Abstract
The ATP-dependent PIM1 protease, a Lon-like protease localized in the mitochondrial matrix, is required for mitochondrial genome integrity in yeast. Cells lacking PIM1 accumulate lesions in the mitochondrial DNA (mtDNA) and therefore lose respiratory competence. The identification of a multicopy suppressor, which stabilizes mtDNA in the absence of PIM1, enabled us to characterize novel functions of PIM1 protease during mitochondrial biogenesis. The synthesis of mitochondrially encoded cytochrome c oxidase subunit I (CoxI) and cytochrome b (Cob) is impaired in pim1 mutants containing mtDNA. PIM1-mediated proteolysis is required for the translation of mature COXI mRNA. Moreover, deficiencies in the splicing of COXI and COB transcripts, which appear to be restricted to introns encoding mRNA maturases, were observed in cells lacking the PIM1 gene. Transcripts of COXI and COB genes harboring multiple introns are degraded in the absence of PIM1. These results establish multiple, essential functions of the ATP-dependent PIM1 protease during mitochondrial gene expression.
Keywords: Mitochondria, ATP-dependent proteolysis, PIM1 protease, translation, RNA processing, RNA stability, cytochrome c oxidase, cytochrome b
Many cellular processes are under the control of ATP-dependent proteases that ensure cellular homeostasis and allow the adaptation to changes in environmental conditions. In eukaryotic cells, the 26S proteasome, a multicatalytic proteolytic complex localized in the cytosol, mediates the energy-dependent degradation of most cellular proteins (Coux et al. 1996; Hilt and Wolf 1996; Baumeister and Lupas 1997). Other than the 26S proteasome, ATP-dependent proteases have only been identified in organelles of endosymbiotic origin, such as mitochondria and chloroplasts, which harbor independent proteolytic systems (Adam 1996; Langer and Neupert 1996; Rep and Grivell 1996; Suzuki et al. 1997). The ATP-dependent proteases of mitochondria fulfill crucial functions during the biogenesis of the organelle, as they are required for the maintenance of the respiratory competence in yeast. However, their physiological substrates have not been described until now.
Two ATP-dependent proteases have been identified in the mitochondrial inner membrane and were termed AAA proteases (Leonhard et al. 1996) as their subunits contain a highly conserved domain characteristic for the AAA family of ATPases (Kunau et al. 1993; Confalonieri and Duguet 1995). Yme1p, an integral inner membrane protein facing the intermembrane space, is the solely identified subunit of the i-AAA protease (Thorsness et al. 1993). Proteolysis by Yme1p is required for the maintenance of respiratory competence of the cells at elevated temperatures and for the formation of a reticulated network of mitochondria (Thorsness et al. 1993; Campbell et al. 1994). The m-AAA protease is composed of multiple copies of Yta10p and Yta12p, integral inner membrane proteins that are homologous to Yme1p but expose their catalytic sites to the mitochondrial matrix (Arlt et al. 1996). Cells lacking Yta10p or Yta12p display deficiencies in the assembly of respiratory chain complexes (Guélin et al. 1994; Tauer et al. 1994; Tzagoloff et al. 1994). Both AAA proteases mediate the degradation of nonassembled inner membrane proteins (Arlt et al. 1996; Guélin et al. 1996). How these AAA proteases are involved in the biogenesis of the respiratory chain and in the maintenance of mitochondrial morphology, however, is still unknown.
The ATP-dependent PIM1 protease controls the selective turnover of proteins in the mitochondrial matrix space (Suzuki et al. 1994; van Dyck et al. 1994). Misfolded polypeptides are degraded by PIM1 protease in cooperation with the mitochondrial Hsp70 system that stabilizes substrate polypeptides against aggregation (Wagner et al. 1994). Overexpression of PIM1 restores the respiratory competence of Δyta10Δyta12 mutants, suggesting a functional overlap with the m-AAA protease (Rep et al. 1996a). Similar to the m-AAA protease, PIM1 protease forms an high molecular weight, presumably homo-oligomeric complex whose assembly depends on its intrinsic ATPase activity (Wagner et al. 1997). Yeast cells lacking the PIM1 gene lose intact mitochondrial DNA (mtDNA) (Suzuki et al. 1994; van Dyck et al. 1994). As essential components of the respiratory chain are encoded by the mitochondrial genome, pim1 mutants are respiratory deficient. Electron dense particles, most likely consisting of aggregated polypeptides, were observed in mitochondria of Δpim1 mutants (Suzuki et al. 1994). It was therefore speculated that the loss of mtDNA in the absence of PIM1 may be caused by the accumulation of misfolded polypeptides (Grivell 1995). Alternatively, one may envision regulatory functions of PIM1 protease in mtDNA metabolism.
Proteins homologous to PIM1 are present in bacteria and mitochondria of human and plant cells and comprise the family of Lon-like proteases (Goldberg 1992; Gottesman and Maurizi 1992; Maurizi 1992). Functional conservation of Escherichia coli Lon protease with PIM1 has recently been demonstrated in yeast (Teichmann et al. 1996). The respiratory competence of cells lacking PIM1, that is, the integrity of mtDNA, can be maintained by expression of E. coli Lon protease. Although complementation depended on the proteolytic activity of the Lon protease, a mutant variant with reduced enzymatic activity, LonK362A protease, was able to substitute for PIM1 protease (Teichmann et al. 1996). Apparently, a low proteolytic activity of a Lon-like protease is sufficient to maintain the respiratory competence of the cells. Substitution of Lon protease for PIM1 was found to occur at 30°C but not when cells were grown at 36°C indicating functional differences between the proteases (Teichmann et al. 1996).
