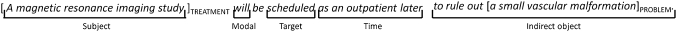Abstract
Objective
A supervised machine learning approach to discover relations between medical problems, treatments, and tests mentioned in electronic medical records.
Materials and methods
A single support vector machine classifier was used to identify relations between concepts and to assign their semantic type. Several resources such as Wikipedia, WordNet, General Inquirer, and a relation similarity metric inform the classifier.
Results
The techniques reported in this paper were evaluated in the 2010 i2b2 Challenge and obtained the highest F1 score for the relation extraction task. When gold standard data for concepts and assertions were available, F1 was 73.7, precision was 72.0, and recall was 75.3. F1 is defined as 2*Precision*Recall/(Precision+Recall). Alternatively, when concepts and assertions were discovered automatically, F1 was 48.4, precision was 57.6, and recall was 41.7.
Discussion
Although a rich set of features was developed for the classifiers presented in this paper, little knowledge mining was performed from medical ontologies such as those found in UMLS. Future studies should incorporate features extracted from such knowledge sources, which we expect to further improve the results. Moreover, each relation discovery was treated independently. Joint classification of relations may further improve the quality of results. Also, joint learning of the discovery of concepts, assertions, and relations may also improve the results of automatic relation extraction.
Conclusion
Lexical and contextual features proved to be very important in relation extraction from medical texts. When they are not available to the classifier, the F1 score decreases by 3.7%. In addition, features based on similarity contribute to a decrease of 1.1% when they are not available.
Keywords: Natural language processing, clinical informatics, medical records systems, computerized, semantic relations
Background and significance
Medical discharge summaries and progress notes contain a variety of medical concepts and relations. The ability to recognize relations between medical concepts (problems, treatments, or tests) enables the automatic processing of clinical texts, resulting in an improved quality of patient care. To address this important aspect of knowledge mining from electronic medical records (EMR), the 2010 i2b2/VA NLP Challenge1 considered a task of relation extraction from EMRs.
The organizers of the 2010 i2b2 Challenge have provided two sets of discharge summaries and progress notes: (1) a training set in which medical concepts and relations between them were annotated; and (2) a testing set in which relation annotations were initially withheld. The training set consists of 349 documents, 27 837 concepts, and 5264 relations. The test set consists of 477 documents, 45 009 concepts, and 9069 relations. These annotations are called gold standard data. Additionally, medical problem concepts have annotations that indicate whether a medical problem is present, absent, associated with someone else, hypothetical, conditional, or possible. The eight types of relations between concepts are illustrated in box 1.
Box 1. The set of relations defined in the 2010 i2b2 relation challenge.
TrIP: A certain treatment has improved or cured a medical problem (eg, ‘infection resolved with antibiotic course’)
TrWP: A patient's medical problem has deteriorated or worsened because of or in spite of a treatment being administered (eg, ‘the tumor was growing despite the drain’)
TrCP: A treatment caused a medical problem (eg, ‘penicillin causes a rash’)
TrAP: A treatment administered for a medical problem (eg, ‘Dexamphetamine for narcolepsy’)
TrNAP: The administration of a treatment was avoided because of a medical problem (eg, ‘Ralafen which is contra-indicated because of ulcers’)
TeRP: A test has revealed some medical problem (eg, ‘an echocardiogram revealed a pericardial effusion’)
TeCP: A test was performed to investigate a medical problem (eg, ‘chest x-ray done to rule out pneumonia’)
PIP: Two problems are related to each other (eg, ‘Azotemia presumed secondary to sepsis’)
We developed a method capable of automatically identifying these relations. Our method operates in two modes: (1) when the concepts mentioned in the clinical texts are already identified (either manually or by another system); and (2) when our method needs to also identify the concepts that are arguments of the relations, as well as the assertions associated with the medical concepts.
