Abstract
Objective
Prescription drugs can be associated with adverse effects (AEs) that are unrecognized despite evidence in the medical literature, as shown by rofecoxib's late recall in 2004. We assessed whether applying information mining to PubMed could reveal major drug–AE associations if articles testing whether drugs cause AEs are over-represented in the literature.
Design
MEDLINE citations published between 1949 and September 2009 were retrieved if they mentioned one of 38 drugs and one of 55 AEs. A statistical document classifier (using MeSH index terms) was constructed to remove irrelevant articles unlikely to test whether a drug caused an AE. The remaining relevant articles were analyzed using a disproportionality analysis that identified drug–AE associations (signals of disproportionate reporting) using step-up procedures developed to control the familywise type I error rate.
Measurements
Sensitivity and positive predictive value (PPV) for empirical drug–AE associations as judged against drug–AE associations subject to FDA warnings.
Results
In testing, the statistical document classifier identified relevant articles with 81% sensitivity and 87% PPV. Using data filtered by the statistical document classifier, base-case models showed 64.9% sensitivity and 42.4% PPV for detecting FDA warnings. Base-case models discovered 54% of all detected FDA warnings using literature published before warnings. For example, the rofecoxib–heart disease association was evident using literature published before 2002. Analyses incorporating literature mentioning AEs common to the drug class of interest yielded 71.4% sensitivity and 40.7% PPV.
Conclusions
Results from large-scale literature retrieval and analysis (literature mining) compared favorably with and could complement current drug safety methods.
Keywords: Data mining, drug safety, machine learning, statistical algorithm, biomedical literature, literature mining
Background
The U.S. Food and Drug Administration (FDA) requires pharmaceutical manufacturers to demonstrate that their products are efficacious in premarketing clinical trials, while not causing major adverse effects (or AEs—unintentional noxious effects or diseases caused by drugs taken at normal doses).1 However, this process is usually conducted in selected populations and typically lacks statistical power for detecting many AEs.2 To aid this process, researchers have analyzed large AE databases, including the FDA's Adverse Events Reporting System (AERS), using numerous statistical learning algorithms. Often, drug safety researchers assess drug–AE associations in three stages: screening of disproportionality analyses to detect those drugs that co-occur with particular diseases in a statistically significant fashion; initial assessment and investigation from a biological and clinical perspective; and in-depth investigation to confirm or reject signals using expert opinion, literature reviews, and other data from randomized controlled trials (RCTs) and epidemiologic studies.3 4 Agencies and researchers have had some success in using these methods to identify and regulate harmful drugs.5 However, the process has been imperfect, as illustrated by the late recall of rofecoxib in 2004, which was approved for marketing in 1999, but was unexpectedly shown to increase the risk of myocardial infarction in a clinical trial in 2004 and in later epidemiologic studies.6 7
Current use of literature reviews and expert opinion to discover or confirm drug–AE relationships4 5 8 implies that systematically analyzing the medical literature could complement AE discovery techniques and provide useful information to regulatory agencies at all stages of the investigative process detailed above. This additional input could help prioritize the tens of thousands of drug–AE associations under consideration by the FDA and other regulatory authorities. One approach might involve conducting systematic reviews using search strategies that have been developed for extracting articles relevant to drug safety,9 but using individual literature reviews to confirm or accept potentially millions of drug–AE hypotheses raises practical difficulties and increases the type I error rate. However, analyzing the literature within the disproportionality framework described above could overcome such challenges; we hypothesize that scientists will show greater interest in true drug–AE pairs as evidenced by published clinical trials, reviews, and studies of biological mechanisms. Supporting this hypothesis, biomedical researchers have used similar analyses of the medical literature to elucidate biological pathways.10 11 If true, disproportionality analyses similar to those used in analyzing AERS should reveal important drug–AE pairs for which there exist relatively more published reports when compared to other drugs and other AEs. However, this analysis faces two major challenges. First, a naive analysis might include many articles that do not test whether a drug causes a disease, including articles in which the drug treats a disease. Second, model performance needs to be judged against a plausible set of true positive and true negative results, because any predictive process is subject to excessive false positives or false negatives (or both).
We aimed to prototype a process for collecting and analyzing relevant literature while minimizing false positive and false negative drug–AE associations. We first improved the data collection process by excluding irrelevant articles using supervised statistical learning techniques that automatically filter citations using MEDLINE indexing terms.12 13 We then used statistically valid machine learning algorithms (from an emerging area of statistical analysis related to simultaneous inference) to identify significant drug–AE links from the several thousands of such pairs while controlling the overall probability of errors.14–18 Finally, we evaluated whether the literature mining process might have detected drug–AE associations that were the subjects of FDA warnings. We used the entire literature in some analyses and simulated prospective analyses by restricting the data to literature available prior to the warnings.