In the present study, we took advantage of the temperature-sensitive growth defect of Δpim1 cells expressing E. coli LonK362A protease and isolated a multicopy suppressor that preserves mtDNA integrity in a pim1 null mutant. The respiratory competence of these cells remains impaired demonstrating a direct involvement of PIM1 protease in the biogenesis of the respiratory chain. Further analysis revealed deficiencies in the synthesis of mitochondrially encoded cytochrome b (Cob) and subunit I of the cytochrome c oxidase (CoxI). PIM1 function is required for the translation of mature COXI mRNA and the stability of COXI and COB transcripts containing multiple introns. Furthermore, pim1 mutants harboring mtDNA show deficiencies in the splicing of COXI and COB pre-mRNAs. Thus, the expression of mitochondrially encoded COXI and COB genes and thereby the assembly of respiratory chain complexes is under the proteolytic control of the ATP-dependent PIM1 protease.
Results
A pim1 mutant harboring intact mtDNA is respiratory deficient
Defects in the integrity of mtDNA result in respiratory deficiency in yeast, as essential components of respiratory chain complexes are mitochondrially encoded. The requirement of PIM1 protease for the maintenance of mtDNA prevents, therefore, a characterization of its role in mitochondrial biogenesis. Genetic approaches, such as a search for multicopy suppressors of the pim1 null mutant phenotype, are hardly applicable because of the lack of mtDNA in these cells. To circumvent this problem, a Δpim1 strain was employed which expresses E. coli LonK362A protease in mitochondria (Δpim1/LON; Teichmann et al. 1996). The expression of Lon protease confers respiratory competence to the cells at 30°C but not at 36°C (Fig. 1; Teichmann et al. 1996). To investigate the function of PIM1 protease, we performed a genetic screen for multicopy suppressors rescuing the conditional growth phenotype of Δpim1/LON cells. This search led to the identification of an extragenic suppressor (YEp13–SUP) that restored the growth of Δpim1/LON cells on nonfermentable carbon sources at 36°C, that is, the integrity and expression of mtDNA (Δpim1/LON/SUP) (Fig. 1).
Figure 1.
Respiratory deficiency of Δpim1 cells carrying intact mtDNA. Wild-type cells (WT), Δpim1 cells (Δpim1), and Δpim1 cells complemented with the E. coli Lon protease (Δpim1/LON), the suppressor gene (Δpim1/SUP), or both (Δpim1/LON/SUP), were grown in glucose-containing selective medium. Cells were harvested in exponential phase, spotted onto YEPG (rich medium containing 3% glycerol), and incubated at 30°C and 36°C for 7 and 10 days, respectively. MtDNA integrity (ρ+, ρ−) of various strains was examined by testing the respiratory competence of a diploid strain generated by mating with a ρ0 PIM1+ strain. (ρ+) Wild-type mtDNA; (ρ−) mutant mtDNA carrying deletions.
To examine whether the suppressor alone stabilizes mtDNA in the absence of PIM1 protease, the PIM1 gene was disrupted in a haploid wild-type strain previously transformed with the rescuing plasmid YEp13–SUP (Δpim1/SUP). In contrast to pim1 null mutants, Δpim1/SUP cells maintained mtDNA as demonstrated by crossing of these cells with a wild-type strain totally devoid of mtDNA. The resulting diploids were able to grow on nonfermentable carbon sources demonstrating the presence of intact mtDNA in Δpim1/SUP cells (data not shown). Although maintaining mitochondrial genome integrity, the suppressor did, however, not provide respiratory competence to Δpim1 cells lacking Lon protease (Fig. 1). Thus, independent of its role in stabilizing the mitochondrial genome, PIM1 function is required for the maintenance of the respiratory competence of the cells.
The rescuing plasmid, YEp13–SUP, contained a 5.4-kb insert from the right arm of chromosome IV bearing the genes SLU7 (Frank and Guthrie 1992), YDR087c, encoding a protein of unknown function, and SSS1 (Esnault et al. 1993). Overexpression of Sss1p alone was sufficient to maintain mtDNA in the absence of PIM1 protease. Disruption of the PIM1 gene in haploid cells expressing Sss1p from a multicopy plasmid did not impair the integrity of mtDNA (data not shown). Sss1p has originally been identified as a multicopy suppressor of the temperature-sensitive sec61-2 mutant (Esnault et al. 1993). It represents a subunit of Sec61p-complexes mediating the translocation of secretory proteins across the membrane of the endoplasmic reticulum (ER) (Esnault et al. 1994; Panzner et al. 1995; Finke et al. 1996). Therefore, an indirect effect on mtDNA metabolism seems likely. It should be noted, however, that a link between mitochondrial function and the ER was also suggested by studies on the yeast signal recognition particle (SRP) (Stirling and Hewitt 1992). The deletion of SRP subunits in yeast results in slow growing cells that are respiratory deficient, an observation whose functional significance remains to be demonstrated. In any case, the stabilization of mtDNA in pim1-null mutants overexpressing Sss1p enabled us to define novel functions of PIM1 protease in mitochondria.
