Abstract
Objectives
To provide an overview and tutorial of natural language processing (NLP) and modern NLP-system design.
Target audience
This tutorial targets the medical informatics generalist who has limited acquaintance with the principles behind NLP and/or limited knowledge of the current state of the art.
Scope
We describe the historical evolution of NLP, and summarize common NLP sub-problems in this extensive field. We then provide a synopsis of selected highlights of medical NLP efforts. After providing a brief description of common machine-learning approaches that are being used for diverse NLP sub-problems, we discuss how modern NLP architectures are designed, with a summary of the Apache Foundation's Unstructured Information Management Architecture. We finally consider possible future directions for NLP, and reflect on the possible impact of IBM Watson on the medical field.
Keywords: Natural language processing; Introduction; clinical NLP, knowledge bases; machine learning; predictive modeling; statistical learning; privacy technology
Introduction
This tutorial provides an overview of natural language processing (NLP) and lays a foundation for the JAMIA reader to better appreciate the articles in this issue.
NLP began in the 1950s as the intersection of artificial intelligence and linguistics. NLP was originally distinct from text information retrieval (IR), which employs highly scalable statistics-based techniques to index and search large volumes of text efficiently: Manning et al1 provide an excellent introduction to IR. With time, however, NLP and IR have converged somewhat. Currently, NLP borrows from several, very diverse fields, requiring today's NLP researchers and developers to broaden their mental knowledge-base significantly.
Early simplistic approaches, for example, word-for-word Russian-to-English machine translation,2 were defeated by homographs—identically spelled words with multiple meanings—and metaphor, leading to the apocryphal story of the Biblical, ‘the spirit is willing, but the flesh is weak’ being translated to ‘the vodka is agreeable, but the meat is spoiled.’
Chomsky's 1956 theoretical analysis of language grammars3 provided an estimate of the problem's difficulty, influencing the creation (1963) of Backus-Naur Form (BNF) notation.4 BNF is used to specify a ‘context-free grammar’5 (CFG), and is commonly used to represent programming-language syntax. A language's BNF specification is a set of derivation rules that collectively validate program code syntactically. (‘Rules’ here are absolute constraints, not expert systems' heuristics.) Chomsky also identified still more restrictive ‘regular’ grammars, the basis of the regular expressions6 used to specify text-search patterns. Regular expression syntax, defined by Kleene7 (1956), was first supported by Ken Thompson's grep utility8 on UNIX.
Subsequently (1970s), lexical-analyzer (lexer) generators and parser generators such as the lex/yacc combination9 utilized grammars. A lexer transforms text into tokens; a parser validates a token sequence. Lexer/parser generators simplify programming-language implementation greatly by taking regular-expression and BNF specifications, respectively, as input, and generating code and lookup tables that determine lexing/parsing decisions.
While CFGs are theoretically inadequate for natural language,10 they are often employed for NLP in practice. Programming languages are typically designed deliberately with a restrictive CFG variant, an LALR(1) grammar (LALR, Look-Ahead parser with Left-to-right processing and Rightmost (bottom-up) derivation),4 to simplify implementation. An LALR(1) parser scans text left-to-right, operates bottom-up (ie, it builds compound constructs from simpler ones), and uses a look-ahead of a single token to make parsing decisions.
The Prolog language11 was originally invented (1970) for NLP applications. Its syntax is especially suited for writing grammars, although, in the easiest implementation mode (top-down parsing), rules must be phrased differently (ie, right-recursively12) from those intended for a yacc-style parser. Top-down parsers are easier to implement than bottom-up parsers (they don't need generators), but are much slower.
The limitations of hand-written rules: the rise of statistical NLP
Natural language's vastly large size, unrestrictive nature, and ambiguity led to two problems when using standard parsing approaches that relied purely on symbolic, hand-crafted rules:
NLP must ultimately extract meaning (‘semantics’) from text: formal grammars that specify relationship between text units—parts of speech such as nouns, verbs, and adjectives—address syntax primarily. One can extend grammars to address natural-language semantics by greatly expanding sub-categorization, with additional rules/constraints (eg, ‘eat’ applies only to ingestible-item nouns). Unfortunately, the rules may now become unmanageably numerous, often interacting unpredictably, with more frequent ambiguous parses (multiple interpretations of a word sequence are possible). (Puns—ambiguous parses used for humorous effect—antedate NLP.)
Handwritten rules handle ‘ungrammatical’ spoken prose and (in medical contexts) the highly telegraphic prose of in-hospital progress notes very poorly, although such prose is human-comprehensible.
The 1980s resulted in a fundamental reorientation, summarized by Klein13:
Simple, robust approximations replaced deep analysis.
Evaluation became more rigorous.
Machine-learning methods that used probabilities became prominent. (Chomsky's book, Syntactic Structures14 (1959), had been skeptical about the usefulness of probabilistic language models).
Large, annotated bodies of text (corpora) were employed to train machine-learning algorithms—the annotation contains the correct answers—and provided gold standards for evaluation.
This reorientation resulted in the birth of statistical NLP. For example, statistical parsing addresses parsing-rule proliferation through probabilistic CFGs15: individual rules have associated probabilities, determined through machine-learning on annotated corpora. Thus, fewer, broader rules replace numerous detailed rules, with statistical-frequency information looked up to disambiguate. Other approaches build probabilistic ‘rules’ from annotated data similar to machine-learning algorithms like C4.5,16 which build decision trees from feature-vector data. In any case, a statistical parser determines the most likely parse of a sentence/phrase. ‘Most likely’ is context-dependent: for example, the Stanford Statistical Parser,17 trained with the Penn TreeBank18—annotated Wall Street Journal articles, plus telephone-operator conversations—may be unreliable for clinical text. Manning and Scheutze's text provides an excellent introduction to statistical NLP.19
Statistical approaches give good results in practice simply because, by learning with copious real data, they utilize the most common cases: the more abundant and representative the data, the better they get. They also degrade more gracefully with unfamiliar/erroneous input. This issue's articles make clear, however, that handwritten-rule-based and statistical approaches are complementary.
NLP sub-problems: application to clinical text
We enumerate common sub-problems in NLP: Jurafksy and Martin's text20 provides additional details. The solutions to some sub-problems have become workable and affordable, if imperfect—for example, speech synthesis (desktop operating systems' accessibility features) and connected-speech recognition (several commercial systems). Others, such as question answering, remain difficult.