Methods
Data sources and document retrieval strategies
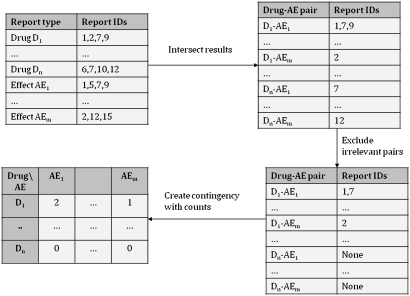
Our primary data were publicly available PubMed records from the National Library of Medicine (NLM). Although PubMed appears dissimilar to AERS, these records also contained drug–AE pairs: articles mentioning a drug could be associated with all diseases in the citation to create a set of drug–AE pairs. We tabulated these reports into a contingency table, in which each cell ij contained the total number of pairs containing Drugi and AEj2 5 (figure 1 illustrates the entire process for hypothetical data).
Figure 1.
Contingency table creation. This figure illustrates contingency table creation using hypothetical data. First, we retrieve all citations involving selected drugs and adverse effects (AEs). Drug–AE pairs are constructed by intersecting the different reports. We then use a statistical document filter to remove irrelevant pairs. Finally, the list of drug–AE pairs is tabulated into a table in which the cells contain the total number of appropriate drug–AE mentions.
We randomly selected a sample of widely used drugs (along with a positive control—rofecoxib) and obtained all PubMed articles mentioning the drug (119 026 articles on September 5, 2009). We obtained data on potential AEs from PubMed using a search strategy based on NLM's Medical Subject Headings (MeSH) index, which comprehensively describes human diseases within PubMed.19 We divided MeSH terms related to human diseases into categories based on human organ systems. For example, articles mentioning ‘eye’ or ‘eye diseases’ or ‘ocular physiology’ were classified under ‘eye diseases.’ The final strategy grouped 2461 terms into 55 categories; in addition, these terms implicitly encompassed lower-level terms in the MeSH hierarchical structure. Further granularity was possible, but many diseases formed logical groups and collapsing categories may reduce the false negative rate of disproportionality analyses.20 In addition, this classification scheme may measure scientific interest more accurately by combining clinical and basic laboratory research for similar topics. We assigned each article retrieved for drugs to one or more AE categories and tabulated counts for each drug–AE combination (figure 1). As described below, we adjusted these article counts by removing articles that did not test whether the drug caused the AE. The corrected count in each cell of the table was used to test the hypothesis of independence between the drug and the AE. We also used NLM's Structured Product Labels database to obtain each drug's major AE warnings and approved therapeutic indications.21
Using statistical document classification to exclude irrelevant articles
We reduced bias by limiting the input data to relevant articles that test whether a drug and an AE are causally related. Prior work suggested that up to 98% of search results are irrelevant to AEs22; including such articles would skew results toward false positive drug–AE linkages. For example, if an article described patients whose dyslipidemia was treated with simvastatin who subsequently developed rhabdomyolysis, an unfiltered search would have yielded one correct drug–AE pair (simvastatin–rhabdomyolysis) and one incorrect drug–AE pair (simvastatin–dyslipidemia). Researchers manually exclude irrelevant articles in systematic reviews, but this approach was not feasible on a large scale. A strategy retaining articles that contained several key terms (including ‘toxicity,’ ‘contraindications,’ and ‘poisoning’) had excellent sensitivity (98%) but 3% positive predictive value (PPV).22 To counter these difficulties, we developed two automated methods for excluding treatment indications and other irrelevant drug–disease pairs.
First, we excluded FDA-approved treatment indications noted in NLM's Structured Product Labels database.21 This step may have excluded instances where the therapy paradoxically increased the risk of its treatment target, such as hormone replacement therapy and ischemic heart disease.23 24 Of note, the hormone replacement therapy case was atypical because it was widely used to prevent ischemic heart disease prior to a definitive RCT being conducted. We assumed that pre-marketing clinical trials had sufficient power for determining efficacy. (We will relax this assumption in future work and use the statistical classifier below to eliminate purely therapeutic articles because we believe that literature mining could provide value by aggregating information on post-marketing studies conducted in broader populations that do not meet RCT inclusion criteria.)
Second, we adapted document classification methods from other biomedical domains and used suggestive MeSH indexing terms to exclude irrelevant articles.12 19 22 25 26 NLM has devoted a great deal of time to assigning MeSH terms and subheadings to the vast majority of PubMed articles based on reviews by multiple indexers.19 Retrieved PubMed articles contained readily available information for statistically modeling drug–AE relationships, despite the fact that drugs and AEs were indexed separately. For example, if an article included a disease index term modified by the subheading ‘chemically induced’ and a drug index term modified by the subheading ‘adverse effects,’ the article likely tested whether the drug caused that disease. As a result, MeSH indexing terms were used to exclude off-label treatment indications, incidental mentions, and instances in which the drug was used to treat the AE of another drug.
