Abstract
Objective
This paper describes natural-language-processing techniques for two tasks: identification of medical concepts in clinical text, and classification of assertions, which indicate the existence, absence, or uncertainty of a medical problem. Because so many resources are available for processing clinical texts, there is interest in developing a framework in which features derived from these resources can be optimally selected for the two tasks of interest.
Materials and methods
The authors used two machine-learning (ML) classifiers: support vector machines (SVMs) and conditional random fields (CRFs). Because SVMs and CRFs can operate on a large set of features extracted from both clinical texts and external resources, the authors address the following research question: Which features need to be selected for obtaining optimal results? To this end, the authors devise feature-selection techniques which greatly reduce the amount of manual experimentation and improve performance.
Results
The authors evaluated their approaches on the 2010 i2b2/VA challenge data. Concept extraction achieves 79.59 micro F-measure. Assertion classification achieves 93.94 micro F-measure.
Discussion
Approaching medical concept extraction and assertion classification through ML-based techniques has the advantage of easily adapting to new data sets and new medical informatics tasks. However, ML-based techniques perform best when optimal features are selected. By devising promising feature-selection techniques, the authors obtain results that outperform the current state of the art.
Conclusion
This paper presents two ML-based approaches for processing language in the clinical texts evaluated in the 2010 i2b2/VA challenge. By using novel feature-selection methods, the techniques presented in this paper are unique among the i2b2 participants.
Keywords: Medical informatics, natural language processing, decision-support systems, clinical, machine learning, clinical informatics, biomedical informatics, pediatrics, e-prescribing, human factors, medical informatics
Introduction
Electronic medical records (EMR), such as hospital discharge summaries, contain a wealth of information only expressed in natural language. Automated methods for extracting information from these records must be able to recognize medical concepts in text, addressing lexical, syntactic, and semantic ambiguity. Furthermore, to perform any automated reasoning on this information, the context of these concepts must be understood. We address a contextual property critical to reasoning: the doctor's belief status (or assertion) of the patient's medical problem. In the 2010 i2b2/VA challenge1 data, a doctor could qualify a particular problem as being present, absent, possible, hypothetical, conditional on another factor, or associated with a someone else. This information must be obtained from the textual context and is difficult for simple rule-based approaches to extract. Alternatively, this information is obtained by a supervised machine-learning approach that uses several natural-language-processing (NLP) components which are both domain-independent and tailored to medical information extraction. These components are combined in an automated feature selection framework to extract medical concepts and classify the assertion for each medical problem.
Background and significance
Research in medical text mining can generally be segmented into two domains: biomedical literature (primarily medical journals and other scholarly research) and clinical notes (hospital discharge summaries, progress reports, or other notes written by doctors).2 Biomedical NLP tends to focus on extracting proteins, genes, pathways, and other biomedical relations.3–9 In clinical NLP, the focus is on building profiles of individual patients10 11 by extracting a broad class of medical conditions (eg, diseases, injuries, and medical symptoms) and responses (eg, diagnoses, procedures, and drugs),1 12–14 with the goal of developing applications that improve patient care.15–18
Despite their differences, biomedical NLP and clinical NLP take advantage of similar resources and text-processing techniques. These resources include medical knowledge sources, such as the unified medical language system (UMLS) Metathesaurus,19 20 as well as numerous medical ontologies.21–23 An important text-processing tool for both biomedical NLP and clinical NLP is MetaMap,24 which links textual references to medical concepts of any semantic type into the UMLS Metathesaurus. MedLEE12–14 is a system that extracts medical information from clinical text, such as a patient's medical problems and their corresponding certainty level and past history. MedLEE was adapted to biomedical literature in the BioMedLEE system4 5 for extracting biomedical entities and relations. Furthermore, techniques for open-domain text processing have been adapted to the medical domain. For instance, the GENIA tagger25 performs part-of-speech tagging, lemmatization, phrase chunking, and entity recognition. Although GENIA was trained on biomedical literature, it is more suitable for clinical text than other NLP tools, which are commonly trained on newswire. While this paper focuses on two tasks from clinical NLP, we make use of several resources intended for biomedical NLP. Additionally, our work provides an automatic method for determining which of these resources are valuable for a given data set—for example, the data provided by the 2010 i2b2/VA challenge.
