Abstract
Objective
Concept extraction is a process to identify phrases referring to concepts of interests in unstructured text. It is a critical component in automated text processing. We investigate the performance of machine learning taggers for clinical concept extraction, particularly the portability of taggers across documents from multiple data sources.
Methods
We used BioTagger-GM to train machine learning taggers, which we originally developed for the detection of gene/protein names in the biology domain. Trained taggers were evaluated using the annotated clinical documents made available in the 2010 i2b2/VA Challenge workshop, consisting of documents from four data sources.
Results
As expected, performance of a tagger trained on one data source degraded when evaluated on another source, but the degradation of the performance varied depending on data sources. A tagger trained on multiple data sources was robust, and it achieved an F score as high as 0.890 on one data source. The results also suggest that performance of machine learning taggers is likely to improve if more annotated documents are available for training.
Conclusion
Our study shows how the performance of machine learning taggers is degraded when they are ported across clinical documents from different sources. The portability of taggers can be enhanced by training on datasets from multiple sources. The study also shows that BioTagger-GM can be easily extended to detect clinical concept mentions with good performance.
Keywords: Natural language processing, medical informatics, medical records systems, computerized
Introduction
Concept extraction is a process to identify phrases referring to concepts of interests in unstructured text. In clinical text processing,1 2 specific concept extraction tasks include, for example, detection of phrases referring to disorders, for example, ‘… prior history of tonsil cancerDisorder’ (an example taken from the paper by Ogren et al3). Concept extraction is a subtask of information extraction (IE) that facilitates automated acquisition of structured information from text, and it has been studied across multiple domains, including news articles and biological research literature.4–6 Over the last decade, machine learning methods have achieved excellent performance in concept extraction.7–10 The excellent performance is attributed in part to effective learning algorithms, such as conditional random field (CRF)11 and support vector machine (SVM).8 12 Other factors, such as appropriate tokenization of input text and selection of features encoding properties of tokens, are also important in each application domain.13–15 In the clinical domain, concept extraction has been a critical component of text processing systems and it has been actively studied.16–20 In contrast, application of machine learning for clinical concept extraction appears to be rather recent.21 22 The reason for the deferred application may be because phrases extracted from clinical text need to be normalized to fine-grained concepts, such as those defined in SNOMED CT23 and the Unified Medical Language System (UMLS).24 Matured extraction systems based on dictionaries and hand-coded rules would have an advantage over machine learning for name recognition, as the former approach simultaneously tackles phrase extraction and normalization, and can handle rare concepts, while allowing for case-by-case error correction. Meanwhile, we also believe the deferred application could be due, at least in part, to the difficulty in making clinical text available to the research community because of the privacy and confidentiality issues with clinical data.
Over the last several years, annotated de-identified clinical text has become available to the research community through the i2b2 shared-task workshops on clinical natural language processing (NLP).25–27 The corpus made available in the 2010 challenge is annotated with phrases referring to three concept types: problem, test, and treatment. The corpus consists of four document sets: three sets of discharge summaries from three different institutions and one set of progress notes from one of the three institutions. This corpus is of great interest not only for the rare availability of annotated clinical text, but also for the multiple data sources. Generally, machine learning models improve as the training data size increases, and there can be advantages in gathering data from multiple sources. However, machine learning is sensitive to heterogeneity in the data, such as different vocabularies and writing styles, and there may be disadvantages in combining data from different sources.
To study the portability of machine learning taggers for concept extraction across institutions and document types, we trained and tested taggers on the four data sets made available in the 2010 i2b2/VA Challenge. We also examined how the performance of taggers improves as the size of the training data set increases to determine if additional annotated documents made available in the future would improve system performance further. These experiments were carried out by adapting an existing tagging system, BioTagger-GM, which we originally developed for gene/protein name extraction in the biology domain.
Background
Concept extraction as sequence labeling
Concept extraction based on machine learning is usually formulated as a sequence labeling problem, that is, labeling of tokens (words and punctuations) in an order, where assigned labels indicate demarcation of target concepts. A commonly used set of labels is {B, I, O},28 indicating each token being the beginning of a concept phrase (B), inside of a concept phrase (I), or outside of a concept phrase (O). Powerful machine learning algorithms for sequence labeling include hidden Markov model (HMM),7 maximum entropy (MaxEnt) tagger,29 CRF,11 and SVM.8 12 Notably, efficient implementations of these algorithms are available in downloadable software packages. In this labeling problem, labels of adjacent tokens are dependent on each other. For example, in the 2010 i2b2/VA Challenge corpus, the phrases ‘chest pain,’ ‘chest x-ray,’ and ‘chest tube’ are annotated, respectively, as problem, test, and treatment, and thus the label of ‘chest’ should be predicted along with that of the succeeding token. A machine learning tagger trained with the aforementioned algorithms tries to find the most likely sequence of labels, given a sequence of tokens. Those algorithms are powerful in that they not only model dependencies among nearby labels, but also accommodate rich overlapping/redundant features of tokens, such as a token string (eg, ‘hypertension’) with its prefixes (eg, ‘hyper-’) and suffixes (eg, ‘-ion’). A tagger exploiting generic features can yield good performance,30 yet customized tokenization, features based on domain knowledge, hand-coded post-processing rules, and other fine-tuning can help improve performance.31
Clinical concept extraction using machine learning
In the clinical domain, Wang et al22 developed an annotated corpus and evaluated a concept extraction system based on a combination of a CRF tagger, an SVM classifier, and a MaxEnt classifier. Ten target concept types were defined based on SNOMED CT. A corpus of 311 admission summaries from an intensive care unit was annotated with these concepts. Initially, concept phrases in text were detected using a CRF tagger, and extracted phrases were re-classified into the 10 concept types using SVM and MaxEnt classifiers. Final concept types of phrases were voted among the three systems. The reported F scores of the CRF tagger range from 0.0 (organism, which only had 36 instances in the corpus) to 0.873 (substance, which had 2449 instances). The overall F score of the CRF tagger was 0.799 and that of the system further exploiting SVM and MaxEnt classifiers was 0.823.