The data from the i2b2 Challenge consist of discharge summaries and progress notes. The records contain semi-structured fields for age, dates relating to admittance, discharge, and treatment, as well as fields for the treatments the patient has undergone. In addition, the discharge summaries contain a written chronological listing of descriptions about the patient's condition, the treatments they underwent, and the tests that were performed. Similarly, the progress notes provide the doctor's description of the patient's current condition, the care provided, and follow-up care to be administered. The relations listed in box 1 are automatically extracted from these sections of the clinical notes through machine learning techniques.
Machine learning-based techniques are not new to medical informatics. The strength of these techniques depends on the features that they use. In this article we show that by customizing the features that capture the context of relations as well as the expression of relations, very promising results are obtained.
Materials and methods
Strategy for extracting relations from EMRs
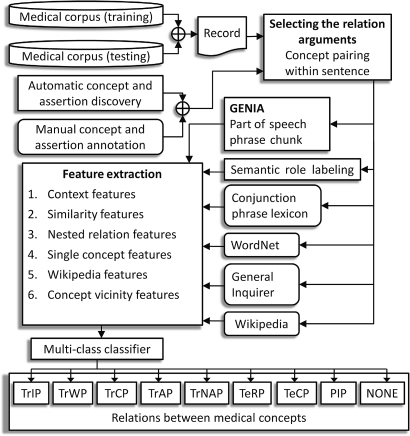
The problem of relation discovery was cast as a multi-class classification problem (defined in Aly2). The classifier considers a pair of medical concepts in text and decides which type of relation exists between them, if any. As illustrated in figure 1, both the training and testing of the relation discovery system is achieved by considering one EMR at a time. In addition, each sentence from the EMR is pre-tokenized and pairs of concepts from the sentence are considered.
Figure 1.
Architecture of the method for identifying relations in clinical texts.
We considered six classes of features as illustrated in figure 1. Feature extraction benefits from several knowledge sources, including a semantic role labeler (SRL), a part of speech (POS) tagger, as well as a phrase chunk parser provided by the GENIA tagger.3 In addition, we have used WordNet,4 the lexico-semantic database encoding a majority of English nouns, verbs, adjectives, and adverbs as well as Wikipedia, a collaboratively written comprehensive encyclopedia containing both structured and unstructured knowledge. We have also used the General Inquirer lexicon5 to detect negative and positive polarity for words.
The multi-class classifier was implemented using the support vector machine (SVM) implementation called LibLINEAR.6 Early testing showed SVMs outperforming other classifiers, including logistic regression and Naïve Bayes. LibLINEAR is an extension of LibSVM7 restricted to a linear kernel to achieve significant speed gains. We used cross-validation on the training set to tune LibLINEAR's parameters. The regularization parameter, C, was set to 0.5. The termination parameter, epsilon, was set to 0.5. Finally, the class weight associated with ‘no relation’ was set to 0.025 to decrease the significance of those concept pairs which had no relation. The weight for other classes was 1.0.
Feature extraction for relation discovery in EMRs
For each pair of concepts from a sentence, we extracted six classes of features to discover possible relations. For example, given sentence S1, there are three possible concept pairs: CP1: (ceftriaxone, azithromycin), CP2: (ceftriaxone, MRSA), and CP3: (azithromycin, MRSA). Using all six classes of features, the task of the system is to extract the two relations present: R1: (ceftriaxone, TrAP, MRSA) and R2: (azithromycin, TrAP, MRSA).
S1: The patient was treated initially with [ceftriaxone]TREATMENT and [azithromycin]TREATMENT based on his history of [MRSA]PROBLEM.
Sentence S1 is representative of the data in both the training and testing sets. That is, the data consist of sentences from EMRs which have been annotated with medical concepts and their types. During training the relation types associated with pairs of concepts are known and used as the outcome label for each pair. In the example above, the labels for CP1, CP2, and CP3 would be: NONE, TrAP, and TrAP, respectively.
The input to the SVM classifier consists of one sparse real-valued feature vector for every pair of concepts from a sentence. The vector representation of a candidate contains one vector element for every pairing of a feature type and one of that feature type's possible values. The vector element is set to one if that combination of feature and value were extracted. The feature vector encodes the six classes of features listed in figure 1 used for discovering the eight relations of interest and presented in the following sections.