In the 2010 i2b2/VA challenge paradigm, extraction of medical concepts involves finding textual expressions of three semantic types: medical problems, tests, and treatments. This is most closely related to the NLP task of named entity recognition (NER). NER systems identify spans of text that belong to a semantic class, such as person, location, or disease. In medical text, this has been applied to tasks such as recognizing diseases, drugs, and proteins, as discussed above. Similar to advances in open-domain NER, early approaches to entity recognition in medical texts were primarily rule-based,3 relying on finely tuned heuristics and lexicons, whereas recent approaches focus on supervised machine-learning models,26 27 which use statistical techniques to handle many types of (often noisy) information. Machine-learning methods have the additional benefit of not needing domain experts to craft rules. For these reasons, we perform concept extraction using two machine-learned classifiers: conditional random fields28 (CRF) for boundary detection and support vector machines29 (SVM) for three-way classification of the concept's type.
The assertion of a medical problem is a classification of the existence, absence, or uncertainty of a problem. A statement such as ‘[his dyspnea] resolved’ implies the problem is absent, while the statement ‘Doctors suspect [an infection of the lungs]’ suggests the problem is possible. This is closely related to the NLP tasks of negation detection and hedge detection because it includes the detection of both negated and uncertain medical problems. The most widely used medical negation detection system is NegEx,30 though more exist.31 NegEx uses a rule-based algorithm that combines negation lexicons with regular expressions matched against the context of terms from UMLS. NegEx is capable of annotating three types of negation status: negated, possible, and actual. In contrast, hedge-detection systems identify instances of uncertainty in natural language. Often this is done at the sentence level,32 but other systems exist that ‘scope’ the uncertainty to a span within a sentence.33 Most recent approaches to hedge detection are supervised.34 We perform a six-way classification of belief status at the concept level (specifically for medical problems), which effectively encompasses all of negation detection and borrows from hedge detection by indicating uncertain medical problems or those that may develop. We incorporate several linguistic features, including NegEx, into a supervised SVM model.
Other submissions to the 2010 i2b2/VA challenge used a range of approaches, including rule-based methods and a variety of ML-based classifiers. However, almost all the top submissions relied on similar ML methods. Seven of the top 10 submissions to the concept task used CRF classifiers or a similar sequence classifier, including our own (the remaining three submissions did not report their methods). Eight of the top 10 submissions to the assertion task used SVM classifiers, including our own (only one did not, while the remaining submission did not report their methods). Many submissions used additional classifiers and strategies for combining the results of multiple classifiers, but it is not clear whether these methods would provide substantial gains for other submissions. As a result of these findings, the focus of our postsubmission research has been the selection of resources and features that maximize a classifier's performance.
Materials and methods
Task description
The 2010 i2b2 challenge1 data consist of 826 discharge summaries and progress notes, split into 349 train and 477 test documents. The documents are annotated by medical professionals familiar with their use. The data contain 72 846 medical concepts (27k train, 45k test). Each concept is classified as a problem (eg, disease, injury), test (eg, diagnostic procedure, lab test), or treatment (eg, drug, preventive procedure, medical device). Medical problems are assigned an assertion type (belief status) among: present, absent, possible, hypothetical, conditional, or associated with someone else. The distribution of assertion types is far from uniform: 69% of all problems are considered present, 20% absent, <5% for possible and hypothetical, and <1% for conditional and associated with someone else. Additionally, the data contain a third set of annotations, relations between concepts, not described in this paper. The data have already been sentence-segmented and tokenized. Automated methods are then required to identify concept start and end tokens, classify the concept's type, and then classify the assertion type for problems. In the official submission, assertion classification was performed only on manually annotated concepts.
Feature selection
Instead of using every possible feature for our classifier, or manually selecting our set of features through trial and error, we use an automated feature selection approach to finding the best set of features. Because our feature set is chosen automatically, we refer to our approach as having a flexible architecture. Given a new task, or simply new data, we can automatically determine a new set of features so long as the new task operates on the same type of input. For example, classifying a concept's type (problem, test, treatment) and a concept's assertion type (present, absent, etc) both operate on the concept level. In both of these tasks, we made largely the same set of features available to the feature selector.