To implement a statistical document classifier, we first constructed training data using 678 randomly selected articles (which comprised 1599 drug–AE pairs). We determined the outcome variable (each drug–AE pair's relevance) by reviewing the associated article's title, abstract, and full text (if the relationship between drug and harm was unclear in the abstract); we created 54 independent variables related to how the drug and disease in question were described in associated MeSH terms and subheadings. We then modeled relevance as a function of these numerous independent variables (MeSH data). We used a shrinkage and selection method for regression called Lasso that minimized the usual sum of squared errors, with a bound on the sum of the absolute values of the coefficients.27–29 This method shrank coefficients of less important variables to zero, resulting in fewer independent variables with better predictive power. We developed and tested several Lasso-based supervised learning models for predicting relevance.28 The shrinkage factor was determined by a tenfold cross-validation of the entire training data. Using coefficients derived from the Lasso procedure, we considered drug–AE pairs with an estimated probability of relevance greater than 0.5 to be relevant, after testing revealed that this threshold minimizes the error rate. We verified the method by randomly selecting three-fourths of the coded articles to derive an empirical model, while calculating performance characteristics on the remaining coded test data (the remaining one-fourth of the data). We compared the predicted scores with the actual scores to obtain average performance characteristics for 20 simulations: sensitivity (% of relevant articles correctly predicted, also known as recall), PPV (% of predicted relevant articles that are actually relevant, also known as precision), and the error rate (% of articles that are correctly classified as relevant or irrelevant). Finally, we calculated predicted relevance scores for uncoded drug–AE pairs by applying coefficients derived using training data to extracted MeSH variables. We retained those articles tagged as relevant and created a contingency table using predicted counts for drug–AE article pairs (figure 1).
To test the robustness of the Lasso document classifier described above, we evaluated two additional statistical methods and two non-statistical filters. (Of note, as with the Lasso model, each of these models used data in which FDA-approved treatment indications were removed.) We developed one model that used standard (linear) logistic regression to model relevance as a function of the same MeSH-derived independent variables discussed in relation to Lasso. However, this method did not discard less important variables. Also, we applied the gradient-boosting method (GBM), a non-parametric tree-based prediction approach based on boosting.28 Boosting improved the classifier (in this case, classification trees) by optimally combining a sequence of classifiers, each of which were iteratively built to give more weight to the training observations that were misclassified in previous classifiers. We evaluated these models' performance using the same metrics given for the Lasso method. We also applied a non-statistical filter (adapted from earlier research),21 which classified articles as relevant if any of three terms (‘adverse effects,’ ‘chemically induced,’ and ‘toxicity’) were present anywhere in the article—regardless of whether the terms were linked to drugs or diseases of interest. Finally, we used minimally filtered data from which only FDA-approved treatment indications had been removed. We computed performance statistics for the non-statistical filters based on the entire sample because their development did not involve training procedures.
Statistical identification of drug–AE associations
Our statistical analysis compared the observed number of drug–AE literature citations with the expected count under the null hypothesis of independence of drugs and AEs, while controlling type I error rates. As in prior drug toxicity analyses and illustrated in figure 1, we constructed a contingency table of drugs versus AEs with n×m cells for the n drugs and m AEs. Each cell contained the count of predicted relevant citations mentioning both the drug and the AE. The observed count xi,j was the number of events observed for the (i,j)th cell. The expected count was Npi.p.j, where pi. and p.j denoted the marginal probabilities of observing the drug and the AE under the hypothesis of independence and N was the total count of AEs for the sample of drugs. Accordingly, the p value for the (i,j)th cell was P(X≥xi,j) where X was a Poisson random variable with mean Npi.p.j, which was calculated using standard Poisson or normal distributional approximations; this resulted in 2090 hypotheses and p values (from 38 drugs×55 possible AEs).
While we aimed to retrieve the greatest number of true signals of disproportionate reporting (SDRs—true drug–AE associations), we also needed to limit the number of false drug–AE associations. Using the common 5% level of significance (α), we expected to retrieve 105 SDRs (even in the absence of true SDRs) from 2090 hypotheses. However, we controlled overall error rates by adapting methods that have emerged under the label of Simultaneous Inference; these methods effectively analyzed a large number of genomics microarray data while controlling familywise type I error rates and false discovery rates.14–18 30 After ordering all p values from largest to smallest, step-down methods find the smallest p value violating or greater than the error measure threshold and reject all null hypotheses with smaller p values. Step-up procedures work in an opposite manner.15 17 Dalal and Mallows proved that valid step-up procedures exist and demonstrated the existence and monotonicity of a sequence of error measure thresholds. They computed this sequence such that the probability that the total count of false positive drug–AE pairs did not exceed some criterion k is greater than or equal to 1−α.14 We applied the above procedure with k=1 and α=0.01 to the drug–AE contingency table to obtain positive drug–AE associations.18
We visualized these analyses using a heat map of the p values; heat maps have been used successfully in diverse areas including genetic cluster analysis and clustering of graphical user interface elements.31 32
Evaluating empirical findings against a reference standard
In our base-case analysis, we compared positive drug–AE associations to a reference set of true associations obtained from the ‘Warnings’ section of Structured Product Labels and also from FDA Enforcement Reports; we excluded three of a total of 2090 drug–AE hypotheses whose labeling information was not definitive. Although a perfect reference set was unavailable in the absence of complete knowledge of various medications' biological effects, associations considered by drug manufacturers and regulatory authorities to be strong were a plausible proxy.4 21 33 True negatives and false positives were difficult to identify because purportedly safe drugs might be found to carry excess risk later. However, drug–AE associations that were absent from product labels from well-studied drugs that have been marketed for at least 6 years were considered to be plausible true negatives.