In the account below, we mention clinical-context issues that complicate certain sub-problems, citing recent biomedical NLP work against each where appropriate. (We do not cover the history of medical NLP, which has been applied rather than basic/theoretical; Spyns21 reviews pre-1996 medical NLP efforts.)
Low-level NLP tasks include:
Sentence boundary detection: abbreviations and titles (‘m.g.,’ ‘Dr.’) complicate this task, as do items in a list or templated utterances (eg, ‘MI [x], SOB[]’).
Tokenization: identifying individual tokens (word, punctuation) within a sentence. A lexer plays a core role for this task and the previous one. In biomedical text, tokens often contain characters typically used as token boundaries, for example, hyphens, forward slashes (‘10 mg/day,’ ‘N-acetylcysteine’).
Part-of-speech assignment to individual words (‘POS tagging’): in English, homographs (‘set’) and gerunds (verbs ending in ‘ing’ that are used as nouns) complicate this task.
Morphological decomposition of compound words: many medical terms, for example, ‘nasogastric,’ need decomposition to comprehend them. A useful sub-task is lemmatization—conversion of a word to a root by removing suffixes. Non-English clinical NLP emphasizes decomposition; in highly synthetic languages (eg, German, Hungarian), newly coined compound words may replace entire phrases.22 Spell-checking applications and preparation of text for indexing/searching (in IR) also employ morphological analysis.
Shallow parsing (chunking): identifying phrases from constituent part-of-speech tagged tokens. For example, a noun phrase may comprise an adjective sequence followed by a noun.
Problem-specific segmentation: segmenting text into meaningful groups, such as sections, including Chief Complaint, Past Medical History, HEENT, etc.23
Haas24 lists publicly available NLP modules for such tasks: most modules, with the exception of cTAKES (clinical Text Analysis and Knowledge Extraction System),25 have been developed for non-clinical text and often work less well for clinical narrative.
Higher-level tasks build on low-level tasks and are usually problem-specific. They include:
1. Spelling/grammatical error identification and recovery: this task is mostly interactive because, as word-processing users know, it is far from perfect. Highly synthetic phrases predispose to false positives (correct words flagged as errors), and incorrectly used homophones (identically sounding, differently spelled words, eg, sole/soul, their/there) to false negatives.
2. Named entity recognition (NER)26 27: identifying specific words or phrases (‘entities’) and categorizing them—for example, as persons, locations, diseases, genes, or medication. An common NER task is mapping named entities to concepts in a vocabulary. This task often leverages shallow parsing for candidate entities (eg, the noun phrase ‘chest tenderness’); however, sometimes the concept is divided across multiple phrases (eg, ‘chest wall shows slight tenderness on pressure …’).
The following issues make NER challenging:
Word/phrase order variation: for example, perforated duodenal ulcer versus duodenal ulcer, perforated.
Derivation: for example, suffixes transform one part of speech to another (eg, ‘mediastinum’ (noun) → ‘mediastinal’ (adjective)).
Inflection: for example, changes in number (eg, ‘opacity/opacities)’, tense (eg, ‘cough(ed)’), comparative/superlative forms (eg, ‘bigger/biggest)’).
Synonymy is abundant in biomedicine, for example, liver/hepatic, Addison's disease/adrenocortical insufficiency.
Homographs: polysemy refers to homographs with related meanings, for example, ‘direct bilirubin’ can refer to a substance, laboratory procedure, or result. Homographic abbreviations are increasingly numerous28: ‘APC’ has 12 expansions, including ‘activated protein C’ and ‘adenomatous polyposis coli.’
3. Word sense disambiguation (WSD)29–31: determining a homograph's correct meaning.
4. Negation and uncertainty identification32–34: inferring whether a named entity is present or absent, and quantifying that inference's uncertainty. Around half of all symptoms, diagnoses, and findings in clinical reports are estimated to be negated.35 Negation can be explicit, for example, ‘Patient denies chest pain’ or implied—for example, ‘Lungs are clear upon auscultation’ implies absence of abnormal lung sounds. Negated/affirmed concepts can be expressed with uncertainty (‘hedging’), as in ‘the ill-defined density suggests pneumonia.’ Uncertainty that represents reasoning processes is hard to capture: ‘The patient probably has a left-sided cerebrovascular accident; post-convulsive state is less likely.’ Negation, uncertainty, and affirmation form a continuum. Uncertainty detection was the focus of a recent NLP competition.36
5. Relationship extraction: determining relationships between entities or events, such as ‘treats,’ ‘causes,’ and ‘occurs with.’ Lookup of problem-specific information—for example, thesauri, databases—facilitates relationship extraction.
Anaphora reference resolution37 is a sub-task that determines relationships between ‘hierarchically related’ entities: such relationships include:
Identity: one entity—for example, a pronoun like ‘s/he,’ ‘hers/his,’ or an abbreviation—refers to a previously mentioned named entity;
Part/whole: for example, city within state;
Superset/subset: for example, antibiotic/penicillin.
6. Temporal inferences/relationship extraction38 39: making inferences from temporal expressions and temporal relations—for example, inferring that something has occurred in the past or may occur in the future, and ordering events within a narrative (eg, medication X was prescribed after symptoms began).
7. Information extraction (IE): the identification of problem-specific information and its transformation into (problem-specific) structured form. Tasks 1–6 are often part of the larger IE task. For example, extracting a patient's current diagnoses involves NER, WSD, negation detection, temporal inference, and anaphoric resolution. Numerous modern clinical IE systems exist,40–44 with some available as open-source.25 44 45 IE and relationship extraction have been themes of several i2b2/VA NLP challenges.46–49 Other problem areas include phenotype characterization,50–52 biosurveillance,53 54 and adverse-drug reaction recognition.55
The National Library of Medicine (NLM) provides several well-known ‘knowledge infrastructure’ resources that apply to multiple NLP and IR tasks. The UMLS Metathesaurus,56 which records synonyms and categories of biomedical concepts from numerous biomedical terminologies, is useful in clinical NER. The NLM's Specialist Lexicon57 is a database of common English and medical terms that includes part-of-speech and inflection data; it is accompanied by a set of NLP tools.58 The NLM also provides a test collection for word disambiguation.59
Some data driven approaches: an overview
Statistical and machine learning involve development (or use) of algorithms that allow a program to infer patterns about example (‘training’) data, that in turn allows it to ‘generalize’—make predictions about new data. During the learning phase, numerical parameters that characterize a given algorithm's underlying model are computed by optimizing a numerical measure, typically through an iterative process.
