Abstract
Background
Open-source clinical natural-language-processing (NLP) systems have lowered the barrier to the development of effective clinical document classification systems. Clinical natural-language-processing systems annotate the syntax and semantics of clinical text; however, feature extraction and representation for document classification pose technical challenges.
Methods
The authors developed extensions to the clinical Text Analysis and Knowledge Extraction System (cTAKES) that simplify feature extraction, experimentation with various feature representations, and the development of both rule and machine-learning based document classifiers. The authors describe and evaluate their system, the Yale cTAKES Extensions (YTEX), on the classification of radiology reports that contain findings suggestive of hepatic decompensation.
Results and discussion
The F1-Score of the system for the retrieval of abdominal radiology reports was 96%, and was 79%, 91%, and 95% for the presence of liver masses, ascites, and varices, respectively. The authors released YTEX as open source, available at http://code.google.com/p/ytex.
Keywords: Hepatitis; visualization of data and knowledge; languages; computational methods; advanced algorithms; natural-language processing; distributed systems; agents; software engineering: architecture; developing and refining EHR data standards (including image standards); data models, data exchange; communication; integration across care settings (inter- and intra-enterprise); detecting disease outbreaks and biological threats; translational research—application of biological knowledge to clinical care; linking the genotype and phenotype; natural-language processing; delivering health information and knowledge to the public; monitoring the health of populations
Introduction
The rich clinical data stored in the electronic medical record are important to clinical-decision support, comparative effectiveness research, and epidemiological and clinical research studies.1 2 The electronic medical record stores much of the relevant information in the form of unstructured free text. Automated document classification and information-extraction techniques are the keys to accessing the clinical data locked in unstructured text.
Methods for automated document classification include rule-based and machine-learning techniques.3 4 In the rule-based approach, experts manually define classification rules. In the machine-learning approach, algorithms construct classifiers automatically using training data. Clinical natural language processing (NLP) systems annotate syntactic structure and semantic content within clinical text, and typically store annotations in a hierarchical data structure.5–7 In contrast, rule and machine-learning classifiers typically operate on ‘flat’ feature vectors. Converting between the hierarchical document representation output by NLP systems and the flat feature space required by classifiers is one of the most time-consuming and labor-intensive tasks in the development process, and one of the most important: the power of machine-learning algorithms depends on the construction of a feature representation that makes learning tractable.8 9 Our goal was to extend an open-source clinical NLP system to simplify feature extraction and the development of rule and machine-learning based document-classification systems.
Background
Historically, clinical NLP systems were built for specific healthcare systems, focused on specific goals, and involved a large implementation effort. Recently, open-source clinical NLP systems based on modular frameworks have become available, dramatically lowering the amount of resources and level of expertise needed to develop effective clinical document-classification systems. These include the clinical Text Analysis and Knowledge Extraction System (cTAKES), the Medical Knowledge Analysis Tool (MedKAT/P), Health Information Text Extraction (HITEx), and the Cancer Text Information Extraction System (caTIES).10–13 The open-source WEKA data-mining toolkit has been used in conjunction with these systems to develop machine-learning based text classifiers.12 14 15
These systems annotate syntactic structures such as sections, sentences, phrases (chunks), tokens (words), and their part-of-speech; perform named entity recognition and map spans of text to concepts from a controlled vocabulary or ontology; and identify the negation context of named entities. Different combinations of text annotations may be appropriate for different classification tasks, and finding the optimal feature representation is critical to classifier development.16–18
The application motivating this study was the automation of document classification in the Veterans Aging Cohort Study (VACS), an ongoing, prospective cohort study that follows HIV-infected and demographically similar HIV-uninfected veterans receiving medical care at eight Veterans Health Administration (VHA) facilities.19 Central to many VACS projects are medical chart reviews, the objective of which is to extract a well-defined set of information from a specific subset of medical records. Automated document classification can facilitate this process by identifying the reports relevant to the chart review. We sought to deploy and extend an open-source clinical NLP system and develop a methodology for the rapid implementation of document classifiers to facilitate VACS chart reviews.