We now describe the three types of feature selection we perform. In each case, the feature sets are scored using cross-validation on the 2010 i2b2/VA training data. Additionally, for features that take parameters, we allow the feature selector to choose among a set of reasonable values. For simplicity, we refer to parameterized features below simply as features.
Greedy forward
Also known as additive feature selection, this method takes a ‘greedy’ approach by always selecting the best feature to add to the feature set. At each iteration, each unused feature is tested in combination with the current selected feature set. The feature corresponding to the highest scoring set is then added to the selected feature set. The algorithm terminates when no new features improve the score.
Greedy forward/backward
Also known as floating forward feature selection,35 this is an extension of greedy forward selection that greedily attempts to remove features from the current feature set after a new feature is added. Intuitively, over time, some features may become redundant or even harmful after new features are added.
Genetic algorithm
Based on biological natural selection, this non-greedy heuristic feature selection technique can overcome local maxima over many generations of genetic crossover and mutation. In this setting, features are analogous to genes, feature sets are analogous to genomes, and the classifier's score using a given feature set is referred to as the feature set's ‘fitness.’ We keep a beam (or ‘population’) of the 500 fittest feature sets found so far. In the crossover step, two feature sets are chosen proportional to their fitness rank and form an ‘offspring.’ Any feature both parents have in common is automatically included in the child feature set. Features in only one parent have a 50% chance of being in the child. The mutation step can then add or remove features from the child. There is a 0.5n+1 chance of having n mutations. For each mutation, a feature is chosen at random from all parameterized features. If that feature is in the child, it is removed, otherwise it is added. The child is then scored and added to the beam if fit. Many termination conditions are possible, but we allow the algorithm to run for several days and take the highest-performing feature set.
External resources
We use numerous external resources to derive features. These resources include UMLS,19 MetaMap,24 NegEx,30 GENIA,25 WordNet,36 PropBank,37 the General Inquirer,38 and Wikipedia. We provide a detailed description of each of these resources in the online supplement.
Concept extraction
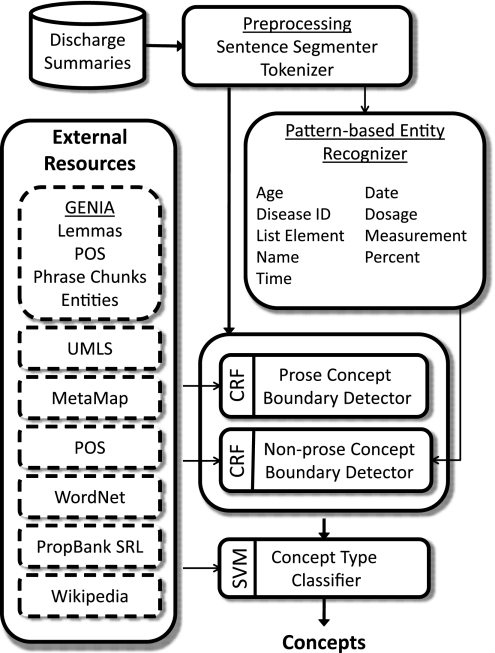
The overall architecture of our concept extraction approach is shown in figure 1. Each discharge summary in the dataset is provided with tokenization and sentence boundaries. We use regular expressions to recognize nine entity types that support concept extraction: names, ages, dates, times, IDC-9 identifiers, percents, measurements, dosages, and list elements. Each sentence is then categorized as being prose or non-prose using a simple heuristic. Sentences that end with a colon are assumed to be section headers and are not considered prose. A sentence is considered prose if it ends with a period or question mark, or if it consists of at least five tokens, less than half of which may be punctuation. Otherwise, it is considered non-prose.
Figure 1.
Architecture of our concept-extraction approach.
We then detect concept boundaries (start and end tokens) using two CRF classifiers39: one CRF for prose sentences; the other CRF for non-prose sentences. This allows us to use separate features for each classifier; each set of features reflects the different problems faced when extracting concepts in prose and non-prose text.
For concept extraction, we used only greedy forward feature selection. The feature selector primarily chose lexical and pattern-entity features for non-prose concepts, along with MetaMap features. For prose concepts, a wide variety of features commonly used in NLP were chosen, including the four annotations provided by GENIA. The lists of features chosen by the feature selector for each CRF classifier is shown in box 1.