Defective synthesis of mitochondrially encoded CoxI and Cob in pim1 mutants
Seven subunits of respiratory complexes and one mitochondrial ribosomal subunit are encoded by mtDNA in yeast (Tzagoloff and Myers 1986; Grivell and Schweyen 1989; Costanzo and Fox 1990). To investigate the essential role of PIM1 for respiration, mitochondrially encoded proteins were labelled with [35S]methionine in Δpim1 cells that maintain mtDNA because of the expression of the E. coli Lon protease (Δpim1/LON), the suppressor (Δpim1/SUP), or both (Δpim1/LON/SUP) (Fig. 2A). Labeling of mitochondrially encoded proteins occurred with similar efficiencies in wild-type and Δpim1/LON cells, but incorporation of [35S]methionine was less efficient in Δpim1/SUP cells. The suppressor did not affect mitochondrial translation as indicated by the identical patterns of proteins synthesized in mitochondria of Δpim1/LON and Δpim1/LON/SUP cells (Fig. 2A).
Figure 2.
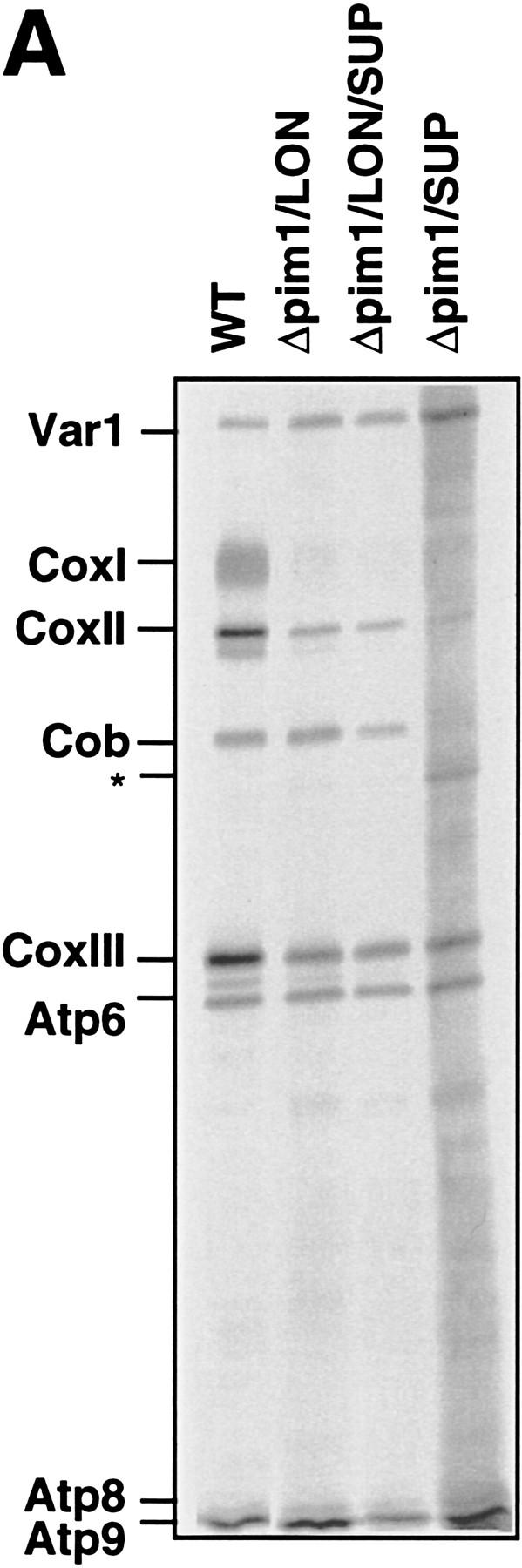
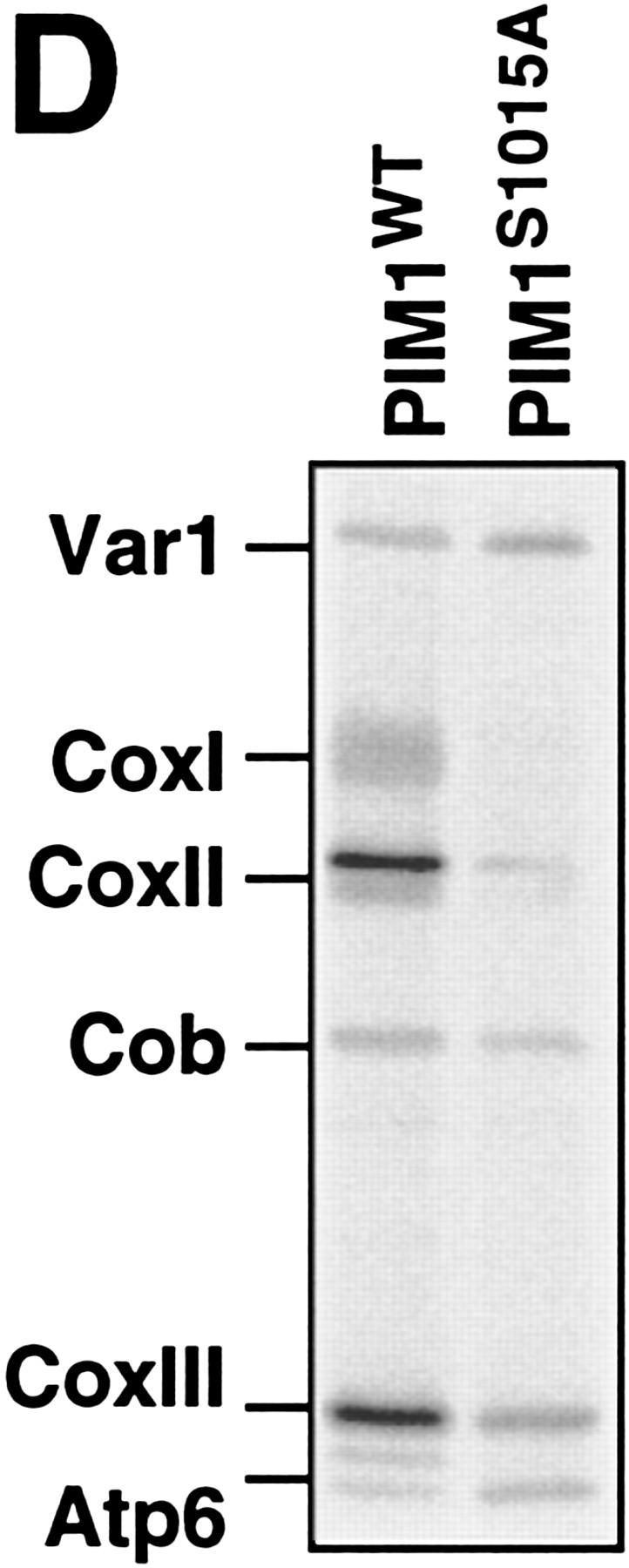
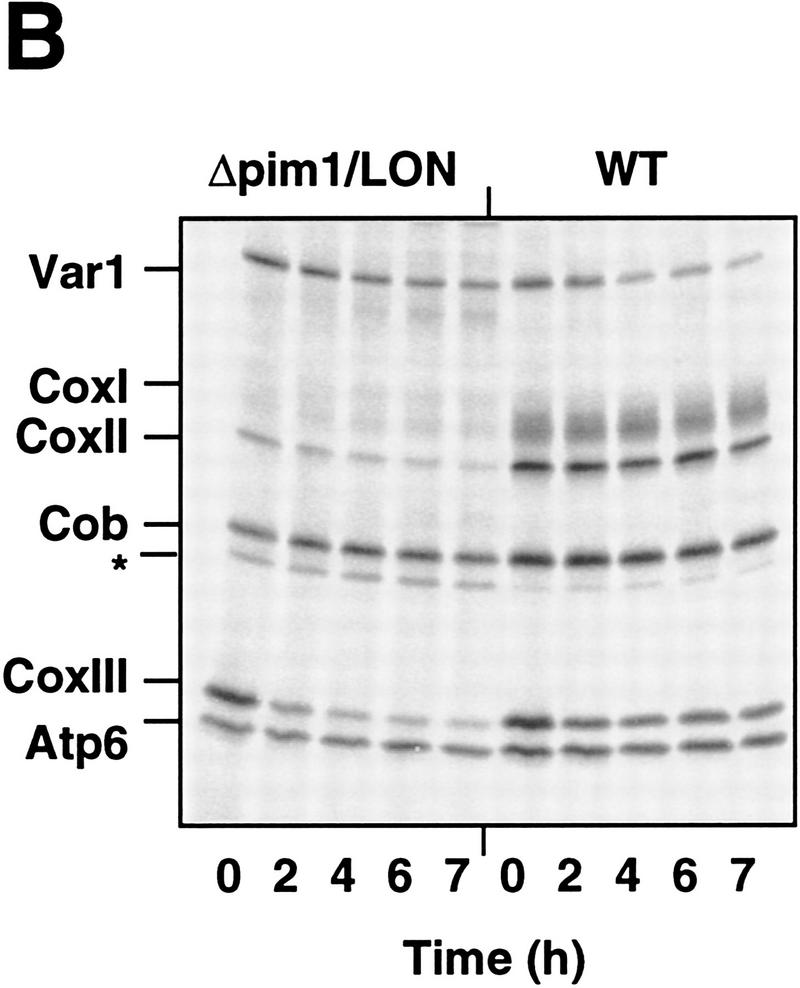
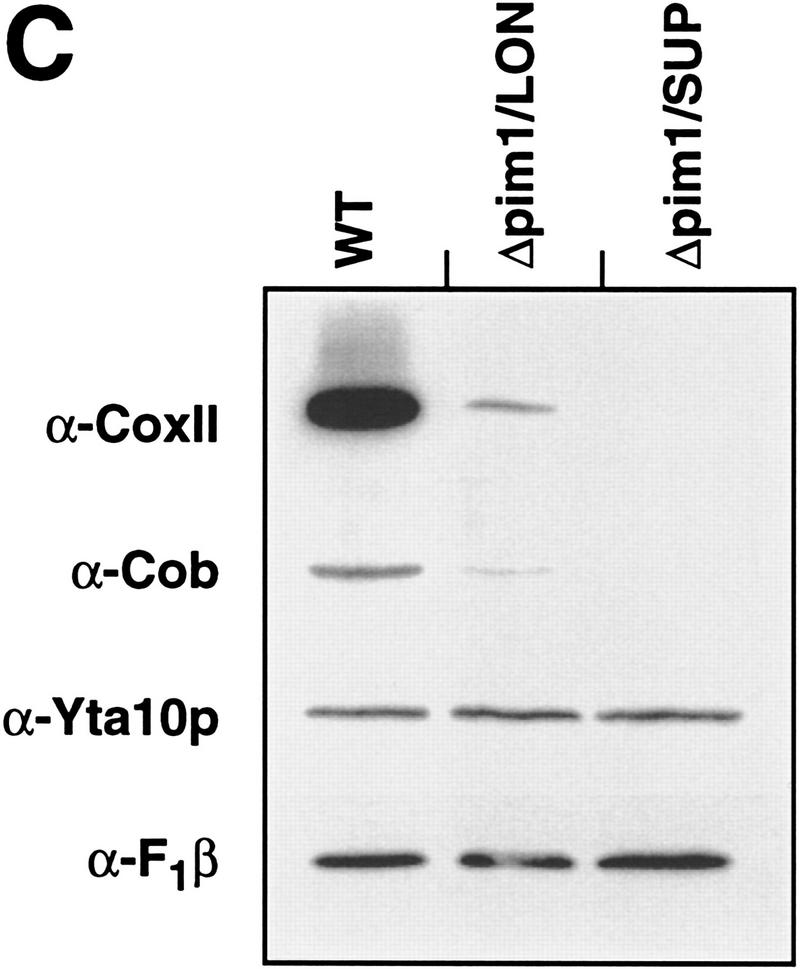
Requirement of PIM1 protease for the synthesis of CoxI and Cob. (A) Synthesis of mitochondrially encoded proteins in vivo. Mitochondrial translation products were labeled with [35S]methionine in the presence of cycloheximide for 10 min (WT, Δpim1/LON, Δpim1/LON/SUP) or 30 min (Δpim1/SUP) at 30°C in vivo as described in Materials and Methods and analyzed by SDS-PAGE. The translation efficiency was reduced in Δpim1/SUP cells. A band marked with an asterisk (*) is not strain-specific and was also detected in ρ0 strains. (CoxI, CoxII, CoxIII) Subunits