Abstract
Objective
The authors' goal was to develop and evaluate machine-learning-based approaches to extracting clinical entities—including medical problems, tests, and treatments, as well as their asserted status—from hospital discharge summaries written using natural language. This project was part of the 2010 Center of Informatics for Integrating Biology and the Bedside/Veterans Affairs (VA) natural-language-processing challenge.
Design
The authors implemented a machine-learning-based named entity recognition system for clinical text and systematically evaluated the contributions of different types of features and ML algorithms, using a training corpus of 349 annotated notes. Based on the results from training data, the authors developed a novel hybrid clinical entity extraction system, which integrated heuristic rule-based modules with the ML-base named entity recognition module. The authors applied the hybrid system to the concept extraction and assertion classification tasks in the challenge and evaluated its performance using a test data set with 477 annotated notes.
Measurements
Standard measures including precision, recall, and F-measure were calculated using the evaluation script provided by the Center of Informatics for Integrating Biology and the Bedside/VA challenge organizers. The overall performance for all three types of clinical entities and all six types of assertions across 477 annotated notes were considered as the primary metric in the challenge.
Results and discussion
Systematic evaluation on the training set showed that Conditional Random Fields outperformed Support Vector Machines, and semantic information from existing natural-language-processing systems largely improved performance, although contributions from different types of features varied. The authors' hybrid entity extraction system achieved a maximum overall F-score of 0.8391 for concept extraction (ranked second) and 0.9313 for assertion classification (ranked fourth, but not statistically different than the first three systems) on the test data set in the challenge.
Keywords: Natural language processing; machine learning; named entity recognition; clinical text processing; NLP; ethical study methods; Text generation from discrete data; Classical experimental and quasi-experimental study methods (lab and field); Developing and refining EHR Data standards (including image standards); Controlled terminologies and vocabularies; Human–computer interaction and human-centered computing; Surveys and needs analysis; Qualitative/ethnographic field study; policy; Legal; historical; Automated learning; Translational research—application of biological knowledge to clinical care; Uncertain reasoning and decision theory; Linking the genotype and phenotype; languages; computational methods; Advanced algorithms; discovery; text and data mining methods; Knowledge representations, Knowledge bases
Introduction
In 2010, the Center of Informatics for Integrating Biology and the Bedside (i2b2) at Partners Health Care System and Veterans Affairs (VA) Salt Lake City Health Care System organized a challenge in natural-language processing (NLP) for clinical data. The challenge had three tiers: (1) extracting clinical concepts from natural language text, to include medical problems, tests, and treatments; (2) classifying assertions made about medical problems; and (3) identifying the relations among medical problems, tests, and treatments. The data set used in the challenge included discharge summaries and some progress notes obtained from three institutions: Partners HealthCare, Beth Israel Deaconess Medical Center, and University of Pittsburgh Medical Center. The organizers manually annotated 826 clinical notes, which served as a gold standard for three tasks in the challenge. For the challenge, 349 annotated clinical notes were used as a training set, and the remaining 477 annotated notes were used as a test set to evaluate the performance of participating systems. In this paper, we describe a novel hybrid system that combines machine learning (ML) and rule-based methods to accurately extract clinical entities and their assertions, and it was ranked second in the concept extraction task of the i2b2/VA challenge. In addition, as part of this work, we investigated the effects of features and ML algorithms on clinical entity recognition, and we conducted a manual analysis to compare the ML-based approach with existing NLP systems that use dictionaries for entity recognition.
Background
Narrative text is the primary communication method in the medical domain. Much of the important patient information is only found in clinical notes in electronic medical records. NLP technologies offer a solution to convert free text data into structured representations. Over the last two decades, there have been many efforts to apply NLP technologies to clinical text. The Linguistic String Project,1 2 the Medical Language Extraction and Encoding System (MedLEE)3–5 and SymText/MPlus6–8 are a few of the earliest NLP systems developed for clinical domain. More recently, open source clinical NLP systems such as cTAKES9 and HiTEX10 have also been introduced into the community. Most of clinical NLP systems can extract various types of named entities from clinical text and link them to concepts in the Unified Medical Language System (UMLS), such as MetaMap11 and KnowledgeMap.12 After a clinical concept is identified, many applications require determination of its assertion (ie, is a medical condition present or absent?). Among various negation detection algorithms, NegEx13 has arguably been used the most widely and has been incorporated into many systems.