Context features
Context features capture characteristics of the text surrounding the medical concepts that may be arguments of a relation. The context features used by the relation extraction method were:
CF1: Any word between the relation arguments.
CF2: Any POS tag between the relation arguments. POS tags are extracted by GENIA.3
CF3: Any bigram between the relation arguments. A bigram is a string of two consecutive tokens.
CF4 and (CF5): Word preceding first (second) argument of the relation.
CF6 and (CF7): Any of the three words succeeding the first (second) argument of the relation.
CF8: Any concept type used between the relation arguments. Concept types are provided or automatically discovered.
CF9: Indicator of whether a conjunction regular expression matched the string of words between the relation arguments. We describe later in this section how feature CF9 is extracted.
CF10: Sequence of phrase chunk types between the relation arguments. Phrase chunks are recognized by GENIA.3
CF11: String of words between the relation arguments.
CF12 and (CF13): Predicates associated with the first (second) relation argument. Predicates are extracted by a method detailed later in this section.
CF14: Any predicate associated with both relation arguments.
Features CF1–CF7 capture important lexical information about the relation arguments and their context. These features allow the classifier to learn which words are associated with certain types of relations. On the other hand, features CF9–CF11 each capture some aspect of the syntactic context of the relation arguments. Finally, features CF12–CF14 are able to extract semantic information from the text. While many of the teams participating in the i2b2 2010 Challenge utilized similar lexical and syntactic features, the use of semantic parsing was not common.
The conjunction feature CF9 aims to detect when the concepts are mentioned in an EMR through a conjunct (eg, ‘, consistent with’, ‘and question of’, or ‘, possibly as well as’). We found that concepts participating in a conjunction do not often constitute the arguments of the same relation. One of the 11 regular expressions used is shown below:
(,)?((and|or))?((possibly|probably))?including.
The words between two medical concepts are also grouped into different phrase chunks which are utilized by feature CF10. The toolkit from GENIA3 allows us to recognize such phrase chunks and to identify their sequence as a grammatical or syntactic expression of the context. For an example, consider the concepts C1=Bipolar D/O and C2=overdose, and their context shown in sentence S2.
S2: [Bipolar D/O]NP [with]PP [suicide attempt]NP [by]PP [overdose]NP.
In this case, the value of CF10 is ‘PP-NP-PP’.
Features CF12–CF14 use the output from ASSERT,8 an SRL system. SRL results from semantic parsing based on PropBank9 which identifies verbal predicates in text, along with their arguments. Figure 2 shows a sentence annotated with semantic roles discovered by ASSERT. The predicate, or target, in this example is the verb scheduled, and it has four arguments: Subject, Modal, Time, and Indirect object. The sentence is also annotated with a pair of relation arguments being considered, namely a TREATMENT and a PROBLEM. Features CF12 and CF13 extract the target predicates associated with a relation argument. In the example CF12, CF13, and CF14 are all set to scheduled because both the TREATMENT and the PROBLEM participate in one of the roles associated with the verb scheduled.
Figure 2.
Example sentence marked with semantic roles.
Similarity features
Relations that have similar contexts should also have similar relation types. However, conventional lexical features fail to directly capture this. For instance, the context feature CF11 (string of words between the relation arguments) is unable to capture minor lexical variations. To overcome this, we use a sequence similarity metric known as Levenshtein distance.10 This metric calculates the number of additions, deletions, or substitutions of sequence elements needed to convert one sequence to another. For instance, if two word sequences differ by only a single word, then their Levenshtein distance is one (because only one substitution is needed). We use this distance metric on the training data to find other relations (including those of type NONE) which are similar to a query relation. The similarity features then indicate the percentage of similar relations of each relation type. The number of similar relations used varies for each feature.
Each similarity feature corresponds to one of five different sequence types:
ST1: POS tags for the entire sentence extracted by GENIA.3 The 100 most similar relations are used.
ST2: Phrase chunks between the relation arguments extracted by GENIA. The 15 most similar relations are used.
