Abstract
Prostate cancer (PrCa) is the most common non-skin cancer diagnosed among males in developed countries and the second leading cause of cancer mortality, yet little is known regarding its etiology and factors that influence clinical outcome. Genome-wide association studies (GWAS) of PrCa have identified at least 30 distinct loci associated with small differences in risk. We conducted a GWAS in 2782 advanced PrCa cases (Gleason grade ≥ 8 or tumor stage C/D) and 4458 controls with 571 243 single nucleotide polymorphisms (SNPs). Based on in silico replication of 4679 SNPs (Stage 1, P < 0.02) in two published GWAS with 7358 PrCa cases and 6732 controls, we identified a new susceptibility locus associated with overall PrCa risk at 2q37.3 (rs2292884, P= 4.3 × 10−8). We also confirmed a locus suggested by an earlier GWAS at 12q13 (rs902774, P= 8.6 × 10−9). The estimated per-allele odds ratios for these loci (1.14 for rs2292884 and 1.17 for rs902774) did not differ between advanced and non-advanced PrCa (case-only test for heterogeneity P= 0.72 and P= 0.61, respectively). Further studies will be needed to assess whether these or other loci are differentially associated with PrCa subtypes.
INTRODUCTION
Prior to recent genome-wide association studies (GWAS), only three major prostate cancer (PrCa) risk factors were known, namely age, African ancestry and family history. Familial and twin studies suggested the importance of inherited factors for PrCa development (1–2), prognosis and tumor aggressiveness (3). A critical clinical question in PrCa has been to determine who will develop aggressive as opposed to indolent disease, as men with poorly differentiated or advanced-stage tumors are more likely to die from this type of cancer (4–5). Family studies have identified at least three promising regions linked to the aggressive form of PrCa, but these results have not been definitively replicated (6–9).
Over the past 3 years, GWAS have successfully identified at least 30 independent common PrCa susceptibility alleles across the genome, each of which confers a small increase in risk for both advanced and non-advanced PrCa (10–26). There is preliminary evidence that three loci could be associated with the risk for advanced disease alone, namely those on 9q33.2 (harboring the candidate gene, DAB2IP; 24), 22q13.1 (27) and 17p12 (25), but further work is required to confirm these observations. The known PrCa susceptibility loci explain a modest fraction of the 2-fold familial relative risk observed for first-degree relatives of affected individuals (11).
In an attempt to identify additional loci that influence susceptibility to PrCa overall, and, in particular, advanced PrCa, we conducted a genome-wide scan using cases with advanced disease (Gleason grade ≥ 8 or tumor stage C/D) from the National Cancer Institute Breast & Prostate Cancer Cohort Consortium (BPC3). To validate associations observed in the initial GWAS, we performed in silico replication for the most promising markers using data from two previous GWAS conducted in the UK and Australia (11–12) and the Cancer Prostate in Sweden (CAPS) Study (23). We identified a new region associated with the risk of PrCa at 2q37.3 and confirmed a proposed association at 12q13 that had previously not achieved genome-wide significance (12). These loci were associated with both advanced and non-advanced PrCa, and neither locus showed compelling evidence for heterogeneity in effects by tumor type.
RESULTS
For the advanced PrCa GWAS (denoted Stage 1), 2891 advanced PrCa cases and 4592 controls of European ancestry from the seven BPC3 cohorts (see Supplementary Material, Table S1) were genotyped across four genotyping centers, mostly using the Illumina HumanHap610 quad array. After quality control metrics (see Materials and Methods), 571 243 single nucleotide polymorphisms (SNPs) were analyzed in 2782 advanced PrCa cases and 4458 controls. Genotype frequencies in cases and controls were compared using a 1-d.f. trend test within each cohort and combined using fixed-effect meta-analysis (see Materials and Methods). After adjustment for differential population structure, there was little evidence for inflation in the genomic control test statistic (λ1000 = 1.01, Supplementary Material, Fig. S1).
We observed evidence of association with advanced PrCa for the majority of previously reported PrCa loci (Supplementary Material, Table S2). For the confirmed loci, the estimated per-allele odds ratios for advanced PrCa were consistent with those from the original reports in which both advanced and non-advanced PrCa were combined. For 8 of the 30 previously reported regions (2p21, 2p15, 4q22.3, 4q24, 8p21, 13q22.1, 17p12 and 19q13.33 [KLK3]), including the proposed advanced-only region 17p12, we observed no statistically significant association (P≥ 0.06) with advanced PrCa in our prospectively ascertained advanced cases and controls. We also observed no association between the proposed advanced-only marker at 22q13.1 and advanced PrCa after excluding subjects that overlapped with the initial report of that marker (P= 0.39). The failure to observe associations in these regions may be due to limited power, differences in linkage disequilibrium structure (the 5p15, 6q22 and 13q22 regions were identified in a recent GWAS in Japanese men) or differences in observed per-allele odds ratios between advanced and non-advanced disease (28–29).
One previously proposed advanced-only marker on chromosome 9q33.2 was nominally statistically significantly associated with advanced PrCa after removing samples that overlapped with original reports for these loci (rs1571801, P= 1.4 × 10−3; Supplementary Material, Table S2). This SNP was genotyped in the full BPC3 cohort (10 501 cases and 10 831 controls), including both advanced and non-advanced PrCa cases as part of an earlier validation study (30); no evidence for differences in association by Gleason grade (<8 versus ≥8; P= 0.50) or tumor stage (AB versus CD P= 0.62) was found.
