Abstract
We have identified a novel gene, Spalten (Spn) that is essential for Dictyostelium multicellular development. Spn encodes a protein with an amino-terminal domain that shows very high homology to Gα-protein subunits, a highly charged inter-region, and a carboxy-terminal domain that encodes a functional PP2C. Spn is essential for development past the mound stage, being required cell autonomously for prestalk gene expression and nonautonomously for prespore cell differentiation. Mutational analysis demonstrates that the PP2C domain is the Spn effector domain and is essential for Spn function, whereas the Gα-like domain is required for membrane targeting and regulation of Spn function. Moreover, Spn carrying mutations in the Gα-like domain that do not affect membrane targeting but affect specificity of guanine nucleotide binding in known GTP-binding proteins are unable to fully complement the spn− phenotype, suggesting that the Gα-like domain regulates Spn function either directly or indirectly by mediating its interactions with other proteins. Our results suggest that Spn encodes a signaling molecule with a novel Gα-like regulatory domain.
Keywords: Dictyostelium, GTP-binding proteins, development, PP2C, cell-type gene expression
Under the condition of ample food supply, Dictyostelium amoebae live as unicellular organisms. Upon starvation, a developmental program is initiated that leads to the formation of a multicellular structure consisting of a vacuolated stalk supporting a spore mass (Loomis 1982). Multicellularity results from the aggregation of up to 105 cells in response to oscillatory pulses of the chemoattractant cAMP (Chen et al. 1996). As the multicellular aggregate forms, the concentration of extracellular cAMP is thought to rise (Abe and Yanagisawa 1983), which leads to the activation of the transcription factor G-box binding factor (GBF) and the subsequent induction of morphogenesis and cell-type differentiation (Firtel 1995, 1996; Ginsburg et al. 1995; Williams 1995). At this stage, cells differentiate into two major cell types: prespore cells (∼70%) and several subpopulations of prestalk cells (∼30%). The prestalk cells sort to the top of the mound where a tip is formed. The tip extends to form a finger, which falls onto the substratum, producing a migrating slug with a well-established spatial patterning. Prespore cells are localized in the posterior region, whereas the individual prestalk cell types are further organized along the anterior-posterior axis in the anterior 20% of the slug. A third subpopulation of cells with some characteristics of prestalk cells, anterior-like cells (ALCs), is found scattered through the slug (Devine and Loomis 1985; Sternfeld and David 1992). Coordinated morphogenesis involving cell–cell interaction and cell sorting results in the formation of a well-proportioned fruiting body (Firtel 1995; Williams 1995). Although the morphogens cAMP and differentiation inducing factor (DIF) are known to mediate cell-type differentiation, the signaling pathways that control the developmental switch at the mound stage, which leads to cell-type differentiation, are not well understood. A number of proteins, including the transcription factor GBF, the cell-surface signaling molecule LagC, and the serine protease ATP transporter tagB, have been shown or are predicted to be required, at mound stage for further development and morphogenesis (Dynes et al. 1994; Schnitzler et al. 1994, 1995; Shaulsky et al. 1995; Firtel 1996), suggesting a complex regulatory network that is far from being fully elucidated.
Reversible protein phosphorylation is a crucial event in regulating intracellular signaling cascades activated in response to growth factors, morphogens, or chemoattractants. In Dictyostelium, serine/threonine protein kinases, including the cAMP-dependent protein kinase PKA (Mann et al. 1992; Reymond et al. 1995; Firtel 1996), the MAP kinase ERK2 (Segall et al. 1995), and the glycogen synthase kinase-3 (GSK-3) (Harwood et al. 1995), have been found to play key roles during the developmental program. Considerable evidence has established the roles of PKA and ERK2 during aggregation and their requirement for cell-type differentiation (Hopper et al. 1993a,b; Mann and Firtel 1993; Gaskins et al. 1996; Zhukovskaya et al. 1996; Mann et al. 1997). Whereas protein tyrosine phosphatases are known to have pathway-specific regulatory functions in Dictyostelium (Gamper et al. 1995), it is not known whether tightly regulated, pathway-specific protein Ser/Thr phosphatases control developmental decisions. Protein Ser/Thr phosphatases are represented by two distinct families (Barford 1996). The PPP family includes PP1, PP2A, and PP2B, some members of which have been identified in Dictyostelium and shown to be generally required for development (Haribabu and Dottin 1991; Horn and Gross 1996). The PPM family is a large family whose defining member is the mammalian PP2C but which also includes a variety of PP2C-type phosphatases such as ABI1 and KAPP-1 from Arabidopsis (Meyer et al. 1994; Stone et al. 1994; Leung et al. 1997), SpoIIE from Bacillus subtilis (Bork et al. 1996), and Fem2 from Caenorhabditis elegans (Chin-Sang and Spence 1996). The PPM family members are characterized by their absolute requirement of Mg2+/Mn2+ for catalytic activity and their insensitivity to certain phosphatase inhibitors such as microcystin or okadaic acid.
In this work, we describe a novel signaling protein, Spalten (Spn), that contains two distinct domains: a carboxy-terminal active PP2C homologous domain and a heterotrimeric G-protein Gα-subunit-like domain at the amino terminus of the protein separated by a highly charged inter-region. Spn is essential for Dictyostelium development because its disruption results in a morphological arrest at the mound stage and a defect in cell-type differentiation. We show that Spn is maximally expressed at mound stage and is mainly expressed in the prestalk cell population during the later multicellular stages. Spn is required cell autonomously for prestalk-specific gene expression and nonautonomously for prespore cell differentiation. Analysis of the different domains indicates that the phosphatase domain is the effector domain of Spn and the Gα-like domain is required for the appropriate intracellular localization of Spn at the plasma membrane. Point mutations in the Gα-like domain that should affect the nucleotide-binding specificity of a bona fide Gα protein partially disrupt Spn function, suggesting a more complex function for this unusual amino-terminal domain in regulating the function of the PP2C domain. Our results are consistent with Spn containing a novel GTP-binding domain that, like previously characterized GTP-binding proteins, may function as a molecular switch to regulate the function of an effector, in this case a PP2C-type protein phosphatase.
Results
Isolation of the Spn gene by REMI mutagenesis
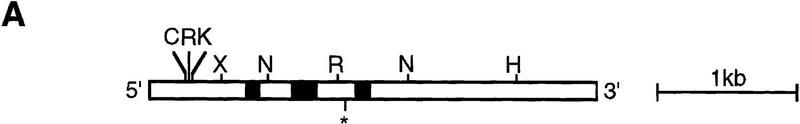
We identified Spn as a developmentally essential gene in a REMI insertional mutagenesis screen for genes required for Dictyostelium differentiation (see Materials and Methods). The inserted vector and 1 kb of surrounding DNA were isolated. The rescued NdeI genomic DNA fragment was used to screen a 12–16 hr developmental λZAP cDNA library. Sequence analysis of the full-length cDNA revealed an ORF of 975 amino acids (Fig. 1B). Sequence comparison of the cDNA and the genomic DNA, amplified by PCR with oligonucleotides at the amino terminus and at the carboxyl terminus of the gene, indicated the presence of three introns (Fig. 1A). To confirm that the phenotype of the REMI mutant (see below) was caused by the plasmid insertion at this locus, the endogenous gene was disrupted by homologous recombination by use of either the rescued plasmid or a disruption construct made by insertion of the Bsr cassette into the cDNA (Materials and Methods; Fig. 1A). In both cases, the gene diruptions, confirmed by Southern blot analysis, displayed the same phenotype as the original REMI mutant (data not shown).
Figure 1.
MAP and amino acid sequence (A) Map of Spn gene. On Spn map, all restriction enzyme sites for ClaI (C), EcoRV (R), KpnI (K), XhoI (X), NdeI (N), and HindIII (H) are shown. The black boxes represent the locations of the three known introns (94, 181, and 105 bp), which were derived from comparison of the sequence of the cDNA and genomic DNA. (*) Insertion site of pUCBsr in the original REMI mutant. (B) Amino acid sequence derived from Spn cDNA. The amino-terminal Gα-like domain and the carboxy-terminal PP2C homologous domain are boxed. The proline, lysine, and glutamic acid-rich region of the IR is shown in boldface letters. (C) Schematic diagram of the protein encoded by Spn cDNA. The Gα-like domain, the IR, and the PP2C domains are indicated.