ST3: Word lemmas for the span beginning two words before the first relation argument up to and including the second word after the second argument. Word lemmas are computed using WordNet.4 The 20 most similar relations are used.
ST4: The concept types for any concepts found in the sentence. The 100 most similar relations are used.
ST5: The shortest dependency path between the arguments. We use the Stanford dependency parser11 to determine this path. The 20 most similar relations are used.
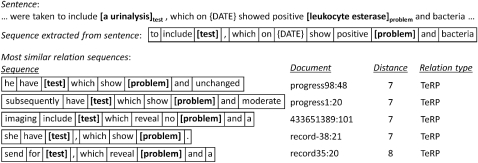
Within the sequences used, any components corresponding to one of the relation arguments are replaced with the argument's concept type. Figure 3 shows the results of searching for sequences similar to a query sequence. Because all of the most similar sequences correspond to the relation TeRP, the values associated with the similarity feature ST3 will help decide that the relation between urinalysis and leukocyte esterase is TeRP.
Figure 3.
Example sequences of word lemmas from electronic medical records displaying high similarity measured by Levenshtein distance.
Nested relations features
We have noticed that relations can be discovered in the text span between the arguments of another relation. For example, when discovering R2 in sentence S1 we call relation R2 to be nested within relation R1. Because of this, our relation discovery classification orders the pairs of concepts by the distance between their argument concepts. The distance is measured in tokens. In this way, relations such as R2 are discovered before relations such as R1. This order of relation discovery leads to the ability to extract features from nested relations (eg, R2) to be used in the discovery of new relations (eg, R1). There is only one nested relation feature. It lists the relation types that are recognized in the text span between the arguments of the relation under consideration.
Single concept features
SCF1 and (SCF2): Any word lemma from the first (second) relation argument. The word lemma is computed using WordNet.4
SCF3 and (SCF4): Any word used to describe the first (second) relation argument.
SCF5 and (SCF6): String of words in first (second) relation argument.
SCF7 and (SCF8): Concept type for first (second) relation argument.
SCF9: Concatenation of assertion types for both relation arguments. Assertion types are either provided or automatically classified.
SCF10 and (SCF11): Any positive or negative semantic category returned by the General Inquirer lexicon5 for the first (second) relation argument.
The features SCF10 and SCF11 are able to detect certain words which may indicate a positive sentiment, such as care, well, and benign, or words carrying a negative sentiment, such as pain, failure, and fever. The General Inquirer lexicon assigns over 200 semantic categories to 9300 English words. We extract the positive and negative categories for any words found in a relation argument.
Wikipedia features
Wikipedia (http://www.wikipedia.org) represents a large collective effort to encode much of human knowledge in an encyclopedic format. While still rapidly growing, it has articles on many medical problems, treatments, and tests of the kind annotated in the i2b2 2010 data set. We found that 93.5% of concepts in the training data contain at least one Wikipedia article title (excluding common words such as the or all from consideration). Several binary features are based on information from Wikipedia's hyperlink structure and category hierarchy:
WF1: Indicates whether neither of the relation arguments contains any substring that may be matched against the title of a Wikipedia article.
WF2: Indicates the absence of Wikipedia links between Wikipedia articles that were retrieved based on the relation arguments.
WF3 and (WF4): Indicates that a Wikipedia link exists from the Wikipedia article pertaining to the first (second) argument of the relation to the Wikipedia article pertaining to the second (first) argument of the relation.
WF5: Indicates that there are Wikipedia links between the Wikipedia articles pertaining to both relation arguments.
WF6: Indicates that both arguments correspond to the same concept type according to their Wikipedia categories. We manually annotated concept types for 26 Wikipedia categories (and their hierarchical descendants).