Li et al21 examined automated extraction of disorder names from clinical text using CRF and SVM. They used different types of annotated clinical documents (outpatient notes, discharge summaries, and inpatient service notes),3 which they partitioned into a training set and a test set containing, respectively, 1265 and 292 disorder names. They reported the best F scores of 0.86 and 0.64, respectively, for a CRF tagger and for an SVM classifier. The SVM classifier they used did not consider the sequence and dependency of token labels, which may explain its significantly low performance compared to the CRF tagger.
In the 2009 i2b2 workshop on NLP challenges,27 the shared-task was to extract selected concepts from discharge summaries to fill in the ‘fields’ for each medication mention in the summaries. These fields were medication (m), dosage (do), mode (mo), frequency (f), duration (du), and reason (r). A set of 696 discharge summaries provided by Partners HealthCare was released to the participants of the shared-task challenge, of which 17 had been annotated by the organizer as examples. The participants' systems were evaluated on separate 251 summaries. The best performing system by Patrick et al32 used a CRF tagger to extract concept names and an SVM classifier to identify fields of the extracted names (table 1). To train these systems, this team annotated 145 longest summaries among the provided data set, assuming that the longer summaries contained more medication information than shorter ones. The workshop organizer used this same annotated corpus to train MaxEnt taggers and reported competitive extraction performance34 (table 1). A system by Li et al that participated in the challenge workshop employed a CRF tagger for name recognition and AdaBoost with decision stumps for field identification.33 They randomly selected 147 summaries among the provided data, and annotated them to train the machine learning models. After the workshop, Doan et al35 used 268 annotated discharge summaries (17 summaries initially provided as examples and 251 summaries provided as the test set) and evaluated SVM taggers12 (table 1).
Table 1.
Performance (F scores) of machine learning taggers on the 2009 i2b2 Challenge corpus
| Authors | Data | Method | F scores | |||||
| M | Do | Mo | F | Du | R | |||
| Patrick et al32 | 145 longest summaries for training and 251 for testing | CRF and SVM | 0.884 | 0.893 | 0.899 | 0.897 | 0.446 | 0.444 |
| Li et al33 | 147 random summaries for training and the same test set as Patrick et al | CRF and AdaBoost with decision stumps | 0.802 | 0.802 | 0.821 | 0.813 | 0.180 | 0.030 |
| Halgrim et al34 | The same data sets as Patrick et al | MaxEnt tagger | 0.841 | 0.898 | 0.933 | 0.932 | 0.515 | 0.471 |
| Doan et al35 | 10-fold cross-validation on 268 summaries | SVM tagger | 0.812 | 0.864 | 0.947 | 0.889 | 0.214 | 0.333 |
| SVM tagger with a rule-based system | 0.923 | 0.927 | 0.954 | 0.944 | 0.496 | 0.484 | ||
These studies have shown the feasibility and potential of machine learning for clinical concept extraction. Additionally, the studies suggest the significance of the corpus size as well as choices of features in using machine learning methods.32–34
BioTagger-GM
BioTagger-GM is a gene/protein name extraction system that we initially developed to exploit BioThesaurus, an extensive thesaurus of gene/protein names compiled and maintained by Liu et al.36 The system has been evaluated in the Gene Mention (GM) shared-task of BioCreative II, a workshop for the evaluation of biomedical IE and text mining.31 It was developed further after the workshop and tested in our evaluation study.13 It has been used for biological literature mining.37 38
BioTagger-GM is essentially a machine learning tagger exploiting features based on dictionary lookup. The system is tuned to gene/protein name detection. It utilizes two large terminology resources, the UMLS24 and BioThesaurus.36 39 It also uses a set of hand-coded rules to correct prevalent errors by machine learning taggers, such as errors due to coordinated gene/protein names, abbreviations in parenthetical expressions, and name boundaries involving tokenization issues. The system that participated in the BioCreative workshop combines outputs from two machine learning taggers, CRF and HMM, and ranked sixth out of the 19 participating teams. After the competition, we extended it to utilize four taggers trained with three algorithms, CRF, maximum entropy Markov model,29 and HMM, and achieved a performance comparable to the system ranked first in the workshop.