Identification of clinically relevant entities from text is an important step for any clinical NLP system. It is a type of Named Entity Recognition (NER) task, which is to locate and classify words/phrases into predefined semantic classes such as person names, locations, and organizations. When mining biomedical literature, researchers have developed various NER methods for biological entities, such as gene/protein names, including rule-based methods that rely on existing biomedical databases/dictionaries,14 and ML-based methods that are trained on available annotated data sets.15 Hybrid approaches, which combine both rule-based and ML methods, have shown good performance on gene-name recognition tasks.16
Despite their success in biomedical literature NER tasks, ML-based methods have not been studied extensively for NER in clinical notes. To the best of our knowledge, there has been no comprehensive study that focuses on ML-based approach for recognition of broad types of clinical entities before the 2010 i2b2/VA challenge. In the 2009 i2b2 NLP challenge, two teams reported ML-based NER methods for clinical text; however, the scope was limited to medication-related entities only.17 18 One relevant study investigated the contribution of syntactic information to semantic categorization of words in discharge summaries using Support Vector Machines (SVM)19; but the study was done on a small data set with 48 clinical notes. In this paper, we describe a systematic investigation on ML-based approaches for recognizing broad types of clinical entities and determining their assertion status, and report a new hybrid clinical entity extraction framework, which achieved good performance in the i2b2/VA NLP challenge.
Methods
ML-based approaches for clinical NER
The i2b2/VA Concept Extraction task is a typical NER task, which requires determination of the boundaries of clinical entities and assignment of their semantic types (problem, test, or treatment). We transformed the annotated data into the ‘BIO’ format,20 in which each word was assigned into a label as follows: B=beginning of an entity, I=inside an entity, and O=outside of an entity. For example, the sentence ‘No active bleeding was observed’ will be labeled as ‘No/O active/B bleeding/I was/O observed/O,’ if ‘active bleeding’ is annotated as an entity. The NER task then becomes a classification task—to assign each word into one of the three labels (B, I, or O) based on the characteristics of each word and its context. As there were three types of entities in the challenge, we defined three different B classes and three different I classes. For example, for medical problems, we defined the B class as ‘B-problem,’ and the I class as ‘I-problem.’ Therefore, we had a total of seven possible labels of classes (including the O class). A multiclass classifier was then built to assign the label for each word. Different ML algorithms and different types of features were investigated in this study.
ML algorithms
Two ML algorithms, Conditional Random Fields (CRF)21 and Support Vector Machines (SVM),22 which have been widely used in biological NER such as gene names,16 23–26 were investigated in this study. For CRF, we used the CRF++ package (http://crfpp.sourceforge.net/), which has been used for various NER tasks.27 28 For SVM, we used TinySVM along with Yamcha (http://chasen.org/∼taku/software/TinySVM) developed at NAIST.29 30 We used a polynomial kernel function with the degree of kernel as two, a context window of two, and the pairwise (one-against-one) strategy for multiclassification, based on reported biomedical NER tasks such as those reported in previous literature.23–25
Types of features
We systematically investigated various types of features that were extracted from the word itself and its context, including:
Word Level Information: Bag-of-word, Orthographic information—such as capitalization of letters in words, and prefixes and suffixes of words;
Syntactic Information: Part of Speech tags obtained using MedPOST31 (http://www.ncbi.nlm.nih.gov/staff/lsmith/MedPost.html);
Lexical and Semantic Information from NLP systems: mainly normalized concepts (eg, UMLS concept unique identifiers) and semantic types identified by NLP systems. Three NLP systems were used: (1) MedLEE; (2) KnowledgeMap; (3) a Dictionary-based Semantic Tagger (DST) developed for this task, which uses vocabularies from public (eg, UMLS) and private (eg, Vanderbilt's problem list) sources and filtered them for medical problems, tests, and treatments;
Discourse Information: Sections in the clinical notes (eg, ‘Current Medications’ section) and Sources of the notes (eg, ‘Partners HealthCare System’), obtained by customized programs developed for the challenge data.
Experiments
We developed and evaluated ML-based NER approaches using the training data set containing 349 annotated clinical notes (see table 1 for the distribution of entities in the data set). A fivefold cross-validation method was used in the evaluation. Optimized parameters for each ML algorithm were determined by the best performance on one fold of test data. To evaluate the effects of different types of features, we started with the baseline method that used bag-of-word features only, and then added additional types of features and reported corresponding results.
Table 1.