We selected the cTAKES, a comprehensive clinical NLP system based on the Unstructured Information Management Architecture (UIMA) that has previously been deployed within the VHA.5 13 17 The cTAKES stores annotations in the UIMA Common Analysis Structure (CAS), a structured object graph superimposed over the unstructured document text. One method for extracting features is the development of a CAS Consumer, a custom software component that accesses the CAS and exports annotations in a user-specified format. However, it is impractical to write software for each set of desired features. The Mayo Weka/UIMA Integration (MAWUI) library provides tools for exporting data from applications based on UIMA for use with the Weka machine-learning environment.15 MAWUI requires the implementation of custom software components by the user to extract features, and thus suffers from the same limitations as the CAS Consumer. Another alternative for the extraction of UIMA annotations is the Common Feature Extraction System (CFE), which enables the declarative extraction of data from the CAS using an XML-based Feature Extraction Specification Language.20 One limitation of the CFE is that it cannot perform aggregate calculations on document features; for example, the CFE cannot output the number of times each term occurs within a document, a commonly used feature in document classification.21 The Automated Retrieval Console (ARC) is a clinical document classification system based on cTAKES.17 The ARC is designed to enable end users with little knowledge of NLP or machine learning to develop document classifiers; it does this by training machine-learning algorithms on various combinations of cTAKES annotations to classify documents. The ARC does not support functionality that more advanced users require, such as rule-based classifier development, manual feature selection, and customizable feature representation. A further limitation is that the gold standard document corpus must be curated within the ARC user interface; this is incompatible with the typical chart review process, which requires the review of thousands of notes and the storage of chart abstraction data in a format amenable to subsequent analyses. A limitation of both the CFE and ARC is their inability to integrate other structured data sources such as administrative data, pharmacy, and laboratory values with the document representation.
To simplify feature extraction and classifier development, we extended cTAKES to store document annotations in a relational database. This approach enabled seamless integration with other structured data sources stored in relational databases; enabled the development of rule-based classifiers using the structured query language (SQL); and enabled the declarative extraction of document annotations for use with WEKA, simplifying the development of machine-learning based classifiers.
Design objectives
We evaluated the ease of document classifier development by applying our system to a document corpus derived from a chart review to screen for and confirm cases of hepatic decompensation in VACS.22 As part of this chart review, trained abstractors reviewed over 13 000 radiology reports from over 395 patients; fewer than 400 reports asserted the presence of a clinical condition indicative of hepatic decompensation. Our objective for this use case was to quickly develop classifiers that accurately identified these reports, thereby dramatically reducing the effort involved in future screens of hepatic decompensation in the VACS study. Abstractors would still have to review the automatically identified reports to extract needed information; therefore, our goal was to develop document classifiers with high recall (sensitivity)—greater than 90%—and acceptable precision (positive predictive value)—greater than 80%.
Methods
We extended the cTAKES pipeline to improve NLP capabilities, simplify feature extraction, and facilitate document classifier development. We constructed a gold standard document corpus of radiology reports suggestive of hepatic decompensation. We then applied the system as follows: (1) developed rule-based classifiers, (2) performed system tuning in which we iteratively improved document annotation by modifying the system configuration, and (3) evaluated machine-learning algorithms for document classification.
YTEX pipeline
The cTAKES is a modular pipeline of annotators that combines rule-based and machine-learning techniques to annotate syntactic constructs, named entities, and their negation context in clinical text. cTAKES uses the OpenNLP Maximum Entropy package for sentence detection, tokenizing, part-of-speech tagging, and chunking; uses the SPECIALIST lexical variant generator for stemming; and uses an algorithm based on NegEx for negation detection.23–25 The cTAKES DictionaryLookup module performs named entity recognition by matching spans of text to entries from a dictionary. We used the cTAKES distribution included with ARC, which is distributed with Unified Medical Language System (UMLS) database tables for use with the DictionaryLookup module.26 The UMLS Metathesaurus unifies over 100 source vocabularies and assigns each term a concept unique identifier (CUI).
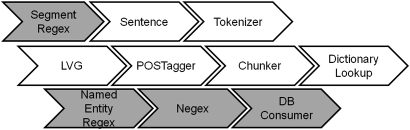
We modified cTAKES as follows: we developed regular-expression-based named entity recognition and section detection annotators (NamedEntityRegex and SegmentRegex); we adapted the latest version of the NegEx algorithm to cTAKES for negation detection (Negex); and we developed a module to store annotations in a relational database (DBConsumer; see figure 1). The annotators we developed are highly configurable; refer to the online appendix for a detailed description of all modifications to the cTAKES pipeline and configurations used in this study.
Figure 1.
Yale clinical Text Analysis and Knowledge Extraction System Extensions (YTEX) pipeline. New annotators developed as part of this study are shaded in gray. DB, database.
cTAKES can annotate demarcated sections from documents that conform to the Clinical Document Architecture format, which is not used in the VHA. To identify document sections, we developed an annotator that identifies section headings and boundaries based on regular expressions.