The features WF1–WF5 rely on the hyperlink structure of Wikipedia. We determine which titles of Wikipedia articles are contained as substrings of relation arguments and check whether these Wikipedia articles link to each other. For example, given the sentence: ‘The [liver enzimes[sic]]TEST come down, no signs of [biliary stasis]PROBLEM’, the arguments are liver enzimes and biliary stasis. The titles of three Wikipedia articles are contained as substrings of these arguments: Liver, Biliary Stasis, and Biliary. The biliary stasis (also called cholestasis) Wikipedia article contains the following text and links: ‘In medicine, cholestasis is a condition where bile cannot flow from the liver to the duodenum.’ Because the Wikipedia article pertaining to biliary stasis links to the Wikipedia article pertaining to liver, the feature WF4 is set to true.
Concept vicinity features
Two concept vicinity features consider concepts which occur immediately preceding or succeeding the relation arguments:
CVF1: Concatenation of the concept types of the first relation argument and the closest preceding concept in the sentence, if one exists.
CVF2: Concatenation of the concept types of the second relation argument and the closest succeeding concept in the sentence, if one exists.
Concepts and their types are either provided by manual annotators or discovered automatically using the approaches described in the next section.
Feature overview
Table 1 contains an overview of which tools and resources are used by each feature.
Table 1.
A listing of all the tools and resources used by each feature
| Class | Features | Tools and resources |
| Context features | CF1, CF3–CF7 | Tokenization |
| CF2 | GENIA part of speech tagger | |
| CF9 | Conjunction lexicon | |
| CF8 | Concepts and concept types | |
| CF10 | GENIA phrase chunker | |
| CF12–CF13 | ASSERT semantic role labeler | |
| Similarity features | ST1 | GENIA part of speech tagger |
| ST2 | GENIA phrase chunker | |
| ST3 | Tokenization, WordNet lemmatization | |
| ST4 | Concepts and concept types | |
| ST5 | Stanford dependency parser | |
| Nested features | All | Annotated relations (training) or discovered relations (testing) |
| Single concept features | SCF1–SCF2 | Tokenization, WordNet lemmatization |
| SCF3–SCF6 | Tokenization | |
| SCF7–SCF8 | Concepts and concept types | |
| SCF9 | Assertion types (manual or automatic) | |
| SCF10–SCF11 | General Inquirer lexicon | |
| Wikipedia features | WF1 | Wikipedia article titles |
| WF2–WF5 | Wikipedia titles and hyperlinks | |
| WF6 | Wikipedia titles and categories | |
| Vicinity features | CV1–CV2 | Concepts and concept types |
Strategy for discovering concepts
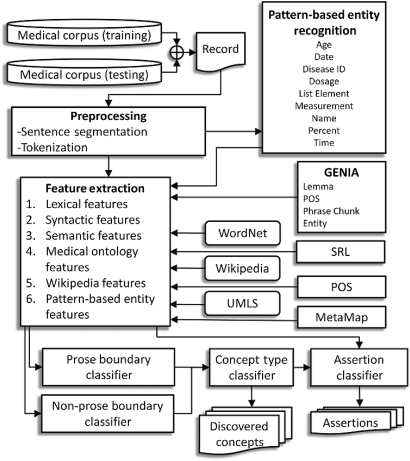
Medical concepts are extracted with supervised machine learning algorithms. Conditional random fields12 (CRF) determine the concept boundaries and SVM13 classify the concept types. CRFs are well suited to sequence classification problems such as the boundary detection problem encountered with concepts, while SVMs perform well on the task of classifying a single instance as is the case for type classification. We chose to separate boundary (concept start and end tokens) and type (problem, test, or treatment) classification in this way because different concept types often share similar contexts.
We use one CRF classifier for text identified as ‘prose’ (grammatical natural language) and another CRF classifier for non-prose (eg, a list of items, a field in the discharge summary). This allows us to use different sets of features for prose and non-prose concept boundary detection. In prose text our classifier relies more on NLP features such as part-of-speech tags and phrase chunks, which assume grammatical sentences. In non-prose text NLP features are less effective and so our boundary classifier relies more on pattern-based contextual features (eg, a nearby measurement, date, or list indicator). The output of both classifiers is given to an SVM to decide if each concept is a problem, test, or treatment. The architecture of our concept extraction method is shown in figure 4.
Figure 4.
Architecture for discovering concepts in text.