Spn encodes a bimodular protein
Comparison of Spn amino acid sequence to the GenBank database by use of the BLAST program revealed two different domains with homologies to distinct gene families (Fig.1C). The amino-terminal portion (residues 98–458) of Spn shares substantial sequence homology with the heterotrimeric Gα-subunit family of GTP-binding proteins in domains required for Gα subunit function. Figure 2A shows the alignment of Spn predicted amino acid sequence with that of several known Gα subunits. The extent of the homology is almost as high as the homology between Gα subunits from distantly related organisms. The Spn Gα-like domain contains the conserved P-loop (GXXXXGKS/T), which is required for GTP-binding (Kjeldgaard et al. 1996). In the other conserved domains, Spn shows strong amino acid sequence homology, but it also possesses some unusual features with potentially conservative substitutions. By computer modeling with the crystallographic coordinates of Gαt, we tried to predict the possible effects of such substitutions (Noel et al. 1993). In the guanine ring-binding motif NKXD, the conserved lysine is replaced by a threonine (Thr 374). Crystallographic data have shown that the guanine ring is sandwiched by Van der Waals interactions involving this particular lysine and a threonine in the carboxy-terminal TCAT box (Noel et al. 1993). The TCAT box is absent in Spn; however, a leucine (Leu 430) is found in the homologous location to the second Thr in the TCAT box. Computer substitution modeling suggests that the combination of Leu 430/Thr 374 may also be able to stabilize the guanine ring, as these two amino acids should be able to form the roof and the floor of the hydrophobic guanine binding pocket similarly. In Gαt, both the Asp of the conserved DXXG box and Thr177 are involved in Mg2+ coordination. In Spn, in the Mg2+ binding domain DXXG, the usually conserved aspartate is replaced by a glycine (position 286), whereas a Lys (Lys 267) replaces the Thr at the equivalent position to Thr177 in Gαt. The computer modeling suggests that the long positively charged sidechain of this Lys places it in a position in which it may mimic the presence of Mg2+ in the Mg2+-binding pocket, opening up the possibility that Mg2+ may not be crucial for Spn intrinsic activity if it is a GTP-binding protein. Another interesting feature of Spn is the presence of several extra domains. Compared with most known heterotrimeric G protein Gα-subunits, Spn has a long amino-terminal extension upstream of the P-loop and several internal insertions. According to our alignment, these internal domains would localize in loop regions of Gαt and, therefore, may not affect the ability of Spn to exhibit a potential Gα-like conformation.
Figure 2.

Sequence and functional analysis of Spn. (A) Analysis of the Gα-like domain sequence. Alignment of the deduced sequence of the Spn Gα-like domain with bona fide members of the Gα-subunit family of GTP-binding proteins. Asterisks (*) show positions of point mutations described in the text. (Bars) Gα-subunit conserved domains mentioned in the text. (D4) Dictyostelium discoideum Gα4 (P34042), (GQ) human Gq (U40038), (D2) D. discoideum Gα2 (P16051), (GT) Bos taurus Gαt (P04695), (GS) B. taurus Gs (G71882), (I1) Rattus norvegicus Gi1 (P10824), (GZ) R. norvegicus Gz (P19627). (B) Amino acid sequence comparison of Spn PP2C-domain with PP2C homologs from S. cerevisiae (Sc) (P35182), human (Hs) (P35813), and C. elegans (Ce) (P49596). (*) Conserved aspartic acids that were mutated into alanine in the mutant SpnD920A/D924A. (C,D) Spn possesses a phosphatase activity. Spn (•) and SpnD920A/D924A (▴) were expressed in Sf9 insect cells as histidine-tagged proteins, purified, and tested for their phosphatase activity on 32P-labeled casein in the presence of 20 mm MgCl2. The release of Pi was followed as a function of time. The data are given as means ±s.d. (n = 3) (C) The Mg2+/Mn2+ requirement for (His)6–Spn activity was tested by incubating (His)6–Spn with 32P-labeled casein in the presence of 20 mm MgCl2, MnCl2, CaCl2, or EDTA. The effect of different inhibitors on Spn phosphatase activity is shown in D. (His)6–Spn was incubated with the substrate in the presence of 20 mm Mg2+ and 50 mm NaF, 10 μm microcystin, or 1 mm vanadate. The amount of released Pi was measured after 30 min incubation. The phosphatase activity was expressed as a percentage of the activity measured in the presence of MgCl2 alone. The graph shows a representative experiment (D).
The carboxy-terminal region (residues 703–975) of Spn shows strong homology to the Ser/Thr phosphatases of the PP2C class (Fig. 2B). The amino acid sequence of this domain in Spn is 30% identical to PTC2 from Saccharomyces cerevisiae, 23% identical to human PP2C, and 25% identical to PP2C from C. elegans. A high similarity was found in the domains required for phosphatase activity according to the crystal structure of human PP2C (Das et al. 1996). The PP2C-homologous domain and the Gα-like domain are separated by an inter-region of ∼240 amino acids rich in lysine, glutamic acid, and proline that shares no homologies with other proteins in the databases. Among the phosphatases of type 2C, Spn is the only one featuring an amino-terminal domain homologous to Gα-subunits, although some PP2C family phosphatases possess amino-terminal targeting or regulatory domains (see Discussion). The presence of a long Gα-subunit-like domain suggests that Spn activity may be regulated differently from the canonical mammalian PP2C proteins.
Spn has serine/threonine phosphatase activity in vitro
To examine whether Spn has a PP2C-like phosphatase activity, amino-terminally (His)6-tagged Spn [(His)6–Spn] expressed in insect cells was purified on Ni2+-agarose beads and its phosphatase activity assayed by use of 32P-labeled PKA-phosphorylated casein as a substrate. (His)6–Spn dephosphorylated 32P-labeled casein in the presence of Mg2+ linearly as a function of time (Fig. 2C). A similar phosphatase activity was also measured in the presence of Mn2+, whereas almost no activity was detected when Mg2+ was replaced by Ca2+ or if EDTA was added to the reaction mixture (Fig. 2D), similar to the properties of other PPM family members. This Mg2+-dependent phosphatase activity was inhibited by addition of 50 mm NaF, but insensitive to treatment with 10 μm microcystin, a potent inhibitor of PP1 and PP2A or 1 mm vanadate, an inhibitor of protein tyrosine phosphatases (Fig. 2D). According to the human PP2C crystal structure, two Mn2+ ions are coordinated through four invariant aspartic acid residues localized in the catalytic site (Das et al. 1996). Mutation of one of these highly conserved residues into alanine leads to an inactive phosphatase protein in both TPD1, a yeast PP2C homolog, and SpoIIE (Barford 1996). For further functional analysis of Spn, two invariant aspartate residues (D920 and D924 in Spn) were changed to alanine. When tested in vitro for its phosphatase activity, the mutated version of Spn, SpnD920A/D924A, was unable to dephosphorylate the substrate (Fig. 2C). These results are consistent with Spn being a member of the PP2C family.
Spn is essential for development
To examine the developmental phenotype of spn null cells, axenically grown cells were washed free of nutrients and plated on non-nutrient Na-KPO4 agar plates. Upon starvation, spn null (spn−) cells aggregated and formed mounds with kinetics similar to those of wild-type cells; however, the null strain failed to continue through morphogenesis (Fig. 3). Instead, at ∼16 hr of development, the mounds disaggregated to form smaller aggregates that eventually produced abnormal looking finger-like structures (Fig. 3D,E).
Figure 3.
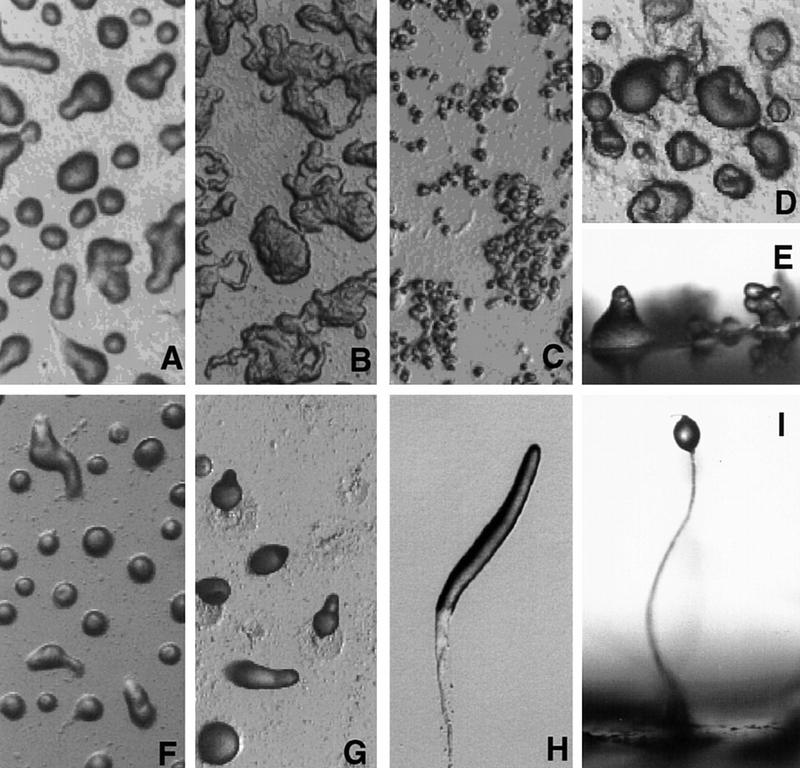
Developmental morphology of spn− cells. Axenically grown cells were washed and plated on non-nutrient NaKPO4 buffered agar plates for development (see Materials and Methods). Pictures of spn− cells (A–E) and wild-type cells (F–I) were taken at different times of development. (A,F) 8 hr; (B,G) 13 hr; (C,H) 16 hr; (E,I) final morphology. (H) Wild-type slug; (I) wild-type fruiting body. Images in D, E, H, and I are at a higher magnification than the other panels.