Dictionary lookup
BioTagger-GM uses dictionary matching to derive the UMLS Metathesaurus and BioThesaurus-based features, where both input text and dictionary entries are first normalized to facilitate flexible matching. Besides intensive tokenization of text, the normalization step includes (a) converting tokens (words) to their base forms according to the UMLS SPECIALIST lexicon, (b) changing letters to lower case, (c) ignoring punctuation marks, and (d) converting digit sequences and Greek letters to 9 and G, respectively. All phrase occurrences, including overlapping phrases, are recorded during dictionary lookup (table 2). For example, tokens constituting phrases in the UMLS Metathesaurus are assigned with features indicating the beginning of a phrase and the inside of a phrase, for example, {UMLS_B_<semantic type>, UMLS_I_<semantic type>}, where <semantic type> is a single UMLS semantic type or a compound semantic type (a concatenation of multiple semantic types) if a concept unique identifier assigned to the phrase is associated with more than one semantic type.
Table 2.
An example of UMLS-based features
|
Each token constituting any phrase found in the UMLS is assigned with a UMLS-based feature. For example, ‘aortic valve stenosis’ known as the type Disease or Syndrome (dsyn) from the UMLS is detected in the example sentence, and the tokens ‘aortic,’ ‘valve,’ and ‘stenosis’ are assigned with UMLS_B_bsyn, UMLS_I_dsyn, and UMLS_I_dsyn. Since ‘aortic valve’ is also recorded as the type Body Part, Organ or Organ Component (bpoc) in the UMLS, ‘aortic’ and ‘valve’ are assigned UMLS_B_bpoc and UMLS_I_bpoc, respectively.
Machine learning
BioTagger-GM uses a CRF implementation of MALLET40 as a baseline tagger. Sentences were tokenized and specified with features characterizing tokens and token occurrences in their contexts. The BioCreative II GM task was a concept extraction with one target type, genes/proteins, and thus the system was designed to assign a set of three labels, {B, I, O}. Besides widely-used features for concept extraction, such as nearby words and part of speech tags within a window size, as stated above, BioTagger-GM incorporates dictionary lookup results.
A second-order CRF model trained using the above features outperformed a first-order CRF model and first- and second-order maximum entropy Markov models trained using the same features. It also yielded better performance than a CRF model trained using an existing gene/protein name extraction system, ABNER,41 and an HMM model.42 Meanwhile, gene/protein names identified by multiple models were more likely to be true gene/protein names. Outputs from these models were combined through voting and improved extraction performance was achieved.43 44
Materials and methods
2010 i2bi2/VA Challenge corpus
The training corpus made available in the 2010 i2b2/VA Challenge consists of four data sets: three sets of discharge summaries from Beth Israel Deaconess Medical Center (BETH), Partners HealthCare (PARTNERS), and University of Pittsburgh Medical Center (UPMCD) and one set of progress notes from University of Pittsburgh Medical Center (UPMCP). For the purpose of concept extraction, the corpus was annotated with three concept types: problem, test, and treatment. For example, phrases frequently annotated as problem are ‘hypertension,’ ‘pain,’ ‘low’ (almost all occurrences in UPMCP as laboratory values, eg, ‘Hgb 10.7 gm/dl Lowproblem’), ‘chest pain,’ and ‘afebrile,’ those frequently annotated as test are ‘creatinine,’ ‘glucose,’ ‘blood pressure,’ ‘BUN,’ and ‘hematocrit,’ and those frequently annotated as treatment are ‘coumadin,’ ‘aspirin,’ ‘lasix,’ ‘lisinopril,’ and ‘protonix.’ The corpus is pre-tokenized, and thus the unit of labeling/annotation is fixed. The statistics of documents, lines, and tokens as well as annotated concept phrases are shown in table 3. The basic statistics of the corpus show some differences in the data sets constituting the corpus. For example, the number of documents in BETH (ie, 73) is smaller than in the others, but the number of lines (ie, 8 727) and the number of tokens (ie, 88 722) are larger. The number of annotated concept phrases in BETH is also larger than that of the others.
Table 3.
Basic statistics of the 2010 i2b2/VA Challenge training corpus
| Set name | Document type | Documents | Lines | Tokens | Total concepts | ||
| Problem | Test | Treatment | |||||
| BETH | Discharge summaries | 73 | 8727 | 88 722 | 4187 | 3036 | 3073 |
| PARTNERS | Discharge summaries | 97 | 7515 | 60 819 | 2886 | 1572 | 1771 |
| UPMCD | Discharge summaries | 98 | 7328 | 62 727 | 2728 | 1217 | 2308 |
| UPMCP | Progress notes | 81 | 7025 | 48 302 | 2167 | 1544 | 1348 |
| Total | 349 | 30 597 | 260 570 | 11 968 | 7369 | 8500 | |
The corpus consists of a set of document files and corresponding annotation files. Text in each file is already tokenized, and lines are roughly sentences, or list items.
Adaptation of BioTagger-GM
We participated in the concept extraction task of the 2010 i2b2/VA Challenge by rapidly adapting BioTagger-GM to clinical text. The hand-coded rules for post-processing error correction in BioTagger-GM were taken out because they were highly tuned to gene/protein name mentions in scientific literature, for example, gene/protein synonyms commonly mentioned in parenthetical expressions. As the system was originally designed for one target concept type, genes/proteins, extraction of the three concept types in the i2b2/VA Challenge was tackled as three independent tasks each with a single target concept type. The performance of BioTagger-GM on the evaluation corpus of the i2b2/VA NLP Challenge is shown in table 4.