To identify additional PrCa risk markers in these previously reported regions, we examined the association between 41 756 high-quality imputed SNPs (Supplementary Methods) within 1cM of any of the published risk markers listed in Supplementary Material, Table S2 and advanced PrCa in the Stage 1 data. We analyzed each imputed SNP twice: once without adjusting for any published marker and once adjusting for the published markers in the region containing the SNP. Supplementary Material, Figure S2 presents the unconditional (dark grey) and conditional (blue) results for each region within 1 cM of the index signal for 30 regions. Of the 22 regions where a previously reported SNP was nominally significantly associated with advanced PrCa in Stage 1, three (3p12.1, 6q22.1 and 11p15.5) exhibited strong evidence that a novel SNP was a better PrCa marker than a previously reported marker (approximate Bayes factor >10; Supplementary Material, Table S3). However, after conditioning on known markers, only 1 of the 22 regions (8q24.21) contained novel markers that were suggestively associated with advanced PrCa (P< 10−4; Supplementary Material, Table S3). This highlights the complex genetic architecture in these regions, which will require additional studies with dense genotyping in large sample sizes to deconstruct (19,31–33).
No novel regions were detected at genome-wide significance in Stage 1 (P< 5 × 10−8). We performed an in silico replication in two existing GWAS (11–12,23) for the most significant 5000 SNPs after removing previously reported SNPs and filtering out correlated SNPs with r2 > 0.2 (see Materials and Methods).
Results from analyses combining both non-advanced and advanced PrCa cases as well as those examining only advanced PrCa cases are presented in Table 1 for the two novel loci that achieved genome-wide significance. A novel region associated with PrCa risk overall was identified at 2q37 (rs2292884; P= 4.3 × 10−8). We also confirmed an association between rs902774 at 12q13 and PrCa (P= 4.7 × 10−9). This SNP was highlighted in a previous report of the GWAS conducted in the UK and Australia (12); despite suggestive evidence for association in the first stage of that study, it did not achieve genome-wide significance in the combined first and second stages (denoted UK1 and UK2+Aus in Table 1). The significant heterogeneity in per-allele odds ratio estimates for rs902774 across the four studies included in this report (P= 2.6 × 10−3) was also driven by the UK2+Aus samples; we observed no significant heterogeneity in this effect among BPC3 studies (P= 0.33; Supplementary Material, Table S4) or among the BPC3, UK1 and CAPS studies (P= 0.11).
Table 1.
Novel SNPs associated with overall and advanced PrCa risk by study and combined
| Overall prostate cancer |
Advanced prostate cancer |
Case-only analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPa | Cytoband/positionb | Study | RAF in controls | Cases/controls | OR (95% CI) | P-valuec | Cases/controls | OR (95% CI) | P-valuec | Advanced/non-advanced | ROR (95% CI)d | P-valuee |
| rs2292884 | 2q37 | BPC3 | 0.25 | 3641/4458 | 1.15 (1.06–1.24) | 8.1 × 10−4 | 2782/4458 | 1.15 (1.06–1.25) | 5.7 × 10−4 | 282/859f | 0.92 (0.74–1.14) | 0.44 |
| A/G | 238107965 | UK1 | 0.25 | 1854/1894 | 1.10 (0.99–1.22) | 0.06 | 617/1894 | 1.13 (0.98–1.32) | 0.10 | 617/1237 | 0.97 (0.83–1.13) | 0.08 |
| UK2+Aus | 0.25 | 3650/3940 | 1.13 (1.05–1.22) | 1.2 × 10−3 | 1084/3940 | 1.08 (0.96–1.21) | 0.19 | 1084/2566 | 1.04 (0.92–1.17) | 0.06 | ||
| CAPS | 0.23 | 1854/898 | 1.17 (1.02–1.34) | 0.02 | 962/898 | 1.19 (1.02–1.39) | 0.03 | 961/893 | 1.04 (0.89–1.20) | 0.65 | ||
| Combined | 1.14 (1.09–1.19) | 4.3 × 10−8 | 1.14 (1.07–1.20) | 7.8 × 10−6 | 1.00 (0.93–1.08) | 0.91 | ||||||
| P-het* | 0.88 | 0.70 | ||||||||||
| rs902774 | 12q13 | BPC3 | 0.17 | 3641/4458 | 1.18 (1.07–1.30) | 1.0 × 10−3 | 2782/4458 | 1.17 (1.07–1.29) | 1.0 × 10−3 | 282/859f | 0.90 (0.70–1.17) | 0.44 |
| G/A | 51560171 | UK1 | 0.14 | 1854/1894 | 1.39 (1.22–1.57) | 2.0 × 10−7 | 617/1894 | 1.49 (1.24–1.78) | 1.4 × 10−5 | 617/1237 | 0.93 (0.78–1.11) | 0.09 |
| UK2+Aus | 0.15 | 3650/3940 | 1.04 (0.95–1.14) | 0.35 | 1084/3940 | 1.12 (0.98–1.28) | 0.10 | 1084/2566 | 0.87 (0.76–1.01) | 0.07 | ||
| CAPS | 0.16 | 1854/898 | 1.24 (1.07–1.44) | 3.9 × 10−3 | 961/898 | 1.25 (1.06–1.48) | 9.6 × 10−3 | 961/893 | 1.01 (0.85–1.19) | 0.94 | ||
| Combined | 1.17 (1.11–1.24) | 4.7 × 10−9 | 1.21 (1.13–1.29) | 1.4 × 10−8 | 0.93 (0.85–1.01) | 0.08 | ||||||
| P-het* | 2.6 × 10−3 | 0.07 | ||||||||||
adbSNP rs number, reference/risk allele. The odds ratio refers to the change in odds per additional copy of the risk allele.
bChromosome and build 36 position.
cTest for trend from logistic regression.
dRatio of advanced PrCa per-allele odds ratio to non-advanced per-allele odds ratio.
eTest for difference in advanced and non-advanced odds ratios.
fFor BPC3, case-only analyses are restricted to advanced cases and non-advanced cases from PLCO that were genotyped as part of the CGEMS PrCa GWAS.