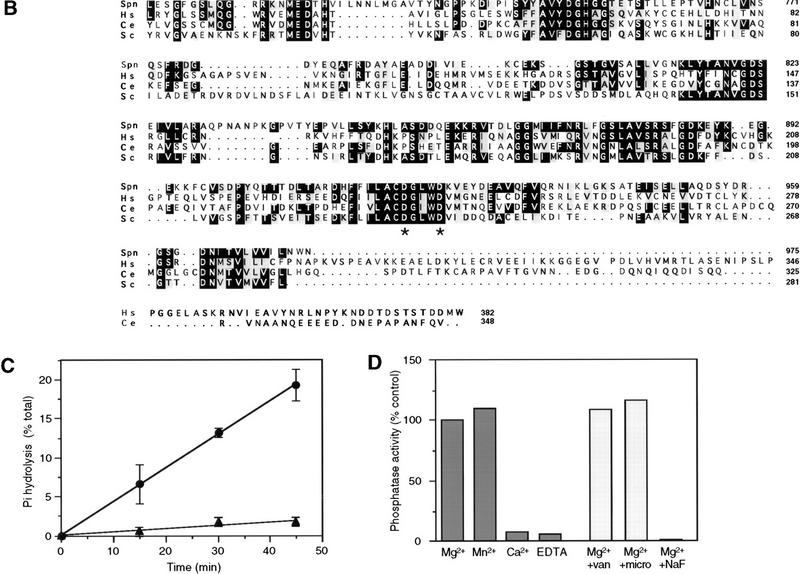
A developmental RNA time course shows that the ∼4-kb Spn mRNA is present at moderate levels during growth. Transcript levels increase during development, peaking at ∼8 hr of development (mound stage) and then decrease gradually during the later stages (Fig. 4A). This transcript is not found in the spn− cells (data not shown). An antibody was raised against the carboxyl terminus domain of Spn (residues 773–975) and used in a Western blot analysis to probe a developmental protein time course. The antibody revealed the presence of an ∼120-kD protein in wild-type cells (Fig. 4B) that is absent in the spn− cells (see below). The protein is present throughout development and increases ∼fourfold at the tipped aggregate stage (12 hr of development), consistent with the mRNA time course. Although Spn is already expressed at the onset of development, the effect of its disruption is manifest visibly only after the cells reach mound stage, when the expression of the protein is more highly induced.
Figure 4.
Gene expression analysis. (A,B) The temporal expression of Spn mRNA (A) and protein (B). Exponentially growing wild-type cells were washed in 12 mm NaKPO4 buffer (pH 6.2) and plated for development on Millipore filters. RNA was isolated at the indicated times of development [(V) vegetative], size-fractionated on a denaturing gel, and probed with a 32P-labeled EcoRV fragment from Spn cDNA (A) as described previously (Mehdy and Firtel 1985). For the Western blot analysis, developed cells were collected at the indicated times and boiled in SDS sample buffer. Equal amounts of protein extracts were separated on an 8% SDS gel and analyzed by Western blot by use of the rabbit polyclonal anti-Spn antibody (B). (C) Expression of developmentally regulated genes is shown. Wild-type and spn− cells were plated for development on Millipore filters or non-nutrient agar plates and RNA was isolated at the times indicated. RNA blots were hybridized with probes for CsA (aggregation-stage gene), GBF (postaggregative gene), LagC (postaggregative gene), ecmA (prestalk), and SP60/cotC (prespore). (D) The effect of cAMP on cell-type specific gene expression is shown. Wild-type and spn− cells were washed, resuspended in NaKPO4 buffer, and starved for 4 hr in suspension. Cells were then stimulated with 300 μm cAMP for 6 hr (Mehdy and Firtel 1985). RNA samples were isolated, size-fractionated on a denaturing gel, and hybridized with ecmA and SP60/cotC probe fragments. (E) The effect of DIF on cell-type specific gene expression is shown. Wild-type and spn− cells were developed on NaKPO4 buffered agar plates for 5 or 11 hr. Cells were then harvested, dissociated, and resuspended in NaKPO4 buffer. Cells were stimulated for 6 hr in shaking culture as indicated with different combinations of 5 nm DIF, 300 μm cAMP, and 0.2 mm Ca2+ as described previously (Jermyn et al. 1987). RNA samples were isolated, size-fractionated on a denaturing gel, and probed with ecmA and SP60/cotC probe fragments.
Spn is required for prestalk and prespore differentiation
After mound formation, a developmental switch occurs that leads to the induction of postaggregative gene expression, morphogenesis, and the initiation of cell-type differentiation (Firtel 1996). As spn− cells failed to develop past the tight mound stage, we investigated the effect of Spn mutation on the expression of developmentally regulated genes (Fig. 4C). The cAMP pulse-induced gene CsA and the gene encoding the transcription factor GBF were used as molecular markers for aggregation stage and early postaggregation gene expression, respectively. In wild-type and spn− cells, CsA transcripts accumulate normally during early development (4–8 hr) and then decrease as the mound forms. However, in spn− cells, CsA expression is reinduced at ∼20 hr of development. The transcription factor GBF plays a central role in the developmental switch, as it controls the expression of some postaggregative genes, including the cell-surface signaling molecule LagC and prespore and prestalk cell-type-specific genes (Schnitzler et al. 1994, 1995). In wild-type cells, the GBF transcript level increases after 4 hr of development, peaks at tipped-mound formation (∼10 hr), and continues to be present thereafter. In spn− cells, the GBF mRNA level decreases dramatically just after mound formation, but is reinduced again at ∼20 hr, the time of the formation of the small tips, as if the developmental program was reinitiated. However, the transcription factor GBF is appropriatly activated in spn− mutant as these cells are able to express LagC, albeit with an abnormal temporal expression pattern, which probably results from the altered pattern of GBF expression.
Neither the prestalk-specific gene ecmA nor the prespore-specific gene SP60/cotC were detectably expressed in spn− cells when the cells were developed on filters. ecmA and SP60/cotC expression was just barely detectable after extended autoradiography when the cells were developed on NaK phosphate agar plates, indicating that Spn is required for both prestalk and prespore differentiation (Fig. 4C). This result is consistent with the morphological phenotype of the mutant and its inability to progress past mound formation. The results of this RNA blot analysis were confirmed by use of ecmA/ and SP60/lacZ constructs: No β-galactosidase staining was obtained in spn− cells containing either reporter construct (data not shown).
Induction of cell-type differentiation is under the control of at least two known morphogens, cAMP and the chlorinated hexaphenone DIF (Kay 1992; Williams 1995; Firtel 1996). We examined the possibility that the spn− phenotype was caused by, in part, an inability to produce these morphogens in sufficient quantities by providing exogenous cAMP and/or DIF under conditions that stimulate the expression of the cell-type-specific genes SP60/cotC and ecmA in wild-type cells (see Materials and Methods). Whereas both the prespore and the prestalk markers were induced in wild-type cells, no expression was detected in spn− cells when stimulated with exogenous cAMP (Fig 4D). To examine the possibility that spn− cells did not produce sufficient DIF, we tested the ability of exogenous DIF combined with cAMP to induce ecmA expression (Jermyn et al. 1987). As described previously (Jermyn et al. 1987; Early and Williams 1988; Mehdy and Firtel 1995), cAMP induces the expression of both the prestalk and prespore genes in suspension but addition of DIF to cultures containing cAMP results in a significant enhancement of prestalk gene expression and a repression of prespore gene expression. In spn− cells, such treatment did not induce the expression of the cell-type-specific markers ecmA and SP60/cotC to any level comparable with that of wild-type cells (Fig. 4E).
Cell-autonomous and nonautonomous functions of Spn in controlling cell-type differentiation
To determine whether Spn functions autonomously, chimeras of Act15/lacZ reporter-tagged spn− cells and untagged wild-type cells (in a ratio of 1:3, respectively) were stained at different stages of development. The chimeric organisms developed with wild-type morphology and timing. In early development, spn− cells were found scattered in the mound, but seemed to be excluded from the emerging tip (Fig. 5A–D). At first finger and slug stages, mutant cells were found in the very posterior of the developing organism. During culmination, spn− cells were transiently found in the developing stalk and, later on, mainly in the spore mass of the fruiting body. Thus, although spn− cells are unable to progress past mound stage when developed on their own, they participate in development, albeit poorly, when mixed with wild-type cells, suggesting a partially cell nonautonomous defect. Similar experiments were conducted with spn− mutant carrying either the prespore- (SP60/lacZ) or prestalk-specific (ecmAO/lacZ) reporters. In chimeric organisms, no ecmAO/lacZ expression was detectable, indicating that Spn function is cell autonomous for prestalk cell differentiation. However, when SP60/lacZ expression was examined, staining could be detected during culmination, mainly in the spore mass of the fruiting body (Fig. 5E,F). It is clear that the SP60/cotC expression defect is not fully complemented because the staining appears only during culmination, whereas SP60/cotC should be expressed starting in the tight aggregate in wild-type organisms (Haberstroh and Firtel 1990). In contrast to spn− cells developed alone, spn− cells in chimeras were able to differentiate spores and express the spore-specific marker SpiA (Fig. 5G,H). The above results suggest that the Spn requirement for prespore/spore cell differentiation is cell nonautonomous. To examine this furter, we made chimeras with a mutant strain, pslA null cells, which does not detectably express prespore-specific genes and produces a fruiting body that contains vacuolated stalk cells but lacks any prespore or spore cells (H. Yasukawa, S. Mohanty, and R.A. Firtel, in prep.). In pslA−:spn−/SP60/lacZ chimeras, the spn− cells also expressed SP60/cotC and formed spores (data not shown), suggesting that the prestalk cells could induce prespore/spore cell differentiation in spn− cells.
Figure 5.