Distribution of different types of entities and assertions in the training and test sets in the Center of Informatics for Integrating Biology and the Bedside/Veterans Affairs natural-language processing challenge
| Concepts (N=72 846) | Assertions (N=30 518) | ||||||||||
| Data set | Problem | Treat | Test | All | Present | Absent | Possible | Conditional | Hypothetical | Not associated with the patient | All |
| Training —349 | 11 968 | 8500 | 7369 | 27 837 | 8052 | 2535 | 535 | 103 | 651 | 92 | 11 968 |
| Test—477 | 18 550 | 13 560 | 12 899 | 45 009 | 13 025 | 3609 | 883 | 171 | 717 | 145 | 18 550 |
ML-based approaches for assertion classification
The assertion classification task is to assign one of the six possible assertion labels (‘Present,’ ‘Absent,’ ‘Possible,’ ‘Conditional,’ ‘Hypothetical,’ and ‘Not associated with the patient’) to each medical problem. We developed a multiclass classifier based on SVM using the LIBLINEAR package,32 which can train on large-scale data sets very efficiently, and we systematically evaluated the contributions of different types of features for the assertion task using the 349 training notes (see table 1 for the distribution of assertions). For each medical problem, we chose a window of context, from which features were extracted. The window size was optimized based on the performance on training set, and it was set to 10 words on the left and five words on the right. Similar types of features used in Concept Extraction task were applied to this task as well. For features from NLP systems, we used only MedLEE's outputs as features because of its good performance in the concept extraction task. One notable feature was the ‘certainty’ information identified by MedLEE. In addition, direction (left vs right) and distance (eg, second word on the left) information of words in the window was also considered. The evaluation of the contributions of different types of features was carried out on the training set, using a fivefold cross-validation. We also conducted systematic parameter selection for the type of solver, cost parameter (C), and tolerance of termination criterion (Epsilon).
Hybrid NLP system for concept extraction and assertion classification
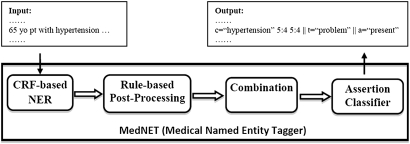
To participate in the i2b2/VA challenge, we further developed a novel hybrid NLP system for clinical concept extraction and assertion classification, building on the top of the ML-based approaches described above. We named the hybrid system Medical Named Entity Tagger (MedNET), which consists of four components in a pipeline: (1) the CRF-based NER module that uses optimized parameters and feature sets as revealed by the training data; (2) a postprocessing program that uses heuristic rules to correct possible errors and further improve the performance; (3) a combination module that optimizes the results from multiple classifiers; and (4) the assertion classifier based on SVM. The first three components form the NER system for concept extraction task, and the last component is for assertion classification. Figure 1 shows an overview of the MedNET system. As components 1 and 4 have been described above, we focus on the two rule-based modules about postprocessing and combination in this section.
Figure 1.
Architecture of the Medical Named Entity Tagger, a hybrid system for clinical Named Entity Recognition (NER). CRF, conditional random fields.
Postprocessing module
Based on a manual review of the errors in the ML-based NER module, a set of heuristic rules were developed and implemented as a postprocessing program. Some rules are intended to fix false negatives, such as abbreviations that are missed by the system. Some rules are used for disambiguation, such as to determine the correct type of a semantically ambiguous entity based on its context. For example, a medication term is usually classified a ‘Treatment’ (eg, ‘Tacrolimus 3 mg twice daily’). However, when it refers to drug levels in blood, it should be classified as ‘Test’ (eg, ‘Tacrolimus level 10.0 ng/ml’). Therefore, we could develop a rule such as ‘IF contextual words around a medication name include keywords such as “level,” THEN overwrite the semantic type of the medication term as “TEST”.’
Combination module
As previous NER studies reported improved performance when multiple classifiers were combined,16 we developed a module to combine the results from four individual NER classifiers, which used different sets of features: three of them used all other features and inputs from one of the NLP systems (MedLEE, KnowledgeMap, or DST), and the fourth one used all other features and merged inputs from all three NLP systems. The combination module works in a two-step way: first, it generates the ‘intersection’ of clinical entities extracted by all three individual classifiers, in other words, only clinical entities extracted by all three classifiers constitute the result; second, it takes a ‘union’ operation between the intersection from step 1 and outputs from the fourth classifier, which means that any entity generated by the first step or by the fourth classifier will be included. The intention is to improve the performance by detecting possible missed false negatives by the fourth classifier alone.