I, II, and III of cytochrome c oxidase, respectively; (Cob) cytochrome b; (Atp6, Atp8, Atp9) subunits 6, 8, and 9 of the F0F1–ATPase, respectively. (B) Degradation of newly synthesized CoxII and CoxIII in Δpim1/LON cells. Mitochondrially encoded polypeptides were synthesized in vivo for 30 min in the presence of [35S]methionine. After addition of cold methionine (10 mm), cells were further incubated for the indicated time periods and then analyzed by SDS-PAGE. (C) Steady-state levels of CoxII and Cob in mitochondria lacking PIM1 protease. Mitochondria were isolated from wild-type, Δpim1/LON, and Δpim1/SUP cells, subjected to SDS-PAGE and analyzed by Western blotting with polyclonal antisera directed against CoxII (α-CoxII), Cob (α-Cob), and Yta10p (α-Yta10p) and the β-subunit of the F1–ATPase (α-F1β) as gel loading controls. (D) Dependence of CoxI synthesis on PIM1-mediated proteolysis. Labeling of mitochondrial translation products was performed for 10 min at 30°C in Δpim1/LON cells expressing wild-type PIM1 or PIM1S1015A from multicopy plasmids. Wild-type and mutant protease accumulated at similar levels in mitochondria.
Newly synthesized ATP synthase subunits 6, 8, and 9 (Atp6, Atp8, and Atp9) and the ribosomal subunit Var1 accumulated at similar levels in wild-type and in Δpim1 cells containing mtDNA (Fig. 2A). In contrast, labeling of CoxI protein was strongly impaired in these cells (Δpim1/LON; Δpim1/SUP; Fig. 2A). CoxI did not accumulate at high levels in Δpim1/LON cells even when labeling was performed for longer time periods (Fig. 2B). CoxII and CoxIII, however, were synthesized in Δpim1/LON mitochondria, but