In general, learning can be supervised—each item in the training data is labeled with the correct answer—or unsupervised, where it is not, and the learning process tries to recognize patterns automatically (as in cluster and factor analysis). One pitfall in any learning approach is the potential for over-fitting: the model may fit the example data almost perfectly, but makes poor predictions for new, previously unseen cases. This is because it may learn the random noise in the training data rather than only its essential, desired features. Over-fitting risk is minimized by techniques such as cross-validation, which partition the example data randomly into training and test sets to internally validate the model's predictions. This process of data partitioning, training, and validation is repeated over several rounds, and the validation results are then averaged across rounds.
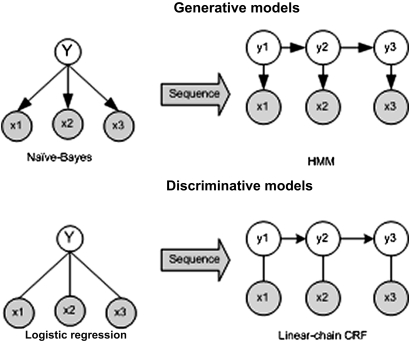
Machine-learning models can be broadly classified as either generative or discriminative. Generative methods seek to create rich models of probability distributions, and are so called because, with such models, one can ‘generate’ synthetic data. Discriminative methods are more utilitarian, directly estimating posterior probabilities based on observations. Srihari60 explains the difference with an analogy: to identify an unknown speaker's language, generative approaches would apply deep knowledge of numerous languages to perform the match; discriminative methods would rely on a less knowledge-intensive approach of using differences between languages to find the closest match. Compared to generative models, which can become intractable when many features are used, discriminative models typically allow use of more features.61 Logistic regression and conditional random fields (CRFs) are examples of discriminative methods, while Naive Bayes classifiers and hidden Markov models (HMMs) are examples of generative methods.
Some common machine-learning methods used in NLP tasks, and utilized by several articles in this issue, are summarized below.
Support vector machines (SVMs)
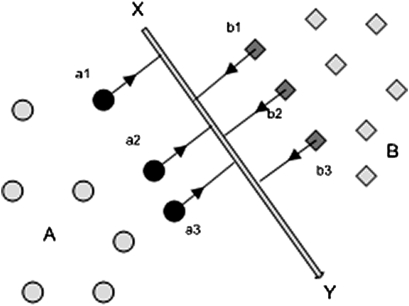
SVMs, a discriminative learning approach, classify inputs (eg, words) into categories (eg, parts of speech) based on a feature set. The input may be transformed mathematically using a ‘kernel function’ to allow linear separation of the data points from different categories. That is, in the simplest two-feature case, a straight line would separate them in an X–Y plot: in the general N-feature case, the separator will be an (N−1) hyperplane. The commonest kernel function used is a Gaussian (the basis of the ‘normal distribution’ in statistics). The separation process selects a subset of the training data (the ‘support vectors’—data points closest to the hyperplane) that best differentiates the categories. The separating hyperplane maximizes the distance to support vectors from each category (see figure 1).
Figure 1.
Support vector machines: a simple 2-D case is illustrated. The data points, shown as categories A (circles) and B (diamonds), can be separated by a straight line X–Y. The algorithm that determines X–Y identifies the data points (‘support vectors’) from each category that are closest to the other category (a1, a2, a3 and b1, b2, b3) and computes X–Y such that the margin that separates the categories on either side is maximized. In the general N-dimensional case, the separator will be an (N−1) hyperplane, and the raw data will sometimes need to be mathematically transformed so that linear separation is achievable.
A tutorial by Hearst et al62 and the DTREG online documentation63 provide approachable introductions to SVMs. Fradkin and Muchnik64 provide a more technical overview.
Hidden Markov models (HMMs)
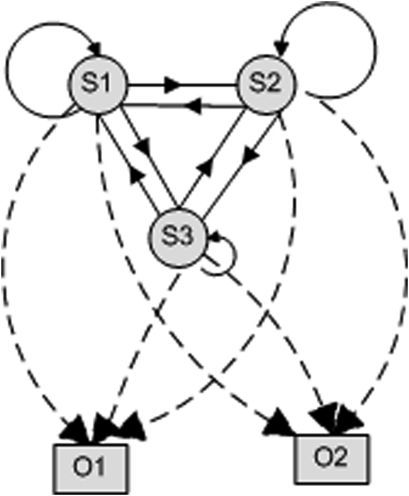
An HMM is a system where a variable can switch (with varying probabilities) between several states, generating one of several possible output symbols with each switch (also with varying probabilities). The sets of possible states and unique symbols may be large, but finite and known (see figure 2). We can observe the outputs, but the system's internals (ie, state-switch probabilities and output probabilities) are ‘hidden.’ The problems to be solved are:
Figure 2.
Hidden Markov models. The small circles S1, S2 and S3 represent states. Boxes O1 and O2 represent output values. (In practical cases, hundreds of states/output values may occur.) The solid lines/arcs connecting states represent state switches; the arrow represents the switch's direction. (A state may switch back to itself.) Each line/arc label (not shown) is the switch probability, a decimal number. A dashed line/arc connecting a state to an output value indicates ‘output probability’: the probability of that output value being generated from the particular state. If a particular switch/output probability is zero, the line/arc is not drawn. The sum of the switch probabilities leaving a given state (and the similar sum of output probabilities) is equal to 1. The sequential or temporal aspect of an HMM is shown in figure 3.
Inference: given a particular sequence of output symbols, compute the probabilities of one or more candidate state-switch sequences.
Pattern matching: find the state-switch sequence most likely to have generated a particular output-symbol sequence.
Training: given examples of output-symbol sequence (training) data, compute the state-switch/output probabilities (ie, system internals) that fit this data best.
B and C are actually Naive Bayesian reasoning extended to sequences; therefore, HMMs use a generative model. To solve these problems, an HMM uses two simplifying assumptions (which are true of numerous real-life phenomena):
The probability of switching to a new state (or back to the same state) depends on the previous N states. In the simplest ‘first-order’ case (N=1), this probability is determined by the current state alone. (First-order HMMs are thus useful to model events whose likelihood depends on what happened last.)