Evaluation and results
Evaluation of features and algorithms for ML-based NER using the training data
The evaluation was carried out using a script provided by i2b2/VA organizers, which can report micro-averaged Precision, Recall, and F-measure for all concepts using both exact and inexact matching methods.33 The exact matching criterion requires that the starting and ending offsets of a concept have to be exactly the same as those in the gold standard, while inexact matching refers to cases where their offsets are not exactly the same as those in gold standard, but they overlap with each other. The best F-measure was 0.8475, achieved by CRF algorithm, with all the features used. Table 2 shows the results of CRF-based NER approach on the training data using fivefold CV when different sets of features were used, as well as the best result from SVM-based NER approach. The reported numbers are averages of the results from fivefold CV. For SVM, two parameters including the degree of the polynomial kernel function (D) and the cost (C) used in the optimization step of learning have to be optimized. We explored various combinations of values for the two parameters using the baseline features and noticed that the model performance was more stable among different values of C when D was set to 2. Therefore, D=2 and C=0.1 were chosen as the parameters for SVM learning in this study (see online only appendix (www.jamia.org) for detailed results from SVM experiments with different parameters and combinations of features).
Table 2.
Results of conditional random fields (CRF)-based Named Entity Recognition module on the training set (using a fivefold cross validation), when different sets of features were used
| Feature set | Preision | Recall | F-measure |
| CRF—Baseline (bag-of-word) | 0.8338 | 0.7162 | 0.7705 |
| CRF—Baseline+POS | 0.8318 | 0.7466 | 0.7868 |
| CRF—Baseline+orthographic | 0.8315 | 0.7364 | 0.7811 |
| CRF—Baseline+prefix & suffix | 0.8524 | 0.7473 | 0.7963 |
| CRF—Baseline+(POS+Orthographic+prefix & suffix) | 0.8303 | 0.7719 | 0.8000 |
| CRF—Baseline+(POS+Orthographic+prefix & suffix)+MedLEE | 0.8642 | 0.8119 | 0.8372 |
| CRF—Baseline+(POS+Orthographic+prefix & suffix)+KnowledgeMap | 0.8557 | 0.7957 | 0.8245 |
| CRF—Baseline+(POS+Orthographic+prefix & suffix)+DST | 0.8601 | 0.8040 | 0.8311 |
| CRF—Baseline+(POS+Orthographic+prefix & suffix)+(MedLEE+KnowledgeMap+DST) | 0.8708 | 0.8243 | 0.8469 |
| CRF—Baseline+(POS+Orthographic+prefix & suffix)+(MedLEE+KnowledgeMap+DST)+source§ion | 0.8717 | 0.8246 | 0.8475 |
| Support Vector Machines—best result | 0.8507 | 0.8153 | 0.8326 |
Baseline, words in context window with indications of left or right; DST, Dictionary-based Semantic Tagger; MedLEE, Medical Language Extraction and Encoding System; POS, part of speech tags of context words.
Evaluation of features for assertion classification using the training data set
The best result for assertion classification was 0.9398 (F-measure), when all the features were used. Table 3 shows the contributions of different types of features for the SVM-based assertion classification.
Table 3.
Results of assertion classifier on training set (fivefold cross validation), when different types of features were used
| Feature set | F-measure |
| Baseline (words in context window, plus direction: left or right) | 0.9132 |
| Baseline+Bigram | 0.9161 |
| Baseline+Bigram+Distance | 0.9225 |
| Baseline+Bigram+Distance+POS | 0.9233 |
| Baseline+Bigram+Distance+POS+Certainty | 0.9248 |
| Baseline+Bigram+Distance+POS+Certainty+Source and Section | 0.9367 |
| Baseline+Bigram+Distance+POS+Certainty+Source and Section+Concept and Semantic | 0.9398 |
Baseline, words in context window with indications of left or right; Bigram, bi-grams identified within the context window; Certainty, certainty information identified by Medical Language Extraction and Encoding System; Concept and Semantic, Unified Medical Language System concept unique identifiers and semantic types of words in the context window, identified by MedLEE; Distance, distance between the feature word and the target word—for example, third word on the left; POS, part of speech tags of context words; Source and Section, source of the note and section where the target word occurs, identified by a customized program.
Evaluation of the MedNET system using the test data set
The performance of MedNET was evaluated on the independent test set (477 notes), and the overall class-level F-measure based on exact matching was used for the rankings in the challenge. MedNET achieved a best class-level F-measure of 0.8391 (highlighted in table 4), which was ranked second among 22 participating teams in the i2b2/VA challenge. The detailed results by semantic type are shown in table 4 as well.
Table 4.