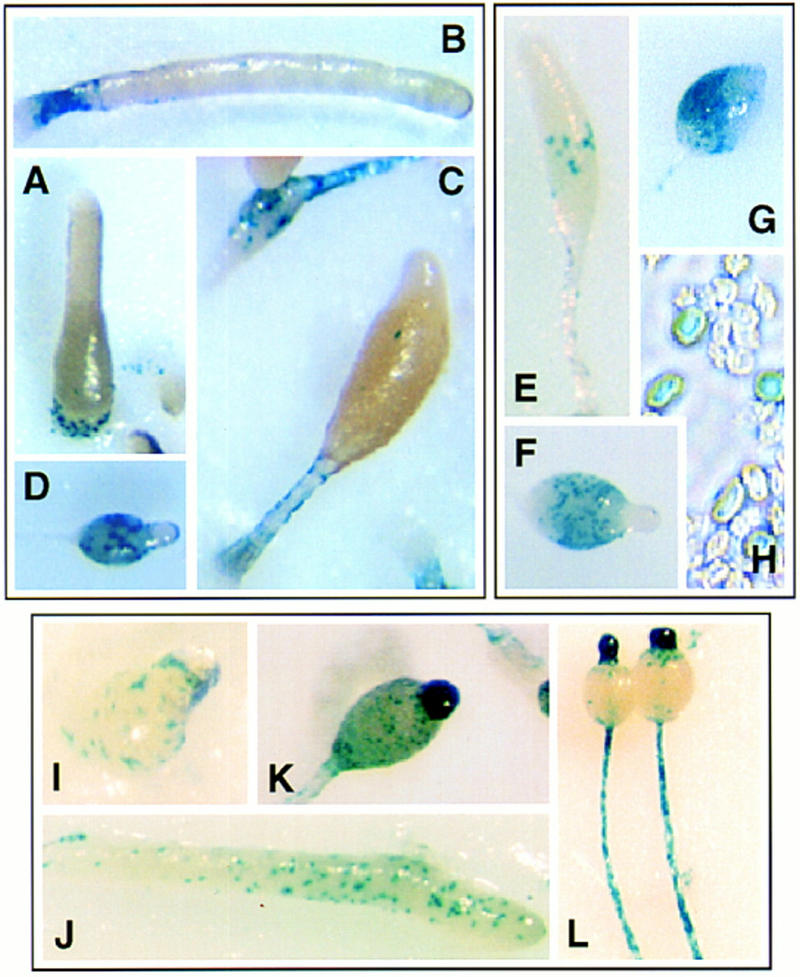
Chimeric organism analysis and spatial expression of Spn. spn− cells carrying the reporter constructs Act15/lacZ (A–D), SP60/lacZ (E,F), and SpiA/lacZ (G,H) were allowed to coaggregate with wild-type cells (1:3 ratio spn−/wild-type cells) and form chimeric organisms. Aggregates were stained at different developmental stages as described in Materials and Methods. (A) First finger; (B) slug; (C,E) culmination, (D,F,G) fruiting body; (H) spores. The Spn promoter region was used to drive the expression of the reporter gene lacZ. Wild-type cells carrying the expression construct pSpn/lacZ were allowed to develop on Millipore filters and histochemically stained at different stages of development for β-gal activity (see Materials and Methods). (I) First finger; (J) slug; (K) culminant; (L) fruiting body.
Spn is expressed in ALCs and prestalk cells during multicellular development
To determine the spatial pattern of Spn expression, we cloned the 4-kb region upstream of Spn. As this region included the carboxyl terminus of the upstream gene, we expect that it contains the full-length promoter (pSpn). This was used to drive the expression of lacZ in wild-type cells (Fig. 5I–L; Materials and Methods). During growth, when expression of Spn is low, staining was very faint and restricted to a small fraction of the cells (data not shown). At the mound and slug stages, pSpn/lacZ-expressing cells were found scattered throughout the organism. In the early culminant, stained cells were still distributed throughout the organism, whereas in late culminants, the β-gal staining was primarily localized in the tip and the stalk of the differentiating fruiting body. pSpn/lacZ expressing cell distribution coincides with the distribution of ALCs in the mound and slug and both ALCs and prestalk cells during culmination (Sternfeld and David 1982; Jermyn and Williams 1991).
The phosphatase domain is the effector domain of Spn
To gain insight about the function of Spn during Dictyostelium development, the full-length protein was overexpressed from the Spn promoter in either wild-type or spn− cells. For the overexpression studies, the promoter region was reduced to 1 kb of upstream sequences (ΔpSpn). This promoter exhibited the same spatial and temporal pattern of expression as that of the 4-kb promoter (data not shown). Western blot analysis of the stable transformants indicated an approximately fivefold increase in the level of expression of the protein (data not shown; see below). Overexpression of Spn did not affect the growth rate or size of vegetative cells (data not shown).
Overexpression of Spn complemented the null phenotype with the formation of wild-type-looking fruiting bodies after 24 hr of development (Fig. 6A). However, overexpression of Spn carrying the double aspartate mutation in the PP2C domain, SpnD920A/D924A, which exhibits an extremely low catalytic activity, did not rescue the null phenotype. This strongly indicates that the phosphatase activity of Spn is required for development to proceed (Fig. 6E). Overexpression of wild-type Spn or SpnD920A/D924A in wild-type cells did not affect development (data not shown). To confirm the requirement of the PP2C domain, this domain was overexpressed in both backgrounds. The PP2C domain overexpression construct, ΔpSpn/PP2C, was made as an in-frame fusion of the PP2C domain with the first 94 amino acids of Spn and lacked the Gα-like and the inter-region (IR) domains. Overexpression of the phosphatase domain partially rescued the null phenotype with formation of short, small, abnormal-looking fruiting bodies (Fig. 6D) containing spores (data not shown). A similar phenotype was observed when the PP2C domain was overexpressed in wild-type cells, indicating that a fivefold overexpression of the PP2C domain alone resulted in abnormal development (Fig. 6H). Overexpression of the PP2C domain alone carrying the double aspartate mutation D920A,D924A did not complement the null phenotype, nor did it alter wild-type development (data not shown). These observations support the conclusions that the phosphatase activity is essential for development but that the normal function of Spn requires the additional domains of the protein.
Figure 6.
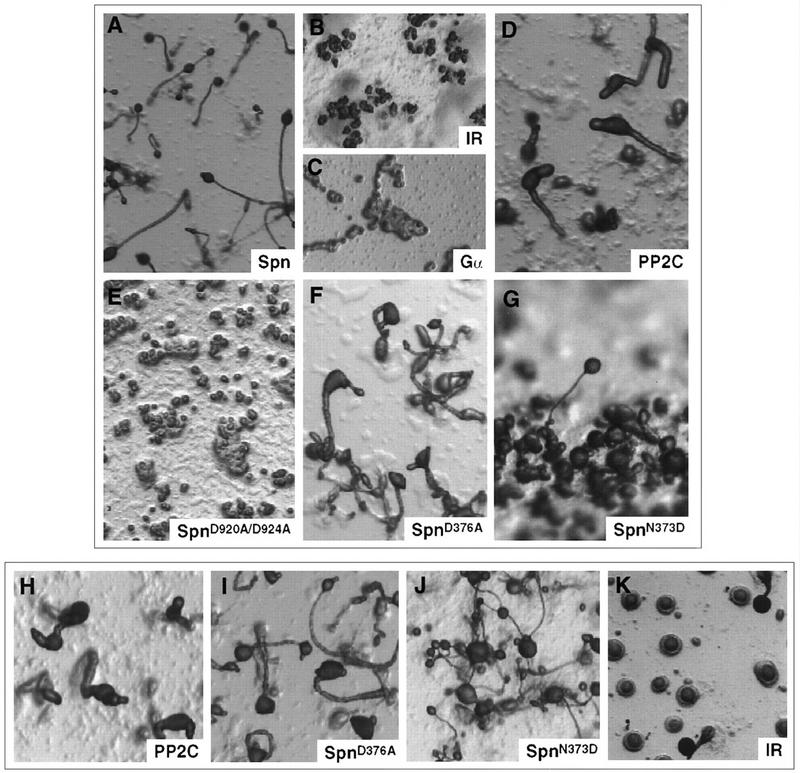
Phenotypic analysis of overexpression of Spn and mutant Spn protein. Wild-type cells (H–K) and spn− cells (A–G) carrying the following constructs were washed and plated for development on non-nutritive agar plates. (A) ΔpSpn/Spn; (B,K) ΔpSpn/IR; (C) ΔpSpn/Gα; (D,H) ΔpSpn/PP2C; (E) ΔpSpn/SpnD920A/D924A; (F,I) ΔpSpn/SpnD376A; (G,J) ΔpSpn/SpnN373D. The pictures represent the final stage of development of the different strains.
The Gα-like domain is required for wild-type Spn function
To examine the potential role of the Gα-like domain and the IR, wild-type cells and spn− cells were transformed with ΔpSpn/Gα and ΔpSpn/IR. Neither domain complemented the null phenotype (Fig. 6B,C). Whereas overexpression of the Gα-like domain did not have any detectable effect in wild-type cells, overexpression of the IR domain resulted in a dominant-negative phenotype with most aggregates arresting at the mound stage (Fig. 6K). To further characterize the function of the Gα-like domain and to test the possibility that a GDP/GTP switch may regulate Spn function, we introduced amino acid substitutions in the conserved guanine ring-binding domain that has been shown to be required for GTP-binding of bona fide GTPases, including p21ras and Goα (Schmidt et al. 1996; Yu et al. 1997). Figure 2A shows the various mutations that were created by site directed mutagenesis. Both the D376A and N373D mutations are expected to alter the GTP-binding specificity. The mutant-overexpressing constructs were transformed into wild-type and spn− cells. In both backgrounds, overexpression of SpnD376A led to the formation of abnormal-looking fruiting bodies (Fig. 6F,I). Overexpression of SpnN373D in spn− cells partially complemented the null phenotype, as most of the mounds did not form fruiting bodies (Fig. 6G). The fruiting bodies were very small compared with those of control wild-type cells. In the wild-type background, overexpression of the same construct led to the formation of very small-sized fruiting bodies (Fig. 6J). This mutational study strongly supports the idea that GTP-binding is required for proper Spn function in vivo.