To assess the predictive validity over time, we tested drug–AE associations using only data available until a certain point in time. For example, rofecoxib would have been considered to have a true association with heart disease by the beginning of 2004 if the literature collected until the end of 2003 indicated this association. However, the relative paucity of literature in earlier years precluded precise measurement prior to 1990. Thus, we censored those results in which drug–AE associations became known to regulatory agencies by 1990 and were recognized by our algorithms using data available by 1990. However, if the drug–AE association was known prior to 1990, but our processes identified it using literature available after 1990, we counted this as a ‘delayed true positive.’ After censoring, we calculated the percentage of true SDRs that would have been detected prior to or concurrent with an FDA warning.
In the base-case analysis described above, we restricted our analysis to articles in which the exact drug was mentioned. However, drugs often share both benefits and AEs with members of the same class.34–36 In a secondary analysis, we incorporated these ‘class effects’ by revising our search methods to capture all articles from each drug class. For example, the base-case analysis captured only articles that explicitly mention atenolol, while the secondary analysis incorporated articles mentioning any member of its class (β-blockers). After excluding irrelevant drug–AE article mentions as before, we incorporated all class-based drug–AE article counts into a contingency table after weighting them appropriately. We empirically tested class weights (which have unknown importance) by varying the weight assigned to the class-based drug–AE article pairs from 0 (the base-case scenario without class effects) to 1 (where drugs assume the characteristics of their class). We selected the optimal weight by choosing the weight associated with the highest F measure (the harmonic mean of PPV and sensitivity with each weighted equally).37
We also conducted sensitivity analyses on the initial data-filtering Lasso step by examining whether data transformed by alternate document classifiers performed better at discovering positive and negative SDRs. We applied our standard SDR algorithm to the four contingency tables created using the alternate document classifiers (logistic, GBM, non-statistical filtering, and minimal filtering). We compared sensitivity and PPV for each alternate table to the base-case (Lasso) results. As in the primary analysis, we also calculated whether true drug–AE associations could be detected using only literature available at the time the FDA warning was issued. We further conducted sensitivity analyses on the statistically permissible false positive rate by varying k between 0 and 10 and α between 0.1 and 0.001. We conducted all analyses using the R statistical package v 2.9.
Results
Data characteristics and filtering
For the primary analysis, we retrieved 119 026 unique articles and 228 920 drug–disease mentions involving at least one of the included drugs (table 1). After removing FDA-approved therapeutic indications, we retained 123 587 drug–disease mentions. For the sensitivity analysis, we also retrieved 293 454 unique articles and 834 437 drug–disease mentions pertaining to at least one of the included drug classes; 460 498 remained after removing FDA-approved therapeutic indications.
Table 1.
Data characteristics
| Sample characteristic | n |
| Unique drugs | 38 |
| Unique diseases | 55 |
| Primary analysis | |
| Median publication year | 2000 |
| Publication year (range) | (1959–2009) |
| Unique articles | 119 026 |
| Drug article mentions | 155 655 |
| Total drug–disease mentions | 228 920 |
| Excluding primary treatment indications | 123 587 |
| Final, excludes all irrelevant drug–disease mentions | 15 565 |
| Including class effects (for sensitivity analyses): | |
| Median publication year | 1998 |
| Publication year (range) | (1949–2009) |
| Unique articles | 293 454 |
| Drug article mentions | 552 498 |
| Total drug–disease mentions | 834 437 |
| Excluding primary treatment indications | 460 498 |
| Final, excludes all irrelevant drug–disease mentions | 50 891 |
We tested the performance of several document classifiers in removing other irrelevant drug–disease mentions. (table 2). Lasso (10.2% error, 81.2% sensitivity, and 87% PPV) and GBM (10.2% error, 79.7% sensitivity, and 88.3% PPV) performed equally well, although Lasso had better sensitivity and GBM had better PPV. The logistic filter had the poorest error rate of the three statistical filters, but performed substantially better than the non-statistical and minimal filters. Based on Lasso's extensive record in the published literature and its good performance here, we used the Lasso-based method to remove irrelevant drug–disease mentions in base-case analyses. We retained the other filters for performing sensitivity analyses.
Table 2.
Comparison of document relevance filters on training data (N=1599 abstracts)
| Filter | Sensitivity (%) | PPV (%) | Error (%) |
| Minimal filtering | 100.0 | 32.9 | 67.1 |
| Non-statistical filtering | 95.6 | 46.1 | 38.3 |
| Logistic algorithm* | 82.6 | 79.4 | 12.6 |
| Lasso* | 81.2 | 87.0 | 10.2 |
| GBM* | 79.7 | 88.3 | 10.2 |
Results of 20 random trials on test data. Lasso corresponds to the base-case statistical model.
GBM, gradient-boosting method; PPV, positive predictive value.