degraded upon further incubation of the cells in pulse chase experiments (Fig. 2B). Defects in cytochrome c oxidase assembly in the presence of limited concentrations of CoxI presumably result in the proteolysis of nonassembled CoxII and CoxIII (McEwen et al. 1986). Notably, the analysis of cell extracts by Western blotting revealed the presence of CoxII in low amounts in Δpim1/LON but not in Δpim1/SUP cells (Fig. 2C). This finding is consistent with the pattern of growth on nonfermentable carbon sources at 30°C (see Fig. 1) and suggests the presence of low but functionally significant levels of CoxI in Δpim1/LON cells.
Interestingly, synthesis of Cob occurred at wild-type levels in Δpim1/LON cells, whereas it was defective in mitochondria lacking a Lon-like protease (Δpim1/SUP; Fig. 2A). Consistently, Cob protein was not detectable in Δpim1/SUP cells upon Western blotting but accumulated, although at reduced levels, in Δpim1/LON cells (Fig. 2C). The presence of a Lon-like protease with reduced enzymatic activity in Δpim1 mitochondria is apparently sufficient to maintain the expression of mitochondrially encoded Cob, but not the efficient synthesis of CoxI. The impaired assembly of the Cox complex in Δpim1/LON cells may indirectly cause slow degradation of newly synthesized Cob, thereby explaining the reduced amount of Cob in these cells. Similar observations have previously been reported for other respiratory chain subunits (Rep and Grivell 1996). Taken together, these results point to a requirement of PIM1 protease for the synthesis of CoxI and Cob and thereby explain the respiratory deficiency of Δpim1 cells containing mtDNA.