Box 1. Features for concept extraction.
-
Non-prose boundary
NF1. uncased word
NF2. pattern-based entity
NF3. uncased prev word
NF4. prev word POS
NF5. 3-token POS context
NF6. MetaMap type
NF7. MetaMap CUI
-
Concept type
TF1. uncased words
TF2. 4-char prefix
TF3. Prev lemma
TF4. next lemma
TF5. uncased prev bigram
TF6. SRL pred.+arg type
TF7. UMLS concept type
TF8. Wiki. concept type
-
Prose boundary
PF1. Word lemma
PF2. Prev word
PF3. Uncased prev word
PF4. 2-char suffix
PF5. prev POS
PF6. 1-token POS context
PF7. UMLS concept parents
PF8. MetaMap type
PF9. GENIA lemma
PF10. GENIA entity type
PF11. GENIA phrase chunk
PF12. Prev GENIA POS
PF13. Prev GENIA lemma
PF14. Prev GENIA phrase chunk
PF15. Next GENIA lemma
After detecting the concept boundaries, our approach classifies each concept as a problem, treatment, or test. We use a single SVM classifier40 for all concepts, prose and non-prose, and employ the same greedy feature-selection technique. The selected features are shown in box 1.
The feature selector for boundaries chose from a set of 125 features, choosing seven for non-prose concept boundaries and 15 for prose concept boundaries. For concept type, a total of 222 features were available to the feature selector (most of which were developed for assertion classification), of which eight were chosen. For features that can take non-numeric values (eg, NF1 can be any word, while TF1 can be many words for a given concept), we expand these features into N binary features, where N is the number of values seen for the feature in the training data. This results in large, sparse feature vectors that can be problematic for some machine-learning techniques, but are easily handled by SVMs and CRFs.
Assertion classification
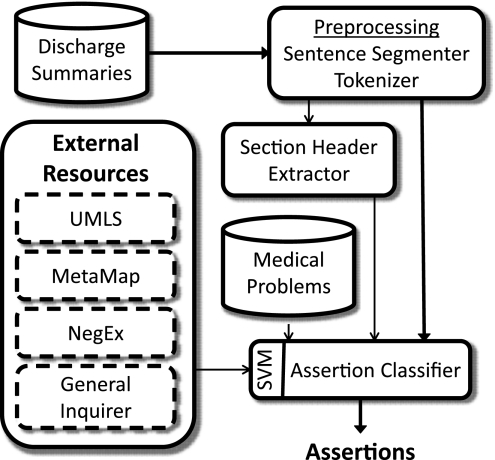
The belief status (or assertion type) of a medical problem is determined by a single SVM classifier.40 Problems are categorized as present, absent, possible, hypothetical, conditional, or associated with someone else. The overall architecture of our assertion classification approach is shown in figure 2, and our features are shown in box 2. We describe only the features chosen by the feature selector. The feature selector chose an optimal set of 27 of the 396 available features (the same set available for concept type plus eight features based on the output of the concept type classifier and 166 features based on significant n-grams developed after the original submission and described below).
Figure 2.
Architecture of our assertion-classification approach.
Box 2. Features for assertion classification.