The probability of generating a particular output in a particular state depends only on that state.
These assumptions allow the probability of a given state-switch sequence (and a corresponding observed-output sequence) to be computed by simple multiplication of the individual probabilities. Several algorithms exist to solve these problems.65 66 The highly efficient Viterbi algorithm, which addresses problem B, finds applications in signal processing, for example, cell-phone technology.
Theoretically, HMMs could be extended to a multivariate scenario,67 but the training problem can now become intractable. In practice, multiple-variable applications of HMMs (eg, NER68) use single, artificial variables that are uniquely determined composites of existing categorical variables: such approaches require much more training data.
HMMs are widely used for speech recognition, where a spoken word's waveform (the output sequence) is matched to the sequence of individual phonemes (the ‘states’) that most likely produced it. (Frederick Jelinek, a statistical-NLP advocate who pioneered HMMs at IBM's Speech Recognition Group, reportedly joked, ‘every time a linguist leaves my group, the speech recognizer's performance improves.’20) HMMs also address several bioinformatics problems, for example, multiple sequence alignment69 and gene prediction.70 Eddy71 provides a lucid bioinformatics-oriented introduction to HMMs, while Rabiner72 (speech recognition) provides a more detailed introduction.
Commercial HMM-based speech-to-text is now robust enough to have essentially killed off academic research efforts, with dictation systems for specialized areas—eg, radiology and pathology—providing structured data entry. Phrase recognition is paradoxically more reliable for polysyllabic medical terms than for ordinary English: few word sequences sound like ‘angina pectoris,’ while common English has numerous homophones (eg, two/too/to).
Conditional random fields (CRFs)
CRFs are a family of discriminative models first proposed by Lafferty et al.73 An accessible reference is Culotta et al74; Sutton and McCallum75 is more mathematical. The commonest (linear-chain) CRFs resemble HMMs in that the next state depends on the current state (hence the ‘linear chain’ of dependency).
CRFs generalize logistic regression to sequential data in the same way that HMMs generalize Naive Bayes (see figure 3). CRFs are used to predict the state variables (‘Ys’) based on the observed variables (‘Xs’). For example, when applied to NER, the state variables are the categories of the named entities: we want to predict a sequence of named-entity categories within a passage. The observed variables might be the word itself, prefixes/suffixes, capitalization, embedded numbers, hyphenation, and so on. The linear-chain paradigm fits NER well: for example, if the previous entity is ‘Salutation’ (eg, ‘Mr/Ms’), the succeeding entity must be a person.
Figure 3.
The relationship between Naive Bayes, logistic regression, hidden Markov models (HMMs) and conditional random fields (CRFs). Logistic regression is the discriminative-model counterpart of Naive Bayes, which is a generative model. HMMs and CRFs extend Naive Bayes and logistic regression, respectively, to sequential data (adapted from Sutton and McCallum73). In the generative models, the arrows indicate the direction of dependency. Thus, for the HMM, the state Y2 depends on the previous state Y1, while the output X1 depends on Y1.
CRFs are better suited to sequential multivariate data than HMMs: the training problem, while requiring more example data than a univariate HMM, is still tractable.
N-grams
An ‘N-gram’19 is a sequence of N items—letters, words, or phonemes. We know that certain item pairs (or triplets, quadruplets, etc) are likely to occur much more frequently than others. For example, in English words, U always follows Q, and an initial T is never followed by K (though it may be in Ukrainian). In Portuguese, a Ç is always followed by a vowel (except E and I). Given sufficient data, we can compute frequency-distribution data for all N-grams occurring in that data. Because the permutations increase dramatically with N—for example, English has 26^2 possible letter pairs, 26^3 triplets, and so on—N is restricted to a modest number. Google has computed word N-gram data (N≤5) from its web data and from the Google Books project, and made it available freely.76
N-grams are a kind of multi-order Markov model: the probability of a particular item at the Nth position depends on the previous N−1 items, and can be computed from data. Once computed, N-gram data can be used for several purposes:
Suggested auto-completion of words and phrases to the user during search, as seen in Google's own interface.
Spelling correction: a misspelled word in a phrase may be flagged and a correct spelling suggested based on the correctly spelled neighboring words, as Google does.
Speech recognition: homophones (‘two’ vs ‘too’) can be disambiguated probabilistically based on correctly recognized neighboring words.
Word disambiguation: if we build ‘word-meaning’ N-grams from an annotated corpus where homographs are tagged with their correct meanings, we can use the non-ambiguous neighboring words to guess the correct meaning of a homograph in a test document.
N-gram data are voluminous—Google's N-gram database requires 28 GB—but this has become less of an issue as storage becomes cheap. Special data structures, called N-gram indexes, speed up search of such data. N-gram-based classifiers leverage raw training text without explicit linguistic/domain knowledge; while yielding good performance, they leave room for improvement, and are therefore complemented with other approaches.
Chaining NLP analytical tasks: pipelines
Any practical NLP task must perform several sub-tasks. For example, all of NLP sub-problems section′s low-level tasks must execute sequentially, before higher-level tasks can commence. Since different algorithms may be used for a given task, a modular, pipelined system design—the output of one analytical module becomes the input to the next—allows ‘mixing-and-matching.’ Thus, a CRF-based POS tagger could be combined with rule-based medical named-entity recognition. This design improves system robustness: one could replace one module with another (possibly superior) module, with minimal changes to the rest of the system.
This is the intention behind pipelined NLP frameworks, such as GATE77 and IBM (now Apache) Unstructured Information Management Architecture (UIMA).78 UIMA's scope goes beyond NLP: one could integrate structured-format databases, images, and multi-media, and any arbitrary technology. In UIMA, each analytical task transforms (a copy of) its input by adding XML-based markup and/or reading/writing external data. A task operates on Common Analysis Structure (CAS), which contains the data (possibly in multiple formats, eg, audio, HTML), a schema describing the analysis structure (ie, the details of the markup/external formats), the analysis results, and links (indexes) to the portions of the source data that they refer to. UIMA does not dictate the design of the analytical tasks themselves: they interact with the UIMA pipeline only through the CAS, and can be treated as black boxes: thus, different tasks could be written in different programming languages.