After applying the Lasso document classifier, we converted 15 565 predicted relevant drug–AE article pairs (and 9133 unique articles) into a contingency table with 2087 cells; each cell contained the count of relevant articles mentioning a drug–AE hypothesis. After using the Lasso filtering process to exclude irrelevant class–disease mentions, we converted the remaining 50 891 class–AE mentions into contingency tables that are used in the disproportionality analyses below.
Identification of drug–AE associations using disproportionality analyses
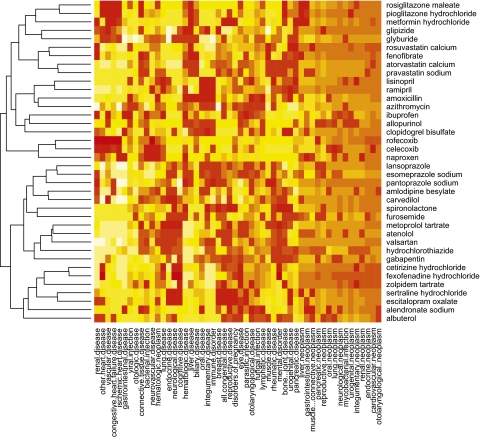
We used a heat map to visualize the p values for each cell where we clustered the drugs (harms) according to their similar p value profiles across harms (drugs) and represented cells with high and low p values along a spectrum of yellow to red (figure 2). These data were drawn from literature published until 2009 but did not incorporate class effects. The graphical representation of hierarchical clustering (a dendrogram) for drugs is shown on the left axis. Several notable features emerged. First, many drug classes were grouped together accurately using only their distribution in the AE literature. For example, rosiglitazone maleate and pioglitazone hydrochloride were known to be members of the same class of drugs but could also be grouped together using their distribution in the AE literature. We saw other appropriate grouping including the COX-2 inhibitors celecoxib and rofecoxib. Our successful grouping of chemicals using literature characteristics alone gives credence to our hypothesis and method. Second, the heat map demonstrated many associations, as seen in the numerous cells with lower p values. Many corresponded to known associations such as rosiglitazone maleate and congestive heart failure. The remainder of this section describes the results of our process for selecting the most significant associations while minimizing false positive associations.
Figure 2.
Heat map representation of contingency table. Each cell represents the probability of the observed count exceeding the expected count under the assumption of independence of adverse effects (AEs) and drugs. p Values were represented using a spectrum of low (red) to high (yellow). Drugs were clustered together on the basis of their similarity across harms. Harms were clustered together on the basis of their similarity across drugs.
We then calculated SDRs for the entire sample (table 3). The base-case scenario, which assumes that class effects were unimportant (class weight=0), revealed 64.9% sensitivity and 42.4% PPV, and a 4.6% overall error rate. Of the detected true positive associations, after censoring 13 drug–AE associations for which no determination can be made, 54% (20/37) were present using the literature available at the time of the safety alert.
Table 3.
Primary analysis
| Article weight* (%) | TP | FP | FN | TN | Sensitivity (%) | PPV (%) | Censored | Positive before FDA | Positive after FDA | F measure |
| 0† | 50 | 68 | 27 | 1942 | 64.9% | 42.4% | 13 | 20 | 17 | 51.3% |
| 10 | 51 | 70 | 26 | 1940 | 66.2% | 42.1% | 13 | 23 | 15 | 51.5% |
| 20 | 55 | 80 | 22 | 1930 | 71.4% | 40.7% | 15 | 24 | 16 | 51.9% |
| 30 | 57 | 94 | 20 | 1916 | 74.0% | 37.7% | 19 | 25 | 13 | 50.0% |
| 40 | 59 | 105 | 18 | 1905 | 76.6% | 36.0% | 19 | 25 | 15 | 49.0% |
| 50 | 58 | 121 | 19 | 1889 | 75.3% | 32.4% | 20 | 26 | 12 | 45.3% |
| 60 | 58 | 134 | 19 | 1876 | 75.3% | 30.2% | 22 | 26 | 10 | 43.1% |
| 70 | 59 | 146 | 18 | 1864 | 76.6% | 28.8% | 22 | 26 | 11 | 41.8% |
| 80 | 62 | 154 | 15 | 1856 | 80.5% | 28.7% | 22 | 26 | 14 | 42.3% |
| 90 | 62 | 165 | 15 | 1845 | 80.5% | 27.3% | 24 | 26 | 12 | 40.8% |
| 100 | 62 | 168 | 15 | 1842 | 80.5% | 27.0% | 24 | 26 | 12 | 40.4% |
There are 2087 potential AE associations in the sample, of which 77 were the subject of an FDA warning. Drug–AE associations are considered positive before the FDA if the association is detected using literature published before or concurrent with the FDA warning. A drug–AE association was considered positive after the FDA warning if the association is detected using literature published after the FDA warning. Positive drug–AE associations for which timing with respect to FDA warnings could not be definitively determined are censored. For all models, the number of statistically allowable false positives (k) and the significance threshold (α) are set to 1 and 0.01, respectively.