Detailed results by semantic type in the best run for the Center of Informatics for Integrating Biology and the Bedside/Veterans Affairs Concept Extraction task on the test data set
| Category | Exact matching | Inexact matching | ||||
| Recall | Precision | F-measure | Recall | Precision | F-measure | |
| Overall—concept | 0.8298 | 0.8828 | 0.8555 | 0.8974 | 0.9546 | 0.9251 |
| Overall—class | 0.8139 | 0.8658 | 0.8391 | 0.8951 | 0.9317 | 0.9130 |
| Problem | 0.8403 | 0.8746 | 0.8571 | 0.9158 | 0.9539 | 0.9345 |
| Treatment | 0.8202 | 0.8801 | 0.8491 | 0.8882 | 0.9535 | 0.9197 |
| Test | 0.8248 | 0.8980 | 0.8598 | 0.8804 | 0.9570 | 0.9171 |
For the Assertion Classification task, our system achieved an F-measure of 0.9313 on the test set, which was ranked fourth in the competition, but with no statistically significant difference with the first ranked system that achieved an F-measure of 0.9362.
Discussion
Concept extraction and assertion determination are two fundamental tasks for successful application of clinical NLP. Current clinical NLP systems typically use rule- and/or knowledge-based approaches for entity recognition, instead of statistical learning methods. Our ML-based approaches in the i2b2/VA challenge achieved good results, finishing second overall in the entity recognition and fourth overall in the assertion task (though statistically similar to the top three entries). Further investigations are needed to reveal the strengths and weaknesses of this approach to clinical NLP. Below, we summarize some interesting findings and challenges to ML-based clinical entity extraction.
ML-based NER for clinical text
For biological NER tasks (eg, genes), some studies reported better results using CRF,26 while others showed that the SVM also achieved a high performance.23 Keerthi et al34 conducted some experiments and demonstrated that CRF and SVM were similar in performance when identical features were used. In our experiments, CRF outperformed SVM with equivalent features. Additional feature and kernel optimization for the SVM may improve its performance.35 However, it also indicates the complexity of SVM parameter optimization.
The contribution of different features to system performance varied (table 2). Lexical and semantic features derived from dictionary-based NLP systems improved the performance largely. Thus, we conclude that medical knowledge bases are crucial to clinical NER, which is similar to findings from gene/protein NER tasks.16 Prefix and suffix information improved the performance reasonably. But improvements from part-of-speech and orthographic features were less important than that seen in gene/protein NER.36 37 More orthographic clues are observed in gene/protein names (eg, numbers in gene names) while prefixes and suffixes are often observed in medical words (eg, ‘-itis’ in ‘tonsillitis’). Unexpectedly, section information did not add much value to the performance, and sometimes even reduced the performance (in SVM experiments; see appendix). Our observation revealed that many sections contained a mixture of entities with different semantic types; therefore, section information may not be a useful predictor for semantic types. For example, one may expect that a ‘Medication’ section should contain medications (a ‘treatment’), but we also often observed medical problems in ‘Medication’ sections, such as ‘…take as needed for pain.’ In addition, a simple section detection program was used, which did not normalize similar section headers (eg, ‘medications:’ or ‘medication list:’) into one normalized concept. Application of more advanced section tagging programs such as SecTag38 to this task may improve the contribution of section information. In general, more features almost always improve the NER performance.
The relative contribution from different dictionary-based NLP systems varied. While the best result combined MedLEE, KnowledgeMap, and DST, MedLEE generated the best result when used in isolation. F-measures on the test data set were 0.8354, 0.8130, and 0.8259 respectively, when only MedLEE, KnowledgeMap, or DST was used. Based on our observation, we believe such differences result primarily from the underlying semantic lexicon sources implemented. MedLEE is designed to extract only clinically relevant findings. Its output includes semantic types similar to those in the i2b2/VA challenge, such as ‘problem,’ ‘labtest,’ and ‘procedure.’ In addition, all semantic lexicons in MedLEE were manually reviewed, and they are highly accurate and specific to their corresponding semantic types. In contrast, KnowledgeMap is a general purpose concept indexing tool that used the entire UMLS metathesaurus, which provides good coverage but could be noisy and imprecise. The DST system was built specifically for the i2b2/VA challenge by selecting concepts and semantic types from UMLS and other sources that are relevant to the problem/test/treatment only. The UMLS semantic types were then regrouped into self-defined semantic categories, such as ‘medication,’ ‘labtest,’ and ‘disease.’ Results showed that it reached reasonable performance.