To establish the dependence of CoxI synthesis on the proteolytic activity of PIM1, a proteolytically inactive mutant form of the protease was employed (PIM1S1015A): Replacement of the conserved serine 1015 by alanine abolishes the proteolytic activity of PIM1 but does not affect the overall protein stability nor the ATP-dependent assembly of the homo-oligomeric protease (Rep et al. 1996b; Wagner et al. 1997). Wild-type and mutant protease were expressed in Δpim1/LON cells and mitochondrial protein synthesis was analyzed (Fig. 2D). Labeling of CoxI occurred in Δpim1/LON cells harboring active PIM1 protease, but CoxI was hardly detectable in Δpim1/LON mitochondria in the presence of proteolytically inactive PIM1. Thus, PIM1-mediated proteolysis is required for the synthesis of CoxI in mitochondria.
PIM1 protease is required for intron-containing pre-mRNA stability and for translation of COXI mRNA
The defective synthesis of CoxI and Cob in pim1 mutants could result from impaired transcription or translation, or might reflect deficiencies in the stability or processing of the corresponding transcripts. As both genes harbor introns (Costanzo and Fox 1990; Pel and Grivell 1993, 1995), pre-mRNA splicing defects must also be considered.
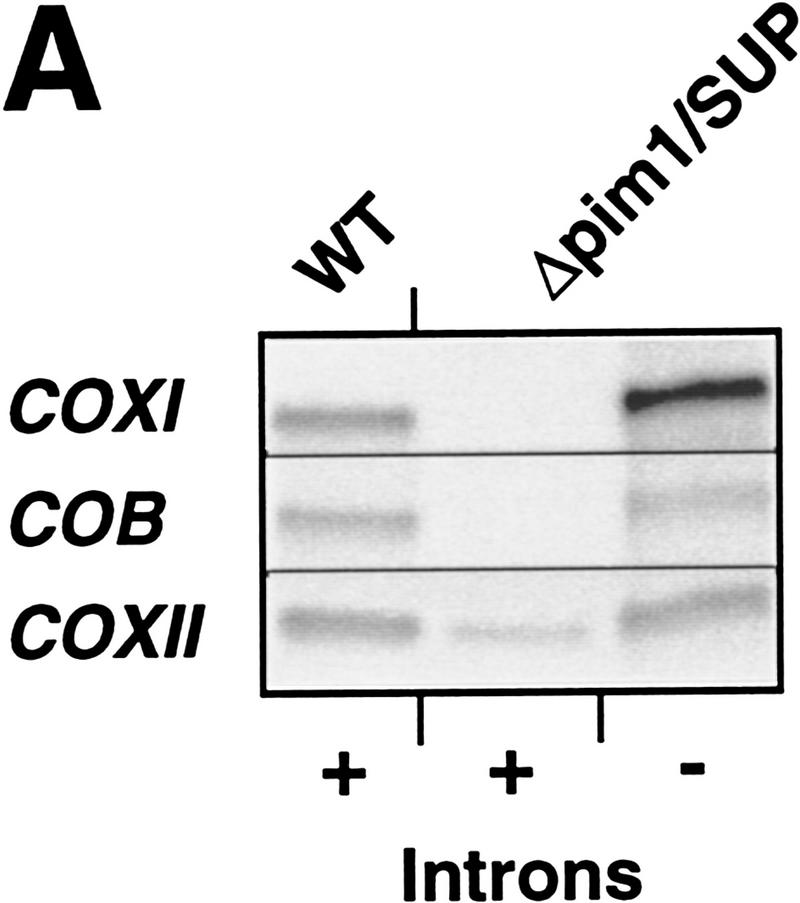
We investigated the possibility of the PIM1 function being related to the presence of introns in the COXI and COB gene. Δpim1/LON and Δpim1/SUP cells were converted to ρ0 mutants and strains devoid of mitochondrial introns were derived by cytoduction (Conde and Fink 1976; Berlin et al. 1991). This procedure allows the introduction of new mitochondrial information in a parent (cytoductant) that has conserved its nuclear genotype. Synthesis of CoxI occurred with similar efficiencies in wild-type and Δpim1/LON cells carrying intronless mtDNA (Fig. 3). Thus, in the presence of the E. coli Lon protease, removal of introns is sufficient to allow synthesis of Cob and CoxI. However, efficient synthesis of Cob but not CoxI was observed in cells carrying SUP but not LON (Fig. 3). These cells were respiratory deficient and did not grow on nonfermentable carbon sources (data not shown). Thus, efficient expression of a COXI gene lacking introns still depends on PIM1 or LON, suggesting defects in CoxI translation or mRNA stability in cells devoid of a Lon-like protease.
Figure 3.

Mitochondrial protein synthesis in Δpim1 cells carrying intronless mtDNA. Δpim1/LON and Δpim1/SUP cells devoid of mitochondrial introns were generated by cytoduction. Mitochondrial translation products were labeled with [35S]methionine for 10 min (WT; Δpim1/LON) or 30 min (Δpim1/SUP) at 30°C in vivo and analyzed by SDS-PAGE. The difference in the electrophoretic mobility of Var1 reflects gene polymorphism in the different mitochondrial genomes (Butow et al. 1985).