Weights to articles directly mentioning the drugs of interest are always 1. Weights for articles mentioning members of the same class as the drugs of interest are varied between 0 and 1.
Base-case model.
FDA, U.S. Food and Drug Administration warning; F measure, harmonic mean of PPV and sensitivity; FN, false negative; FP, false positive; TN, true negative; TP, true positive; PPV, positive predictive value.
Sensitivity generally rose and PPV fell as class effects were assigned greater weight and specific information about the drugs was given less importance (table 3). Selection of a specific weight depended on the trade-off between PPV and sensitivity. Using the F measure, the optimal choice corresponded to a 20% class weight (71.4% sensitivity, 40.7% PPV; see row 3 of table 3). Of true positive associations detected using this weighting scheme, and after censoring 15 drug–AE associations for which no determination could be made, 60% (24/40) were present using the literature available at the time of the safety alert. Notably, adding class effects increased the number of censored drugs by discovering several additional drug–AE associations using earlier literature.
We linked general cardiovascular diseases with rofecoxib (using literature available by 2001) and with celecoxib (using literature available by 2002). These results persisted with and without class effects. Then again, the FDA warned consumers in 2001 that rosiglitazone potentially exacerbated or unmasked congestive heart failure,38 but this association was only evident in base-case models using literature released prior to 2003.
Sensitivity analyses
We tested the robustness of the Lasso-derived document filter by comparing base-case results with those obtained using data derived from alternate filters (table 4). Using unfiltered data yielded slightly higher sensitivity (67.5% vs >60%) but substantially worse PPV (12.8% vs >40%) in all comparisons with statistical filters. All performed similarly in detecting drug–AE associations using literature published prior to the FDA warning. Overall, the three statistical filters performed similarly, suggesting that switching filters would not have improved the results. Of note, the base-case (Lasso) model that included class effects (table 3) had superior sensitivity and PPV when compared to the non-statistical and minimal filters in table 4. This suggests that the statistical relevance filtering did not remove valuable information, despite the great reduction in the number of included drug–AE article mentions (122 598 to less than 16 000 for all statistical filters).
Table 4.
Evaluating document classification strategies in detecting drug–AE associations
| Filter | Drug–AE article mentions | Sensitivity (%) | PPV (%) | Censored | Positive before FDA | Positive after FDA |
| Minimal filtering | 122 598 | 67.5% | 12.8% | 17 | 21 | 14 |
| Non-statistical filtering | 36 613 | 66.2% | 22.0% | 13 | 21 | 17 |
| Logistic algorithm | 15 372 | 66.2% | 42.9% | 13 | 20 | 18 |
| Lasso* | 15 505 | 64.9% | 42.4% | 13 | 20 | 17 |
| GBM | 14 145 | 62.3% | 41.4% | 13 | 20 | 15 |
Base-case document-filtering strategy.
AE, adverse effects; FDA, U.S. Food and Drug Administration warning; GBM, gradient-boosting method; PPV, positive predictive value.
In addition, key results (including the rofecoxib–cardiovascular disease association) persisted despite varying key model parameters k and α (the number of allowable false positives and the significance threshold, respectively) across wide ranges.
Discussion
Our methods for collecting, filtering, and analyzing the medical literature showed promise for detecting early SDRs among a large set of drug–AE hypotheses. We combined several recently developed approaches that improved PPV while maintaining reasonable sensitivity rates, including detecting the link between rofecoxib and cardiovascular diseases using literature published several years prior to rofecoxib's recall. Although researchers often referenced previously published literature to discover or confirm SDRs, prior work primarily analyzed AE databases, healthcare claims data, and clinical trials. Our results support the hypothesis that analyzing the medical literature within a disproportionality framework could supplement current methods for discovering drug–AE relationships.