Table 4.
Performance of BioTagger-GM on the test corpus of the i2b2/VA Challenge
| TP | FP | FN | Precision | Recall | F score | |
| Problem | 14 551 | 2365 | 3999 | 0.860 | 0.784 | 0.821 |
| Test | 9814 | 1555 | 3085 | 0.863 | 0.761 | 0.809 |
| Treatment | 10 279 | 1681 | 3281 | 0.859 | 0.758 | 0.806 |
| Micro average | 34 644 | 5601 | 10 365 | 0.861 | 0.770 | 0.813 |
FN, false negative; FP, false positive; TP, true positive.
The performance of BioTagger-GM is owing to BioThesaurus and the UMLS Metathesaurus, as well as the powerful machine learning algorithms as reported in our previous study.13 In the current task, instead of using BioThesaurus, we used a collection of clinical terms extracted from discharge summaries for a clinical vocabulary viewer.45 The UMLS Metathesaurus was used just as in BioTagger-GM. We employed the first-order CRF model for this task, instead of the second-order model, since the former yielded superior performance in our preliminary tests. Two slight variations of this baseline tagger were considered during our participation in the challenge workshop. As one variant, we supplemented the section header, for example, ‘problem,’ ‘procedure,’ or ‘indication,’ as an additional feature. As the second variant, we adjusted the context window size to derive features encoding nearby tokens. The window size has previously been tuned for gene/protein name recognition in biological literature, where an asymmetric window size yielded better performance, but we considered a wider, symmetric context window. Outputs from these three systems were combined through voting. Improvement though system combination, however, was limited. In the current study, therefore, we only used baseline first-order CRF taggers.
Experiment
We designed our experiment to examine the portability of machine learning systems for concept extraction using BioTagger-GM and the training corpus of the 2010 i2b2/VA Challenge workshop (BETH, PARTNERS, UPMCD, and UPMCP). Our first question was how well a set of annotated phrases in one data set covers concept phrases annotated in another data set. Therefore, we initially examined concept extraction based on dictionary lookup using a dictionary compiled from each data set. In order to overlook minor variations of concept phrases, word tokens were lower-cased and special symbols and determiners (‘a,’ ‘an,’ and ‘the’) were ignored during lookup. The performance was measured by the widely-used F score (F1 score), that is, the harmonic mean of precision (positive predictive value) and recall (sensitivity).
Next, using BioTagger-GM, we trained and evaluated CRF taggers on different combinations of the data sets. Taggers were trained on individual data sets, combinations of any two data sets (eg, BETH and PARTNERS), combinations of any three data sets (eg, BETH, PARTNERS, and UPMCD), and the combination of all four data sets (ie, the training corpus). The trained taggers were evaluated on each of the four data sets. When the training set(s) and the test data set did not overlap (eg, the training data is the combination of BETH, PARTNERS, and UPMCD, and the test data is UPMCP), the entire data sets were used for training and testing. When there was an overlap (eg, training data set consists of BETH, PARTNERS, and UPMCD, and test data set is BETH), 10-fold cross-validation was conducted, and averaged F scores were reported.
Results
Table 5 shows the results from dictionary lookup where the dictionary was compiled from the training document sets. The F scores vary, but they are all low. For example, regarding problem concepts (table 5A), the F scores range from 0.347 (when the lookup approach was evaluated on PARTNERS using a dictionary compiled from UPMCP) to 0.651 (on UPMCP using a dictionary compiled from UPMCP, where 10-fold cross-validation was used because the evaluation set and the dictionary source were the same).
Table 5.
Dictionary lookup using dictionaries compiled from corpora
| Dictionary source | ||||
| BETH | PARTNERS | UPMCD | UPMCP | |
| a. Results for problem concepts | ||||
| Documents | ||||
| BETH | 0.479 (0.532/0.439) | 0.413 (0.530/0.339) | 0.447 (0.785/0.312) | 0.379 (0.519/0.298) |
| PARTNERS | 0.484 (0.664/0.381) | 0.407 (0.437/.0383) | 0.435 (0.747/0.307) | 0.347 (0.462/0.278) |
| UPMCD | 0.542 (0.703/0.441) | 0.443 (0.540/0.375) | 0.500 (0.535/0.483) | 0.401 (0.483/343) |
| UPMCP | 0.567 (0.753/0.454) | 0.461 (0.645/0.359) | 0.518 (0.738/0.399) | 0.651 (0.637/0.670) |
| b. Results for test concepts | ||||
| Documents | ||||
| BETH | 0.518 (0.406/0.733) | 0.497 (0.530/0.339) | 0.447 (0.785/0.312) | 0.463 (0.443/0.463) |
| PARTNERS | 0.409 (0.324/0.555) | 0.413 (0.320/0.622) | 0.435 (0.747/0.307) | 0.435 (0.404/435) |
| UPMCD | 0.349 (0.253/0.561) | 0.528 (0.530/0.528) | 0.344 (0.251/0.581) | 0.457 (0.407/523) |
| UPMCP | 0.466 (0.379/0.607) | 0.445 (0.458/0.432) | 0.518 (0.738/0.399) | 0.516 (0.396/0.756) |
| c. Results for treatment concepts | ||||
| Documents | ||||
| BETH | 0.514 (0.473/0.570) | 0.469 (0.609/0.382) | 0.458 (0.509/0.417) | 0.409 (0.550/0.325) |
| PARTNERS | 0.468 (0.498/0.442) | 0.433 (0.419/0.462) | 0.471 (0.510/0.437) | 0.298 (0.429/0.228) |
| UPMCD | 0.523 (0.593/0.469) | 0.514 (0.705/0.404) | 0.537 (0.525/0.552) | 0.356 (0.622/0.453) |
| UPMCP | 0.480 (0.508/0.455) | 0.463 (0.621/0.369) | 0.487 (0.561/0.431) | 0.567 (0.509/0.658) |
The three table sections, 4A, 4B, and 4C, show F scores (precisions/recalls) for concept extraction using dictionary lookup for the three target concept types, where the dictionaries are compiled from concepts annotated in the training document sets. For instance, the second cell in the top row, 0.413 (0.530/0.339), indicates that the F score of dictionary lookup is 0.413 when the dictionary of phrases annotated in PARTNERS, is evaluated on BETH (based on phrase occurrences, not unique phrases).