AF1. Section name
AF2. Sentence contains pattern-based entity
AF3. Other concepts in sentence
AF4. Previous assertion type in sentence
AF5. Previous assertion type within 5 tokens
AF6. Previous assertion type plus words between
AF7. Previous assertion in doc is hypothetical
AF8. NegEx modifier
AF9. Concept is in unified medical language system
AF10. Concept detected by MetaMap with score ≥800
AF11. Inquirer word with category ‘If’ before concept
AF12. Next word's part-of-speech
AF13. Unigrams in concept
AF14. Next word
AF15. Previous word
AF16. Uncased next word
AF17. Uncased previous word
AF18. Uncased previous bigram
AF19. Uncased previous bigram if contains a stopword
AF20. Unigrams in sentence
AF21. ANG(P, sent, 10) most significant is absent
AF22. ANG(F, sent, 10) most significant is associated with someone else
AF23. ANG(P, sent, 3) most significant is hypothetical
AF24. ANG(F, sent, 3) types with log value ≤−100
AF25. ANG(P, sent, 3) types with value ≤−1
AF26. ANG(F, 5 tokens, 3) types with log value ≤−2
AF27. ANG(F, 5 tokens, 3) n-grams with log value ≤−2
ANG (x, y, z) is an assertion n-gram feature, where x is the metric (P for pointwise mutual information, F for Fisher), y the context, and z the minimum count
After we preprocess the discharge summaries as in concept extraction, we partition the document into sections and associate each section with its header. We assume that sentences ending with a colon are section headers. The uncased section name is then used as a feature (AF1) for all problems in that section. Common section names that are useful in assertion classification are ‘allergies,’ ‘family history,’ and ‘infectious disease.’ Another feature derived from preprocessing uses the pattern-based entities discussed in concept extraction. This feature (AF2) indicates if one of these entities is in the current sentence. This is a good indicator of medical problems which are present, as the entities often refer to dosages given for the treatment of some problem.
Five features (AF3–7) capture information about other concepts in the context. AF3 simply returns the other concepts in the sentence, while AF4–7 deal specifically with the assertion type of previous concepts. Since assertions are classified based on their order in the document, only the types of the previous problems are available to the classifier.
We use a NegEx feature (AF8) to indicate the negation word associated with the medical problem. Additional medical features indicate if the problem was found in UMLS (AF9) or MetaMap (AF10), as the distribution of assertion types for problems found within these resources differs from that of the documents.
We use the General Inquirer's category information to better understand the context of a medical problem. We only use the ‘If’ category, which indicates uncertainty words such as ‘unexpected,’ ‘hesitant,’ or ‘suspicious.’ This feature (AF11) only looks at the five previous tokens.
We use eight lexical features to capture the words both inside and outside the concept. Features representative of the concept, such as the words within the concept (AF13), are important because different medical problems have different assertion type distributions. Some concepts are by their very nature absent (eg, ‘afebrile’ means without fever, and all 213 mentions are marked as absent). To gather lexical context, we use the word on either side of the concept (AF14–19) as well as every word in the sentence (AF20).
Since lexical features create a high-dimensionality problem for the classifier, we developed a method to use only the most significant words and phrases in a problem's context. This is sometimes referred to as statistical feature selection, and differs from automated feature selection in that it filters features based on their statistical significance instead of the final output of the classifier on an evaluation set. We use two statistical measures: pointwise mutual information (PMI) and Fisher exact test. Both measures provide a scoring method for words and phrases based on how often they co-occur with problems of a specific assertion type in the training data. For instance, the phrase ‘family history’ is strongly correlated with a problem associated with someone else, ‘on exertion’ is highly correlated with a conditional, and ‘no evidence of’ is highly negatively correlated with present. While these n-grams are captured in the lexical features previously described, by removing statistically insignificant n-grams the feature can be given a more appropriate weight. This provides our approach with additional flexibility, as the n-grams could be extracted directly from any training set.
We allow the feature selector to choose between various parameters for our significant n-gram features: (1) the choice of metric (PMI or Fisher), (2) context window size, (3) the minimum count an n-gram must have in the training set to be considered, and (4) various score thresholds. This provides further motivation for automated feature selection: manually evaluating well over a hundred parameterizations would prove too costly. Automated feature selection learns the best parameterization relative to the other features being used by the classifier. The feature selector chose several parameterized features using both statistical measures, a range of windows from the entire sentence to a five-word span around the concept, minimum counts of three and 10, as well as scores that capture both highly positively and negatively correlated n-grams. A total of seven features (AF21–27) based on these n-grams were chosen.
Results
Table 1 shows the results of our concept extraction approach as submitted to the 2010 i2b2/VA challenge, where we placed ninth. The best, mean, and median results across all 22 submissions are shown as well. Additionally the table shows the breakdown into boundary detection (with both prose and non-prose) and type classification. The largest source of error was boundaries. Most boundary errors were partial, involving missed or incorrectly added words, especially at the beginning of the concept where the i2b2 guidelines specified only certain types of modifiers. The submission's inexact score was 89.25, meaning over half of all errors were still partial matches.
Table 1.