For concept discovery features, we use a combination of off-the-shelf external resources and in-house tools to provide a range of features to the classifier. Features from external resources combine medical knowledge from MetaMap,14 UMLS,15 and GENIA3; open-domain knowledge from Wikipedia and WordNet4; as well as SRL.9 Additionally we use a variety of lexical features to capture word context and pattern-based entities. Our pattern-based entity recognizer uses regular expressions to identify names, ages, dates, times, list elements, percentages, measurements, dosages, and ICD-9 disease IDs.
Strategy for classifying assertions
Assertion types for medical problems (eg, conditional for ‘dyspnea on exertion’) are determined using a single SVM.13 For features, we primarily rely on lexical features from the words of the medical problem and in the problem's sentential context along with features based on the General Inquirer, similar to the relation features described above. Additionally, we use a section header feature to gather extra-sentential context.
Experiment for relation extraction using manually annotated concepts and assertions
The first experiment is similar to the submission for the i2b2 2010 relation identification task, by using manually annotated concept and assertion annotations. This allows us to evaluate the relation extraction method independently. The training set consists of 349 discharge summaries and progress notes, containing 5264 relations, while the test set was 477 medical records with 9069 relations.
Experiment for relation extraction using automatically discovered concepts and assertions
In this experiment we consider a more realistic setting in which manual concepts and assertions have not been provided. We use the strategy for discovering concepts and the strategy for classifying assertions discussed previously. The approach for discovering concepts has previously been evaluated in the i2b2 2010 Challenge and achieved an F1 score of 79.59 for extracting concepts and their types. Similarly, the assertion system achieved an accuracy of 92.75% for assigning assertion types. These methods are reported in Roberts et al.16
There is an inherent bias that results from training and making predictions on the same data. Moreover, because the relation extraction system requires annotations of concepts and assertions, we had to create similar models of training and testing for concept discovery and assertion classification prior to training the relation extraction system. This was achieved by performing stacked learning.17 The training data are split into 10 partitions and the system that annotates one partition uses a model trained on the remaining partitions. In this way, the concept and assertion models can provide annotations for all of the training data, which can then be used by the relation extraction strategy.
Results
The relation extraction method was evaluated in two ways: (M1) by measuring the correctness of the relation type identified between two concepts; and (M2) by measuring only the correctness of identifying whether a relation is present between two concepts. The scoring metric is the F1 measure. The F1 measure is defined as F1=2×P×R/(P+R), where P is the precision, and R is the recall. Precision measures the fraction of automatically discovered relations which were correct over all the identified relations. Recall measures the fraction of relations that were identified over all relations that exists and should be identified in the text. We have considered two metrics: M1 to evaluate the relation types, and M2 to evaluate the relation arguments alone. M1 and M2 are evaluated using the F1 measure. Table 2 illustrates the results for our method during the 2010 i2b2 Challenge.
Table 2.
Results of the relation identification method operating on gold standard data
| Evaluation | P | R | F1 |
| Relation type (M1) | 72.0 | 75.3 | 73.7 |
| Relation (M2) | 78.8 | 82.4 | 80.5 |
| TrIP relation type | 56.2 | 29.8 | 38.9 |
| TrWP relation type | 27.8 | 3.5 | 6.2 |
| TrCP relation type | 54.2 | 56.5 | 55.3 |
| TrAP relation type | 70.7 | 81.4 | 75.7 |
| TrNAP relation type | 43.2 | 19.9 | 27.2 |
| PIP relation type | 66.4 | 72.6 | 69.4 |
| TeRP relation type | 82.5 | 90.6 | 86.4 |
| TeCP relation type | 45.6 | 59.4 | 51.6 |
P, precision; R, recall.