Unfiltered literature was too statistically noisy to allow accurate signal detection.22 However, our use of a two-stage filter for removing treatment indications and other irrelevant articles limited the dataset to an enriched set of articles that tested whether drugs cause potential AEs. To our knowledge, this was the first validated filter for retrieving articles related to particular drug–AE pairs. This literature-based method discovered true drug–AE associations with greater than 70% sensitivity and 40% PPV. Furthermore, we detected numerous associations prior to FDA warning, suggesting that literature mining did not simply provide a lagging indicator of widely known drug–AE associations. These results persisted in several sensitivity analyses. Statistical learning tools using data such as AERS reported higher sensitivity rates for dozens of true positive drug–AE associations. However, these tools often detected thousands of additional drug–disease pairs of unclear importance, suggesting a lower PPV than a literature mining approach.5 39
In accord with several prior studies, we gauged performance against a gold standard of drug–AE associations that were presumed to be true. In the absence of complete knowledge of a medication's true biological effects, some expert judgment is always required. In contrast to other studies that chose to use alternate gold standard drug–AE associations,5 39 we chose to use FDA warnings for the set of drugs and AEs under consideration for several reasons. First, although imperfect, the FDA decision-making process requires review of prior clinical trials, biological evidence, data mining results, and expert judgment, along with substantial documentation.4 21 33 This certainly makes their decision-making process superior to our own judgment regarding historical drug–AE associations (although we believe that this work will aid their processes in the future). Second, prior gold standard lists did not overlap with our dataset of drug–AE associations (and these all required some expert judgment as well). Finally, FDA warnings appear to strongly influence drug sales and policy,40 making them a de facto standard in the US market, and a respected source elsewhere. Given these reasons, FDA warnings served as a plausible gold standard for this study and for future work. Our methods carried several limitations. First, we relied entirely on PubMed records, which had been regarded as insufficient for literature reviews.9 However, PubMed offered a large sample of available literature and was substantially more accessible than data sources such as EMBASE. Second, literature-based statistical learning methods may only duplicate AERS analyses and discovered drug–AE associations may already be under consideration. However, the early retrieval of rofecoxib toxicity suggested that literature-based methods might prove complementary. Furthermore, researchers currently reference the literature when evaluating statistical analyses of AE databases, RCTs, and epidemiologic data. Therefore, signals from the literature could improve regulatory decision-making by providing additional statistically robust information to all stages of the investigative process. Third, MEDLINE indexing typically requires several months, which delayed data retrieval. Fourth, we weighed all articles equally regardless of quality. It is possible that giving greater weight to high-quality meta-analyses41 and large RCTs41 would have changed the results. We plan to explore additional weighting schemes in future research. Fifth, only one author reviewed the training set, which may make estimates less generalizable. However, the final results were robust when judged against our reference set, despite any classification errors, suggesting that the problem was relatively small. Finally, our results may only pertain to the small group of included drugs. However, increasing the number of drugs and AEs in the contingency table will increase the number of drug–AE hypotheses; this may aid identification by increasing the relative signal strength of important drug–AE associations.
Drug safety has benefited from systematic reviews and from the application of statistical learning techniques to spontaneously generated AE data. We described an approach for identifying major drug–AE associations that combined large-scale literature analysis with statistical learning techniques. We dramatically improved PPV and sensitivity rates with respect to major known drug–AE associations by applying successive filters and incorporating drug class effects. Regulatory agencies and drug safety researchers may be able to use these techniques to improve decision-making about drug safety. In addition, we anticipate improving drug–AE detection by optimizing model parameters, by applying these methods to additional drugs and data sources (including unpublished literature), and by combining these signals with those obtained from standard spontaneously generated AE data.
Acknowledgments
The authors thank Dana Goldman, PhD (RAND Corporation and University of Southern California), Jay Bhattacharya, MD, PhD (Stanford University), David Madigan, PhD (Columbia University), Michael Lesk, PhD (Rutgers University), and Emmett Keeler, PhD (RAND Corporation) for reviewing earlier versions of this manuscript and providing helpful comments without compensation.
Footnotes
Funding: The RAND Corporation funded this work through an internal grant to Dr Dalal, and had no role in the study.
Competing interests: None.
Contributors: Both Drs Dalal and Shetty had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All significant contributors have been acknowledged.
Provenance and peer review: Not commissioned; internally peer reviewed.
References
- 1.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 1998;279:1200–5 [DOI] [PubMed] [Google Scholar]
- 2.Almenoff JS, Pattishall EN, Gibbs TG, et al. Novel statistical tools for monitoring the safety of marketed drugs. Clin Pharmacol Ther 2007;82:157–66 [DOI] [PubMed] [Google Scholar]
- 3.Hochberg AM, Hauben M. Time-to-signal comparison for drug safety data-mining algorithms vs. traditional signaling criteria. Clin Pharmacol Ther 2009;85:600–6 [DOI] [PubMed] [Google Scholar]
- 4.Hauben M, Noren GN. A decade of data mining and still counting. Drug Saf 2010;33:527–34 [DOI] [PubMed] [Google Scholar]
- 5.Almenoff J, Tonning JM, Gould AL, et al. Perspectives on the use of data mining in pharmaco-vigilance. Drug Saf 2005;28:981–1007 [DOI] [PubMed] [Google Scholar]
- 6.Levesque LE, Brophy JM, Zhang B. The risk for myocardial infarction with cyclooxygenase-2 inhibitors: a population study of elderly adults. Ann Intern Med 2005;142:481–9 [DOI] [PubMed] [Google Scholar]
- 7.Edwards IR. What are the real lessons from Vioxx? Drug Saf 2005;28:651–8 [DOI] [PubMed] [Google Scholar]
- 8.Kelly TN, Bazzano LA, Fonseca VA, et al. Systematic review: glucose control and cardiovascular disease in type 2 diabetes. Ann Intern Med 2009;151:394–403 [DOI] [PubMed] [Google Scholar]
- 9.Higgins JPT, Green S, eds. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. Available from www.cochrane-handbook.org [Google Scholar]
- 10.Aubry M, Monnier A, Chicault C, et al. Combining evidence, biomedical literature and statistical dependence: new insights for functional annotation of gene sets. BMC Bioinformatics 2006;7:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yetisgen-Yildiz M, Pratt W. Using statistical and knowledge-based approaches for literature-based discovery. J Biomed Inform 2006;39:600–11 [DOI] [PubMed] [Google Scholar]
- 12.Tanaka LY, Herskovic JR, Iyengar MS, et al. Sequential result refinement for searching the biomedical literature. J Biomed Inform 2009;42:678–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen AM, Hersh WR, Peterson K, et al. Reducing workload in systematic review preparation using automated citation classification. J Am Med Inform Assoc 2006;13:206–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalal S, Mallows C. Buying with exact confidence. Ann Appl Probab 1992;2:752–65 [Google Scholar]
- 15.Dalal S, Mallows C. Optimal stopping with exact confidence on remaining defects. Technometrics 2008;50:397–406 [Google Scholar]
- 16.Efron B. Large-scale simultaneous hypothesis testing: the choice of a null hypothesis. J Am Stat Assoc 2004;99:96–104 [Google Scholar]
- 17.Finner H, Roters M. Asymptotic comparison of step-down and step-up multiple test procedures based on exchangeable test statistics. Ann Stat 1998;26:505–24 [Google Scholar]
- 18.Lehmann E, Romano J. Generalizations of the familywise error rate. Ann Stat 2005;33:1138–54 [Google Scholar]
- 19.National Library of Medicine Introduction to MeSH—2011, 2009. http://www.nlm.nih.gov/mesh/introduction.html (accessed 12 Nov 2009).