The diagonal cell values are underlined in the table to indicate that they were obtained using 10-fold cross-validation (otherwise, the recall of phrases is trivially 100%).
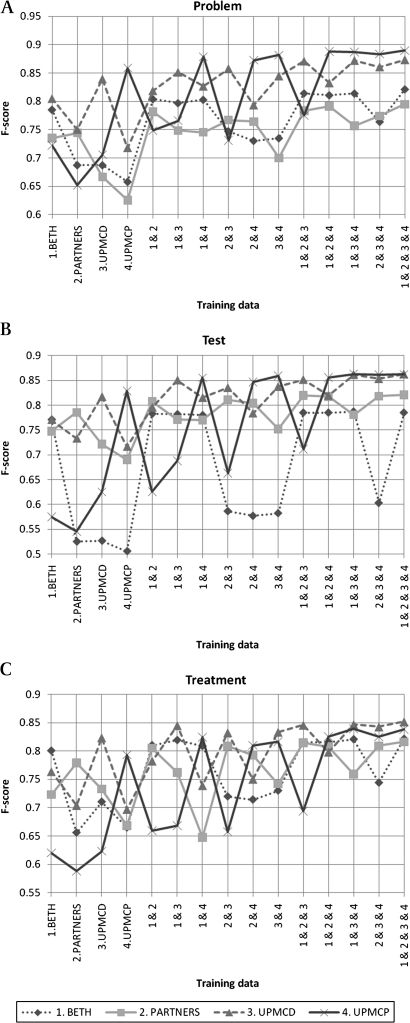
Figure 1 shows the results of the experiments. Across the concept types and the test data sets, taggers achieved good F scores, ranging from 0.787 on BETH for test concepts to 0.890 on UPMCP for problem concepts. Generally, performance of taggers was improved by supplementing an additional data set to the training data. As expected, good F scores could be achieved when text from the same set as the test data was included in the training data. For example, when tested on BETH, a tagger trained on BETH outperformed one trained on PARTNERS. There was relatively large improvement when one additional data set was supplemented. The improvement, however, was limited when more data sets were supplemented further; the impact of supplementing a data set becomes smaller.
Figure 1.
F scores of taggers. Each figure (A, B, and C) shows F scores evaluated for different concept types (problem, test, and treatment). There are four lines in each figure corresponding to the four data sets used as the test corpora of a tagger (1. BETH, 2. PARTNERS, 3. UPMCD, and 4. UPMCP). The horizontal axis indicates a single data set (eg, BETH, the leftmost) or a combined data set (eg, all four combined (1&2&3&4), the rightmost) used as the training corpus of a tagger. For example, in each of the three figures, the 15 diamond marks connected with the dotted line show the F scores of 15 taggers trained on different training corpora (BETH, PARTNERS, …, and all four combined) and tested on BETH. (A). Results for Problem concepts. (B). Results for Test concepts. (C). Results for Treatment concepts.
In the most cases, the best F scores were observed when all four data sets were used for training, except for a few cases (eg, when tested on BETH, the tagger trained on all the four data sets achieved the averaged F score of 0.785, but the tagger trained on three data sets, BETH, UPMCD, and UPMCP, achieved the slightly better averaged F score of 0.787). Even for such exceptional cases, taggers trained on all four data sets achieved the near-best F scores. They performed well on any of the four data sets. Meanwhile, in terms of a specific test data set, near-best F scores can be achieved for taggers trained on the combinations of three or sometimes two data sets. For example, when tested on UPMCP, the tagger trained on the combination of BETH and UPMCP and that trained on the combination of BETH, PARTNERS, and UPMCP achieved F scores of 0.882 and 0.888, respectively, which are close to the 0.890 obtained using all four data sets. In other words, as stated earlier, improvement of taggers by supplementing the fourth (or even third) additional data set to the training data was small, and sometimes none.