The DictionaryLookup algorithm performs named entity recognition by matching spans of document text to word sequences from a dictionary. Some clinical concepts are too complex, have too many lexical variants, or consist of non-contiguous tokens, making them difficult to represent in a simple dictionary. To address this issue, we developed an annotator that uses regular expressions to identify such concepts.
The cTAKES negation-detection algorithm is based on an older version of the NegEx algorithm and has limited support for long-range detection and post-negation triggers. To address these issues, we replaced the cTAKES negation-detection algorithm with an annotator based on the latest version of the Java General NegEx package, which supports long-range detection and post-negation triggers.27
In order to efficiently extract different feature sets from documents annotated with cTAKES, we developed a module that stores cTAKES annotations in a relational database. UIMA annotations are limited in complexity and obey a strict class hierarchy. These restrictions on the structure of UIMA annotations facilitate a high-fidelity relational representation. We used an object-relational mapping tool (Hibernate) to map UIMA annotations to relational database tables using a table-per-subclass strategy; refer to the online appendix for a detailed description of the data model.28 YTEX supports SQL Server, Oracle, and MySQL databases. The effort involved in mapping new or modified annotations to the database is minimal, making this approach applicable to any UIMA annotation.
Storing annotations in a relational database greatly simplifies the development of rule-based classifiers: document feature vectors can be retrieved using SQL queries, and rules can be implemented using SQL ‘case’ statements.
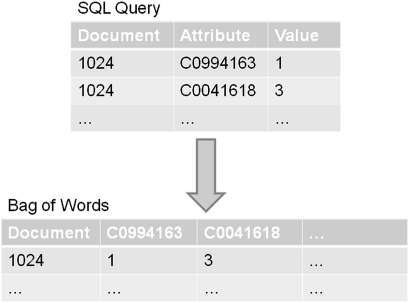
Machine-learning document-classification techniques often employ the ‘bag-of-words’ or ‘term-document matrix’ representation of documents.21 In this representation, documents occupy a feature space with one dimension for each word or term; words may be a word from a natural language or may be a technical identifier. The value of each dimension is typically either an indicator, asserting the presence of the word in the document, or a numeric value, indicating the term frequency. This feature space is typically high-dimensional and sparse, that is, the feature vectors mostly contain zeros. Most statistical packages support specialized file formats for efficient handling and exchange of sparse data sets. To use the bags-of-words document representation with WEKA, we developed a tool for exporting annotations obtained via SQL queries in the WEKA sparse file format. The tool takes as a parameter an SQL query that retrieves instance id, attribute name, and attribute value triples; it executes the query and rotates rows into columns to produce a sparse matrix representation of the data (figure 2). This transformation is similar to the SQL ‘pivot’ operator but differs in that it can create a matrix with an arbitrary number of columns.
Figure 2.
Bag-of-Words Exporter pivots instance id, attribute name, attribute value triples into a sparse matrix.
The generic nature of the tool allows classification on any unit of text: the instance id can refer to a document, sentence, or phrase. The attribute name represents a dimension—for example, a stemmed word or concept identifier; and the attribute value may be numeric or categorical. The tool enables the integration of other relational data sources with document annotation data—for example, administrative, pharmacy, or laboratory data. Refer to the online appendix for sample SQL statements used to export document annotations and administrative data for use with WEKA.
Reference standard document corpus construction
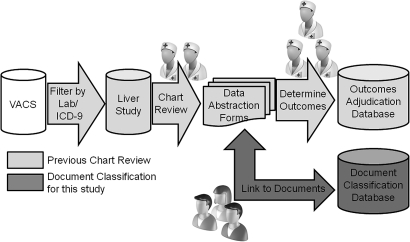
To develop a gold-standard classification of radiographic findings indicative of hepatic decompensation, we used the results of a chart review designed to screen for and confirm cases of hepatic decompensation in the Veterans Aging Cohort Study (VACS) (figure 3). For the chart review, subjects enrolled in VACS were screened for radiographic findings of hepatic decompensation at enrollment by evaluating for suggestive International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) diagnostic codes, and laboratory abnormalities up to 1 year before through 6 months after entry into the cohort to identify possible prevalent cases. Additionally, a random sample of 100 patients who did not screen positive by the above criteria was selected to ensure the absence of hepatic decompensation events. Two trained data abstractors reviewed reports of abdominal ultrasounds, abdominal CT scans, and MRI studies, and recorded onto structured data-collection forms the following information: presence and quantity of ascites (fluid within the peritoneal cavity); presence and location of varices (dilated veins within the esophagus and stomach), and presence, number, and dimensions of liver masses. Two endpoint adjudicators with expertise in chronic liver diseases reviewed data forms and determined whether these outcomes of interest (ie, ascites, varices, liver masses) were present or absent. Disagreement on classification of the finding resulted in a review by a third reviewer to adjudicate the outcome. All findings were recorded in an electronic ‘adjudication database.’
