Shinder V., Peiffer D.A., Gunderson K.L., et al. Increased LIS1 expression affects human and mouse brain development. Nat. Genet. 2009;41:168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zollino M., Lecce R., Fischetto R., Murdolo M., Faravelli F., Selicorni A., Butte C., Memo L., Capovilla G., Neri G. Mapping the Wolf-Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am. J. Hum. Genet. 2003;72:590–597. doi: 10.1086/367925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cody J.D., Carter E.M., Sebold C., Heard P.L., Hale D.E. A gene dosage map of Chromosome 18: a map with clinical utility. Genet. Med. 2009;11:778–782. doi: 10.1097/GIM.0b013e3181b6573d. [DOI] [PubMed] [Google Scholar]
- 75.Martin C.L., Nawaz Z., Baldwin E.L., Wallace E.J., Justice A.N., Ledbetter D.H. The evolution of molecular ruler analysis for characterizing telomere imbalances: from fluorescence in situ hybridization to array comparative genomic hybridization. Genet. Med. 2007;9:566–573. doi: 10.1097/gim.0b013e318149e1fc. [DOI] [PubMed] [Google Scholar]
- 76.Hauge X., Raca G., Cooper S., May K., Spiro R., Adam M., Martin C.L. Detailed characterization of, and clinical correlations in, 10 patients with distal deletions of chromosome 9p. Genet. Med. 2008;10:599–611. doi: 10.1097/GIM.0b013e31817e2bde. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.