As shown in table 2, the results obtained when using the M1 metric are 8.5% lower than the results obtained when M2 was used. M2 measures how well the method is identifying the presence of any relation and acts as an upper bound for M1 which additionally considers the relation type. This indicates that relation type classification accounts for only 32.3% of our total error rate when evaluated by the M1 metric. Therefore, we believe that focusing future work on improving the detection of relations is more important than the type classification of relations. In addition, table 2 details the results that were obtained on each of the eight types of relation. Our method obtained the best results on the TeRP, TrAP, and PIP relations. These are also the most common relation types in the training data, accounting for 83.5% of the relations in the training data and 82.8% in the test data. TeRP occurs 1734 times in the training data. In contrast, the TrWP relation type occurs only 56 times in the training data, TrNAP occurs 106 times, and TrIP occurs 107 times. We believe the small number of training instances is the primary reason for the lower F1 scores for these relations.
Table 3 shows the degradation of the final F1 score evaluated with the metric M1 when a single feature set is removed. The results show that removing context features had the biggest impact. The similarity features, which also capture context, have the next largest impact. We also examined the impact that the removal of each feature set had on the results for individual relation types. The ablation evaluation indicated that the removal of any of the feature sets resulted in a similar degradation across all relation types. We conclude that no feature set impacts only one of the relations we extracted. Moreover, because our features seem to capture similar correlations, we believe that another way of measuring the impact of each feature set could also be considered. Therefore, we performed another evaluation where the relation extraction method used only one set of features, and then expanded that by adding one set at a time.
Table 3.
Degradation to scores for the M1 metric when each feature set was removed from the full system
| Removed feature set | F1 (M1) | Impact |
| Context features | 70.0 | −3.7 |
| Similarity features | 72.6 | −1.1 |
| Nested relation features | 73.2 | −0.5 |
| Single concept features | 73.4 | −0.3 |
| Wikipedia features | 73.4 | −0.3 |
| Concept vicinity features | 73.6 | −0.1 |
Table 4 shows the F1 score enhancement as the different feature sets are made available to our system. We considered each set of features separately and found that the similarity features, when used alone, result in the best F1 score, namely 65.8%. Then we evaluated the system when each of the remaining sets of features was added. We found that the context features added to the similarity features performed best, obtaining an F1 score of 71.3. In the same way we have continued adding features, obtaining the results listed in table 4.
Table 4.
Scores for metric M1 starting with similarity features only, and building up to the full system
| Feature sets used | F1 |
| Similarity features | 65.8 |
| + Context features | 71.3 |
| + Single concept features | 72.8 |
| + Concept vicinity features | 72.7 |
| + Wikipedia features | 73.2 |
| + Nested relation features | 73.7 |
When trained on automatically discovered concepts and assertions, the relation extraction method achieved an F1 score of 48.4, a precision of 57.6 and a recall of 41.7 when using the M1 measure. For the more lenient M2 metric, the method achieved an F1 of 54.2, a precision of 64.6, and a recall of 46.7. These results indicate a serious degradation of the performance of the relation extraction. An approach which attempts to jointly discover concepts and relations simultaneously would likely improve on this performance because the information from relations can inform concept discovery and assertion classification.
Discussion
Our relation extraction method achieved the highest score in the 2010 i2b2 NLP Challenge,1 with an F1 measure of 73.65. The importance of a machine learning approach is emphasized by the fact all submissions in the top 10 evaluated in the challenge used a supervised classifier, rather than only sets of rules. The rationale is driven by the availability of large sets of training data.
Our relation extraction approach made use of many existing NLP tools and knowledge resources, which made this work much easier. The availability of high quality, freely available NLP tools for POS tagging, phrase chunking, and syntactic and semantic parsing enable researchers to focus on new problems of interest in medical informatics.
Our evaluation showed that each set of features benefited the extraction of all types of relations. However, some individual features provide information which is more useful to the extraction of a specific relation. For instance, the Wikipedia category feature WF6 will primarily aid the PiP relation (defined as a relation between two medical problems), because those features rely on the relatedness of two concepts within a semantic class hierarchy. The predicate-based features (CF12–CF14) are useful only to relations which are expressed by a verb such as ‘improved’ (TrIP: treatment improves problem) or ‘revealed’/‘showed’ (TeRP: test revealed problem). There are other features that have similarly limited impact, such as SCF9 which uses assertion types determined by the assertion classification strategy. Since assertion types are only available for problems, this feature helps the extraction of PiP relations the most. The sentiment features SCF10 and SCF11 improve results for relations such as TrIP (treatment improves problem) or TrWP (treatment worsens problem) as those relations incorporate sentiment (improve/worsen).