Figure 3.
Development of the gold standard. ICD, International Classification of Diseases, Ninth Revision; VACS, Veterans Aging Cohort Study.
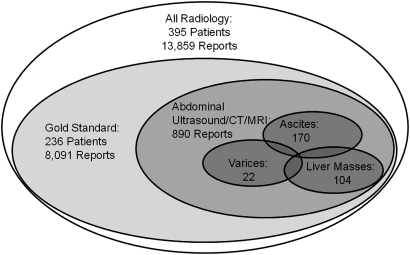
As part of this study, we randomly selected the data-abstraction forms of 236 patients with ICD-9-CM diagnostic codes and/or laboratory abnormalities suggestive of hepatic decompensation and transcribed them to a database. We then linked the abstraction data to the original radiology reports, and defined a gold-standard classification of radiology reports. We labeled radiology reports included in the chart review ‘abdominal radiology reports.’ We assigned additional class labels to these reports indicating the presence of ascites, varices, and/or liver masses based on the data-abstraction forms (figure 4).
Figure 4.
Dimensions of the document corpus.
Rule-based classifier development
We initially classified documents using manually developed rules. These interpretable classifiers allowed us to explore the feature space, optimize feature representations, and understand and rectify NLP errors that caused misclassification. We implemented the rules as SQL case statements, operating on feature vectors retrieved via SQL queries. For example, to identify radiology reports that assert the presence of varices, we focused on named entity annotations that contain CUIs related to varices, and represented documents as vectors with a column for each concept. Refer to the online appendix for sample SQL statements and a list of features we used in classification rules.
The ability to filter, aggregate, and transform document annotations using SQL queries allowed us to easily experiment with different representations of document concepts and their semantic and syntactic context. We found the following feature selection and representation approaches effective: filtering out concepts located within certain document sections; representing the negation status of concepts using a ‘relative negation count’; combining different concepts in a single feature; and using within-sentence concept co-occurrence.
The document section to which a term belongs is an important feature for document classification: for the discrimination of abdominal radiology reports from other radiology reports, terms in the title had far more importance than terms in the document body. For the identification of documents that assert the presence of a clinical condition (ie, ascites, varices, or liver masses), we found that filtering out terms from the clinical history section of documents improved classifier performance.
We combined distinct UMLS concepts under a single feature, thereby reducing the number of features needed and simplifying rule development. For example, the distinct UMLS concepts ‘Ascites’ (C0003962), ‘Peritoneal Fluid’ (C0003964), and intra-abdominal collection (C0401020) could for the purposes of this classification task be grouped under a single feature ‘Ascites.’
For the identification of liver masses, within-sentence co-occurrence was an important feature. For example, the sentence ‘A rounded, echogenic focus is seen in the left lobe of the liver’ contains the terms ‘echogenic focus’ and ‘liver’. We used co-occurrence of these terms within a sentence as a simple heuristic to infer the presence of a liver mass. Knowing that both these terms are in the same document is insufficient to infer the presence of a liver mass.
Concepts can be negated and affirmed within the same document as a result of errors in the negation detection algorithm, or due to deeper semantic content; exclusively considering affirmed or negated terms obscures this information. To address this issue, we represented the negation context of concepts using a ‘relative affirmation count’: the number of times a concept was affirmed minus the number of times it was negated within a document.
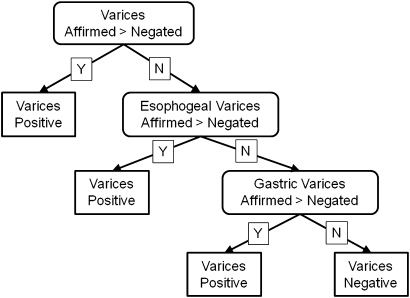
For example, the rule for the classification of varices compares the number of affirmed varices terms to the number of negated varices terms outside of the ‘Clinical History’ section of the document (figure 5). If any particular varices term is affirmed more than negated, the document is classified as ‘varices positive.’ Refer to the online appendix for a description of other rule-based classifiers.
Figure 5.
Varices classification rule. If any one of the varices terms is affirmed more than it is negated, the tree assigns the document the class label ‘varices positive.’
