Abstract
Objective
Information extraction and classification of clinical data are current challenges in natural language processing. This paper presents a cascaded method to deal with three different extractions and classifications in clinical data: concept annotation, assertion classification and relation classification.
Materials and Methods
A pipeline system was developed for clinical natural language processing that includes a proofreading process, with gold-standard reflexive validation and correction. The information extraction system is a combination of a machine learning approach and a rule-based approach. The outputs of this system are used for evaluation in all three tiers of the fourth i2b2/VA shared-task and workshop challenge.
Results
Overall concept classification attained an F-score of 83.3% against a baseline of 77.0%, the optimal F-score for assertions about the concepts was 92.4% and relation classifier attained 72.6% for relationships between clinical concepts against a baseline of 71.0%. Micro-average results for the challenge test set were 81.79%, 91.90% and 70.18%, respectively.
Discussion
The challenge in the multi-task test requires a distribution of time and work load for each individual task so that the overall performance evaluation on all three tasks would be more informative rather than treating each task assessment as independent. The simplicity of the model developed in this work should be contrasted with the very large feature space of other participants in the challenge who only achieved slightly better performance. There is a need to charge a penalty against the complexity of a model as defined in message minimalisation theory when comparing results.
Conclusion
A complete pipeline system for constructing language processing models that can be used to process multiple practical detection tasks of language structures of clinical records is presented.
Keywords: agents, automated learning, classification, clinical, controlled terminologies and vocabularies, designing usable (responsive) resources and systems, discovery, distributed systems, information classification, information extraction, i2b2 challenge, knowledge bases, natural language processing, ontologies, software engineering: architecture, text and data mining methods, 2010 i2b2 challenge
In the clinical domain, there is a large amount of textual data in patients' notes so efficient processing techniques are necessary to make use of this valuable information. Information extraction and classification tools for processing clinical narratives are valuable for assisting clinical staff to quickly find data of relevance to themselves.
In this study the focus is on extraction of medical problems, tests and treatments, classification of assertions made about medical problems and relationships between medical problems, tests and treatments.1 This paper presents an approach for building the required extraction models using both training data and local knowledge resources, including gazetteers of entities, acronyms and abbreviations, and a spelling correction process with methods for resolving unknown words and non-word tokens.
Background
Challenge requirements
The fourth i2b2/VA challenge is a three-tiered challenge that studies:
Extraction of medical problems, tests and treatments in clinical records.
Classification of assertions made on medical problems. Assertion is a context-based attribute of medical concepts to determine what medical problems the note asserts. Each medical problem is re-classified into one of six categories of assertions (ordered by their priorities): associated with someone else, hypothetical, conditional, possible, absent and present.
Classification of a relationship between a pair of concepts that appear in the same sentence where at least one concept is a medical problem. These relations are: treatment improves problem (TrIP) or worsens problem (TrWP), treatment causes problem (TrCP), treatment is administered (TrAP) or not administered because of problem (TrNAP); test reveals problem (TeRP), test conducted to investigate problem (TeCP) and problem indicates problem (PIP).
Corpus description
The i2b2 challenge corpus is composed of 1653 clinical records provided by Partner Healthcare, which is divided into training and testing data. The training set contains 349 manually annotated notes (gold standard) and 827 raw records. The size of testing data is 477 records, which is approximately 1.5 times larger than gold standard data. The ground truths of testing data were also released after each submission closed (concept, assertion and relation) so the computations for the next tier of the challenge could use the true data rather than each team using their own erroneous annotations.
Lexical semantic resources
For clinical Natural Language Processing (NLP) research, ontologies and lexical semantic resources, such as unified medical language system (UMLS) and the systematic nomenclature of medicine—clinical terms (SNOMED–CT) are now available.2 3 UMLS, with the largest available medical lexicon, integrates and distributes key terminology, classification and coding standards and associated resources to promote the creation of more effective and interoperable biomedical information systems and services, including electronic health records. SNOMED–CT is a standardized healthcare terminology including comprehensive coverage of diseases, clinical findings, therapies, procedures and outcomes.