*P-value for test of heterogeneity of effect across study.
RAF, risk allele frequency; OR, per-allele (multiplicative) odds ratio; CI, confidence interval.
The per-allele odds ratios for advanced and non-advanced PrCa were not significantly different for either of these loci (case-only test for heterogeneity P > 0.08; Table 1). However, this heterogeneity test is biased towards the null since aggressive and non-aggressive PrCa cases were combined to reach genome-wide significance. Additional studies will be needed to evaluate aggressive- and non-aggressive-specific effects.
We examined the association between these two genome-wide significant SNPs and PrCa in three non-European populations: African Americans (1071 cases and 1074 controls), Latinos (1043 cases, 1057 controls) and Japanese (1033 cases, 1042 controls) (see Materials and Methods). No significant association was observed for either of these markers in any one of these three non-European populations (Supplementary Material, Table S5 and Fig. S3), which may be due to low power or differences in allele frequencies or linkage disequilibrium. However, the direction of association between the 2q37 SNP rs2292884 and PrCa risk was consistent across all four populations, and a meta-analysis of the African American, Latino and Japanese results suggested this SNP was also associated with PrCa in these non-European populations (Supplementary Material, Table S5; P= 7.1 × 10−3). This region will need to be further evaluated in larger sample sizes of both European and non-Europeans.
Eight additional markers located in or near known risk regions achieved genome-wide significance (P< 5 × 10−8) in analyses combining Stage 1 and the in silico replication studies (Table 2). By design, the linkage disequilibrium between these markers and known markers was low (r2 < 0.2). After conditioning on known risk alleles, the statistical significance for the new markers dropped by three to eight orders of magnitude. None remained genome-wide significant after conditioning, with the exception of rs651164 at 6q25.3, which had been identified as a possible PrCa marker in a previous report (11). Several recombination hotspots separate rs651164 and the previously reported rs9364554 (12), which lie over 252 kb apart (Supplementary Material, Fig. S4); consequently, the linkage disequilibrium between these two markers is low (r2 = 0.00 and |D'| = 0.16 in the HapMap CEU sample). This suggests that the 6q25.3 region contains two statistically independent markers of PrCa risk, rs651164 and rs9364554. Also of note, rs7629490 at 3p11 remained strongly associated with the risk of PrCa (P= 1.2 × 10−7) after conditioning on the previously reported PrCa risk marker rs2660753 (12). The SNP rs7629490 is located 130 kb centromeric from rs2660753. Although these two SNPs are in a large block of limited recombination (spanning 320 kb from rs1370041 at chr3:87,171,855 to rs4858957 at chr3:87,491,848; Supplementary Material, Fig. S4), they are weakly correlated (r2 = 0.05; |D'| = 0.38). Further fine-mapping studies are required to identify the mechanism underlying the statistical associations between PrCa and multiple, weakly correlated markers in these regions.
Table 2.
Genome-wide significant markers located in known regionsa
| Marginal analysis |
Conditional analysis |
|||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Allelesb | Cytoband | Pos | Gene | OR (95% CI) | P-value | OR (95% CI) | P-value |
| rs7629490c | C/T | 3p11 | 87324187 | – | 1.15 (1.10–1.19) | 1.9 × 10−10 | 1.06 (1.04–1.09) | 1.2 × 10−7 |
| rs651164d | G/A | 6q25 | 160501364 | SLC22A1 | 0.85 (0.81–0.89) | 1.4 × 10−13 | 0.87 (0.83–0.91) | 2.2 × 10−10 |
| rs13252298e | A/G | 8q24 | 128164338 | – | 0.85 (0.81–0.89) | 4.5 × 10−12 | 0.89 (0.85–0.95) | 4.3 × 10−6 |
| rs6999921e | G/A | 8q24 | 128510110 | – | 0.81 (0.75–0.87) | 7.4 × 10−9 | 0.90 (0.83–0.97) | 7.0 × 10−3 |
| rs1447293e | T/C | 8q24 | 128541502 | – | 1.14 (1.09–1.18) | 3.1 × 10−9 | 1.00 (0.95–1.04) | 0.84 |
| rs11228594f | A/G | 11q13 | 68779663 | – | 0.87 (0.83–0.91) | 1.2 × 10−9 | 0.90 (0.86–0.95) | 2.4 × 10−5 |
| rs7940107f,g | G/A | 11q13 | 68784346 | – | 1.20 (1.12–1.28) | 4.7 × 10−8 | 1.06 (1.02–1.11) | 3.6 × 10−3 |
| rs742134h | A/G | 22q13 | 41848219 | BIK | 1.20 (1.15–1.33) | 5.6 × 10−9 | 1.16 (1.01–1.23) | 5.9 × 10−6 |
aUnless otherwise specified, results are from combined Stage 1 and in silico replication studies, including both advanced and non-advanced cases.
bReference allele/risk allele; the odds ratio is the change in odds per additional copy of the risk allele.
cConditional analyses adjusted for rs2660753.
dConditional analyses adjusted for rs9364554.
eConditional analyses adjusted for rs7841060, rs16901979, rs620861, rs6983267, rs4242382.
fConditional analyses adjusted for rs10896449.
gResults from advanced-only analysis.
hConditional analyses adjusted for rs5759167.