System tuning
To improve classifier performance, we performed multiple iterations of system tuning: (1) we generated document annotations with YTEX; (2) we classified documents using rule-based classifiers; (3) we manually examined all misclassified documents, and modified rules to resolve misclassification errors where necessary; (4) we reconfigured YTEX to rectify NLP errors; (5) we forwarded incorrectly labeled radiology reports to endpoints arbitrators (VLR and ZD-S) who reviewed these documents and updated document labels and patient adjudication databases.
We found that many classification errors were due to problems in named entity recognition (NER) and negation detection. To address these issues, we reconfigured the NER and negation-detection modules: we added entries to the dictionary used by the DictionaryLookup module; we configured regular expressions for use with the NamedEntityRegex module; and we modified the list of negation triggers used by the NegEx module. Refer to the online appendix for a detailed description of the regular expressions, dictionary entries, and negation triggers used for this study.
Upon evaluation of misclassified documents, we noticed that lexical variants of clinical concepts needed for classification were not included in the UMLS. For example, ultrasounds were often denoted with the term ‘echogram,’ which is not contained in the UMLS. We added additional entries to the YTEX UMLS dictionary to identify these concepts.
Some clinical concepts consisted of non-contiguous tokens, making them difficult to capture in a dictionary. For example, the following phrases were used to note the presence of ascites in radiology reports: ‘fluid is noted in the subhepatic area’, ‘free fluid around the liver is noted’, or ‘free fluid in the perihepatic region’. In these examples, the term ‘fluid’ is separated from the term ‘liver’ or ‘hepatic’ by several variable words. We configured regular expressions to identify these concepts.
Evaluation of machine-learning algorithms
Although the accuracy of the rule-based classifiers was satisfactory, we explored whether machine-learning algorithms could improve classification accuracy by using additional features that we overlooked, or features that could not easily be used in simple rule-based classifiers. For example, radiology reports that asserted the presence of hepatocellular carcinoma often asserted the presence of liver masses; machine-learning algorithms may leverage such associations to improve classification accuracy. We trained and evaluated the following machine-learning algorithms: decision trees (C4.5 algorithm), machine-learning analogs of rule-based classifiers29 30; random forests, ensembles of decision trees31; and SVMs, which have been successfully applied in document classification.3 32–34
To test whether system tuning and feature representation improved classifier performance, we evaluated classifiers against different representations of the document corpus:
baseline: this dataset represents the annotations generated by the un-tuned pipeline;
simple: this dataset employs a bag of affirmed terms document representation, which ignores document section and negated terms;
rich: this dataset uses the rich document feature representation that leverages the syntactic and negation context of named entities as described above.
We exported the document corpus in the WEKA sparse file format, split the corpus into a training set and a held-out test set, performed cross-validation on the training set, selected the optimal algorithm, and performed a final evaluation of classifier accuracy against the held-out test set. We used the cross-validation results to estimate classifier accuracy for varices, as we did not have enough reports for a held-out test set. Refer to the online appendix for a detailed description of the different corpus representations and machine learning process.
The datasets we exported had over 4000 features. For feature selection, we ranked features from the training set by mutual information and evaluated classifier performance using a 4‐fold cross-validation on the top n features, with n varying between 1 and 500. Accuracy peaked with fewer than 500 features for all classification tasks. We then performed a 4‐fold cross-validation 25 times with the optimal algorithm and number of features on the training set to generate empirical distributions for the information retrieval metrics specificity, precision, recall, and F1-Score with which we assess classifier performance. These are defined as follows35:
specificity: TN/(TN+FP);
precision (positive predictive value): P=TP/(TP+FP);
recall (sensitivity): R=TP/(TP+FN);
F1-score: (2*P*R)/(P + R);
TP: true positives (classified as positive when in fact positive);
FP: false positives (classified as positive when in fact negative);
TN: true negatives (classified as negative when in fact negative);
FN: false negatives (classified as negative when in fact positive).
Results
Cross-validation results
Classifiers trained on the tuned dataset that employed the rich feature representation performed significantly better than classifiers trained on the untuned dataset, and the dataset based on a simple feature representation (table 1). An exception was varices, in which classifiers trained on the simple feature representation performed best; this difference was however not statistically significant (p=0.0157). These results show that tuning NER, negation detection, and optimizing the feature representation significantly improved classifier performance.
Table 1.