Besides the clinical resources (UMLS and SNOMED–CT), a general English lexicon, MOBY, was used for lexical verification and misspelling correction in the processing pipeline.4
Related work
A variety of methods and systems has been implemented in the clinical domain to extract information from free text. Friedman et al5 developed the medical language extraction and encoding (MedLEE) system, which used a domain-specific vocabulary and semantic grammar to process clinical narrative reports. It was initially used to participate in an automated decision-support system, and to allow natural language queries. MedLEE was then adapted to automatically identify the concepts in clinical documents, map the concepts to semantic categories and semantic structures.6 The final semantic representation of each concept contained information on status, location and certainty of each concept instance. Haug et al7 introduced symbolic text processor (SymText), a natural language understanding system for chest x-ray reports. SymText processes each sentence in a document independently with syntactic and probabilistic semantic analysis. Bayesian networks are used in SymText to determine the probability that a disease is present in the patient.
An early combined classifier approach in biomedical named entity recognition (NER) proposed a two-state model in which boundary recognition and term classification are separated into two phases.8 In each classification phase, different feature sets were selected independently, which is more efficient for each task. A comparative study between two classical machine learning methods, conditional random fields (CRF) and support vector machines (SVM) for clinical named entity recognition shows that the CRF outperformed SVM in clinical NER.9
When extracting information from narrative text documents, the context or assertion of the concepts extracted play a critical role.10 The NegEx algorithm of Chapman et al11 implements dictionaries of pre-UMLS and post-UMLS phrases that are indicative of negation to identify positive and negative assertions. NegEx uses a rule-based method and heuristics to limit the scope of indicative phrases. The challenge's assertion classification is an extension of previous system designed by Uzuner et al,12 in new specification of an uncertainty assertion divided into values of hypothetical, conditional and possible. A combination of machine learning and rule-based approaches are utilized in the system of Uzuner et al.12 One of these approaches extends the rule-based NegEx algorithm to capture alter-association in addition to positive, negative and uncertain assertions; the other employs SVM to present a machine learning solution to assertion classification.
For the relationship classification task, there are many definitions of relationships between concepts in which each system classifies different relationship types. In general, relevant features are extracted from the text and are usually selected on the basis of the experimental results and intuition, or by statistical techniques.13 First, by experience and intuition, we designed feature sets that were expected to have a strong correlation with the target classification. Forward selection was applied by sequentially adding each feature set to the model and evaluating its performance. The feature set is retained if a better result is achieved otherwise it is discarded before the next cycle is repeated.
The closest research related to our methods is the information extraction system for the clinical notes of Wang,14 in which a clinical corpus was annotated for clinical named entities and relationships based on the SNOMED–CT. The work of Wang14 used CRF and SVM machine learners to create an information extraction system of multiple classifiers rather than a single classifier in his NER strategy.15 16
In comparison with these information extraction systems, our work is an adaptation based on the specific requirements of the i2b2 challenge with three levels of classification. We have developed a pipeline system for clinical NLP, which includes a proofreading process, with gold standard reflexive validation and correction. The information extraction system is the combination of a machine learning approach and a rule-based approach. Furthermore, a post-processing step was implemented to refine the results. CRF and SVM are two classic machine-learning approaches used, but in this case with some new feature sets introduced (dictionary; abbreviation, acronym, misspelling expansion; text to SNOMED–CT convertor and medication extraction system results).17 18 A detailed description and use of the feature sets is explained in the next section.
Methods
Model construction strategy
The important system requirement is to generate models that can be used to interpret the real world phenomena of the language structures and clinical knowledge in the text. The system also enables the optimal classifier from a set to be assessed in different applications. The required extraction models could be built using training data and local knowledge resources. For the challenge, two machine learners adopted were CRF and SVM. The CRF learner was used for concept annotation and assertion classification while the SVM was designed to identify the relationships between two entities.
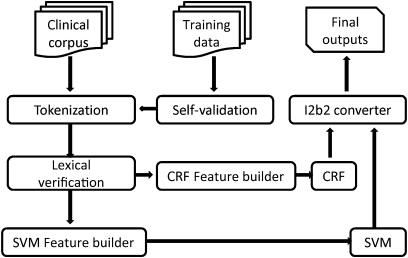
Figure 1 shows the system architecture in which the first processing step is self-validation of the training data (reflexive validation). The gold standard was created by the i2b2 organizers by manual annotation, which usually contains minor errors and inconsistencies. The gold standard can be corrected for inconsistencies between annotations by using a reflexive validation process, which we also denote as ‘100% train and test’. This involves using 100% of the training set to build a model and then testing on the same set. With this self-validation process, more than 100 errors in the training data were detected. The three most frequent error types in concept annotation are: (1) missing modifier (any, some); (2) including punctuation (full stop, comma, hyphen); (3) missing annotation (false negative). As theoretically all data items used for training should be correctly identifiable by the model, any errors represent either inconsistencies in annotations or weaknesses in the computational linguistic processing. The former faults identify training items that are rejected, and the latter gives indications of where to concentrate efforts to improve the preprocessing system. This process improved scores of the order of 0.5%.