The Gα-like domain is required for targeting Spn to the plasma membrane
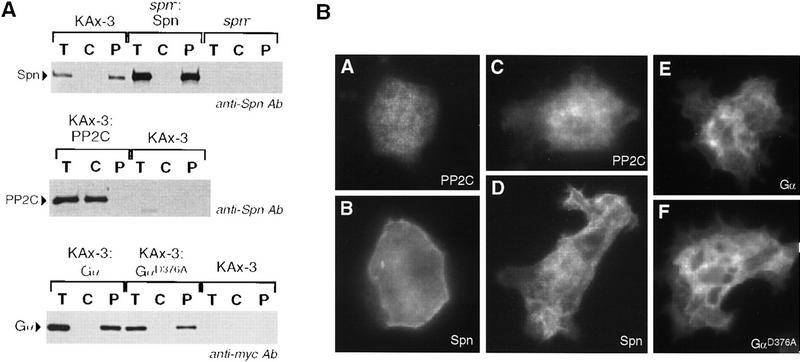
Subcellular fractionation was used to examine the distribution of Spn in wild-type cells. Cytosolic and pellet fractions were separated by high-speed centrifugation by use of lysates from 8 hr developed cells (loose mound stage). Western blot analysis indicated that Spn was found in the particulate fraction (Fig. 7A). A similar subcellular distribution was obtained in wild-type cells or spn− cells overexpressing Spn or a myc epitope-tagged Spn (ΔpSpn/Spn–myc) (Fig. 7A; data not shown). Next, we examined the subcellular distribution of myc epitope-tagged versions of the PP2C-domain, the Gα-like domain, and the mutant Gα-like domain GαD376A expressed in wild-type cells. Gα–myc and GαD376A–myc displayed the same distribution as Spn–myc, whereas PP2C–myc was found predominantly in the cytosolic fraction. Most G-protein α subunits are modified by palmitoylation and/or myristoylation on cysteine and glycine, respectively, at the amino terminus of the protein. Spn does not contain a cysteine or myristoylation consensus sequence (MGXXXS) at the amino terminus of the protein, but we cannot exclude an internal palmitoylation site. As both constructs ΔpSpn/Gα–myc and ΔpSpn/PP2C–myc contain the first 94 amino acids of Spn, the amino-terminal extension is probably not solely responsible for the subcellular localization of Spn.
Figure 7.
Spn localizes to the plasma membrane. (A) Subcellular fractionation of Spn is shown. Wild-type and spn− cells carrying ΔpSpn/Spn–myc, ΔpSpn/Gα-myc, ΔpSpn/GαD376A–myc, ΔpSpn/PP2C–myc, or control cells were plated for development on NaKPO4 buffered agar plates and left to develop for 8 hr. Cells were then harvested in 20 mm triethanolamine at pH 7.5, and lysed through a 3 μm Nuclepore filter. Nuclei and intact cells were removed by a 800g centrifugation, and the remaining supernatant was then centrifuged at 100,000g to separate the cytosol from the particulate fraction. The pellet was resuspended in the original volume of buffer. Aliquots of supernatant taken before the 100,000g centrifugation (T) and of cytosol (C) and pellet fraction (P) taken after the 100,000g centrifugation were separated by SDS-PAGE and analyzed by Western blot with either anti-myc or anti-Spn antibodies. (B) Subcellular localization of Spn is examined by indirect immunofluorescence. Wild-type cells carrying ΔpSpn/PP2C–myc (A,C), ΔpSpn/Spn–myc (B,D), ΔpSpn/Gα–myc (E), or ΔpSpn/GαD376A–myc (F) were starved for 3 hr in NaKPO4 buffer and stimulated for 2 hr with 300 μm cAMP to induce the expression of the various constructs. Cells were then fixed in MeOH (A,B) or paraformaldehyde (C–F) and treated as described in the Materials and Methods.
Indirect immunofluorescence was used to visualize the subcellular distribution of Spn–myc, PP2C–myc Gα–myc, and GαD376A–myc in stable transformants with an anti-myc monoclonal antibody. Cells were fixed after 3 hr starvation in NaKPO4 buffer and subsequent stimulation with high cAMP for 2 hr. Such conditions induced the expression from the Spn promoter (data not shown). Both Spn–myc and the myc-tagged Gα-like domain were observed in a nonuniform distribution at the periphery of the cells in the cortical region, primarily in regions of the plasma membrane that may coincide with membrane ruffles (Fig. 7B). However, the PP2C–myc exhibited a cytosolic staining, supporting the subcellular fractionation results.
Taken together, these data are consistent with a function for the Gα-like domain in the targeting of Spn to the plasma membrane. However, the results also suggest that the role of the Gα-like domain is probably not restricted to this particular function because mutations that are known to alter GTP-binding activity in G proteins results in an Spn protein that is unable to fully complement the spn− phenotype but, at least for the case of GαD376A–myc, does not affect Spn subcellular localization.
Discussion
Spn is a bimodular protein having two distinct functional domains
The Spn amino acid sequence predicts a novel signaling protein that contains two distinct functional domains: a novel Gα-like domain and a domain encoding a PP2C-type serine–threonine phosphatase, a member of the PPM serine-threonine phosphatase family. In addition, Spn has a long (∼240 amino acid) IR that is rich in proline, lysine, and glutamic acid, which exhibits dominant phenotypes when overexpressed, suggesting a specific, but yet undefined function for this domain. Our biochemical and mutational analysis demonstrate that the Spn carboxy-terminal domain encodes a PP2C activity. Because the null phenotype can be partially rescued by overexpression of the PP2C domain alone, the intracellular function of Spn during development is likely to reside mainly in its phosphatase activity. This idea is supported by the fact that inactivation of the phosphatase domain by a double point mutation abrogates the ability of the mutant Spn from complementing the null phenotype.
Recently, a number of PP2C homologs have been identified in different species that are involved in various signaling cascades: PP2C homologs in Schizosaccharomyces pombe and S. cerevisiae are negative regulators of stress response pathways (Maeda et al. 1994; Gaits et al. 1997); the Arabidopsis PP2C-like protein phosphatases ABI1 and ABI2 are required for proper cellular response to the plant hormone abscisic acid (Leung et al. 1997); the PP2C homolog Fem-2 is involved in male sex determination in C. elegans (Chin-Sang and Spence 1996); and the B. subtilis SpoIIE phosphatase regulates sporulation by dephosphorylating SpoIIA, an antitranscription factor (Bork et al. 1996). In some of these proteins, like Spn, the PP2C domain is associated with an amino-terminal functional domain. For example, ABI1 contains a putative Ca2+ binding EF hand, whereas KAPP, another Arabidopsis PP2C, consists of a phosphatase domain fused to an amino-terminal kinase interacting domain. SpoIIE also features a long amino-terminal extension upstream of the PP2C-domain that contains 10 membrane-spanning regions (Stone et al. 1994; Bork et al. 1996; Leung et al. 1997).
A particularly intriguing characteristic of Spn is the presence of a domain with strong homology to Gα subunits. Whereas the PP2C domain is the Spn effector domain and alone can complement the null, although poorly, our data clearly indicate that the Gα-like domain is necessary for the proper function of the protein and may act as a regulatory domain. By subcellular fractionation experiments and indirect immunofluorescence, we have shown that the Gα-like domain is necessary for the targeting of the protein to particular regions of the plasma membrane. We expect that a combination of the inappropriate localization and the lack of proper regulation of the phosphatase activity when the PP2C domain is expressed on its own contributes to the inability of this domain alone to fully rescue the null phenotype. Many heterotrimeric G-proteins function at the plasma membrane as molecular switches to transduce information from a transmembrane receptor to an appropriate effector and to regulate a large number of cellular responses (Gilman 1987; Bourne et al. 1990; Simon et al. 1991). A number of lines of evidence suggest that the Spn amino-terminal Gα-like domain may be a very novel form of a GTP-binding protein. The amino acid sequence comparison would strongly suggest that this domain of Spn is very related to Gα proteins and may either have evolved from one or may have a common ancestor. Sequence comparison of the highly conserved domains of bona fide Gα subunits shows some differences in key residues that have prescribed functions in controlling GTP binding and hydrolysis (see Results). It is, however, highly unlikely that this domain interacts with Gβγ subunits. Our analysis of these residues through projection onto the crystal structure of Gαt suggests that some of these amino acid changes might serve the same function as those in heterotrimeric Gα protein subunits (Noel et al. 1993). Indirectly supporting the model that the Spn amino-terminal domain functions as a GTP-mediated switch is the fact that amino acid substitutions in Spn that would abrogate the GTP-binding function of Gα protein subunits result in a loss of the ability of the expressed protein to fully complement the null phenotype. This also suggests that the amino-terminal domain functions as more than just a targeting domain, as these mutant proteins also target to the membrane. Our data suggest that this domain functions to control the PP2C-like activity either directly or by controlling Spalten’s interaction with its substrate or another regulatory protein.