Classifier parameters, information retrieval scores, and probability mean of the simple/baseline F1-score, and how they differ from rich F1-scores
| Task | Dataset | Classifier | Features | Specificity | Precision | Recall | F1-score | p Value |
| Abdominal | Rich | Tree | 10 | 0.997 | 0.996 | 0.997 | 0.997 | |
| Radiology | Baseline | Svm | 75 | 0.994 | 0.960 | 0.928 | 0.944* | 0 |
| Reports | Simple | Svm | 30 | 0.991 | 0.938 | 0.939 | 0.938* | 0 |
| Ascites | Rich | Tree | 1 | 0.983 | 0.928 | 0.939 | 0.932 | |
| Baseline | Tree | 1 | 0.976 | 0.895 | 0.893 | 0.893* | 0 | |
| Simple | Tree | 1 | 0.962 | 0.855 | 0.989 | 0.916* | 0.0047 | |
| Liver masses | Rich | Tree | 2 | 0.979 | 0.830 | 0.781 | 0.800 | |
| Baseline | Tree | 8 | 0.975 | 0.782 | 0.696 | 0.728* | 0 | |
| Simple | Random forest | 125 | 0.970 | 0.710 | 0.528 | 0.595* | 0 | |
| Varices | Rich | Tree | 1 | 0.995 | 0.894 | 0.94 | 0.911 | |
| Baseline | Svm | 4 | 0.992 | 0.863 | 0.905 | 0.871* | 0.0033 | |
| Simple | Tree | 1 | 0.995 | 0.904 | 1.000 | 0.946 | 0.0157 |
Significant at 0.01 level.
On the rich data set, classifiers achieved optimum performance with only the features used by our rule-based classifiers; additional features did not add predictive power to the classifier. The decision trees ‘learned’ from the rich dataset were similar or identical to the rule-based classifiers. Because of their similarity to machine-learned trees, we did not explicitly evaluate the performance of the rule-based classifiers.
On the rich dataset, simple decision trees using few features achieved optimal performance. For the identification of abdominal radiology reports and liver masses, classifiers trained on other datasets required more features and the more complex random forest and SVM algorithms to attain optimal performance.
Performance on test set
For each classification task, we selected the best classifier and evaluated it against the held-out test set (table 2).
Table 2.
Classifier performance on test set
| Classification task | Specificity | Precision | Recall | F1-score |
| Abdominal radiology reports | 0.997 | 0.950 | 0.976 | 0.963 |
| Ascites | 0.981 | 0.896 | 0.915 | 0.905 |
| Liver masses | 0.982 | 0.735 | 0.862 | 0.794 |
Discussion
We achieved the goal of rapidly developing accurate document classifiers to facilitate the hepatic decompensation chart review. The extensions we developed greatly simplified the development of rule and machine-learning based document classifiers, allowing us to complete classifier development in a total of five man days. This included developing rule-based classifiers, tuning the NLP system, and training and evaluating machine-learning classifiers. The system we developed classified radiology reports from eight VHA sites with high accuracy; we were able to meet the goals of >90% recall and >80% precision for the classification of abdominal radiology reports and varices; the system, however, failed to meet these goals for the classification of reports that assert the presence of liver masses. To achieve these results for this use case, we tuned named entity recognition and negation detection, and explored various feature combinations. Interpretable rule-based classifiers simplified tuning the NLP pipeline and exploring feature representations.
Through an examination of misclassified documents, we recognized issues where further work is required. Common causes of misclassification included lack of temporal context, lack of location context, and lack of pronominal anaphora or co-reference resolution; refer to the online appendix for a detailed discussion and examples of misclassification. These issues are the focus of active research in the clinical NLP field36–41; we will address these issues in future work.
Tuning named entity recognition (NER) and negation detection significantly improved classifier performance: this is demonstrated by the relative performance of classifiers evaluated on annotations derived from the tuned and untuned pipelines (rich vs baseline). We improved NER by adding lexical variants of clinical concepts to the dictionary, and by using regular expressions to identify clinical concepts. We used the NegEx algorithm for negation detection, and updated the default negation triggers. The resulting system was optimized for classification tasks specific to this study. Evaluations of cTAKES estimate the F1-score of NER to be 0.82413; thus, in general, it may be necessary to tune NER for specific classification tasks. Our tuning approach is generally applicable, and can be used to optimize NER and negation detection for other problem domains.
Choosing an optimal feature representation significantly improved classification performance. Text classification systems have used combinations of words, phrases, word sense, UMLS/SNOMED concepts, and others; no single feature representation is optimal for all document classification tasks.16–18 42 Finding the optimal representation for a given classification task requires exploration and experimentation with multiple feature representations. Domain knowledge can be incorporated in the feature representation via feature selection, and by combining multiple features to create new variables.42 We simplified this process by storing document annotations in a relational database, allowing us to efficiently explore the feature space and optimize the feature representation.