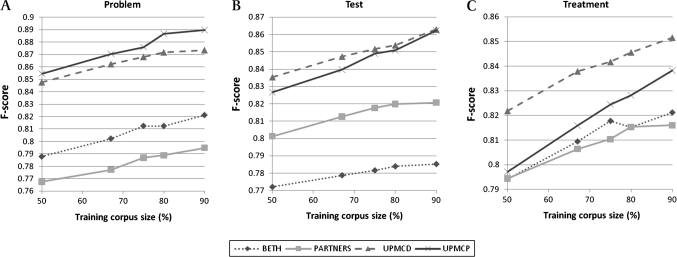
The observation above raised the question if it was the case that the performance of taggers almost reached the limit after combining the four data sets, or if it was the case that further improvement could be expected when text from the same sources could be supplemented. We were unable to answer this question directly by supplementing additional clinical text. Therefore, we varied the training data size (conducted n-fold cross-validation with varying n) to observe the trajectory of the F scores (figure 2). In figure 2, for most of the concept types and the test data sets, the F scores appear to improve steadily, without being tapered off, as the training data size increases. This result suggests that tagger performance is likely to improve as more data become available.
Figure 2.
F scores of taggers trained on different sizes of training corpora. Cross-validation tests (m×n-fold cross-validation tests) were conducted using the entire data sets for (m,n)=(5,2), (4,3), (3,4), (2,5), and (1,10). In these tests, since the corpus was partitioned into n subsets, the sizes of the training corpus were, respectively, 50%, 67%, 75%, 80%, and 90%.
Discussion
Our experiments show that BioTagger-GM, originally developed for gene/protein name extraction, can be adapted for clinical concept extraction. In fact, despite the relatively limited resources we could afford during our participation in the 2010 i2b2/VA shared-task challenge, the resulting taggers performed reasonably well as demonstrated in this paper. Meanwhile, lessons have been learned from our participation in the shared-task challenge as discussed below.
Although it has been well known and also observed in past studies in the clinical domain,32 33 46 we confirmed the significance of the data resources in developing machine learning models, including large and high quality corpora and dictionary resources. Implementations of powerful machine learning algorithms have been available in downloadable software packages, for example, MALLET.40 Provided with an annotated corpus, one can readily develop an acceptable concept extraction system. Improved performance in a particular domain can be achieved using concept dictionaries as exemplified by BioTagger-GM. In retrospect, BioTagger-GM has an advantage in rapid adaptation because dictionary lookup results are simply encoded as one type of feature for machine learning, and the adaptation process was primarily to replace BioThesaurus with a set of phrases collected in the clinical domain. The system may be further refined though fine-tuned features, hand-coded post-processing rules, system ensemble, and their effective combination as seen in past shared-task systems and post shared-task systems in the biology domain.13–15 Meanwhile, intensive system tuning can be a time-consuming process, and the gain could be smaller as the performance improves. Moreover, some ad hoc solutions, such as hand-coded error correction rules, can be highly specific not only to the application domain, but also to the development corpora and annotation guidelines. When we ported BioTagger-GM to the clinical domain, we observed no improvement or degradation of the performance for hand-coded error correction rules that we created for gene/protein name extraction. It is important to consider the trade-off between intensive system tuning and system portability.
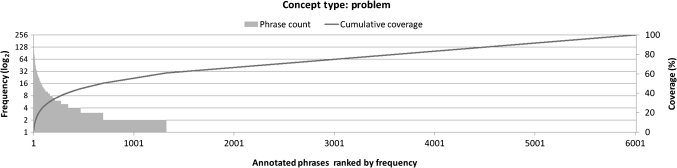
The current study showed that a clinical concept tagger may be ported to other institutions/document types, while the degradation of tagger performance can vary. When ported, taggers trained on BETH appear to perform relatively well, and we believe this is owing to the large concept vocabulary used in the data set. In clinical text, concept names frequently appear in lists27 and/or in coordination, for example, ‘a history of type 2 diabetesproblem, high cholesterolproblem, and coronary artery diseaseproblem.' For such name occurrences, there is little contextual clue for concept extraction, and we believe the performance of the system is highly dependent on the concept vocabulary in the training corpus and/or additional thesauri the system has access to. The distribution of annotated concept phrases follows Zipf's law as seen in figure 3, and there would always be many unseen concept phrases for a concept tagger. In creating an additional annotated corpus to improve the performance of concept extraction, it would be efficient to review clinical text using diverse concept phrases with higher concept occurrences. In practice, when a fixed number of documents are annotated, selecting longer documents that are likely to contain more concept phrases may be effective.32
Figure 3.
Frequency of concept phrases annotated as Problem. The horizontal axis represents distinctive Problem phrases ranked from 1 to 6009 by their frequencies. The bar graphs show the frequencies of each phrases in log2 scale (left vertical axis) and the line graph shows the cumulative percentage of phrase frequency over all Problem phrase occurrences (left vertical axis). The phrase ranked first is ‘hypertension.’ It is annotated 147 times, which constitutes 1.2% of all Problem phrase occurrences (11 968). Nearly 80% of Problem phrases appear only once in the corpus.
The significance of the vocabulary/dictionary does not mean contextual clues were not helpful for concept extraction, since we could observe certain helpful phrases near annotated phrases. For example, as the contextual indications of the type problem, we observed ‘history of’ and ‘evidence of’ to the left of phrase occurrences, and ‘secondary to’ and ‘consistent with’ to the right. There were clues indicative of more than one concept type, such as ‘patient/he/she underwent’ as the left context for test and treatment, and ‘showed’ and ‘revealed’ as the right context for problem and test. Concept extraction is indeed a complex problem involving multiple clues present in text. Powerful machine learning algorithms that could exploit many overlapping clues (features), such as CRF, would be desirable in this task.