Spn regulates prestalk cell differentiation
At mound formation, pathways are activated that regulate subsequent morphogenesis and cell-type differentiation in Dictyostelium (Firtel 1995; Williams 1995). In this work, we describe a novel activator of the developmental program that is essential for the unicellular-multicellular transition that occurs at the mound stage. Cells lacking Spn fail to undergo morphogenesis and do not induce cell-type-specific genes. However, spn− cells induce the earliest stages of the developmental transition at the mound stage, including the expression of GBF and the early postaggregative gene LagC, which itself is required for cell-type-specific gene expression (Dynes et al. 1994). The expression of these genes and the aggregation-stage, cAMP pulse-induced gene CsA are reinduced later in development as the spn− mounds dissipate and reform tiny aggregates with tips.
Our data clearly demonstrate that Spn is required for cell-type differentiation because spn− cells are effectively unable to express cell-type specific genes during multicellular development or in suspension in response to cAMP. Analysis of chimeric organisms indicates a cell-autonomous requirement of Spn for the expression the prestalk-specific marker ecmA. Although ecmA expression can be induced by treatment with cAMP and the morphogen DIF in wild-type cells in cell suspension, spn− cells do not respond to these morphogens, suggesting a defect in the earliest stages of the prestalk induction pathway. However, although the null mutant does not express the prespore marker SP60/cotC when developed on filters or in suspension, the prespore marker is induced in chimeras with wild-type cells. This strongly suggests that Spn functions to control a cell nonautonomous pathway for prespore cell differentiation and may be required directly or indirectly for the production of an intercellular developmental signal. However, wild-type cells do not effectively induce the prespore pathway in spn− cells until later in multicellular differentiation. Because Spn is expressed very early in development, we cannot exclude a possible cell-autonomous role of this protein in prespore cell differentiation. The spn− defect can also be partially rescued by codevelopment with pslA− cells, which are unable to induce the prespore pathway (H. Yasukawa, S. Mohanty, and R.A. Firtel, in prep.). The ability of pslA− cells to rescue prespore gene expression in spn− cells favors the model that a developmental signal triggering prespore differentiation together with cAMP might be provided by the prestalk cell population. This is consistent with Spn being expressed in ALCs and prestalk/stalk cells during multicellular stages. In spn− cells, prespore differentiation might not occur because of the absence of prestalk cells and, thus, the prestalk-mediated signaling molecule. However, we cannot exclude the possibility that any cell type could function to complement the spn− defect. Recently, a prespore/spore-inducing factor that works on culmination-stage cells to induce spore formation has been defined (Anjard et al. 1998). The relationship of this factor to our proposed prespore/spore-inducing signaling molecule is not known.
Model for Spn function
The physiological substrate of Spn has not been identified; however, dephosphorylation of Spn target is apparently a key event for morphogenesis to proceed. Considering the cell autonomous effect of Spn null mutation on prestalk cell differentiation, Spn is expected to function directly in ALCs or prestalk cells, consistent with its pattern of expression. By antagonizing the activity of a specific protein kinase, Spn may either directly activate a pathway essential for prestalk differentiation or inhibit a negative regulator of such pathway (Fig. 8). Recently, we identified in a second-site suppressor screen, a gene encoding a novel, putative serine/threonine kinase whose diruption in spn− cells allows the double knockout mutant to form fruiting bodies and differentiate spores (L. Aubry and R.A. Firtel, unpubl.). It is likely that Spn and this novel kinase regulate the same pathway by controlling the activity of a common substrate. Whereas a possible cell-autonomous effect of Spn on prespore differentiation cannot be completely excluded (see above), we favor the model presented in Figure 8, in which prestalk cell signaling is required for prespore differentiation in vivo and Spn’s primary function is to control the induction of the prestalk pathway.
Figure 8.
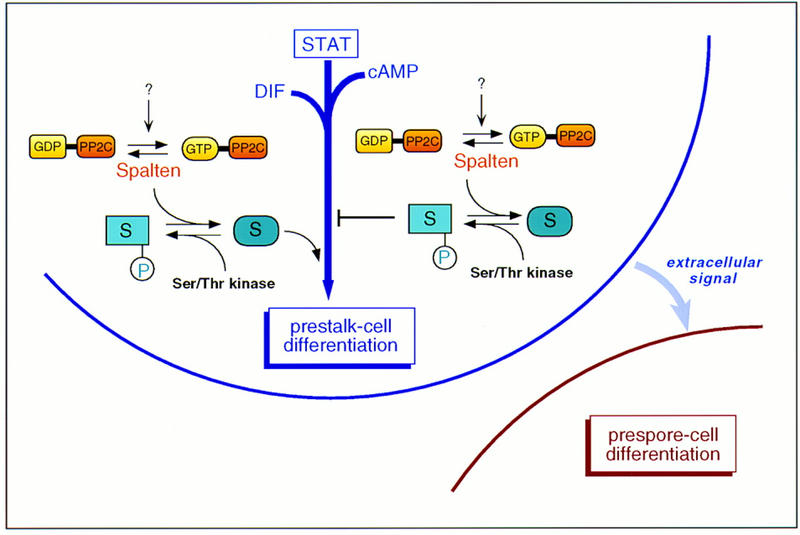
Model for Spn function. The results indicate that Spn is required for prestalk cell differentiation and would be part of a complex network, including the Dictyostelium STAT (Kawata et al. 1997) and the signaling molecules DIF and cAMP (see text for details). Under the appropriate signal, it is possible that the Gα-like domain acts as a molecular switch allowing the activation of the phosphatase domain. Once active, Spn, by antagonizing the effect of a Ser/Thr kinase, may either activate a pathway required for the prestalk differentiation process or inhibit a negative regulator responsible for a block of this pathway.
A variety of proteins have been implicated in the progression past mound stage, suggesting the existence of a complex regulatory network to control this particular transitional stage, including the transcription factor GBF, LagC, the ubiquitin conjugating enzyme UBC, and the cAMP receptor cAR2 (Saxe et al. 1993; Dynes et al. 1994; Schnitzler et al. 1994, 1995; Clark et al. 1997). Spn is a novel component of this integrated network whose functional analysis should allow further understanding of the mechanisms that regulate Dictyostelium development and may be a member of a new family of GTP-regulated molecular switches.
Materials and methods
Cell culture and differentiation
All of the experiments were carried out with KAx-3 as the parental Dictyostelium strain. The cells were grown in suspension in HL5 medium containing 5 μg/ml of blasticidin or 15 μg/ml of G418 as required (Clark et al. 1997; Nellen et al. 1987). Clonal selection of overexpressing strains was done by plating onto G418-containing DM plates in association with Escherichia coli (Hughes et al. 1992). Knockout strains were cloned by plating onto SM-agar plates in association with Klebsiella aerogenes. Developmental phenotypes were studied after plating cells on nonnutrient, Na/KPO4-buffered agar plates.
Insertional mutagenesis
Insertional mutagenesis was performed as described previously (Kuspa and Loomis 1992; Clark et al. 1997) with the following modifications. The plasmid pUCBsr, carrying the blasticidin S resistance gene bsr (Sutoh 1993), was linearized with BamHI and electroporated into KAx-3 cells along with the restriction enzyme DpnII. Transformants were selected in blasticidin-containing HL5 and plated for clonal isolation onto SM-agar plates in association with K. aerogenes. The mutants with abnormal developmental phenotypes were kept for future study, including spn null mutant, which is the subject of the present report. Part of Spn genomic DNA flanking the integrated plasmid was isolated as an NdeI fragment as described (Kuspa and Loomis 1992). This 1-kb fragment was used to screen a 12–16 hr developmental λZAP cDNA library (Schnitzler et al. 1994). A cDNA of ∼3.4 kb containing the entire Spn ORF was obtained. The phenotype of the REMI mutant was recapitulated by use of the original rescued plasmid or a gene-disruption construct made by use of the cDNA (see below).
Plasmid constructs
Spn gene-disruption construct was made by insertion in the 3′ EcoRV site in the cDNA of a ∼1.4-kb fragment containing the Bsr resistance cassette (Fig.1). The promoter region of Spn was obtained from the original REMI mutant by isolation of the 5-kb region of genomic DNA upstream of the site of insertion of pUCBsr after digesting the genomic DNA with XbaI and SpeI. The promoter region (∼4 kb) was subcloned as such (pSpn) or reduced to 1 kb upstream of the ATG (ΔpSpn) in the EXP4+ Dictyostelium expression vector (Dynes et al. 1994) lacking the actin promoter and used for overexpression analysis to drive the expression of Spn, the Gα-like domain, and the PP2C domain and for β-galactosidase staining experiments, to drive the expression of the reporter gene lacZ. The ΔpSpn/Gα construct encompasses Spn amino acid sequence from residue 1 to 458. For ΔpSpn/PP2C and pSpn/lacZ constructs, the PP2C domain (residue 773–975) and the lacZ gene were subcloned in the ClaI site of the cDNA in frame with the ATG after PCR amplification to create the appropriate subcloning site. ΔpSpn/Spn–myc, ΔpSpn/Gα–myc, ΔpSpn/PP2C–myc were made similarly after addition by PCR of a (myc)2-tag at the carboxyl terminus of the protein. The GST–ΔPP2C construct was obtained by subcloning of the carboxy-terminal region of Spn (∼600 bp) in-frame to glutathione S-transferase (GST) into the pGEX-KG expression vector (Guan and Dixon 1991). The (His)6–Spn construct was created by subcloning the full-length cDNA into FASTBAC vector (GIBCO) in-frame with the amino-terminal polyhistidine tag contained in the vector. All of the constructs that required PCR amplification were verified by sequencing.