DISCUSSION
We conducted the GWAS of PrCa using 2782 cases with advanced PrCa (Gleason grade ≥ 8 or stage C/D) and 4458 controls of European ancestry nested in seven prospective cohort studies, followed by in silico replication in two previous GWAS (11–12,23). We confirmed a previously proposed PrCa marker (rs902774) at 12q13 and identified a new PrCa marker (rs229884) at 2q37. Our findings are consistent with the expectation that further GWAS will continue to discover additional low penetrance, common alleles associated with the risk for PrCa (34).
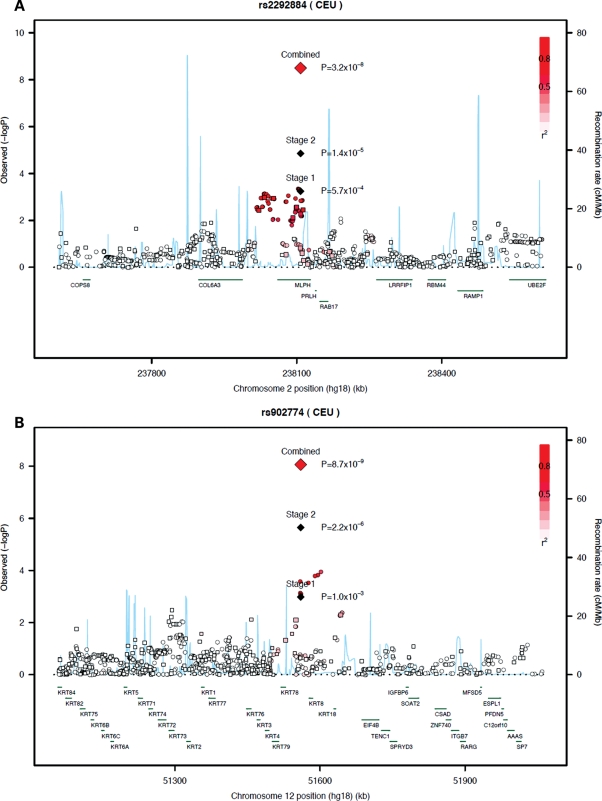
At 2q37, rs2292884 is a missense SNP (Arg347His) located in exon 9 of the melanophilin gene (MLPH) (Fig. 1A). This protein is a member of the exophilin subfamily of Rab effector proteins. MLPH forms a ternary complex with the small Ras-related GTPase Rab27A and myosin Va; this complex functions as a tether for pigment-producing melanosomes to bind with actin cytoskeleton in melanocytes. Mouse models have demonstrated that mutations in MLPH are required for visible pigmentation in the hair and skin (35). A homozygous mutation in the MLPH gene has been associated with the human affliction Griscelli syndrome type 3, a hypopigmentaton disorder (36). Other genes nearby on chromosome 2q37 include prolactin releasing hormone (PRLH) and RAB17, which is a member of the Ras oncogene family that regulates membrane trafficking. The aforementioned candidate genes in this region have not been previously implicated in PrCa carcinogenesis. The SNP rs2292884 was associated with overall PrCa risk at a conventional level for genome-wide statistical significance (P< 5 × 10−8). This provides reassurance that this result is not a false positive due to multiple testing. However, given the modest odds ratio and the fact that the P-value is near the significance threshold, replication of this association in future, large studies will be important.
Figure 1.
Regional association plots of two PrCa loci. (A and B) Plots show the genomic regions of association with overall PrCa risk on chromosome 2q37 (A) and 12q13 (B). Shown are the –log10 association P-values of genotyped (square) and imputed (circle) SNPs in 2782 advanced PrCa cases and 4458 controls in Stage 1. The –log10 association P-values for the index SNP in Stage 1, Stage 2 and combined is shown (diamond). The intensity of red shading indicates the strength of LD with the index SNP. Also shown are the SNP build 36 coordinates in kilobases (kb), recombination rates in centimorgans (cM) per megabase (Mb) (in blue) and genes in the region (in green).
The 12q13 rs902774 SNP lies in the type II keratin family cluster (Fig. 1B), which encodes a set of genes that contribute to the formation of cytoplasmic filaments in epithelial cells. These filaments maintain cellular integrity and function in signal transduction and cellular differentiation. The closest gene (7 kb telomeric) is KRT8, which has a role in apoptosis (37). Other genes nearby include EIF4B, which plays an important role in the metabolism of RNA and is necessary for cell proliferation and survival (38), and TENC1, which is a focal adhesion molecule that regulates cell mobility. The nearby genes have yet to be implicated in PrCa.