Figure 1.
Language processing architecture. CRF, conditional random field; SVM, support vector machine.
As specified by the challenge, each sentence was on separate lines and tokens were split using a white-space tokenizer. Lexical verification for each token included expansion of abbreviations, acronyms, checking against gazetteers and the lexical resources of UMLS, MOBY and SNOMED–CT, then resolving misspellings and unknown words. All the results of this process were saved in a lexicon management system for later use in feature generation. The lexicon management system is a system developed to store the accumulated lexical knowledge of our laboratory and contains categorizations of spelling errors, abbreviations, acronyms and a variety of non-tokens. It also has an interface that supports rapid manual correction of unknown words with a high accuracy clinical spelling suggestor plus the addition of grammatical information and the categorization of such words into gazetteers.17
After lexical verification, seven feature sets were prepared to train a CRF model to identify the named entities classes of problems, tests and treatments. For assertion classification, three approaches (rule-based, CRF and SVM) were tested and the best method was the CRF with only four feature sets. SVM classified relationships between entities using local context feature and semantic feature sets. All these feature sets were sent to corresponding CRF and SVM feature generators. Finally, when the results from CRF, SVM were computed, the i2b2 converter generated the outputs according to the format required in the challenge.
A 10-fold cross-validation method was used for model building with data from the gold standard, which contained 349 annotated records. After performing different experiments with a variety of features and linguistic processes, the model was generated from the full set of training data minus the erroneous annotations with the optimal feature set.
Concept annotation
For the concept annotation task, the best performance was obtained from seven feature sets that were used for each unigram in the training of the CRF:
Three-word context window: the selection of window size was a separate experiment (size varied from three to seven), the results showing that the three-word window size was optimal in both performance and model complexity.
Lemma, part of speech, chunk from the GENIA tagger. The GENIA tagger analyzes English sentences and outputs the base forms, part-of-speech tags, chunk tags. The tagger is specifically tuned for biomedical text and is a useful preprocessing tool to extract information from biomedical documents.19 Lemma is the base form of a word (lemma of ‘chosen’ is ‘choose’). Traditional grammar classifies words based on eight parts of speech: the verb, the noun, the pronoun, the adjective, the adverb, the preposition, the conjunction and the interjection. Chunk is any pair or group of words that are commonly found together, or in close proximity, in this case, GENIA groups words into noun phrase, verb phrase, etc….
Gazetteers and lexical resources: name of gazetteer (ne-drug, ne-disorder, ne-person, ne-location, etc…) or dictionary (UMLS, MOBY, SNOMED–CT) where the word is classified.
Abbreviation, acronym expansion and misspelling correction. After the lexical verification process, each unrecognized word may require to have expansion or correction. This transformation should not be made in the original corpus because it would affect the offsets of tokens that will lead to retrieval of incorrect annotations. Corrections, when used as a feature, will support the model in learning correct forms of misspelt words (‘medicla’ and ‘medcial’ refer to the same word ‘medical’) and variations of abbreviations (‘amnt’ and ‘amt’ are both ‘amount’), and multiple acronyms of the same term (‘ABG’, ‘ABGs’ are both ‘arterial blood gases’).
Number tag: for each token that is recognized as a number, a feature with value ‘number’ is added to distinguish it from word tokens.
TTSCT: this feature is the result of parsing the text to identify SNOMED–CT's concepts using the TTSCT service. TTSCT was developed so that SNOMED–CT concepts could be identified in free text narrative and to annotate them with the clinical reference term. The performance of TTSCT is approximately 69% on a test corpus.17 Its improvement is ongoing research conducted by the authors.
Medication extraction system's results: this is the system the authors used for i2b2 2009 challenge in which medication annotation was shown to contribute to recognizing treatments.
Other feature sets were used during the experiment process but they did not improve the F-score. These feature sets are morphology and finite state automata annotation.