The schema for a particular CAS is developer-defined because it is usually problem-specific. (Currently, no standard schemas exist for tasks such as POS tagging, although this may change.) Definition is performed using XMI (XML Metadata Interchange), the XML-interchange equivalent of the Unified Modeling Language (UML). XMI, however, is ‘programmer-hostile’: it is easier to use a commercial UML tool to design a UML model visually and then generate XMI from it.79
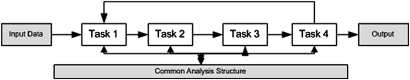
In practice, a pure pipeline design may not be optimal for all solutions. In many cases, a higher-level process needs to provide feedback to a lower-level process to improve the latter's accuracy. (All supervised machine- learning algorithms, for example, ultimately rely on feedback.) Implementing feedback across analytical tasks is complicated: it involves modifying the code of communicating tasks—one outputting data that constitutes the feedback, the other checking for the existence of such data, and accepting them if available (see figure 4). New approaches based on active learning may help select cases for manual labeling for construction of training sets.80 81
Figure 4.
A UIMA pipeline. An input task is sequentially put through a series of tasks, with intermediate results at each step and final output at the end. Generally, the output of a task is the input of its successor, but exceptionally, a particular task may provide feedback to a previous one (as in task 4 providing input to task 1). Intermediate results (eg, successive transformations of the original bus) are read from/written to the CAS, which contains metadata defining the formats of the data required at every step, the intermediate results, and annotations that link to these results.
Also, given that no NLP task achieves perfect accuracy, errors in any one process in a pipeline will propagate to the next, and so on, with accuracy degrading at each step. This problem, however, applies to NLP in general: it would occur even if the individual tasks were all combined into a single body of code. One way to address it (adopted in some commercial systems) is to use alternative algorithms (in multiple or branching pipelines) and contrast the final results obtained. This allows tuning the output to trade-offs (high precision versus high recall, etc).
A look into the future
Recent advances in artificial intelligence (eg, computer chess) have shown that effective approaches utilize the strengths of electronic circuitry—high speed and large memory/disk capacity, problem-specific data-compression techniques and evaluation functions, highly efficient search—rather than trying to mimic human neural function. Similarly, statistical-NLP methods correspond minimally to human thought processes.
By comparison with IR, we now consider what it may take for multi-purpose NLP technology to become mainstream. While always important to library science, IR achieved major prominence with the web, notably after Google's scientific and financial success: the limelight also caused a corresponding IR research and toolset boom. The question is whether NLP has a similar breakthrough application in the wings. One candidate is IBM Watson, which attracted much attention within the biomedical informatics community (eg, the ACMI Discussion newsgroup and the AMIA NLP working group discussion list) after its ‘Jeopardy’ performance. Watson appears to address the admittedly hard problem of question-answering successfully. Although the Watson effort is impressive in many ways, its discernible limitations highlight ongoing NLP challenges.
IBM Watson: a wait-and-see viewpoint
Watson, which employs UIMA,82 is a system-engineering triumph, using highly parallel hardware with 2880 CPUs+16 TB RAM. All its lookup of reference content (encyclopedias, dictionaries, etc) and analytical operations use structures optimized for in-memory manipulation. (By contrast, most pipelined NLP architectures on ordinary hardware are disk-I/O-bound.) It integrates several software technologies: IR, NLP, parallel database search, ontologies, and knowledge representation.
A Prolog parser extracts key elements such as the relationships between entities and task-specific answers. In a recent public display, the task was to compete for the fastest correct answer in a series of questions against two human contestants in the popular US-based television show, ‘Jeopardy.’ During training with a Jeopardy question-databank, NLP is also used to pre-process online reference text (eg, encyclopedia, dictionaries) into a structure that provides evidence for candidate answers, including whether the relationships between entities in the question match those in the evidence.83 The search, and ranking of candidate answers, use IR approaches.
A challenge in porting Watson's technology to other domains, such as medical question answering, will be the degree to which Watson's design is generalizable.
Watson built its lead in the contest with straightforward direct questions whose answers many of the audience (and the skilled human contestants) clearly knew—and which a non-expert human armed with Google may have been able to retrieve using keywords alone (albeit slower). As pointed out by Libresco84 and Jennings,85 Watson was merely faster with the buzzer—electronics beats human reaction time. For non-game-playing, real-world question answering scenarios, however, split-second reaction time may not constitute a competitive advantage.
For harder questions, Watson's limitations became clearer. Computing the correct response to the question about which US city (Chicago) has two airports, one named after a World War II battle (Midway), the other after a World War II hero (O'Hare), involves three set intersections (eg, the first operation would cross names of airports in US cities against a list of World War II battles). Watson lacked a higher-level strategy to answer such complex questions.
Watson's Prolog parser and search, and especially the entire reference content, were tuned/structured for playing Jeopardy, in which the questions and answers are one sentence long (and the answer is of the form ‘what/who is/are X?’). Such an approach runs the risk of ‘over-fitting’ the system to a particular problem, so that it may require significant effort to modify it for even a slightly different problem.
IBM recently conducted a medical diagnosis demonstration of Watson, which is reported in an Associated Press article.86 Demonstrations eventually need to be followed by evaluations. Earlier medical diagnosis advice software underwent evaluations that were rigorous for their time, for example, Berner et al87 and Friedman et al,88 and today's evaluations would need to be even more stringent. The articles from Miller and Masarie89 and Miller90 are excellent starting points for learning about the numerous pitfalls in the automated medical diagnosis domain, and IBM may rediscover these:
Medico-legal liability: ultimately the provider, not software, is responsible for the patient.
Reference-content reliability: determining the reliability of a given unit of evidence is challenging. Even some recent recommendations by ‘authorities’ have become tainted (eg, in psychiatry) with subsequent revelations of undisclosed conflict of interest.
The limited role of NLP and unstructured text in medical diagnosis: it is unclear that accurate medical diagnosis/advice mandates front-end NLP technology: structured data entry with thesaurus/N-gram assisted pick-lists or word/phrase completion might suffice. Similarly, diagnostic systems have used structured, curated information rather than unstructured text for prioritizing diagnoses. Even this information requires tailoring for local prevalence rates, and continual maintenance. Unstructured text, in the form of citations, is used mainly to support the structured information.