Storing annotations in a relational database also greatly simplified the development of both rule and machine-learning based classifiers. The relational representation inherently supports the rule-based document classification approach: we implemented classification rules as SQL case statements, operating on feature vectors retrieved via SQL queries. To support machine-learning approaches, we developed highly configurable tools to extract document features from the database in a bag-of-words representation for use with the WEKA toolkit.
We applied the lessons learned from this study to other chart reviews within VACS. The most laborious step of this study was the construction of a gold standard document corpus: this required the transcription of chart abstraction data from paper forms to a database, and linking these data to specific radiology reports. In general, the information captured as part of medical chart reviews is insufficient to construct a gold-standard corpus: the structured data produced by chart reviews typically synthesizes findings from multiple notes in the patient chart. However, in order to automate document classification and information extraction, notes must be linked to the information extracted from them. To address this issue, we have developed databases that integrate the chart-review data-abstraction process with manual document annotation, yielding gold standard corpora that we can use to develop document classifiers. We have developed databases for chart reviews to confirm cases of cancer from pathology, progress, and radiology notes; to study cases of community acquired pneumonia from microbiology, radiology, and progress notes; and to identify homeless veterans from progress notes. Future work will focus on developing document classifiers to assist these studies.
Conclusions
cTAKES is a comprehensive clinical NLP system that serves as a foundation for the development of clinical document classification and information-extraction systems. The Yale cTAKES Extensions (YTEX) simplify feature extraction and the development of rule and machine-learning based classifiers. We have released YTEX as open source.43
We used these tools to develop document classifiers that identify radiology reports with findings suggestive of hepatic decompensation. YTEX enabled us to efficiently explore the feature space; create a feature representation that leverages domain knowledge, the syntactic structure of the documents, and the negation context of concepts; and quickly develop rule and machine-learning based classifiers. In the future, we will apply these tools to identify reports relevant to medical chart reviews performed as part of other VACS studies.
Footnotes
Funding: Yale School of Medicine (VG). VA grant HIR 08-374 HSR&D: Consortium for Health Informatics (CB, MS). VA Office, Academic Affiliations, Information Research & Development (Medical Informatics Fellowship Program) (JW, CB, AJ). National Institute on Alcohol Abuse and Alcoholism (U10 AA 13566) (AJ, PI; CB, FK). National Institute of Allergy and Infectious Diseases (K01 AI 070001; VLR).
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Bates DW, Kuperman GJ, Wang S, et al. Ten commandments for effective clinical decision support: making the practice of evidence-based medicine a reality. J Am Med Inform Assoc 2003;10:523–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Justice AC, Erdos J, Brandt C, et al. The veterans affairs healthcare system: a unique laboratory for observational and interventional research. Med Care 2006;44(8 Suppl 2):S7–12 [DOI] [PubMed] [Google Scholar]
- 3.Joachims T. Text Categorization With Support Vector Machines: Learning With Many Relevant Features. Computer Science Department of the University of Dortmund;1998, Research Reports of the unit no. VIII (AI), Report 23 137–42 [Google Scholar]
- 4.Hayes PJ, Weinstein SP. CONSTRUE/TIS: a system for content-based indexing of a database of news stories. Proceedings of the The Second Conference on Innovative Applications of Artificial Intelligence. AAAI Press, 1991:49–64 [Google Scholar]
- 5.UIMA. http://uima.apache.org/
- 6.Cunningham H, Maynard D, Bontcheva K, et al. An architecture for development of robust HLT applications. 40th Annual Meeting of the Association for Computational Linguistics, Proceedings of the Conference. 2002:168–75 [Google Scholar]
- 7.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001:17–21 [PMC free article] [PubMed] [Google Scholar]
- 8.Langley P, Simon HA. Applicatons of machine learning and rule induction. Comm ACM 1995;38:55–64 [Google Scholar]
- 9.Wilcox A. Automated Classification of Medical Text Reports. PhD thesis. New York: Columbia University, 2000 [Google Scholar]
- 10.Crowley RS, Castine M, Mitchell K, et al. caTIES: a grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research. J Am Med Inform Assoc 2010;17:253–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. J Biomed Inform 2009;42:937–49 [DOI] [PubMed] [Google Scholar]