Assertion classification
For the assertion classification task, the organizers had provided the ground truth for concept annotations so the boundaries of problem entities could be used as a feature to indicate concepts that needed to be re-classified. Three different methods (rule-based, CRF, SVM) were designed and tested, all of them based on the same ideas of using a manual lexicon as a key feature to classify the assertions made about medical problems. The first step was to implement the rule-based method with a very small lexicon for each class (absent 27 words; possible 26 words; conditional seven words; hypothetical 13 words; not associated eight words; total 81 words). The lexicon data was identified by manually reading sentences in the training data (approximately 1% of each class over 11 967 sentences) and selecting words representative of the assertion classes. The rule-based method can be summarized in three steps: (1) for each sentence in the corpus, generate a list of problems, sorted by their order of appearance in the sentence; (2) the class of each problem will be the name of lexicon which the closest word preceding that problem belongs to; (3) if we cannot find any lexicon word before problem in the sentence, the default type will be present. For example, with the sentence ‘This is very likely to be an asthma exacerbation’, likely is the closest lexicon word (possible lexicon) before problem an asthma exacerbation then assertion of this problem will be possible. Additional heuristics such as backward scanning for lexicon words before a problem until we meet another problem, finding lexical words up to three words after problem for special instances have been added to the rule-based method.
With an F-score performance from the rule-based method of 90.73%, we considered that a statistical approach based on the same idea would produce a better performance. Consequently, the rule-based method was converted to a statistical method for both CRF and SVM tests. However, only the CRF generated a higher score than the rule-based method (92.37%) while the score of the SVM dropped significantly (81.77%). Four feature types were created for the assertion CRF and SVM:
Bag of words with a context window size of three words. With this window size, the model automatically learns the heuristic of finding lexical words up to three words after problem for special instances.
Lexicon type: keep track of current lexicon type for each token in the sentence. At the beginning of sentence, this feature is set as present (default value) and will be updated to corresponding lexicon type whenever it meets a word in that lexicon. Consequently, every token will come with a feature indicating the closest lexicon type before it and so the basic idea from rule-based method has been embedded into the statistical approach.
Negation identifier: indicate whether a concept has been negated or not. If a problem is negated, it is more likely to be an absent problem.
Problem boundary: indicates boundaries of the problem concept that need to be re-classified.
The assertion of each problem was based on information in the sentence it belonged to. Generally, the assertion is decided by the nearest word in a lexicon before a problem, in which the name of the lexicon becomes the type of assertion. Priorities of assertion types were also considered, in which a new assertion type is assigned only if it has higher ranking than the current type. If there was no word in any lexicon, the default assertion type was assigned as present, and there was no lexicon compiled for the present assertion.
Relationship identification
First, the system needs to generate all possible combinations of problem with other concepts within each sentence, and then pass each pair of concepts and the sentence into an SVM. There were nine features used in the SVM to classify the relationships between medical concepts, consisting of local context features and semantic features:
- Local context features:
- a. Three words before the first concept.
- b. Three words after the second concept.
- c. Words between the two concepts.
- d. Words inside of each concept.
- Semantic features:
- a. The type of each concept from the ground truth.
- b. The assertion type of the problem concept.
- c. Concept types between two concepts.
- d. Medication extraction result.
- e. Lexicon type.
Results
Table 1 shows the performance on the concept annotation task with bag of words used as the only feature set in the baseline model. In the baseline model and training model, 10-fold cross validation has been used to select optimal feature sets.
Table 1.
Final scores for concept annotation for the challenge test set, 10-fold cross-validation of the training set and baseline
| Entity type | Training | Testing | Recall (test), recall (train), (baseline) | Precision (test), precision (train), (baseline) | F-score (test), F-score (train), (baseline) |
| Problem | 11 983 | 18 550 |
|
|
|
| Test | 7380 | 12 899 |
|
|
|
| Treatment | 8515 | 13 560 |
|
|
|
| Overall | 27 878 | 45 009 |
|
|
|
Table 2 shows results on assertion classification using CRF developed from rule-based methods.
Table 2.
Scores for challenge test data and the training set for assertion classification
| Assertion type | Training | Testing | Recall (test), recall (train) | Precision (test), precision (train) | F-score (test), F-score (train) |
| Absent | 2535 | 3609 |
|
|
|
| Not associated | 92 | 145 |
|
|
|
| Conditional | 103 | 171 |
|
|
|
| Hypothetical | 651 | 717 |
|
|
|
| Possible | 535 | 883 |
|
|
|
| Present | 8051 | 13 025 |
|
|
|
| Overall | 11 967 | 18 550 |
|
|
|
For the relationship identification results in table 3, the baseline is produced from an SVM classifier with basic feature sets: three words before and after concepts, words between concepts, words inside of each concept and the types of concepts.
Table 3.