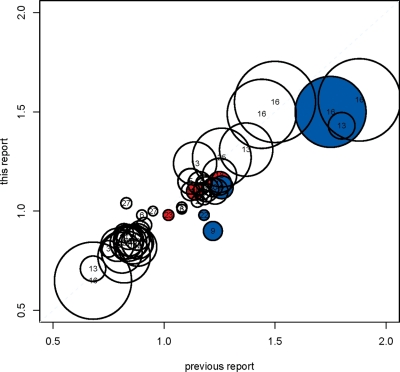
It is notable that although our Stage 1 with 2782 cases is the largest GWAS of advanced PrCa to date and our replication sample contained an additional 2662 advanced PrCa cases, we identified no loci that were primarily associated with advanced PrCa. Moreover, the estimated odds ratios for previously identified risk alleles in our GWAS are similar to those from the original reports, even though our GWAS consisted of advanced PrCa cases and controls drawn from prospective cohort studies, while previous studies have included both advanced and non-advanced cases and controls, mostly ascertained retrospectively (Fig. 2). Since we had over 90% power to detect a marker solely associated with advanced PrCa with a minor allele frequency of 40% and a per-allele odds ratio of 1.18 (and over 50% power to detect an odds ratio of 1.13; Supplementary Methods Fig. S4), this suggests that—unlike breast cancer, where there are unique markers associated with estrogen-receptor positive and negative tumors (39)—there are currently no known common markers differentially associated with advanced or non-advanced PrCa. There are several possible reasons for this observation. It may be that the determinants of aggressive disease occur later in the pathogenesis of PrCa, and germline variants primarily play a role in the initiation of disease; or it may be that current definitions of aggressive disease, although clinically useful, do not reflect etiologic differences in prostate tumors; or it may be that the clinical heterogeneity in diagnosis may be greater than the small effect of any individual locus; or it may be that other distinctions among tumors not evaluated here (such as molecular tumor markers) are associated with genetic heterogeneity.
Figure 2.
Comparison of the observed per-allele odds ratio in the BPC3 advanced PrCa GWAS (Stage 1) versus previously reported per-allele odds ratios for 51 of the 57 markers in Supplementary Material, Table S2. (For six markers, a per-allele odds ratio for overall PrCa could not be calculated from published information.) The reference/risk alleles are defined as in Supplementary Material, Table S2. The radius of the plotted point is proportional to the −log10 P-value in the BPC3 GWAS and the center of each point is marked by the region number (Supplementary Material, Table S2). The red highlighted SNPs were originally published as advanced PrCa markers; the blue highlighted SNPs were published in a GWAS conducted among Japanese men (22).
GWAS of PrCa have identified many regions in the genome harboring susceptibility alleles that confer risk for PrCa, but these studies have not conclusively identified regions that only confer risk of clinically relevant, advanced PrCa, as measured by advanced Gleason score or stage. The PrCa susceptibility loci identified to date may principally influence early stages of tumor development rather than disease progression based on current observations. Larger studies will be needed to identify germline variants with modest effects that influence Gleason score, stage and survival after diagnosis with PrCa.
MATERIALS AND METHODS
Samples
The advanced PrCa cases included in Stage 1 were drawn from seven cohorts in the USA and Europe (Supplementary Material, Table S1 and Supplementary Methods). Cases were men of European ancestry identified through cancer registries or by self-report and confirmed through medical record review. Clinical characteristics were abstracted from medical records. Advanced PrCa was defined as having either high histologic grade (Gleason score ≥ 8) or extra-prostatic extension (Stage C/D). Controls were men without a diagnosis of PrCa and of European ancestry.
The Stage 2 in silico replication included subjects of European ancestry from two previously reported GWAS of PrCa (11–12,23,24) (Supplementary Methods). The first GWAS consisted of two stages, with cases and controls drawn from the UK (12) (UK1) and both the UK and Australia (11) (UK2+Aus), respectively. For the second, cases and controls were taken from the CAPS study (23). Non-advanced cases from the BPC3 that were genotyped as part of the Cancer Genetic Markers of Susceptibility GWAS (19) were also included in the overall PrCa analyses. To explore associations in other populations, non-European cases and controls were taken from the Multi-Ethnic Cohort, scanned as part of an ongoing GWAS of PrCa funded by the NHGRI's GENEVA program (Supplementary Methods).
Ethics statement
Each of the participating studies obtained an informed consent from study participants and approval from their respective institutional review boards for this study.
Genotyping and quality control assessment
All of the subjects from four BPC3 cohorts (1239 cases and 1188 controls), most of the subjects from a fifth (656 cases and 409 controls) and cases from the two remaining cohorts (249 and 437) were genotyped specifically for this study using Illumina Human 610-Quad Beadchips (Illumina, Inc). The remaining cases and controls were genotyped previously (Supplementary Methods). De novo genotyping was performed at four centers using genomic samples extracted from blood and buccal specimens. Cases and controls were randomly distributed across genotyping plates. Genotypes on the Illumina platform were called using GenCall algorithm as implemented in GenomeStudio version 2009.1 (Illumina, Inc.). GenCall quality metrics, sample heterozygosity, marker and sample completion rates, minor allele frequencies and departures from Hardy–Weinberg equilibrium were used to filter under-performing SNPs and samples; these filters removed SNPs with genotyping differences across arrays (Supplementary Methods). Subjects with evidence of significant non-European ancestry and population structure were also excluded (Supplementary Methods Fig. S2).
The UK Stage I samples were genotyped using the Illumina Human Hap550 platform, while the UK and Australian Stage II samples were genotyped using a custom Infinium iSelect. CAPS samples were genotyped using the Affymetrix 500k gene chip. Quality control procedures for these studies have been reported previously (11–12,23). The non-European cases and controls from the MEC were genotyped on the Illumina 610 platform.