- 12.Zeng QT, Goryachev S, Weiss S, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural-language-processing system. BMC Med Inform Decis Mak 2006;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mark H, Eibe F, Geoffrey H, et al. The WEKA data mining software: an update. SIGKDD Explor 2009;11:8 [Google Scholar]
- 15.Savova GK, Ogren PV, Duffy PH, et al. Mayo clinic NLP system for patient smoking status identification. J Am Med Inform Assoc 2008;15:25–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conway M, Doan S, Kawazoe A, et al. Classifying disease outbreak reports using n-grams and semantic features. Int J Med Inform 2009;78:E47–58 [DOI] [PubMed] [Google Scholar]
- 17.D'Avolio LW, Nguyen TM, Farwell WR, et al. Evaluation of a generalizable approach to clinical information retrieval using the automated retrieval console (ARC). J Am Med Inform Assoc 2010;17:375–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilcox A, Hripcsak G. Medical text representations for inductive learning. Proc AMIA Symp 2000:923–7 [PMC free article] [PubMed] [Google Scholar]
- 19.Justice AC, Dombrowski E, Conigliaro J, et al. Veterans Aging Cohort Study (VACS): Overview and description. Med Care 2006;44(8 Suppl 2):S13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sominsky I, Coden A, Tanenblatt M. CFE—A System for Testing, Evaluation and Machine Learning of Uima Based Applications. Morocco: LREC, 2008 [Google Scholar]
- 21.Salton G, McGill M. Introduction to Modern Information Retrieval. New York: McGraw Hill, 1983 [Google Scholar]
- 22.Lo Re, III V, Bidwell GM, Lim JK, et al. A method to identify and confirm hepatic decompensation events in a Multicenter Cohort Study. Pharmacoepidemiology and Drug Safety; In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LVG. http://lexsrv3.nlm.nih.gov/LexSysGroup/Projects/lvg/2010/index.html
- 24.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 25.openNLP MaxEnt. http://maxent.sourceforge.net/
- 26.National Library of Medicine UMLS reference manual. http://www.ncbi.nlm.nih.gov/books/NBK9676/ (accessed 19 May 2011).
- 27.Solti I. Negex: Negation Identification for Clinical Conditions, 2009. http://code.google.com/p/negex/ [Google Scholar]
- 28.Hibernate Inheritance Mapping. http://docs.jboss.org/hibernate/core/3.3/reference/en/html/inheritance.html
- 29.Apte C, Damerau F, Weiss SM. Automated learning of decision rules for text categorization. ACM Trans Inform Syst 1994;12:233–51 [Google Scholar]
- 30.Quinlan JR. C4.5: Programs for Machine Learning. San Francisco: Morgan Kaufmann Publishers, 1993. ISBN: 9781558602380. [Google Scholar]
- 31.Breiman L. Random forests. Mach Learn 2001;45:5–32 [Google Scholar]
- 32.Yang H, Spasic I, Keane JA, et al. A text mining approach to the prediction of disease status from clinical discharge summaries. J Am Med Inform Assoc 2009;16:596–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patrick J, Li M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J Am Med Inform Assoc 2010;17:524–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dumais S, Platt J, Heckerman D, et al. Inductive learning algorithms and representations for text categorization. Proceedings Of The Seventh International Conference On Information And Knowledge Management. Bethesda, MD: ACM, 1998:148–55 [Google Scholar]
- 35.van Rijsbergen CJ. Information Retrieval. 2nd edn London: Butterworth, 1979 [Google Scholar]
- 36.Zhou L, Hripcsak G. Temporal reasoning with medical data—a review with emphasis on medical natural language processing. J Biomed Inform 2007;40:183–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasperin C, Briscoe T. Statistical Anaphora Resolution in Biomedical Texts. Manchester, UK: Coling 2008 Organizing Committee, 2008:257–64 [Google Scholar]
- 38.Mykowiecka A, Marciniak M, Kupść A. Rule-based information extraction from patients’ clinical data. J Biomed Inform 2009;42:923–36 [DOI] [PubMed] [Google Scholar]
- 39.Savova G, Bethard S, Styler W, et al. Towards temporal relation discovery from the clinical narrative. AMIA Annu Symp Proc 2009;2009:568–72 [PMC free article] [PubMed] [Google Scholar]
- 40.Hahn U, Romacker M, Schulz S. MedSynDiKATe—design considerations for an ontology-based medical text understanding system. Proc AMIA Symp 2000:330–4 [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts A, Gaizauskas R, Hepple M, et al. Building a semantically annotated corpus of clinical texts. J Biomed Inform 2009;42:950–66 [DOI] [PubMed] [Google Scholar]
- 42.Wilcox AB, Hripcsak G. The role of domain knowledge in automating medical text report classification. J Am Med Inform Assoc 2003;10:330–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garla V. YTEX: Yale cTAKES Extensions, 2010. http://code.google.com/p/ytex/ [DOI] [PMC free article] [PubMed] [Google Scholar]