Results for concept extraction
| System | Score |
| Best i2b2 submission | 85.23 |
| Our i2b2 submission | 79.59 |
| Median i2b2 submission | 77.78 |
| Mean i2b2 submission | 73.56 |
| SubSystem | Score |
| Boundary | 83.17 |
| (Prose) | 83.45 |
| (Non-prose) | 81.79 |
| Type | 95.49 |
| Total | 79.59 |
Table 2 shows the current status of our assertion classification approach using different feature-selection methods, compared against our original submission to the 2010 i2b2/VA challenge, and the best, mean, and median results. There are two differences between the approach described in this paper and our submission. First, greedy forward (GF) feature selection was used to choose features in the original submission. Second, the n-gram correlation features were not used. We report the improved scores using three feature-selection ‘pipelines’: greedy forward/backward (GFB) alone, then the output of the GFB selector being used as the seed set for a genetic algorithm (GA), and finally adding another GFB on the GA's output to remove any spurious features and quickly find additional features that improve results. Again, all features were chosen using cross-validation on the training data, while the results shown in table 2 are evaluated on the test data (our cross-validation F-measure was 95.3). As can be seen in the results, these additions are significantly better than our original submission.
Table 2.
Results for assertion classification
| System | Score |
| GFB+GA+GFB | 93.94 |
| GFB+GA | 93.93 |
| GFB | 93.84 |
| Best i2b2 submission | 93.62 |
| Our i2b2 submission (GF) | 92.75 |
| Median i2b2 submission | 91.96 |
| All features | 90.67 |
| Mean i2b2 submission | 86.18 |
| Our best with automatic concepts | 73.67 |
| Class | Precision | Recall | F1 |
| Absent | 95.93 | 93.41 | 94.65 |
| Associated with someone else | 91.47 | 81.38 | 86.13 |
| Conditional | 72.86 | 29.82 | 42.32 |
| Hypothetical | 92.17 | 87.03 | 89.53 |
| Possible | 81.63 | 58.89 | 68.42 |
| Present | 94.39 | 98.00 | 96.17 |
GA, genetic algorithm; GF, greedy forward; GFB, greedy forward/backward.
Table 2 also shows the results on a per-class basis. Generally speaking, performance is relative to the number of examples in the training data. The notable exception is the ‘associated with someone else’ class, which had a specific feature (AF22) that was targeted at this class. Similarly, the hypothetical class, which has roughly the same number of manual annotations as the possible class, performs better due to its targeting feature (AF23). Equivalent features for conditional and possible were not as helpful, suggesting there are fewer overt lexical cues for these classes.
For comparison, we also report results for using all features (instead of the feature selector) as well as the results when using automatically detected concepts. Clearly, using all 396 features impairs the performance significantly. This stands to emphasize that feature selection is an important process, as over 90% of the features devised were either redundant with other features or too noisy to improve held-out results. In regards to the experiment using automatically generated concepts, we found that assertion classification is relatively independent of concept extraction (ie, the problems that were correctly extracted were not necessarily the problems whose assertions were correctly classified), as the result of 73.67 is very close to the 74.76 F-measure (79.59%×93.94%) expected by independence. However, the 73.67 result could be improved to (or exceed) 74.76 if the assertion classifier were trained on the automatic concepts instead of the manually annotated concepts.
Discussion
Automated feature selection permits the consideration of a significant number of features, derived from a large set of resources. The goal of feature selection is to find a near-optimal subset of features for a given task. Here we consider three separate contributions made by employing feature-selection methods: (1) more advanced feature-selection algorithms improve feature choice; (2) feature selection can choose the best parameters for highly parameterized features; and (3) feature selection allows for an empirical evaluation on the value of individual resources.
Enhancements in our automated feature selection improved assertion classification. While the genetic algorithm did not offer a significant improvement (0.1%), it was able to find several useful features. We feel that on more difficult tasks, this genetic algorithm will prove more valuable. Given the diminishing returns of adding new features and resources, it is unlikely that adding more features and resources will significantly improve assertion classification on this dataset. This also limits the genetic algorithm's effectiveness. The greedy forward/backward algorithm did not improve the results using the original features, but it was crucial when the statistically significant n-gram features were used. With fewer features, more relaxed n-gram features prove useful, but as other features are added, new parameterizations prove more effective. Without GFB's ability to prune features, the n-gram features add only 0.4%, but using GFB this increases to 1.1%. Thus, automated feature selection and highly parameterized features are mutually beneficial.