Despite our success we believe that the results on automatic relation extraction on EMRs can be further improved. During development we considered incorporating several medical ontologies to increase our coverage of medical entities. While we did not see significant benefits from these knowledge bases, it is possible they could be more intelligently mined for knowledge. In addition, our approach to relation extraction primarily treated each relation independently, with the exception of the nested relation features. We believe that an approach which attempts to jointly infer all relations from a sentence would see improved results. We will be addressing these issues in our future work.
Conclusion
We have developed a state of the art method that automatically extracts relations between medical concepts. We found that relation extraction benefits from lexical, syntactic, and semantic context features. In addition, knowledge sources such as Wikipedia proved to improve relation extraction by providing information about whether two concepts are strongly associated. We also performed a more realistic evaluation of the method when using automatically discovered concepts and assertions which showed that the quality of concept discovery substantially impacts the quality of relation extraction. This makes us believe that jointly learning systems for concept and relation discovery is a good direction for future work.
Acknowledgments
The authors would like to thank the i2b2 2010 organizers and the VA for providing an invaluable dataset without which this work would not have been possible. We would also like to thank the reviewers for their helpful comments.
Footnotes
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Uzuner O, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aly M. Survey on Multiclass Classification Methods. Technical report, California Institute of Technology, 2005 [Google Scholar]
- 3.Kulick S, Bies A, Liberman M, et al. Integrated annotation for biomedical information extraction. In: Proc. of the Human Language Technology Conference and the Annual Meeting of the North American Chapter of the Association for Computational Linguistics (HLT/NAACL), 2004 [Google Scholar]
- 4.Fellbaum C. WordNet: An Electronic Lexical Database. MA: MIT press Cambridge, 1998 [Google Scholar]
- 5.Stone PJ, Dunphy DC, Smith MS, et al. The General Inquirer: a Computer Approach to Content Analysis. MA: MIT Press Cambridge, 1966 [Google Scholar]
- 6.Fan RE, Chang KW, Hsieh CJ, et al. LIBLINEAR: a library for large linear classification. J Mach Learn Res 2008;9:1871–4 [Google Scholar]
- 7.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology 2011;2:27:1–27:27 [Google Scholar]
- 8.Pradhan S, Ward W, Hacioglu K, et al. Shallow semantic parsing using support vector machines. Proceedings of HLT/NAACL-2004, 2004:1–8 [Google Scholar]
- 9.Palmer M, Gildea D, Kingsbury P. The proposition bank: an annotated corpus of semantic roles. Comput Ling 2005:71–106 [Google Scholar]
- 10.Levenshtein VI. Binary codes capable of correcting deletions, insertions, and reversals. Sov Phys Dokl 1966;10:707–10 [Google Scholar]
- 11.de Marneffe M, Manning CD. The stanford typed dependencies representation. In: Coling 2008: Proceedings of the workshop on Cross-Framework and Cross-Domain Parser Evaluation. Manchester, United Kingdom: Association for Computational Linguistics, 2008:1–8 [Google Scholar]
- 12.McCallum AK. Mallet: a machine learning for language toolkit. 2002. http://mallet.cs.umass.edu
- 13.Joachims T. SVMLight: support vector machine. SVM-Light Support Vector Machine. University of Dortmund, 1999:4 http://svmlight.joachims.org/ [Google Scholar]
- 14.Aronson AR. Effective mapping of biomedical text to the UMLS metathesaurus: the MetaMap program. Proc AMIA Symp 2001:17–21 [PMC free article] [PubMed] [Google Scholar]
- 15.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system. Methods Inf Med 1993;32:281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts K, Harabagiu SM. A flexible framework for deriving assertions from electronic medical records. J Am Med Inform Assoc 2011;18:568–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolpert DH. Stacked generalization. Neural networks 1992;5:241–59 [Google Scholar]