Scores for challenge test, training set and a baseline model for relation classification
| Entity type | Training | Testing | Recall (test), recall (train), (baseline) | Precision (test), precision (train), (baseline) | F-score (test), F-score (train), (baseline) |
| PIP | 1239 | 1986 |
|
|
|
| TrWP | 56 | 143 |
|
|
|
| TrAP | 1422 | 2487 |
|
|
|
| TrNAP | 106 | 191 |
|
|
|
| TrCP | 296 | 444 |
|
|
|
| TrIP | 107 | 198 |
|
|
|
| TeCP | 303 | 588 |
|
|
|
| TeRP | 1733 | 3033 |
|
|
|
| Overall | 5262 | 9070 |
|
|
|
PIP, problem indicates problem; TeCP, test conducted to investigate problem; TeRP, test reveals problem; TrAP, treatment is administered problem; TrCP, treatment causes problem; TrIP, treatment improves problem; TrNAP, treatment not administered because of problem; TrWP, treatment worsens problem.
The micro averaged F-measure for the system outputs achieved high performance in all three tasks of the competition over 42 teams: equal second on relationship identification, equal third on concept annotation and in the first 10 on assertion classification.
Discussion
Concept annotation
Overall, the best F-score is over 83%, approximately 6% higher than the bag of words baseline. Treatment still has the lowest score but the difference to problem (0.12%) and test (1.31%) is less significant. This occurs because:
There are many ways that treatment can be represented in clinical notes (drug name; drug name with dose; drug name with details in brackets, multiple drug names separated by hyphens, etc).
Misspelling of drug names.
Many unseen drug names.
In contrast, the performance for test annotation is highest although it has the smallest frequency of the three entity types. The reason is there are fewer varieties of test expressions so that the model can learn them more effectively.
The training results are nearly 1.5% better than the challenge test result of 81.79%. This is a loss due to unseen data, in which the total number of concepts in the test set is more than one and a half times greater than the training data.
Assertion classification
As shown in table 2, the best performance was obtained by using CRF methods. The explanation for this result is:
The sequence of words in the sentence and especially before each concept is important in deciding the assertion made about the medical problems.
In the CRF method, the sequence of tokens and their features is a key factor to training the model. While for the SVM, only the word itself could be used as a feature and so the sequence contributed little to the classification result.
The most popular classes (present, absent) have the highest performance with the F-score greater than 91% in both training and challenge test data sets. The lowest F-scores were in the scarce types (conditional, not associated) due to a lack of training examples and small number of words used in their respective lexicon feature, especially conditional, which performed the worst with just under 30% for the F-score.
Relationship identification
In the specification of the relation classification task, there is no need to indicate if two concepts do not have a relationship. However in the SVM model, no relation was also treated as a type of relationship to enable the classification process. As can be seen from table 3, the higher the frequency of the relation type the better performance it achieved. The three most frequent classes have the highest F-scores: TeRP (84.62%), TrAP (72.89%) and PIP (67.91%); while the smallest type of TrWP had very low F-scores at under 7%.
The challenge test data are nearly double the size for the larger classes of the training data set. This causes approximately a 2.5% drop in F-score due to unseen examples.
Discussion of overall result
Within a limited time, a completed system has been built to cope with all three tasks in the i2b2 challenge and the ability to extend to other practical tasks. The challenge in the multitask test requires a team to distribute its time and workload for each individual task thus assigning less time to a given task than a team entering only one task. So the overall performance evaluation on all three tasks would be more informative for those teams rather than treating each task assessment as independent. This could be done by computing the F-score over all annotations of the three tasks. In this case, our system submission achieved relatively high performance on all three tasks along with having a unique architectural design. This result also demonstrates that the system design is easily adaptable to different types of clinical NLP tasks. Models of different structures can be evaluated and compared based on a combination of model complexity and goodness of performance.
The simplicity of the model developed in this work should be contrasted with the very large feature space of other participants in the challenge who only achieved slightly better performance. We advocate that there is a need to charge a penalty against the complexity of a model as defined in message minimalisation theories.20 Our system performance was accomplished by using classic machine learning algorithms such as CRF and SVM.
Conclusion
In this paper, a complete system for the i2b2 clinical NLP challenge has been presented. The system generates results for all three tasks in the challenge. Furthermore, we also introduced a general NLP system architecture, which is easily adapted to different requirements in clinical information extraction and classification by choosing relevant feature sets.
In future work, more feature sets could be added such as a sentence parse tree. Finally, this system's pipeline will be developed into an experiment management system so that researchers can efficiently select various feature sets from a feature list and run the experiment for multiple NLP tasks.