Conclusion
We showed that BioTagger-GM, a system to train a concept tagger given a domain corpus and terminology resources, can be adapted to the clinical domain and can achieve good performance. BioTagger-GM uses domain knowledge embedded in a terminology resource simply as one type of feature for a machine learning model, and it could readily incorporate information sources suited for a new application domain. We also showed the varying performance in porting a clinical concept tagger to another institution/document type. In general, taggers trained on data from more than one source can perform well, and they tend to be more robust when ported across institutions and/or document types. While the corpus annotated for the 2010 i2b2/VA Challenge is fairly large, performance of machine learning taggers appears to improve further when an additional training corpus is provided. In real-life IE applications, a primary challenge to machine learning-based concept taggers would be normalizing extracted phrases to entries in a control vocabulary, such as SNOMED CT or the UMLS. Despite this and other challenges toward practical application of machine learning in the clinical domain, we believe the increased accessibility of clinical text for researchers will advance the field, and will help managing and mining information in unstructured clinical text.
Acknowledgments
We thank those who developed and made available the machine learning packages, natural language processing tools, terminological resources, and labeled corpora mentioned in this study.
Footnotes
Funding: This study was supported by the National Science Foundation (ABI: 0845523) and the National Institute of Health (R01LM009959A1). De-identified clinical records used in this research were provided by the i2b2 National Center for Biomedical Computing funded by U54LM008748 and were originally prepared for the Shared Tasks for Challenges in NLP for Clinical Data organized by Dr Ozlem Uzuner, i2b2 and SUNY.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform 2009;42:760–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008:128–44 [PubMed] [Google Scholar]
- 3.Ogren P, Savova G, Chute C. Constructing evaluation corpora for automated clinical named entity recognition. In: Proceedings of the Sixth International Language Resources and Evaluation (LREC'08) at Marrakech, Morocco, Paris, France: European Language Resources Association, 2008:3143–50 [Google Scholar]
- 4.Nadeau D, Sekine S. A survey of named entity recognition and classification. Lingvisticae Investigationes 2007;30:3–26 [Google Scholar]
- 5.Krauthammer M, Nenadic G. Term identification in the biomedical literature. J Biomed Inform 2004;37:512–26 [DOI] [PubMed] [Google Scholar]
- 6.Tran N, Luong T, Krauthammer M. Mapping terms to UMLS concepts of the same semantic type. In: AMIA Annu Symp Proc, Bethesda, MD: American Medical Informatics Association, 2007:1136. [PubMed] [Google Scholar]
- 7.Bikel DM, Miller S, Schwartz R, et al. Nymble: a high-performance learning name-finder. In: Proceedings of the fifth Conference on Applied Natural Language Processing at Washington, DC, Stroudsburg, PA: Association for Computational Linguistics, 1997:194–201 [Google Scholar]
- 8.Altun Y, Tsochantaridis I, Hofmann T. Hidden Markov Support Vector Machines. In: Proceedings of the Twentieth International Conference on Machine Learning (ICML-2003) at Washington, DC, Menlo Park, CA: AAAI Press, 2003:3–10 [Google Scholar]
- 9.McCallum A, Li W. Early results for named entity recognition with conditional random fields, feature induction and web-enhanced lexicons. In: Proceedings of the Seventh Conference on Natural Language Learning at HLT-NAACL at Edmonton, Canada, Stroudsburg, PA: Association for Computational Linguistics, 2003:290–4 [Google Scholar]
- 10.Ratinov L, Roth D. Design challenges and misconceptions in named entity recognition. In: Proceedings of the Thirteenth Conference on Computational Natural Language Learning (CoNLL-2009) at Boulder, CO, Stroudsburg, PA: Association for Computational Linguistics, 2009:147–55 [Google Scholar]
- 11.Lafferty J, McCallum A, Pereira F. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. In: Proceedings of ICML 2001 at Williamstown, MA, San Francisco, CA: Morgan Kaufmann Publishers, 2001:282–9 [Google Scholar]
- 12.Kudo T, Matsumoto Y. Chunking with support vector machines. In: Proceedings of the Second Meeting of the North American Chapter of the Association for Computational Linguistics at Pittsburgh, PA, San Francisco, CA: Morgan Kaufmann Publishers, 2001:192–9 [Google Scholar]
- 13.Torii M, Hu Z, Wu CH, et al. BioTagger-GM: a gene/protein name recognition system. J Am Med Inform Assoc 2009;16:247–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leaman R, Gonzalez G. BANNER: an executable survey of advances in biomedical named entity recognition. Pac Symp Biocomput 2008:652–63 [PubMed] [Google Scholar]