To be fair to IBM, NLP technology may conceivably augment web crawler technologies that search for specific information and alert curators about new information that may require them to update their database. Electronic IE technologies might save curation time, but given the medico-legal consequences, and the lack of 100% accuracy, such information would need to be verified by humans.
From an optimistic perspective, the Watson phenomenon may have the beneficial side effect of focusing attention not only on NLP, but also on the need to integrate it effectively with other technologies.
Will NLP software become a commodity?
The post-Google interest in IR has led to IR commoditization: a proliferation of IR tools and incorporation of IR technology into relational database engines. Earlier, statistical packages and, subsequently, data mining tools also became commoditized. Commodity analytical software is characterized by:
Availability of several tools within a package: the user can often set up a pipeline without programming using a graphical metaphor.
High user friendliness and ease of learning: online documentation/tutorials are highly approachable for the non-specialist, focusing on when and how to use a particular tool rather than its underlying mathematical principles.
High value in relation to price: some offerings may even be freeware.
By contrast, NLP toolkits and UIMA are still oriented toward the advanced programmer, and commercial offerings are expensive. General purpose NLP is possibly overdue for commoditization: if this happens, best-of-breed solutions are more likely to rise to the top. Again, analytics vendors are likely to lead the way, following the steps of biomedical informatics researchers to devise innovative solutions to the challenge of processing complex biomedical language in the diverse settings where it is employed.
Footnotes
Funding: This work is funded in part by grants from the National Institutes of Health (R01LM009520, U54HL108460, and UL1RR031980).
Competing interests: None.
Provenance and peer review: Commissioned; internally peer reviewed.
References
- 1.Manning C, Raghavan P, Schuetze H. Introduction to Information Retrieval. Cambridge, UK: Cambridge University Press, 2008 [Google Scholar]
- 2.Hutchins W. The First Public Demonstration of Machine Translation: the Georgetown-IBM System, 7th January 1954. 2005. http://www.hutchinsweb.me.uk/GU-IBM-2005.pdf (accessed 4 Jun 2011). [Google Scholar]
- 3.Chomsky N. Three models for the description of language. IRE Trans Inf Theory 1956;2:113–24 [Google Scholar]
- 4.Aho AV, Sethi R, Ullman JD. Compilers: Principles, Techniques, Tools. Reading, MA: Addison-Wesley, 1988 [Google Scholar]
- 5.Chomsky N. On certain formal properties of grammars. Inform Contr 1959;2:137–67 [Google Scholar]
- 6.Friedl JEF. Mastering Regular Expressions. Sebastopol, CA: O'Reilly & Associates, Inc., 1997 [Google Scholar]
- 7.Kleene SC. Representation of events in nerve nets and finite automata. In: Shannon C, McCarthy J, eds. Automata Studies. Princeton, NJ: Princeton University Press, 1956 [Google Scholar]
- 8.Kernighan B, Pike R. The UNIX Programming Environment. Englewood Cliffs, NJ: Prentice-Hall, 1989 [Google Scholar]
- 9.Levine JR, Mason T, Brown D. Lex & Yacc. Sebastopol, CA: O'Reilly & Associates, Inc., 1992 [Google Scholar]
- 10.Joshi A, Vijay-Shanker K, Weir D. The convergence of mildly context-sensitive grammar formalisms. In: Sells P, Shieber S, Wasow T, eds. Foundational Issues in Natural Language Processing. Cambridge, MA: MIT Press, 1991:31–81 [Google Scholar]
- 11.Clocksin WF, Mellish CS. Programming in Prolog: Using the ISO Standard. 5th edn New York: Springer, 2003 [Google Scholar]
- 12.Warren DS. Programming in Tabled Prolog. 1999. http://www.cs.sunysb.edu/∼warren/xsbbook/node10.html (accessed 1 Jun 2011). [Google Scholar]
- 13.Klein D. CS 294–5: Statistical Natural Language Processing. 2005. http://www.cs.berkeley.edu/∼klein/cs294-5 (accessed 2 Jun 2011). [Google Scholar]
- 14.Chomsky N. Syntactic Structures. The Hague, Netherlands: Mouton and Co, 1957 [Google Scholar]
- 15.Klein D, Manning C. Accurate Unlexicalized Parsing. Proceedings of the 41st Meeting of the Association for Computational Linguistics; 2003. 2003:423–30 http://nlp.stanford.edu/∼manning/papers/unlexicalized-parsing.pdf (accessed 20 Jul 2011). [Google Scholar]
- 16.Quinlan JR. C4.5: Programs for Machine Learning. San Mateo, CA: Morgan Kaufmann Publishers, 1993 [Google Scholar]
- 17.Klein D, Manning C. Stanford Statistical Parser. 2003. http://nlp.stanford.edu/software/lex-parser.shtml (accessed 4 Jun 2011). [Google Scholar]
- 18.University of Pennsylvania Penn Treebank Project. 2011. http://www.cis.upenn.edu/∼treebank/ (accessed 21 May 2011). [Google Scholar]
- 19.Manning C, Schuetze H. Foundations of Statistical Natural Language Processing. Cambridge, MA: MIT Press, 1999 [Google Scholar]
- 20.Jurafsky D, Martin JH. Speech and Language Processing. 2nd edn Englewood Cliffs, NJ: Prentice-Hall, 2008 [Google Scholar]
- 21.Spyns P. Natural language processing in medicine: an overview. Methods Inf Med 1996;5:285–301 [PubMed] [Google Scholar]
- 22.Deleger L, Namer F, Zweigenbaum P. Morphosemantic parsing of medical compound words: transferring a French analyzer to English. Int J Med Inform 2009;78(Suppl 1):S48–55 [DOI] [PubMed] [Google Scholar]
- 23.Denny JC, Spickard A, 3rd, Johnson KB, et al. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc 2009;16:806–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas S. Tools for Natural language processing. 2011. http://ils.unc.edu/∼stephani/nlpsp08/resources.html#tools (accessed 1 Jun 2011). [Google Scholar]
- 25.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aronson A. Effective mapping of biomedical text to the UMLS metathesaurus: The MetaMap Program. Proc AMIA Symp 2001;2001:17–21 [PMC free article] [PubMed] [Google Scholar]