Finally, we discuss the value of external resources as determined by feature selection. For detailed tests of the contribution of each feature, see the online supplement. Both concept extraction and assertion classification benefit from resources such as Wikipedia, GENIA, and MetaMap. The most useful resource across both tasks is MetaMap, which is used by both concept boundary detectors and the assertion classifier. The most used resource in terms of number of features is GENIA, but all seven features are used by the prose concept boundary detector. The task least affected by external resources is the assertion task (see the online-only feature test for more details). The most relevant resource for assertion classification, NegEx, is itself an automated NLP system. Most of the rules within NegEx are redundant with information extracted directly from the training data.
Owing to the large size of the training data, a basic approach composed entirely of word, part-of-speech, and lemmatization features could perform well. We experimented with just these features and obtained a result of 76.61 on concept extraction and 91.50 on assertion classification, both near the median i2b2 submission. Crucially, however, the difference between an average submission and a top performing submission is the careful use of external resources and statistically derived features. By using automated feature-selection techniques, we were therefore able to experiment with a significantly larger number of resources and features in order to maximize their impact on the final results.
Conclusion
We have described automated approaches for extracting medical concepts and classifying assertions of medical problems. In both cases, supervised ML-based methods were used in combination with feature-selection techniques. Both methods were in the top 10 results on the 2010 i2b2 challenge evaluation. Additionally, we describe improvements made to assertion classification which outperform the best submission to the challenge. These improvements combine selecting the most statistically significant words and phrases as well as enhancements to our automated feature selection.
Acknowledgments
The authors would like to thank the i2b2 National Center for Biomedical Computing for organizing the 2010 challenge and providing the data used in these experiments. We would also like to acknowledge the anonymous reviewers for their valuable suggestions.
Footnotes
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Uzuner O, South B, Shen S, et al. i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collier N, Nazarenko A, Baud R, et al. Recent advances in natural language processing for biomedical applications. Int J Med Inform 2006;75:413–17 [DOI] [PubMed] [Google Scholar]
- 3.Tanabe L, Wilber WJ. Generation of a large gene/protein lexicon by morphological pattern analysis. J Bioinform Comput Biol 2004;1:611–26 [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Friedman C. Extracting phenotypic information from the literature via natural language processing. Medinfo 2004;11(Pt 2):758–62 [PubMed] [Google Scholar]
- 5.Lussier Y, Borlawsky T, Rappaport D, et al. PhenoGO: assigning phenotypic context to gene ontology annotation with natural language processing. Pac Symp Biocomput 2006;11:64–75 [PMC free article] [PubMed] [Google Scholar]
- 6.Rindflesch TC, Tanabe L, Weinstein JN, et al. EDGAR: extraction of drugs, genes, and relations from the biomedical literature. Pac Symp Biocomput 2000;5:514–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hristovski D, Friedman C, Rindflesch TC, et al. Exploiting semantic relations for literature-based discovery. AMIA Annu Symp Proc 2006:216–20 [PMC free article] [PubMed] [Google Scholar]
- 8.Duda S, Aliferis C, Miller R, et al. Extracting drug–drug interaction articles from MEDLINE to improve the content of drug databases. AMIA Annu Symp Proc 2005:216–20 [PMC free article] [PubMed] [Google Scholar]
- 9.Rzhetsky A, Iossifov I, Koike T, et al. GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. J Biomed Inform 2004;37:43–53 [DOI] [PubMed] [Google Scholar]
- 10.Heinze DT, Morsch ML, Holbrook J. Mining free-text medical records. Proc AMIA Symp 2001:254–8 [PMC free article] [PubMed] [Google Scholar]