Other molecular biology
Site-directed mutagenesis was performed by use of the Transformer Site-Directed Mutagenesis kit (Clontech). All constructs were sequenced to confirm the amino acid substitutions and the absence of additional mutations.
Cells carrying the constructs pSpn/lacZ, act15/lacZ (Mann and Firtel 1993), SP60/lacZ (Haberstroh and Firtel 1990), ecmA/lacZ (Jermyn and Williams 1991), and spiA/lacZ (Richardson et al. 1994) were subjected to β-galactosidase staining. Cells were spread on nitrocellulose filters laid on nonnutrient agar plates and allowed to develop. Histochemical localization of β-galactosidase activity was determined as described previously (Haberstroh and Firtel,1990; Mann et al. 1994).
RNA and DNA blots were performed by standard techniques (Nellen et al. 1987).
Antibody and Western blot analysis
The GST–ΔPP2C fusion protein was expressed in E. coli BL21(DE3) and used to raise polyclonal anti-Spn antibodies. Anti-Spn-specific antibodies were purified as described in Gamper et al (1995) and used as well as a monoclonal anti-myc antibody (Invitrogen) for Western blot analysis. Proteins were detected by enhanced chemiluminescence (ECL-Amersham).
Purification of Spn and phosphatase assay
Spn was expressed in Sf9 insect cells as a six-histidine amino-terminal tagged protein [(His)6–Spn construct] by use of the FASTBAC kit from GIBCO. Seventy-two hours after infection, cells were harvested by centrifugation, resuspended in buffer A (20 mm Tris at pH 8.0, 0.5 m NaCl, 5 mm MgCl2, 10 mm β-mercaptoethanol) containing 0.5% NP-40 and proteases inhibitors, and lysed by sonication. The lysate was centrifuged at 100,000g to remove debris and the supernatant was incubated with Ni2+–Sepharose beads (Qiagen). The beads were washed three times in buffer A, and (His)6–Spn was eluted with buffer A containing 50 mm imidazole. Samples were subjected to SDS-PAGE and Coomassie staining to verify that (His)6–Spn was the predominant species.
Casein was used as a substrate to assay the phosphatase activity of (His)6–Spn. Casein was phosphorylated with the catalytic subunit of cAMP-dependent PKA and [γ-32P]ATP, purified through a Sephadex G50 column, and used as described in McGowan and Cohen (1988). Reactions were performed in a volume of 50 μl with purified (His)6–Spn in the presence of 20 mm MgCl2, MnCl2, CaCl2, or EDTA.
Subcellular fractionation
Subcellular fractionation was performed on wild-type cells, spn− cells, and overexpressing strains carrying ΔpSpn/Spn–myc, ΔpSpn/Gα–myc, or ΔpSpn/PP2C–myc. Cells (5 × 107) were left to develop for 8 hr on non-nutrient agar plates, harvested by centrifugation, resuspended in 20 mm triethanolamine at pH 7.5 containing proteases inhibitors, and lysed through a 3-μm Nuclepore filter. The lysate was first centifuged at 800g for 5 min to remove nuclei and any intact cells and then at 100,000g for 2 hr to separate soluble and particulate cell fractions. The pellet was resuspended in the original volume of 20 mm triethanolamine. All procedures were performed on ice. Protein samples were subjected to SDS-PAGE and Western blot analysis with either anti-Spn or anti-myc antibodies.
Indirect immunofluorescence microscopy
Cells expressing myc-tagged proteins were washed, resuspended in Na–KPO4 buffer, and starved for 3 hr in suspension. Cells were then stimulated with 300 μm cAMP for 2 hr and left to adhere for 10 min on a coverslip. Cells were fixed in 40 mm Mes–Na at pH 6.5 containing 4% paraformaldehyde, and permeabilized in 0.2% Triton X-100 in Mes–Na buffer. Alternatively, cells were prefixed in 50% MeOH and fixed in 100% MeOH at 0°C in suspension after cAMP treatment. Cells were then incubated with 1.4 μg/ml anti-myc monoclonal antibody (Invitrogen) in PBS for 1 hr, washed in 0.5% BSA containing PBS, and incubated with FITC-labeled anti-mouse antibodies for 1 hr. After washing, cells were observed with a 60× oil-immersion lens on a Nikon Microphot-FX microscope. Images were captured with a Photometrics Sensys camera and IP Lab Spectrum software.
Acknowledgments
We are indebted to Joe Noel (Salk Institute) for assistance in analysis of the structure of the Spn Gα-like domain. We also thank Marie-Helene Ogliastro (UCSD) for her assistance in expression of Spalten in insect cells, Jason Brown (UCSD) for helpful discussions through the course of this work, and Anson Nomura (UCSD) for expert technical assistance. L.A. was supported in part by a Human Frontiers in Science postdoctoral fellowship. This work was supported by U.S. Public Health Service grants to R.A.F. The GenBank nucleotide accession no. for Spn is AF019985.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rafirtel@ucsd.edu; FAX (619) 534-7073.
References
- Abe K, Yanagisawa K. A new class of rapid developing mutants in Dictyostelium discoideum: Implications for cyclic AMP metabolism and cell differentiation. Dev Biol. 1983;95:200–210. doi: 10.1016/0012-1606(83)90018-0. [DOI] [PubMed] [Google Scholar]
- Anjard C, Zeng C, Loomis WF, Nellen W. Signal transduction pathways leading to spore differentiation in Dictyostelium discoideum. Dev Biol. 1998;193:146–155. doi: 10.1006/dbio.1997.8804. [DOI] [PubMed] [Google Scholar]
- Barford D. Molecular mechanisms of the serine/threonine phosphatases. Trends Biochem Sci. 1996;21:407–412. doi: 10.1016/s0968-0004(96)10060-8. [DOI] [PubMed] [Google Scholar]
- Bork P, Brown NP, Hegyi H, Shultz J. The protein phosphatase 2C (PP2C) superfamily: Detection of bacterial homologues. Protein Sci. 1996;5:1421–1425. doi: 10.1002/pro.5560050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR, Sander DA, McCormick F. The GTPase superfamily: A conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Chen MY, Insall RH, Devreotes PN. Signaling through chemoattractant receptors in Dictyostelium. Trends Genet. 1996;12:52–57. doi: 10.1016/0168-9525(96)81400-4. [DOI] [PubMed] [Google Scholar]
- Chin-Sang ID, Spence AM. Caenorhabditis elegans sex-determining protein FEM-2 is a protein phosphatase that promotes male development and interacts directly with FEM-3. Genes & Dev. 1996;10:2314–2325. doi: 10.1101/gad.10.18.2314. [DOI] [PubMed] [Google Scholar]
- Clark A, Nomura A, Mohanty S, Firtel RA. A ubiquitin-conjugating enzyme is essential for development transitions in Dictyostelium. Mol Biol Cell. 1997;8:1989–2002. doi: 10.1091/mbc.8.10.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AK, Helps NR, Cohen PTW, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- Devine KM, Loomis WF. Molecular characterization of anterior-like cells in Dictyostelium discoideum. Dev Biol. 1985;107:364–372. doi: 10.1016/0012-1606(85)90318-5. [DOI] [PubMed] [Google Scholar]
- Dynes J, Clark A, Shaulsky G, Kuspa A, Loomis W, Firtel R. LagC is required for cell-cell interactions that are essential for cell-type differentiation in Dictyostelium. Genes & Dev. 1994;8:948–958. doi: 10.1101/gad.8.8.948. [DOI] [PubMed] [Google Scholar]
- Early VE, Williams JG. A Dictyostelium prespore-specific gene is transcriptionally repressed by DIF in vitro. Development. 1988;103:519–524. doi: 10.1242/dev.103.3.519. [DOI] [PubMed] [Google Scholar]
- Firtel RA. Integration of signaling information in controlling cell-fate decisions in Dictyostelium. Genes & Dev. 1995;9:1427–1444. doi: 10.1101/gad.9.12.1427. [DOI] [PubMed] [Google Scholar]
- ————— Interacting signaling pathways controlling multicellular development in Dictyostelium. Curr Opin Genet Dev. 1996;6:545–554. doi: 10.1016/s0959-437x(96)80082-7. [DOI] [PubMed] [Google Scholar]
- Gaits F, Shiozaki K, Russell P. Protein phosphatase 2C acts independently of stress-activated kinase cascade to regulate the stress response in fission yeast. J Biol Chem. 1997;272:17873–17879. doi: 10.1074/jbc.272.28.17873. [DOI] [PubMed] [Google Scholar]
- Gamper M, Howard PK, Hunter T, Firtel RA. Protein tyrosine phosphatases in Dictyostelium discoideum. Adv Prot Phosphatases. 1995;9:25–49. [Google Scholar]
- Gaskins C, Clark AM, Aubry L, Segall JE, Firtel RA. The Dictyostelium MAP kinase ERK2 regulates multiple, independent developmental pathways. Genes & Dev. 1996;10:118–128. doi: 10.1101/gad.10.1.118. [DOI] [PubMed] [Google Scholar]