To distinguish between these possibilities, mitochondrial RNA (mtRNA) was isolated from wild-type and from Δpim1/SUP cells carrying intronless mtDNA and analyzed by Northern blot hybridization with probes specific for COXI and COB exons (Fig. 4A). With wild-type cells, the probes hybridized with transcripts of ∼2.1 and 2.2 kb, which correspond to mature COXI and COB mRNA, respectively (Fig. 4A). Similarly, mature-sized COXI and COB transcripts were detected in Δpim1/SUP cells harboring intronless mtDNA (Fig. 4A). COXI mRNA accumulated in significantly increased amounts in Δpim1/SUP cells devoid of mitochondrial introns when compared to wild-type cells (Fig. 4A). Nevertheless, CoxI protein was not synthesized in these cells (see Fig. 3) demonstrating the requirement of PIM1 protease for efficient translation of mature COXI transcripts.
Figure 4.
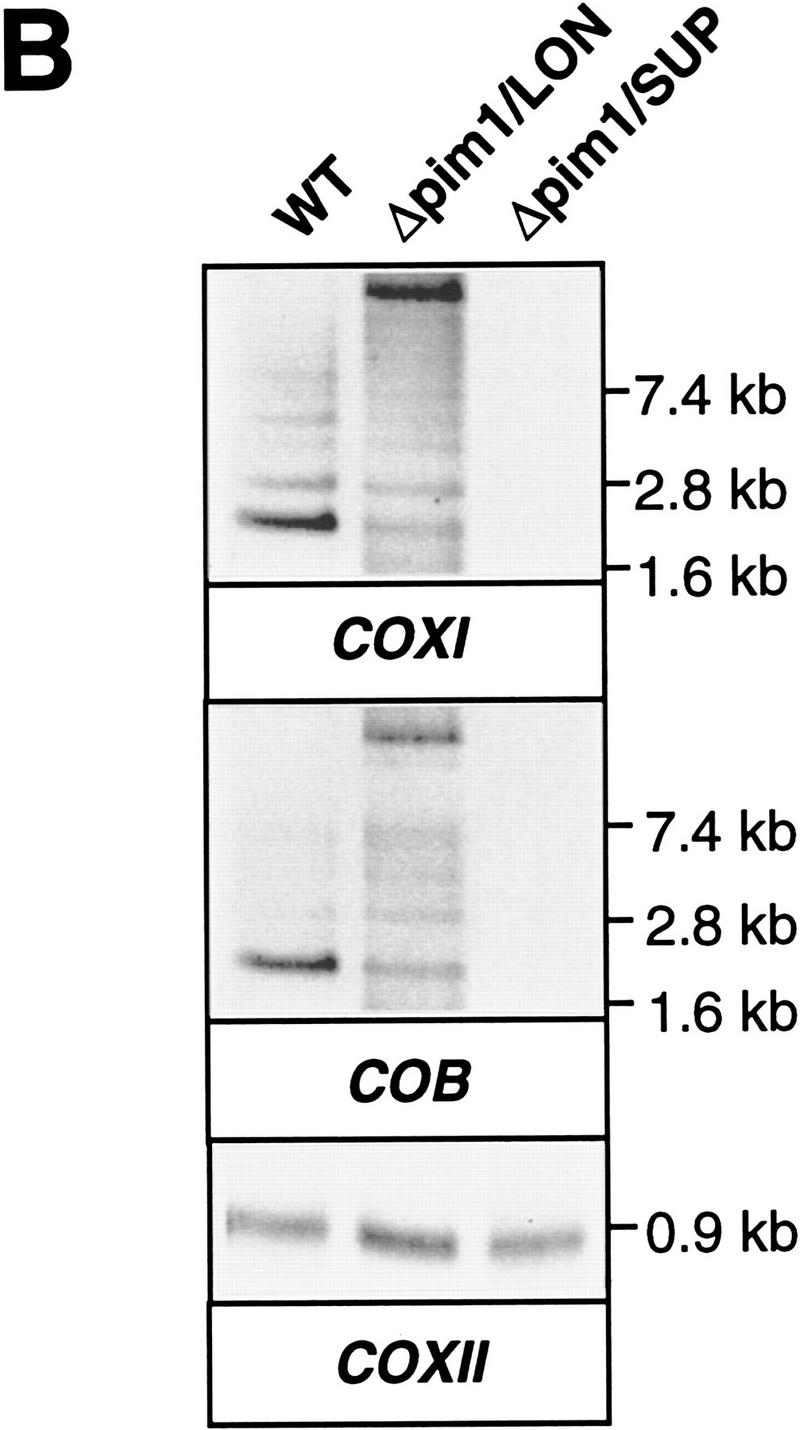
Northern blot analysis of COXI and COB transcripts in mitochondria lacking PIM1 protease. (A) mtRNA was isolated from wild-type (WT) and Δpim1/SUP cells, harboring an intron-containing or an intronless mitochondrial genome (±introns), and analyzed with COXI and COB-specific exon probes and a COXII probe for control as described in Materials and Methods. (B) mtRNA from wild-type (WT), Δpim1/LON, and Δpim1/SUP mitochondria with intron-containing mtDNA was analyzed as in A.