- 20.Pearson RK, Hauben M, Goldsmith DI, et al. Influence of the MedDRA((R)) hierarchy on pharmacovigilance data mining results. Int J Med Inform 2009;78:e97–e103 [DOI] [PubMed] [Google Scholar]
- 21.NLM DailyMed, 2009. http://dailymed.nlm.nih.gov/dailymed/about.cfm (accessed 25 Sep 2009). [Google Scholar]
- 22.Golder S, Loke YK. Search strategies to identify information on adverse effects: a systematic review. J Med Libr Asssoc 2009;97:84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossouw JE, Anderson GL, Prentice RL, et al. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results from the women's health initiative randomized controlled trial. JAMA 2002;288:321–33 [DOI] [PubMed] [Google Scholar]
- 24.Shetty KD, Vogt WB, Bhattacharya J. Hormone replacement therapy and cardiovascular health in the United States. Med Care 2009;47:600–6 [DOI] [PubMed] [Google Scholar]
- 25.Malik R, Franke L, Siebes A. Combination of text-mining algorithms increases the performance. Bioinformatics 2006;22:2151–7 [DOI] [PubMed] [Google Scholar]
- 26.Chen ES, Hripcsak G, Xu H, et al. Automated acquisition of disease drug knowledge from biomedical and clinical documents: an initial study. J Am Med Inform Assoc 2008;15:87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tibshirani R. Regression shrinkage and selection by lasso. J Roy Stat Soc B 1994;58:267–88 [Google Scholar]
- 28.Genkin A, Lewis D, Madigan D. Large-scale bayesian logistic regression for text categorization. Technometrics 2007;49:291–304 [Google Scholar]
- 29.Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat 2001;29:1189–232 [Google Scholar]
- 30.Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol 2002;23:70–86 [DOI] [PubMed] [Google Scholar]
- 31.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 1998;95:14863–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Payne PR, Starren JB. Quantifying visual similarity in clinical iconic graphics. J Am Med Inform Assoc 2005;12:338–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.FDA USFood and drug administration enforcement reports. 2009. http://www.fda.gov/Safety/Recalls/EnforcementReports/default.htm (accessed 5 Sep 2009).
- 34.Christopher-Stine L. Statin myopathy: an update. Curr Opin Rheumatol 2006;18:647–53 [DOI] [PubMed] [Google Scholar]
- 35.Hodel C. Myopathy and rhabdomyolysis with lipid-lowering drugs. Toxicol Lett 2002;128:159–68 [DOI] [PubMed] [Google Scholar]
- 36.Lebovitz HE. Differentiating members of the thiazolidinedione class: a focus on safety. Diabetes Metab Res Rev 2002;18(Suppl 2):S23–9 [DOI] [PubMed] [Google Scholar]
- 37.van Rijsbergen CJ. Information Retrieval. London: Butterworths, 1979 [Google Scholar]
- 38.Singh S, Loke YK, Furberg CD. Long-term risk of cardiovascular events with rosiglitazone: a meta-analysis. JAMA 2007;298:1189–95 [DOI] [PubMed] [Google Scholar]
- 39.Hauben M. A brief primer on automated signal detection. Ann Pharmacother 2003;37:1117–23 [DOI] [PubMed] [Google Scholar]
- 40.Avorn J. Drug warnings that can cause fits–communicating risks in a data-poor environment. N Engl J Med 2008;359:991–4 [DOI] [PubMed] [Google Scholar]
- 41.Mukherjee D, Nissen SE, Topol EJ. Risk of cardiovascular events associated with selective COX-2 inhibitors. JAMA 2001;286:954–9 [DOI] [PubMed] [Google Scholar]