- Gilman AG. G proteins: Transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Ginsburg GT, Gollop R, Yu YM, Louis JM, Saxe CL, Kimmel AR. The regulation of Dictyostelium development by transmembrane signalling J. Euk Microbiol. 1995;42:200–205. doi: 10.1111/j.1550-7408.1995.tb01565.x. [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Haberstroh L, Firtel RA. A spatial gradient of expression of a cAMP-regulated prespore cell type specific gene in Dictyostelium. Genes & Dev. 1990;4:596–612. doi: 10.1101/gad.4.4.596. [DOI] [PubMed] [Google Scholar]
- Haribabu B, Dottin RP. Homology cloning of protein kinase and phosphoprotein phosphatase sequences of Dictyostelium discoideum. Dev Genet. 1991;12:45–49. doi: 10.1002/dvg.1020120109. [DOI] [PubMed] [Google Scholar]
- Harwood A, Plyte S, Woodgett J, Strutt H, Kay R. Glycogen synthetase kinase 3 (GSK-3) regulates cell fate in Dictyostelium. Cell. 1995;80:139–148. doi: 10.1016/0092-8674(95)90458-1. [DOI] [PubMed] [Google Scholar]
- Hopper NA, Anjard C, Reymond CD, Williams JG. Induction of terminal differentiation of Dictyostelium by cAMP-dependent protein kinase and opposing effects of intracellular and extracellular cAMP on stalk cell differentiation. Development. 1993a;119:147–154. doi: 10.1242/dev.119.1.147. [DOI] [PubMed] [Google Scholar]
- Hopper NA, Harwood AJ, Bouzid S, Veron M, Williams JG. Activation of the prespore and spore cell pathway of Dictyostelium: Differentiation by cAMP-dependent protein kinase and evidence for its upstream regulation by ammonia. EMBO J. 1993b;12:2459–2466. doi: 10.1002/j.1460-2075.1993.tb05900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn F, Gross J. A role for calcineurin in Dictyostelium discoideum development. Differentiation. 1996;60:269–275. doi: 10.1046/j.1432-0436.1996.6050269.x. [DOI] [PubMed] [Google Scholar]
- Hughes JE, Podgorski GJ, Welker DL. Selection of Dictyostelium discoideum transformants and analysis of vector maintenance using live bacteria resistant to G418. Plasmid. 1992;28:46–60. doi: 10.1016/0147-619x(92)90035-9. [DOI] [PubMed] [Google Scholar]
- Jermyn KA, Williams JG. An analysis of culmination in Dictyostelium using prestalk and stalk-specific cell autonomous markers. Development. 1991;111:779–787. doi: 10.1242/dev.111.3.779. [DOI] [PubMed] [Google Scholar]
- Jermyn KA, Berks M, Kay RR, Williams JG. Two distinct classes of prestalk-enriched mRNA sequences in Dictyostelium discoideum. Development. 1987;100:745–755. doi: 10.1242/dev.100.4.745. [DOI] [PubMed] [Google Scholar]
- Kawata T, Shevchenko A, Fukuzawa M, Jermyn KA, Totty NF, Zhukovskaya NV, Sterling AE, Mann M, Williams JG. SH2 signaling in a lower eukaryote: A STAT protein that regulates stalk cell differentiation in Dictyostelium. Cell. 1997;89:909–916. doi: 10.1016/s0092-8674(00)80276-7. [DOI] [PubMed] [Google Scholar]
- Kay RR. Cell differentiation and patterning in Dictyostelium. Curr Opin Cell Biol. 1992;4:934–938. doi: 10.1016/0955-0674(92)90121-r. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M, Nyborg J, Clark BC. The GTP binding motif: Variations on a theme. FASEB J. 1996;10:1347–1368. [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF. Development of Dictyostelium discoideum. New York, NY: Academic Press; 1982. [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Mann SKO, Firtel RA. cAMP-dependent protein kinase differentially regulates prestalk and prespore differentiation during Dictyostelium development. Development. 1993;119:135–146. doi: 10.1242/dev.119.1.135. [DOI] [PubMed] [Google Scholar]
- Mann SKO, Yonemoto WM, Taylor SS, Firtel RA. DdPK3, which plays essential roles during Dictyostelium development, encodes the catalytic subunit of cAMP-dependent protein kinase. Proc Natl Acad Sci. 1992;89:10701–10705. doi: 10.1073/pnas.89.22.10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann S, Devreotes P, Eliott S, Jermyn K, Kuspa A, Fechheimer M, Furukawa R, Parent C, Segall J, Shaulsky G, Vardy P, Williams J, Williams K, Firtel R. Cell biological, molecular genetic, and biochemical methods to examine Dictyostelium. In: Celis J, editor. Cell biology: A laboratory handbook. New York, NY: Academic Press; 1994. pp. 412–451. [Google Scholar]
- Mann SKO, Brown JM, Briscoe C, Parent C, Pitt G, Devreotes PN, Firtel RA. Role of cAMP-dependent protein kinase in controlling aggregation and postaggregative development in Dictyostelium. Dev Biol. 1997;183:208–221. doi: 10.1006/dbio.1996.8499. [DOI] [PubMed] [Google Scholar]
- McGowan CH, Cohen P. Protein phosphatase-2C from rabbit skeletal muscle and liver: An Mg2+-dependent enzyme. Methods in Enzymol. 1988;159:416–426. doi: 10.1016/0076-6879(88)59041-9. [DOI] [PubMed] [Google Scholar]
- Mehdy MC, Firtel RA. A secreted factor and cyclic AMP jointly regulate cell-type-specific gene expression in Dictyostelium discoideum. Mol Cell Biol. 1985;5:705–713. doi: 10.1128/mcb.5.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Nellen W, Datta S, Reymond C, Sivertsen A, Mann S, Crowley T, Firtel RA. Molecular biology in Dictyostelium: Tools and applications. Methods Cell Biol. 1987;28:67–100. doi: 10.1016/s0091-679x(08)61637-4. [DOI] [PubMed] [Google Scholar]
- Noel JP, Hamm HE, Sigler PB. The 2.2 Å crystal structure of transducin-α complexed with GTPγS. Nature. 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- Reymond CD, Schaap P, Veron M, Williams JG. Dual role of cAMP during Dictyostelium development. Experientia. 1995;51:1166–1174. doi: 10.1007/BF01944734. [DOI] [PubMed] [Google Scholar]
- Richardson DL, Loomis WF, Kimmel AR. Progression of an inductive signal activates sporulation in Dictyostelium discoideum. Development. 1994;120:2891–2900. doi: 10.1242/dev.120.10.2891. [DOI] [PubMed] [Google Scholar]
- Saxe CL, III, Ginsburg GT, Louis JM, Johnson R, Devreotes PN, Kimmel AR. CAR2, a prestalk cAMP receptor required for normal tip formation and late development of Dictyostelium discoideum. Genes & Dev. 1993;7:262–272. doi: 10.1101/gad.7.2.262. [DOI] [PubMed] [Google Scholar]
- Schmidt G, Lenzen C, Simon I, Deuter R, Cool RH, Goody RS, Wittinghofer A. Biochemical and biological consequences of changing the specificity of p21ras from guanosine to xanthosine nucleotides. Oncogene. 1996;12:87–96. [PubMed] [Google Scholar]
- Schnitzler G, Fischer W, Firtel R. Cloning and characterization of the G-box binding factor, an essential component of the developmental switch between early and late development in Dictyostelium. Genes & Dev. 1994;8:502–514. doi: 10.1101/gad.8.4.502. [DOI] [PubMed] [Google Scholar]
- Schnitzler GR, Briscoe C, Brown JM, Firtel RA. Serpentine cAMP receptors may act through a G protein-independent pathway to induce postaggregative development in Dictyostelium. Cell. 1995;81:737–745. doi: 10.1016/0092-8674(95)90535-9. [DOI] [PubMed] [Google Scholar]
- Segall J, Kuspa A, Shaulsky G, Ecke M, Maeda M, Gaskins C, Firtel R, Loomis W. A MAP kinase necessary for receptor-mediated activation of adenylyl cyclase in Dictyostelium. J Cell Biol. 1995;128:405–413. doi: 10.1083/jcb.128.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulsky G, Kuspa A, Loomis WF. A multidrug resistance transporter serine protease gene is required for prestalk specialization in Dictyostelium. Genes & Dev. 1995;9:1111–1122. doi: 10.1101/gad.9.9.1111. [DOI] [PubMed] [Google Scholar]
- Simon MI, Strathman MP, Gautam N. Diversity of G proteins in signal transduction. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- Sternfeld J, David CN. Fate and regulation of anterior-like cells in Dictyostelium slugs. Dev Biol. 1982;93:111–118. doi: 10.1016/0012-1606(82)90244-5. [DOI] [PubMed] [Google Scholar]
- Stone JM, Collinge MA, Smith RD, Horn MA, Walker JC. Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science. 1994;266:793–795. doi: 10.1126/science.7973632. [DOI] [PubMed] [Google Scholar]
- Sutoh K. A transformation vector for Dictyostelium discoideum with a new selectable marker Bsr. Plasmid. 1993;30:150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- Williams J. Morphogenesis in Dictyostelium: New twists to a not-so-old tale. Curr Opin Genet Dev. 1995;5:426–431. doi: 10.1016/0959-437x(95)90044-h. [DOI] [PubMed] [Google Scholar]
- Yu B, Slepak VZ, Simon MI. Characterization of a Goα mutant that binds xanthine nucleotides. J Biol Chem. 1997;272:18015–18019. doi: 10.1074/jbc.272.29.18015. [DOI] [PubMed] [Google Scholar]
- Zhukovskaya N, Early A, Kawata T, Abe T, Williams J. cAMP-dependent protein kinase is required for the expression of a gene specifically expressed in Dictyostelium prestalk cells. Dev Biol. 1996;179:27–40. doi: 10.1006/dbio.1996.0239. [DOI] [PubMed] [Google Scholar]