- 27.Zou Q, Chu WW, Morioka C, et al. IndexFinder: a method of extracting key concepts from clinical texts for indexing. AMIA Annu Symp Proc 2003:763–7 [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Aronson A, Friedman C. A study of abbreviations in MEDLINE abstracts. Proc AMIA Symp 2002:464–8 [PMC free article] [PubMed] [Google Scholar]
- 29.Rindflesch TC, Aronson AR. Ambiguity resolution while mapping free text to the UMLS Metathesaurus. Proc Annu Symp Comput Appl Med Care 1994:240–4 [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen T. Free NLP Software. 2011. http://www.d.umn.edu/∼tpederse/code.html (accessed 3 Jun 2011). [Google Scholar]
- 31.Weeber M, Mork JG, Aronson AR. Developing a Test Collection for Biomedical Word Sense Disambiguation. Proc AMIA Symp 2001:746–50 [PMC free article] [PubMed] [Google Scholar]
- 32.Chapman W, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 33.Mutalik P, Deshpande A, Nadkarni P. Use of general-purpose negation detection to augment concept indexing of medical documents: a quantitative study using the UMLS. J Am Med Inform Assoc 2001;8:598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y, Lowe HJ. A Novel Hybrid Approach to Automated Negation Detection in Clinical Radiology Reports. J Am Med Inform Assoc 2007;14:304–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapman W, Bridewell W, Hanbury P, et al. Evaluation of negation phrases in narrative clinical reports. Proceedings of the AMIA Fall Symposium; 2001. Washington DC: Hanley & Belfus, Philadelphia, 2001:105–9 [PMC free article] [PubMed] [Google Scholar]
- 36.University of Szeged (Hungary) Conference on Computational Natural Language Learning (CoNLL 2010): Learning to Detect Hedges and their Scope in Natural Text. 2010. http://www.inf.u-szeged.hu/rgai/conll2010st/program.html (accessed 4 May 2010). [Google Scholar]
- 37.Savova GK, Chapman WW, Zheng J, et al. Anaphoric relations in the clinical narrative: corpus creation. J Am Med Inform Assoc 2011;18:459–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao C, Solbrig H, Deepak S, et al. Time-Oriented Question Answering from Clinical Narratives Using Semantic-Web Techniques. Springer, Berlin: Lecture Note on Computer Science, 2011:6496 http://www.springerlink.com/content/67623p256743wv4u/ (accessed 20 Jul 2011). [Google Scholar]
- 39.Hripcsak G, Elhadad N, Chen YH, et al. Using empiric semantic correlation to interpret temporal assertions in clinical texts. J Am Med Inform Assoc 2009;16:220–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taira RK, Johnson DB, Bhushan V, et al. A concept-based retrieval system for thoracic radiology. J Digit Imaging 1996;9:25–36 [DOI] [PubMed] [Google Scholar]
- 41.Sager N, Lyman M, Nhan N, et al. Medical language processing: applications to patient data representation and automatic encoding. Meth Inform Med 1995;34:140–6 [PubMed] [Google Scholar]
- 42.Haug PJ, Ranum DL, Frederick PR. Computerized extraction of coded findings from free-text radiologic reports. Radiology 1990;174:543–8 [DOI] [PubMed] [Google Scholar]
- 43.Christensen L, Haug PJ, Fiszman M. MPLUS: a probabilistic medical language understanding system. Philadelphia, PA: IEEE. Proceedings Workshop on Natural Language Processing in the Biomedical Domain 2002:29–36 http://acl.ldc.upenn.edu/W/W02/W02-0305.pdf (accessed 20 Jul 2011). [Google Scholar]
- 44.Xu H, Friedman C, Stetson PD. Methods for building sense inventories of abbreviations in clinical notes. AMIA Annu Symp Proc 2008:819. [PMC free article] [PubMed] [Google Scholar]
- 45.Christensen L, Harkema H, Irwin J, et al. ONYX: A System for the Semantic Analysis of Clinical Text. Philadelphia, PA: IEEE. Proceedings of the BioNLP2009 Workshop of the ACL Conference 2009. http://www.aclweb.org/anthology/W/W09/W09-1303.pdf (accessed 20 Jul 2011). [Google Scholar]
- 46.Uzuner O, South B, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uzuner O, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008;15:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uzuner O. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc 2009;16:561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chute C. The horizontal and vertical nature of patient phenotype retrieval: new directions for clinical text processing. Proc AMIA Symposium; 2002. Washington DC: American Medical Informatics Association, 2002:165–9 [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L, Friedman C. Extracting phenotypic information from the literature via natural language processing. Stud Health Technol Inform 2004;107:758–62 [PubMed] [Google Scholar]
- 52.Wang X, Hripcsak G, Friedman C. Characterizing environmental and phenotypic associations using information theory and electronic health records. BMC Bioinformatics 2009;10:S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chapman WW, Fiszman M, Dowling JN, et al. Identifying respiratory findings in emergency department reports for biosurveillance using MetaMap. Stud Health Technol Inform 2004;107:487–91 [PubMed] [Google Scholar]
- 54.Chapman WW, Dowling JN, Wagner MM. Fever detection from free-text clinical records for biosurveillance. J Biomed Inform 2004;37:120–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Hripcsak G, Markatou M, et al. Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: a feasibility study. J Am Med Inform Assoc 2009;16:328–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindberg DAB, Humphreys BL, McCray AT. The Unified Medical Language System. Meth Inform Med 1993;32:281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Browne AC, McCray AT, Srinivasan S. The SPECIALIST Lexicon 2000. http://lexsrv3.nlm.nih.gov/LexSysGroup/Projects/lexicon/2003/release/LEXICON/DOCS/techrpt.pdf (accessed 20 Jul 2011).