- 11.Cao H, Hripcsak G, Markatou M. A statistical methodology for analyzing cooccurrence data from a large sample. J Biomed Inform 2007;40:343–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman C, Hripcsak G, Shagina L, et al. Representing information in patient reports using natural language processing and the extensible markup language. J Am Med Inform Assoc 1999;6:76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004;11:392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn U, Romacker M, Schulz S. Creating knowledge repositories from biomedical reports: the MEDSYNDIKATE text mining system. Pac Symp Biocomput 2002;7:338–49 [PubMed] [Google Scholar]
- 16.Babic A. Knowledge discovery for advanced clinical data management and analysis. Stud Health Technol Inform 1999;68:409–13 [PubMed] [Google Scholar]
- 17.Hripcsak G, Bakken S, Stetson PD, et al. Mining complex clinical data for patient safety research: a framework for event discovery. J Biomed Inform 2003;36:120–30 [DOI] [PubMed] [Google Scholar]
- 18.Hripcsak G, Austin JH, Alderson PO, et al. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology 2002;224:157–63 [DOI] [PubMed] [Google Scholar]
- 19.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system. Methods Inf Med 1993;32:281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng Q, Cimino JJ. Automated knowledge extraction from the UMLS. Proc AMIA Symp 1998:568–72 [PMC free article] [PubMed] [Google Scholar]
- 21.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol 2005;6:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith CL, Goldsmith CA, Eppig JT. The mammalian phenotype ontology as a tool for annotating, analyzing, and comparing phenotypic information. Genome Biol 2005;6:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheuermann RH, Ceusters W, Smith B. Toward an ontological treatment of disease and diagnosis. Summit on Translat Bioinforma 2009:116–20 [PMC free article] [PubMed] [Google Scholar]
- 24.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. 2001. http://mmtx.nlm.nih.gov [PMC free article] [PubMed]
- 25.Tsuruoka T, Tateishi T, Kim JD, et al. Developing a robust part-of-speech tagger for biomedical text. Advances in Informations—10th Panhellenic Conference on Informatics 2005:382–92 http://bit.ly/geniatagger [Google Scholar]
- 26.Patrick J, Min L. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J Am Med Infom Assoc 2010;17:524–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou G, Zhang J, Su J, et al. Recognizing names in biomedical texts: a machine learning approach. Bioinformatics 2004;20:1178–90 [DOI] [PubMed] [Google Scholar]
- 28.Lafferty J, McCallum A, Pereira F. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. Proceedings of the International Conference on Machine Learning (ICML) 2001:282–9
- 29.Vapnik V. The Nature of Statistical Learning Theory. Berlin: Springer-Verlang, 1995 [Google Scholar]
- 30.Chapman WW, Bridewell W, Hanbury P, et al. A Simple Algorithm for Identifying Negated Findings and Diseases in Discharge Summaries. Biomed Inform 2001;24:310–10 [DOI] [PubMed] [Google Scholar]
- 31.Gindl S. Negation Detection in Automated Medical Applications. Vienna: Vienna University of Technology, 2006 [Google Scholar]
- 32.Szarvas G. Hedge classification in biomedical texts with weakly supervised selection of keywords. Proceedings of the Association for Computational Linguistics (ACL) 2009:281–9
- 33.Morante R, Daelemans W. Learning the scope of hedge cues in biomedical texts. Proceedings of the Workshop on BioNLP 2009:28–36
- 34.Farkas R, Vencze V, Móra G, et al. The CoNLL-2010 shared task: learning to detect hedges and their scope in natural language text. Proceedings of the Conference on Computational Natural Language Learning: Shared Task 2010:1–12
- 35.Pudil P, Novovičová J, Kittler J. Floating search methods in feature selection. Pattern Recognit Lett 1994;15:1119–25 [Google Scholar]
- 36.Fellbaum C. WordNet: An Electronic Lexical Database. Cambridge, MA: MIT Press, 1998 [Google Scholar]
- 37.Palmer M, Kingsbury P, Gildea D. The proposition bank: an annotated corpus of semantic roles. Computational Linguistics 2005;31:71–106 [Google Scholar]
- 38.Stone PJ, Dunphy DC, Smith MS, et al. The general inquirer: a computer approach to content analysis. MIT studies in comparative politics. 1966.
- 39.McCallum AK. MALLET: a machine learning for language toolkit. 2002. http://mallet.cs.umass.edu
- 40.Fan RE, Change KW, Hsieh CJ, et al. LIBLINEAR: a library for large linear classification. J Mach Learn Res 2008;9:1871–4 [Google Scholar]