The MedNET system provides a generalizable framework for a hybrid approach that combines ML and rule-based methods to improve NER performance. The CRF-based NER module alone achieved an F-measure of 0.8369; CRF module+postprocessing increased it to 0.8380; CRF module+postprocessing+combination further improved the performance to the maximum F-measure of 0.8391. Therefore, both the postprocessing module and combination module improved the overall F-measure very slightly. We found three major types of errors in MedNET: (1) omitted terms (false negatives); (2) incorrect boundaries for extracted terms; and (3) misclassified semantic types. One of the main causes for the first type of errors is unrecognized abbreviations. Boundary and semantic classification errors contribute to both false positives and false negatives. The current postprocessing program implemented very simple rules based on context. Addition of more sophisticated text processing methods, such as abbreviation recognition or supervised word sense disambiguation, may improve performance further. The combination module improved system's performance as well, but not as much as that reported in previous studies on gene names.18 We plan to explore further more advanced combination methods such as building ensembles of classifiers in the next steps.
Assertion classification
Determination of assertion status is an important area of clinical NLP research. Previous efforts have used regular expression patterns13 or linguistic information from sentence parsing.39 Our investigation showed that ML-based approaches for assertion determination were promising. The i2b2/VA assertion task is more challenging than negation determination task (a binary classification problem), since it included six classes. The current task is similar to the classes predicted by ConText, which extends NegEx algorithmically and lexically to include other classes of assertion modifiers.13 Use of the ‘distance’ feature and use of the ‘source/section’ feature both improved results. The ‘Section’ feature primarily benefited a few specific classes of assertion, such as identifying non-patient experiences via a ‘Family History’ section.
The SVM-based classifier for determining ‘absent’ status achieved a similarly high performance (precision 0.9623 and recall 0.9459) as reported in a previous negation study.40 The performances for ‘conditional’ and ‘possible’ classes were much worse than other classes (see appendix for assertion classification results by class). However, we noticed that human determination of ‘conditional’ and ‘possible’ assertions was not always straightforward either. For example, according to the i2b2 gold standard, ‘hypertension’ was assigned a status of ‘possible,’ in the sentence ‘We felt this was likely secondary to hypertension.’ However, a physician in our team would interpret that both ‘this (problem)’ and ‘hypertension’ were ‘present,’ but the relation between them was ‘possible,’ when not considering the context beyond this sentence.
Comparison between ML and dictionary-based systems
Most existing clinical NLP systems rely on vocabularies to recognize clinical entities such as diseases and medications41 in text, probably because of two reasons: (1) the existence of rich clinical knowledge bases and vocabularies, such as the UMLS,42 allowing rapid development of effective systems with little training; and (2) very few annotated data sets of clinical text are available for ML-based NER approaches. It is difficult to directly compare the performance of existing dictionary-based NLP systems with ML using the i2b2/VA annotated data set due to the strictly defined boundaries in the competition (which are not a goal of most dictionary-based NLP systems), and differing scopes of semantic entities. The challenge annotation guideline (https://www.i2b2.org/NLP/Relations/Documentation.php) used syntactic information to determine entity boundaries, instead of considering meanings of clinical entities, which is what dictionary-based NLP systems do. For example, i2b2/VA defined that ‘Up to one prepositional phrase can be annotated together with the preceding concept as one entity.’ In the example ‘defect in lobe of liver,’ only ‘defect in lobe’ can be annotated as an entity. In contrast, systems such as KnowledgeMap and MedLEE are designed to match the ‘best’ concepts (in this case, a more accurate concept), using linguistic structures as a guide instead of a rule. The second issue was that dictionary-based NLP systems usually capture broader types of findings than medical problems, tests, and treatments. Therefore, careful filtering of semantic types from NLP systems' output is required, which is often not straightforward.
Nevertheless, we estimated the performance of MedLEE using the i2b2/VA data set by converting MedLEE's output to offset format required by i2b/VA challenge and filtering MedLEE's outputs to only relevant semantic types. Based on inexact-matching without considering semantic types, MedLEE achieved 0.8733 (recall) and 0.6924 (precision) for extracting i2b2/VA annotated entities. The recall of MedLEE was comparable with some high-performance ML systems in the challenge. When looking into the entities missed by MedLEE, we noticed most of them were abbreviations. The precision of 0.6924 indicated that MedLEE recognized more than 30% additional entities. We randomly selected 50 such entities and reviewed them. About 84% of the additional entities recognized by MedLEE were meaningful clinical findings, such as patient demographics (eg, age and sex), body functions or measures (eg, ‘heart size’), and substance use (eg, smoking or alcohol status). Only 16% were false positives, which often involved ambiguous terms.