Statistical analysis
In Stage 1, we tested associations between each SNP and advanced PrCa separately for each study using a 1-d.f. trend test from a logistic regression, adjusted for the second principal component of genetic covariance in the combined sample, which was the only component of the top 10 nominally significantly associated with PrCa (P= 0.01). For SNPs that were present in at least four studies, the overall estimate of the per-allele odds ratio and the corresponding association test were calculated using fixed-effect meta-analysis. The inflation in the Chi-squared statistic was assessed using the genomic control approach, scaled to a standard sample size of 1000 cases and controls (40).
Five thousand SNPs were chosen for in silico replication in Stage 2 after removing 53 known SNPs associated with PrCa and any correlated markers (r2 > 0.2). The remaining SNPs were ranked according to P-value and further pruned by removing SNPs in LD (r2 > 0.2) with top signals yielding 4976 SNPs. We performed an ancillary GWAS using 2 388 194 imputed SNPs (see Supplementary Methods) and identified 23 additional significant SNPs that were not correlated with any of the selected genotyped SNPs. We included these in the replication set, along with rs4054823, which is reportedly associated with advanced PrCa (41). A total of 4679 SNPs were genotyped or imputed in the replication studies. Logistic regression assuming a log-additive genetic model was performed in each study, using expected allele dosage to account for imputation uncertainty, first comparing all PrCa cases to controls, then comparing only advanced PrCa cases to controls. Fixed-effect meta-analysis was used to combine evidence for association across the replication studies and the Stage 1 studies. Genome-wide significant SNPs (P< 5.0 × 10−8) in known PrCa regions were further evaluated for independent associations by conditioning on known PrCa SNPs (Table 2). Case-only analyses were used to assess whether SNP effects differed for advanced versus non-advanced PrCa.
SUPPLEMENTARY MATERIAL
FUNDING
The BPC3 was supported by the U.S. National Institutes of Health, National Cancer Institute (cooperative agreements U01-CA98233 to D.J.H., U01-CA98710 to M.J.T., U01-CA98216 to E.R., and U01-CA98758 to B.E.H., and Intramural Research Program of NIH/National Cancer Institute, Division of Cancer Epidemiology and Genetics). This work was supported by Cancer Research UK Grants C5047/A7357, C1287/A10118, C5047/A3354, C5047/A10692, C16913/A6135, and C16913/A6835. We would also like to thank the following for funding support: The Institute of Cancer Research and The Everyman Campaign, The Prostate Cancer Research Foundation, Prostate Research Campaign UK, The National Cancer Research Network UK, The National Cancer Research Institute (NCRI) UK. We acknowledge NHS funding to the NIHR Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. We also acknowledge grant support from The National Health and Medical Research Council, Australia (209057, 251533, 450104, 390130), VicHealth, The Cancer Council Victoria, Cancer Council Queensland, The Whitten Foundation, and Tattersall's. The ProtecT study is ongoing and is funded by the Health Technology Assessment Programme (projects 96/20/06, 96/20/99). The ProtecT trial and its linked ProMPT and CAP (Comparison Arm for ProtecT) studies are supported by Department of Health, England; Cancer Research UK grant number C522/A8649, Medical Research Council of England grant number G0500966, ID 75466 and The NCRI, UK. The epidemiological data for ProtecT were generated though funding from the Southwest National Health Service Research and Development. DNA extraction in ProtecT was supported by USA Dept of Defense award W81XWH-04-1-0280, Yorkshire Cancer Research and Cancer Research UK. The bio-repository from ProtecT is supported by the NCRI (ProMPT) study and the Cambridge BMRC grant from NIHR. Financial support for CAPS was provided through a grant from the Swedish Research Council (grant no K2010-70X-20430-04-3 and 70867901), the Swedish Cancer Foundation (grant no 09-0677), the Hedlund Foundation, Söderberg Foundation, Enqvist Foundation, ALF funds from the Stockholm County Council. W.B.I. was supported by National Cancer Institute grants CA112517 and CA133009.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank all the study participants in BPC3, CAPS study and the UK and Australian studies. The authors would like to acknowledge the tremendous contribution of all members of the ProtecT study research group. We should like to acknowledge the NCRN nurses and Consultants for their work in the UKGPCS study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Gronberg H. Prostate cancer epidemiology. Lancet. 2003;361:859–864. doi: 10.1016/S0140-6736(03)12713-4. doi:10.1016/S0140-6736(03)12713-4. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenstein P., Holm N.V., Verkasalo P.K., Iliadou A., Kaprio J., Koskenvuo M., Pukkala E., Skytthe A., Hemminki K. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. doi:10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 3.Hemminki K., Ji J., Forsti A., Sundquist J., Lenner P. Concordance of survival in family members with prostate cancer. J. Clin. Oncol. 2008;26:1705–1709. doi: 10.1200/JCO.2007.13.3355. doi:10.1200/JCO.2007.13.3355. [DOI] [PubMed] [Google Scholar]