- 15.Hsu CN, Chang YM, Kuo CJ, et al. Integrating high dimensional bi-directional parsing models for gene mention tagging. Bioinformatics 2008;24:i286–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010;17:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haug PJ, Koehler S, Lau LM, et al. Experience with a mixed semantic/syntactic parser. In: Proc Annu Symp Comput Appl Med Care, Bethesda, MD: American Medical Informatics Association, 1995:284–8 [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng QT, Goryachev S, Weiss S, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak 2006;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, Schuler K, Savova G. Conditional random fields and support vector machines for disorder named entity recognition in clinical texts. In: Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing at Columbus, OH, Stroudsburg, PA: Association for Computational Linguistics, 2008:94–5 [Google Scholar]
- 22.Wang Y, Patrick J. Cascading classifiers for named entity recognition in clinical notes. In: Proceedings of the Workshop on Biomedical Information Extraction at Borovets, Bulgaria, Stroudsburg, PA: Association for Computational Linguistics, 2009:42–9 [Google Scholar]
- 23.Stearns MQ, Price C, Spackman KA, et al. SNOMED clinical terms: overview of the development process and project status. In: Proc AMIA Symp, Bethesda, MD: American Medical Informatics Association, 2001:662–6 [PMC free article] [PubMed] [Google Scholar]
- 24.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32:D267–70 (Database issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uzuner O, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007;14:550–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uzuner O, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008;15:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramshaw L, Marcus M. Text chunking using transformation-based learning. In: Proceedings of the Third Workshop on Very Large Corpora at Somerset, NJ, Stroudsburg, PA: Association for Computational Linguistics, 1995:82–94 [Google Scholar]
- 29.McCallum A, Freitag D, Pereira F. Maximum entropy Markov Models for Information Extraction and Segmentation. In: Proceedings of ICML 2000 at Stanford, CA, San Francisco, CA: Morgan Kaufmann Publishers, 2000:591–8 [Google Scholar]
- 30.Carpenter B. LingPipe for 99.99 % Recall 1 LingPipe for 99.99 % Recall of Gene Mentions. Paper presented at: the Second BioCreative Challenge Evaluation Workshop at Madrid, Spain, Madrid, Spain: Centro Nacional de Investigaciones Oncológicas, 2007 [Google Scholar]
- 31.Smith L, Tanabe LK, Ando RJ, et al. Overview of BioCreative II gene mention recognition. Genome Biol 2008;9(Suppl 2):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patrick J, Li M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J Am Med Inform Assoc 2010;17:524–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Liu F, Antieau L, et al. Lancet: a high precision medication event extraction system for clinical text. J Am Med Inform Assoc 2010;17:563–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halgrim S, Xia F, Solti I, et al. Extracting medication information from discharge summaries. In: Proceedings of the NAACL HLT 2010 Second Louhi Workshop on Text and Data Mining of Health Documents at Los Angeles, CA, Stroudsburg, PA: Association for Computational Linguistics, 2010:61–7 [Google Scholar]
- 35.Doan S, Xu H. Recognizing medication related entities in hospital discharge summaries using support vector machine. Poster presented at: Coling 2010 at Beijing, China, Stroudsburg, PA: Association for Computational Linguistics, 2010 [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Hu ZZ, Zhang J, et al. BioThesaurus: a web-based thesaurus of protein and gene names. Bioinformatics 2006;22:103–5 [DOI] [PubMed] [Google Scholar]
- 37.Lee CM, Torii M, Hu Z-Z, et al. Evaluating gene/protein name tagging and mapping for article retrieval. In: Proceedings of the Fourth International Symposium on Semantic Mining in Biomedicine (SMBM) at Hinxton, UK, Ed. Collier N, Hahn U, Rebholz-Schuhmann D, Rinaldi F, Pyysalo S. 2010:104–9
- 38.Torii M, Lee CM, Hu Z-Z, et al. Collecting short-long form pairs from MEDLINE specifically for gene/protein entities. Poster presented at: the Fourth International Symposium on Semantic Mining in Biomedicine (SMBM) at Hinxton, UK, 2010
- 39.Liu H, Hu ZZ, Torii M, et al. Quantitative assessment of dictionary-based protein named entity tagging. J Am Med Inform Assoc 2006;13:497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCallum AK. MALLET: A Machine Learning for Language Toolkit. 2002. http://mallet.cs.umass.edu [Google Scholar]
- 41.Settles B. ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text. Bioinformatics 2005;21:3191–2 [DOI] [PubMed] [Google Scholar]
- 42.Alias-i LingPipe. http://alias-i.com/lingpipe
- 43.Torii M, Liu H. At-Least-N Voting Over Biomedical Named Entity Recognition Systems. Paper presented at: the Annual Meeting of the ISMB BioLINK Special Interest Group on Text Data Mining (BioLINK 2008) at Toronto, Canada, 2008 [Google Scholar]
- 44.Kambhatla N. Minority vote: at-least-N voting improves recall for extracting relations. Poster presented at: COLING/ACL 2006 at Sydney, Australia, Stroudsburg, PA: Association for Computational Linguistics, 2006 [Google Scholar]
- 45.Friedman C, Liu H, Shagina L. A vocabulary development and visualization tool based on natural language processing and the mining of textual patient reports. J Biomed Inform 2003;36:189–201 [DOI] [PubMed] [Google Scholar]
- 46.Clark C, Good K, Jezierny L, et al. Identifying smokers with a medical extraction system. J Am Med Inform Assoc 2008;15:36–9 [DOI] [PMC free article] [PubMed] [Google Scholar]