The field of clinical NLP needs to address the issues of trade-offs between model complexity and model accuracy, as creating operational systems will invariably bring to the fore the importance of economic computational models that can be used in restricted environments.
Acknowledgments
The authors would like to give special thanks to members of the Health Information Technologies Research Laboratory for their valuable contributions.
Footnotes
Funding: The 2010 i2b2/VA challenge and the workshop are funded in part by grant no U54-LM008748 on Informatics for Integrating Biology to the Bedside from National Library of Medicine. This challenge and workshop are also supported by resources and facilities of the VA Salt Lake City Health Care System with funding support from the Consortium for Healthcare Informatics Research (CHIR), VA HSR HIR 08-374 and the VA Informatics and Computing Infrastructure (VINCI), VA HSR HIR 08-204. MedQuist, the largest transcription technology and services vendor, generously co-sponsored the 2010 i2b2/VA challenge meeting at AMIA.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Informatics for Integrating Biology and the Bedside (i2b2) NIH-funded National Center for Biomedical Computing (NCBC) based at Partners HealthCare System in Boston, MA; established in 2004. https://www.i2b2.org/NLP (accessed 9 Aug 2010). [Google Scholar]
- 2.Unified Medical Language System (UMLS) U.S National Library of Medicine, National Institutes of Health. http://www.nlm.nih.gov/research/umls (accessed Feb 2010).
- 3.The International Health Terminology Standards Development Organisation (IHTSDO) An international not-for-profit organization based in Denmark. IHTSDO acquires, owns and administers the rights to Sydtematized Nomenclature of Medicine - Clinical Terms (SNOMED-CT) and other health terminologies and related standards. http://www.ihtsdo.org/snomed-ct (accessed Feb 2010).
- 4.The Institute for Language, Speech and Hearing. Grady Ward's Moby; http://icon.shef.ac.uk/Moby (accessed Dec 2009). [Google Scholar]
- 5.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004;11:392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haug PJ, Koehler S, Lau LM, et al. Experience with a mixed semantic/syntactic parser. Proc Annu Symp Comput Appl Med Care 1995:284–8; PMCID: PMC2579100. [PMC free article] [PubMed] [Google Scholar]
- 8.Lee KJ, Hwang YS, Rim HC. Two-phase biomedical NE recognition based on SVMs. Proceedings of ACL Workshop on Natural Language Processing in Biomedicine Sapporo, Japan, 11 July 2003:33–40 [Google Scholar]
- 9.Li D, Kipper-Schuler KC, Savova G. Conditional Random Fields and Support Vector Machines for Disorder Named Entity Recognition in Clinical Texts. BioNLP: Current Trends in Biomedical Natural Language Processing. Ohio, USA: Symposium Proceedings, 2008:94–5 [Google Scholar]
- 10.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearbook Med Inform 2008:128–44 [PubMed] [Google Scholar]
- 11.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 12.Uzuner Ö, Zhang X, Sibanda T. Machine learning and rule-based approaches to assertion classification. J Am Med Inform Assoc 2009;16:109–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haddow B. Using automated feature optimisation to create an adaptable relation extraction system. BioNLP: Current Trends in Biomedical Natural Language Processing. Ohio, USA: Symposium Proceedings, 2008:19–27 [Google Scholar]
- 14.Wang Y. Annotating and Recognizing Named Entities in Clinical Notes. Proceedings of the ACL-IJCNLP Student Research Workshop. Singapore: Suntec, 2009:18–26 [Google Scholar]
- 15.CRF++ Yet another CRF toolkit. Software. http://crfpp.sourceforge.net (accessed 2 Dec 2010).
- 16.Cristianini N, Shawe-Taylor J. An Introduction to Support Vector Machines and other Kernel-Based Learning Methods. Cambridge, UK: Cambridge University Press, 2000 [Google Scholar]
- 17.Patrick J, Wang Y, Budd P. An automated system for conversion of clinical notes into SNOMED clinical terminology. Proceedings of the 5th Australasian Symposium on ACSW frontiers. Ballarat, Victoria, Australia, 30 Jan–2 Feb, 2007;68:219–26 [Google Scholar]
- 18.Patrick J, Li M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J Am Med Inform Assoc 2010;17:524–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GENIA tagger. http://www-tsujii.is.s.u-tokyo.ac.jp/GENIA/tagger (accessed Feb 2010).
- 20.Wallace CS, Dowe DL. Minimum message length and kolmogorov complexity. Computer J 1999;42:270–83 [Google Scholar]