ML-based approaches have a few drawbacks if we want to extend them to general-purpose NLP systems. First, as more types of clinical entities are included, the performance of ML-based may diminish as the number of classes increases. Second, accurate mapping of text to concepts in controlled vocabularies such as UMLS is often more useful for many clinical tasks (eg, search retrieval and decision support), but very challenging, and the degree to which it is helped by accurate boundary detection by ML-based NER systems is unknown. We think such an ML-based method could be complementary to existing dictionary-based NLP systems. It could be integrated with existing NLP systems to help recognize unknown words based on context features (ie, recognize unknown lab test names in lab sections).
Conclusion
In this study, we implemented ML-based approaches for clinical entity recognition and assertion classification, and systematically evaluated the effects of different features and ML algorithms. Our final solution was a novel hybrid clinical NLP system using both ML methods and rule-based components. In the 2010 i2b2/VA NLP challenge, our system achieved a maximum F-score of 0.8391 for concept extraction (ranked second) and 0.9313 for assertion classification (ranked fourth, but not statistically significant different from the top three systems), which indicates that such approaches are very promising.
Supplementary Material
Acknowledgments
We would like to thank the 2010 i2b2/VA challenge organizers for the development of training and test corpora. We also thank Dr Carol Friedman at Columbia University for providing MedLEE for us to use in the challenge.
Footnotes
Funding: This study was partially supported by the grant from the US NIH: NCI R01CA141307, NHGRI U01 HG004603 and 3T15 LM007450-08S1. The MedLEE system was supported by NLM grants LM008635 and LM010016.
Competing interests: None.
Ethics approval: Ethics approval was provided by Vanderbilt University IRB.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Sager N, Friedman C, Chi E, et al. The analysis and processing of clinical narrative. MedInfo 1986:1101–5 [Google Scholar]
- 2.Sager N, Friedman C, Lyman M. Medical Language Processing: Computer Management of Narrative Data. Reading, MA: Addison-Wesley, 1987 [Google Scholar]
- 3.Hripcsak G, Friedman C, Alderson PO, et al. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med 1995;122:681–8 [DOI] [PubMed] [Google Scholar]
- 4.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hripcsak G, Austin JH, Alderson PO, et al. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology 2002;224:157–63 [DOI] [PubMed] [Google Scholar]
- 6.Haug PJ, Koehler S, Lau LM, et al. Experience with a mixed semantic/syntactic parser. Proc Annu Symp Comput Appl Med Care 1995:284–8 [PMC free article] [PubMed] [Google Scholar]
- 7.Fiszman M, Chapman WW, Evans SR, et al. Automatic identification of pneumonia related concepts on chest x-ray reports. Proc AMIA Symp 1999:67–71 [PMC free article] [PubMed] [Google Scholar]
- 8.Haug PJ, Christensen L, Gundersen M, et al. A natural language parsing system for encoding admitting diagnoses. Proc AMIA Annu Fall Symp 1997:814–18 [PMC free article] [PubMed] [Google Scholar]
- 9.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng QT, Goryachev S, Weiss S, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak 2006;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010;17:229–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denny JC, Miller RA, Johnson KB, et al. Development and evaluation of a clinical note section header terminology. AMIA Annu Symp Proc 2008:156–60 [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 14.Hanisch D, Fundel K, Mevissen HT, et al. ProMiner: rule-based protein and gene entity recognition. BMC Bioinformatics 2005;6(Suppl 1):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krauthammer M, Nenadic G. Term identification in the biomedical literature. J Biomed Inform 2004;37:512–26 [DOI] [PubMed] [Google Scholar]
- 16.Torii M, Hu Z, Wu CH, et al. BioTagger-GM: a gene/protein name recognition system. J Am Med Inform Assoc 2009;16:247–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patrick J, Li M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J Am Med Inform Assoc 2010;17:524–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Liu F, Antieau L, et al. Lancet: a high precision medication event extraction system for clinical text. J Am Med Inform Assoc 2010;17:563–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sibanda T, He T, Szolovits P, et al. Syntactically-informed semantic category recognition in discharge summaries. AMIA Annu Symp Proc 2006:714–18 [PMC free article] [PubMed] [Google Scholar]