- 58.Divita G, Browne AC, Rindflesch TC. Evaluating lexical variant generation to improve information retrieval. Proc AMIA Symp 1998:775–9 [PMC free article] [PubMed] [Google Scholar]
- 59.National Library of Medicine Word Sense Disambiguation Collection. 2011. http://wsd.nlm.nih.gov (accessed 2 Jun 2011). [Google Scholar]
- 60.Srihari S. Machine Learning: Generative and Discriminative Models. 2010. http://www.cedar.buffalo.edu/∼srihari/CSE574/Discriminative-Generative.pdf (accessed 31 May 2011). [Google Scholar]
- 61.Elkan C. Log-Linear Models and Conditional Random Fields. 2008. http://cseweb.ucsd.edu/∼elkan/250B/cikmtutorial.pdf (accessed 28 Jun 2011). [Google Scholar]
- 62.Hearst MA, Dumais ST, Osman E, et al. Support vector machines. IEEE Intel Sys Appl 1998;13:18–28 [Google Scholar]
- 63.DTREG Inc An introduction to Support Vector Machines. 2011. http://www.dtreg.com/svm.htm (accessed 4 Jun 2011). [Google Scholar]
- 64.Fradkin D, Muchnik I. Support vector machines for classification. In: Abello J, Carmode G, eds. Discrete Methods in Epidemiology. Piscataway, NJ: Rutgers State University of New Jersey, 2006:13–20 [Google Scholar]
- 65.Viterbi A. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans Inform Theor 1967;13:260–9 [Google Scholar]
- 66.Dempster AP, Laird NM, Rubin DB. Maximum Likelihood from Incomplete Data via the EM Algorithm. J Roy Stat Soc 1977;39:1–38 [Google Scholar]
- 67.Hasegawa-Johnson M. Multivariate-state hidden Markov models for simultaneous transcription of phones and formants. IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP); 2000. Istanbul, Turkey: 2000:1323–26 http://www.isle.illinois.edu/pubs/2000/hasegawa-johnson00icassp.pdf (accessed 20 Jul 2011). [Google Scholar]
- 68.Zhang J, Shen D, Zhou G, et al. Tan C-l. Exploring deep knowledge resources in biomedical name recognition. J Biomed Inform 2004;37:411–22 [DOI] [PubMed] [Google Scholar]
- 69.Sonnhammer ELL, Eddy SR, Birney E, et al. Pfam: Multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res 1998;26:320–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lukashin A, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 1998;26:1107–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eddy SR. What is a hidden Markov model? Nat Biotechnol 2004;22:1315–16 [DOI] [PubMed] [Google Scholar]
- 72.Rabiner LR. A Tutorial on Hidden Markov Models and Selected Applications in Speech Recognition. Proc IEEE 1989;77:257–86 [Google Scholar]
- 73.Lafferty J, McCallum A, Pereira F. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. Proc 18th International Conf on Machine Learning; 2001. 2001:282–9 http://www.cis.upenn.edu/∼pereira/papers/crf.pdf (accessed 20 Jul 2011). [Google Scholar]
- 74.Culotta A, Kulp D, McCallum A. Gene Prediction with Conditional Random Fields. 2005. http://www.cs.umass.edu/∼culotta/pubs/culotta05gene.pdf (accessed 1 Jun 2011). [Google Scholar]
- 75.Sutton C, McCallum A. An Introduction to Conditional Random Fields for Relational Learning. Amherst: University of Massachusetts, 2004 [Google Scholar]
- 76.Franz A, Brants T. Google N-gram database (all our N-grams belong to you). 2011. http://googleresearch.blogspot.com/2006/08/all-our-n-gram-are-belong-to-you.html (accessed 1 Jun 2011). [Google Scholar]
- 77.University of Sheffield Natural Language Group Information Extraction: the GATE pipeline. 2011. http://www.gate.ac.uk/ie (accessed 1 Jun 2011). [Google Scholar]
- 78.Apache Foundation The Unstructured Information Management Architecture. 2011. http://uima.apache.org (accessed 3 Jun 2011). [Google Scholar]
- 79.Apache Foundation Apache UIMA Documentation version 2.3.1. 2011. http://uima.apache.org/documentation.html (accessed 3 Jun 2011). [Google Scholar]
- 80.Thompson C, Califf M, Mooney R. Active learning for natural language parsing and information extraction. In: Conference PotSIML, ed. 1999. http://www.aidanf.net/publications/finn03active.pdf (accessed 20 July). [Google Scholar]
- 81.North American Chapter of the Association for Computational Linguistics Active Learning for NLP. 2020. http://active-learning.net/alnlp2010 (accessed 7 Jun 2011). [Google Scholar]
- 82.Fodor P, Lally A, Ferrucci D. The Prolog Interface to the Unstructured Information Management Architecture. 2011. http://arxiv.org/ftp/arxiv/papers/0809/0809.0680.pdf (accessed 4 Jun 2011). [Google Scholar]
- 83.Lally A, Fodor P. Natural Language Processing With Prolog in the IBM Watson System. Association for Logic Programming (ALP) Newsletter, 2011. http://www.cs.nmsu.edu/ALP/2011/03/natural-language-processing-with-prolog-in-the-ibm-watson-system/ (accessed 20 Jul 2011). [Google Scholar]
- 84.Libresco LA. A Non-Trivial Advantage for Watson. Huffington Post (tech) 2011. http://www.huffingtonpost.com/leah-anthony-libresco/a-nontrivial-advantage-fo_b_825837.html (accessed 20 Jul 2011). [Google Scholar]
- 85.Jennings K. Conversation: Jeopardy! Champ Ken Jennings Washington Post; 2011. http://live.washingtonpost.com/jeopardy-ken-jennings.html (accessed 20 Jul 2011). [Google Scholar]
- 86.Fitzgerald J. 'Jeopardy!'-winning computer delving into medicine. 2011. http://www.physorg.com/news/2011-05-jeopardy-winning-delving-medicine.html (accessed 4 Jun 2011). [Google Scholar]
- 87.Berner ES, Webster GD, Shugerman AA, et al. Performance of four computer-based diagnostic systems. N Engl J Med. 1994;330:1792–6 [DOI] [PubMed] [Google Scholar]
- 88.Friedman CP, Elstein AS, Wolf FM, et al. Enhancement of clinicians' diagnostic reasoning by computer-based consultation: a multisite study of 2 systems. JAMA 1999;282:1851–6 [DOI] [PubMed] [Google Scholar]
- 89.Miller R, Masarie F. The demise of the Greek oracle model of diagnostic decision support systems. Meth Inform Med 1990;29:1–8 [PubMed] [Google Scholar]
- 90.Miller R. Medical diagnostic decision support systems–past, present, and future: a threaded bibliography and brief commentary. J Am Med Inform Assoc 1994;1:8–27 [DOI] [PMC free article] [PubMed] [Google Scholar]