- 4.Albertsen P.C., Hanley J.A., Fine J. 20-year outcomes following conservative management of clinically localized prostate cancer. JAMA. 2005;293:2095–2101. doi: 10.1001/jama.293.17.2095. doi:10.1001/jama.293.17.2095. [DOI] [PubMed] [Google Scholar]
- 5.Bill-Axelson A., Holmberg L., Ruutu M., Haggman M., Andersson S.O., Bratell S., Spangberg A., Busch C., Nordling S., Garmo H., et al. Radical prostatectomy versus watchful waiting in early prostate cancer. N. Engl. J. Med. 2005;352:1977–1984. doi: 10.1056/NEJMoa043739. doi:10.1056/NEJMoa043739. [DOI] [PubMed] [Google Scholar]
- 6.Slager S.L., Schaid D.J., Cunningham J.M., McDonnell S.K., Marks A.F., Peterson B.J., Hebbring S.J., Anderson S., French A.J., Thibodeau S.N. Confirmation of linkage of prostate cancer aggressiveness with chromosome 19q. Am. J. Hum. Genet. 2003;72:759–762. doi: 10.1086/368230. doi:10.1086/368230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slager S.L., Zarfas K.E., Brown W.M., Lange E.M., McDonnell S.K., Wojno K.J., Cooney K.A. Genome-wide linkage scan for prostate cancer aggressiveness loci using families from the University of Michigan Prostate Cancer Genetics Project. Prostate. 2006;66:173–179. doi: 10.1002/pros.20332. doi:10.1002/pros.20332. [DOI] [PubMed] [Google Scholar]
- 8.Witte J.S., Goddard K.A., Conti D.V., Elston R.C., Lin J., Suarez B.K., Broman K.W., Burmester J.K., Weber J.L., Catalona W.J. Genomewide scan for prostate cancer-aggressiveness loci. Am. J. Hum. Genet. 2000;67:92–99. doi: 10.1086/302960. doi:10.1086/302960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witte J.S., Suarez B.K., Thiel B., Lin J., Yu A., Banerjee T.K., Burmester J.K., Casey G., Catalona W.J. Genome-wide scan of brothers: replication and fine mapping of prostate cancer susceptibility and aggressiveness loci. Prostate. 2003;57:298–308. doi: 10.1002/pros.10304. doi:10.1002/pros.10304. [DOI] [PubMed] [Google Scholar]
- 10.Amundadottir L.T., Sulem P., Gudmundsson J., Helgason A., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. doi:10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 11.Eeles R.A., Kote-Jarai Z., Al Olama A.A., Giles G.G., Guy M., Severi G., Muir K., Hopper J.L., Henderson B.E., Haiman C.A., et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. doi:10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eeles R.A., Kote-Jarai Z., Giles G.G., Olama A.A., Guy M., Jugurnauth S.K., Mulholland S., Leongamornlert D.A., Edwards S.M., Morrison J., et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. doi:10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 13.Gudmundsson J., Sulem P., Gudbjartsson D.F., Blondal T., Gylfason A., Agnarsson B.A., Benediktsdottir K.R., Magnusdottir D.N., Orlygsdottir G., Jakobsdottir M., et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. doi:10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudmundsson J., Sulem P., Manolescu A., Amundadottir L.T., Gudbjartsson D., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. doi:10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 15.Gudmundsson J., Sulem P., Rafnar T., Bergthorsson J.T., Manolescu A., Gudbjartsson D., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Blondal T., et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat. Genet. 2008;40:281–283. doi: 10.1038/ng.89. doi:10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gudmundsson J., Sulem P., Steinthorsdottir V., Bergthorsson J.T., Thorleifsson G., Manolescu A., Rafnar T., Gudbjartsson D., Agnarsson B.A., Baker A., et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 2007;39:977–983. doi: 10.1038/ng2062. doi:10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 17.Thomas G., Jacobs K.B., Yeager M., Kraft P., Wacholder S., Orr N., Yu K., Chatterjee N., Welch R., Hutchinson A., et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. doi:10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 18.Yeager M., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. doi:10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 19.Yeager M., Chatterjee N., Ciampa J., Jacobs K.B., Gonzalez-Bosquet J., Hayes R.B., Kraft P., Wacholder S., Orr N., Berndt S., et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. doi:10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng S.L., Stevens V.L., Wiklund F., Isaacs S.D., Sun J., Smith S., Pruett K., Wiley K.E., Kim S.T., Zhu Y., et al. Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiol. Biomarkers Prev. 2009;18:1815–1820. doi: 10.1158/1055-9965.EPI-08-0983. doi:10.1158/1055-9965.EPI-08-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun J., Zheng S.L., Wiklund F., Isaacs S.D., Purcell L.D., Gao Z., Hsu F.C., Kim S.T., Liu W., Zhu Y., et al. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat. Genet. 2008;40:1153–1155. doi: 10.1038/ng.214. doi:10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takata R., Akamatsu S., Kubo M., Takahashi A., Hosono N., Kawaguchi T., Tsunoda T., Inazawa J., Kamatani N., Ogawa O., et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat. Genet. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 23.Hsu F.C., Sun J., Wiklund F., Isaacs S.D., Wiley K.E., Purcell L.D., Gao Z., Stattin P., Zhu Y., Kim S.T., et al. A novel prostate cancer susceptibility locus at 19q13. Cancer Res. 2009;69:2720–2723. doi: 10.1158/0008-5472.CAN-08-3347. doi:10.1158/0008-5472.CAN-08-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duggan D., Zheng S.L., Knowlton M., Benitez D., Dimitrov L., Wiklund F., Robbins C., Isaacs S.D., Cheng Y., Li G., et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J. Natl Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. doi:10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 25.Xu J., Zheng S.L., Isaacs S.D., Wiley K.E., Wiklund F., Sun J., Kader A.K., Li G., Purcell L.D., Kim S.T., et al. Inherited genetic variant predisposes to aggressive but not indolent prostate cancer. Proc. Natl Acad. Sci. USA. 107:2136–2140. doi: 10.1073/pnas.0914061107. doi:10.1073/pnas.0914061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al Olama A.A. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. doi:10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 27.Sun J., Zheng S.L., Wiklund F., Isaacs S.D., Li G., Wiley K.E., Kim S.T., Zhu Y., Zhang Z., Hsu F.C., et al. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009;69:10–15. doi: 10.1158/0008-5472.CAN-08-3464. doi:10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahn J., Berndt S.I., Wacholder S., Kraft P., Kibel A.S., Yeager M., Albanes D., Giovannucci E., Stampfer M.J., Virtamo J., et al. Variation in KLK genes, prostate-specific antigen and risk of prostate cancer. Nat. Genet. 2008;40:1032–1034. doi: 10.1038/ng0908-1032. author reply 1035–1036 doi:10.1038/ng0908-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eeles R.A., Giles G.G., Neal D.E., Muir K., Easton D.F., Consortium f.t.P. Reply to "Variation in KLK genes, prostate-specific antigen and risk of prostate cancer". Nat. Genet. 2008;40:1035–1036. doi:10.1038/ng0908-1035. [Google Scholar]
- 30.Lindstrom S., Schumacher F.R., Siddiq A., Travis R., Campa D., Berndt S., Diver R., Chanock S., Crawford D., Gaziano J.M., et al. Characterizing associations and SNP-environment interactions for GWAS-identified prostate cancer risk markers—results from BPC3. PLoS ONE. 2011;6:e17142. doi: 10.1371/journal.pone.0017142. doi:10.1371/journal.pone.0017142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prokunina-Olsson L., Fu Y.P., Tang W., Jacobs K.B., Hayes R.B., Kraft P., Berndt S.I., Wacholder S., Yu K., Hutchinson A., et al. Refining the prostate cancer genetic association within the JAZF1 gene on chromosome 7p15.2. Cancer Epidemiol. Biomarkers Prev. 2010;19:1349–1355. doi: 10.1158/1055-9965.EPI-09-1181. doi:10.1158/1055-9965.EPI-09-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lou H., Yeager M., Li H., Bosquet J.G., Hayes R.B., Orr N., Yu K., Hutchinson A., Jacobs K.B., Kraft P., et al. Fine mapping and functional analysis of a common variant in MSMB on chromosome 10q11.2 associated with prostate cancer susceptibility. Proc. Natl Acad. Sci. USA. 2009;106:7933–7938. doi: 10.1073/pnas.0902104106. doi:10.1073/pnas.0902104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung C.C., Ciampa J., Yeager M., Jacobs K.B., Berndt S.I., Hayes R.B., Gonzalez-Bosquet J., Kraft P., Wacholder S., Orr N., et al. Fine mapping of a region of chromosome 11q13 reveals multiple independent loci associated with risk of prostate cancer. Hum. Mol. Genet. 2011;20:2869–2878. doi: 10.1093/hmg/ddr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park J.H., Wacholder S., Gail M.H., Peters U., Jacobs K.B., Chanock S.J., Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 2010;42:570–575. doi: 10.1038/ng.610. doi:10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silvers W.K. The Coat Colors of Mice: A Model for Mammalian Gene Action and Interaction. New York: Springer; 1979. [Google Scholar]
- 36.Menasche G., Ho C.H., Sanal O., Feldmann J., Tezcan I., Ersoy F., Houdusse A., Fischer A., de Saint Basile G. Griscelli syndrome restricted to hypopigmentation results from a melanophilin defect (GS3) or a MYO5A F-exon deletion (GS1) J. Clin. Invest. 2003;112:450–456. doi: 10.1172/JCI18264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He T., Stepulak A., Holmstrom T.H., Omary M.B., Eriksson J.E. The intermediate filament protein keratin 8 is a novel cytoplasmic substrate for c-Jun N-terminal kinase. J. Biol. Chem. 2002;277:10767–10774. doi: 10.1074/jbc.M111436200. doi:10.1074/jbc.M111436200. [DOI] [PubMed] [Google Scholar]
- 38.Shahbazian D., Parsyan A., Petroulakis E., Topisirovic I., Martineau Y., Gibbs B.F., Svitkin Y., Sonenberg N. Control of cell survival and proliferation by mammalian eukaryotic initiation factor 4B. Mol. Cell Biol. 2010;30:1478–1485. doi: 10.1128/MCB.01218-09. doi:10.1128/MCB.01218-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraft P., Haiman C.A. GWAS identifies a common breast cancer risk allele among BRCA1 carriers. Nat. Genet. 2010;42:819–820. doi: 10.1038/ng1010-819. doi:10.1038/ng1010-819. [DOI] [PubMed] [Google Scholar]
- 40.de Bakker P.I., Ferreira M.A., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. doi:10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J., Zheng S.L., Isaacs S.D., Wiley K.E., Wiklund F., Sun J., Kader A.K., Li G., Purcell L.D., Kim S.T., et al. Inherited genetic variant predisposes to aggressive but not indolent prostate cancer. Proc. Natl Acad. Sci. USA. 2010;107:2136–2140. doi: 10.1073/pnas.0914061107. doi:10.1073/pnas.0914061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.