- 20.Sang EFTK, Veenstra J. Representing text chunks. Proceedings of the Ninth Conference of the European Chapter of the Association for Computational Linguistics, Association for Computational Linguistics Morristown, NJ, USA, 1999:173–9 [Google Scholar]
- 21.Lafferty JD, McCallum A, Pereira FCN. Conditional random fields: probabilistic models for segmenting and labeling sequence data. Proceedings of the Eighteenth International Conference on Machine Learning. Morgan Kaufmann Publishers Inc, Burlington, MA, USA, 2001:282–9 [Google Scholar]
- 22.Joachims T. Making large-scale SVM learning practical. Advances in Kernel Methods – Support Vector Learning. MIT Press, Cambridge MA, USA, 1998 [Google Scholar]
- 23.Takeuchi K, Collier N. Bio-medical entity extraction using support vector machines. Proceedings of the ACL 2003 workshop on Natural Language Processing in Biomedicine. Vol. 13 Sapporo, Japan: Association for Computational Linguistics, 2003:57–64 [Google Scholar]
- 24.Kazama J, Makino T, Ohta Y, et al. Tuning support vector machines for biomedical named entity recognition. Proceedings of the ACL-02 Workshop on Natural Language Processing in the Biomedical Domain. Vol 3 Phildadelphia, PA: Association for Computational Linguistics, 2002:1–8 [Google Scholar]
- 25.Yamamoto K, Kudo T, Konagaya A, et al. Protein name tagging for biomedical annotation in text. Proceedings of the ACL 2003 Workshop on Natural Language Processing in Biomedicine. Vol 13 Sapporo, Japan: Association for Computational Linguistics, 2003:65–72 [Google Scholar]
- 26.Li D, Kipper-Schuler K, Savova G. Conditional random fields and support vector machines for disorder named entity recognition in clinical texts. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing. Columbus, OH: Association for Computational Linguistics, 2008:94–5 [Google Scholar]
- 27.He Y, Kayaalp M. Biological entity recognition with conditional random fields. AMIA Annu Symp Proc 2008:293–7 [PMC free article] [PubMed] [Google Scholar]
- 28.Song Y, Kim E, Lee GG, et al. POSBIOTM-NER: a trainable biomedical named-entity recognition system. Bioinformatics 2005;21:2794–6 [DOI] [PubMed] [Google Scholar]
- 29.Kudo T, Matsumoto Y. Chunking with support vector machines. Proceedings of the Second Meeting of the North American Chapter of the Association for Computational Linguistics on Language Technologies. Pittsburgh, Pennsylvania: Association for Computational Linguistics, 2001:1–8 [Google Scholar]
- 30.Kudoh T, Matsumoto Y. Use of support vector learning for chunk identification. Proceedings of the 2nd Workshop on Learning Language in Logic and the 4th Conference on Computational Natural Language Learning. Vol 7 Lisbon, Portugal: Association for Computational Linguistics, 2000:142–44 [Google Scholar]
- 31.Smith L, Rindflesch T, Wilbur WJ. MedPost: a part-of-speech tagger for bioMedical text. Bioinformatics 2004;20:2320–1 [DOI] [PubMed] [Google Scholar]
- 32.Fan RE, Chang KW, Hsieh CJ, et al. LIBLINEAR: a library for large linear classification. J Mach Learn Res 2008;9:1871–4 [Google Scholar]
- 33.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keerthi SS. CRF versus SVM-Struct for Sequence Labeling. Technical Report, Yahoo Research, Santa Clara, CA, USA, 2007 [Google Scholar]
- 35.Leopold E, Kindermann J. Text categorization with support vector machines. how to represent texts in input space? Machine Learning 2002;46:423–44 [Google Scholar]
- 36.Zhou G, Shen D, Zhang J, et al. Recognition of protein/gene names from text using an ensemble of classifiers. BMC Bioinformatics 2005;6(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou G, Zhang J, Su J, et al. Recognizing names in biomedical texts: a machine learning approach. Bioinformatics 2004;20:1178–90 [DOI] [PubMed] [Google Scholar]
- 38.Denny JC, Spickard A, 3rd, Johnson KB, et al. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc 2009;16:806–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y, Lowe HJ. A grammar-based classification of negations in clinical radiology reports. AMIA Annu Symp Proc 2005:988. [PMC free article] [PubMed] [Google Scholar]
- 40.Mutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents: a quantitative study using the UMLS. J Am Med Inform Assoc 2001;8:598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010;17:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindberg DA, Humphreys BL, McCra AT. The unified medical language system. Methods Inf Med 1993;32:281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.