COXI and COB transcripts were not detected in Δpim1/SUP cells with an intron-containing mitochondrial genome (Fig. 4A), although transcription proceeded normally in these cells (see below). Apparently, PIM1 protease is required for the stability of COXI and COB transcripts harboring multiple introns. It is, however, conceivable that processing defects in the absence of PIM1 cause the degradation of COB and COXI transcripts, as previous studies revealed a correlation between mRNA processing defects and transcript degradation (Grivell 1995).
PIM1 protease affects COXI and COB pre-mRNA processing
To further analyze a possible role of PIM1 protease in pre-mRNA processing, we again took advantage of the observation that E. coli Lon protease with reduced enzymatic activity is sufficient to maintain the respiratory competence of pim1 null mutant cells at 30°C. RNA was isolated from wild-type and Δpim1/LON mitochondria and subjected to Northern blot analysis with probes specific for exons of COXI or COB (Fig. 4B). In contrast to Δpim1/SUP cells, COXI and COB transcripts accumulated in Δpim1/LON cells. When compared to wild-type cells, however, a significant decrease in the amounts of mature transcripts and an increase of larger precursor transcripts were detected with COXI- and COB-specific probes in Δpim1/LON cells (Fig. 4B). Apparently, the presence of a Lon-like protease with reduced enzymatic activity in mitochondria is sufficient to stabilize COXI and COB transcripts containing multiple introns, but does not allow efficient RNA processing to occur. These results suggest an involvement of PIM1 protease in splicing processes in mitochondria. Notably, despite the presence of reduced levels of mature COB transcripts, Cob synthesis was hardly affected in Δpim1/LON cells (see Fig. 2A).
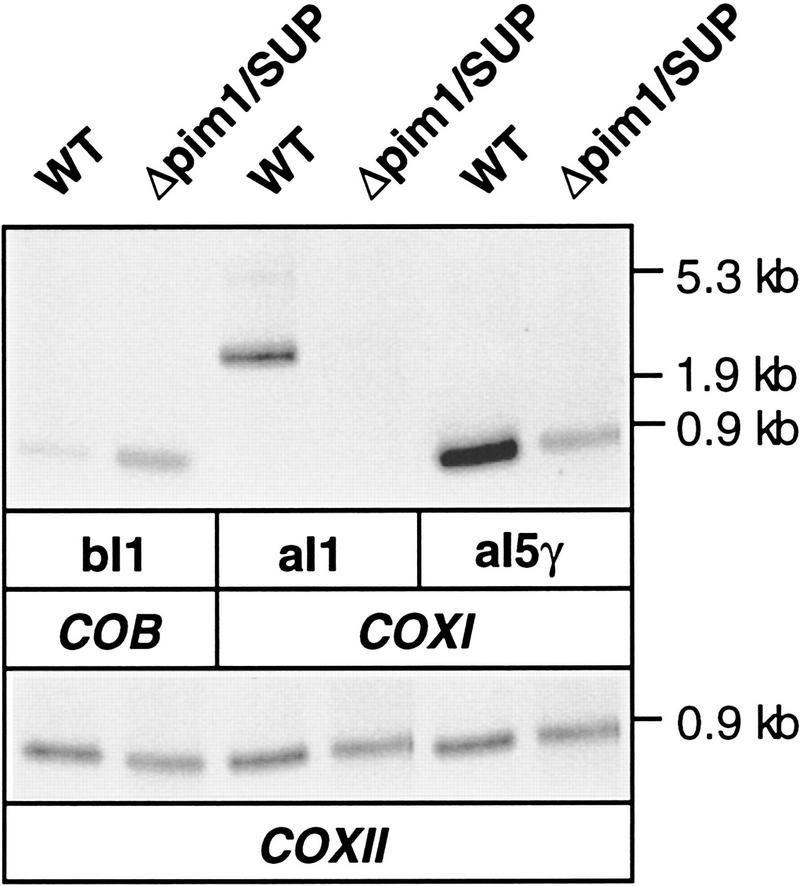
mtRNA of wild-type and Δpim1/SUP cells was analyzed with probes specific for introns of group II. In contrast to group I introns, these introns form stable lariat structures upon splicing and can therefore be detected by Northern blot hybridization (Costanzo and Fox 1990; Perlman 1990). Probes specific for the first intron of COB (bI1) or the last intron of COXI (aI5γ) hybridized to transcripts of ∼0.8 kb from wild-type and Δpim1/SUP mitochondria; these species correspond in size to the excised lariat forms (Fig. 5). Interestingly, excised intron bI1 accumulated at higher levels in Δpim1/SUP cells, most likely indicating an upregulation of transcription because of the impaired synthesis of Cob in these cells. These findings confirm transcription of COB and COXI in Δpim1/SUP cells and demonstrate that PIM1 protease is not required for the splicing of these group II introns.
Figure 5.
Characterization of COXI and COB pre-mRNA processing defects in pim1 mutants by Northern hybridization using intron-specific probes. mtRNA isolated from wild-type (WT) and Δpim1/SUP cells was analyzed with intron-specific COB and COXI probes. DNA probes specific for the group II introns bI1 of COB, aI1 and aI5γ of COXI and COXII, as a control, were employed.
Many mitochondrial introns contain an open reading frame that encodes an mRNA maturase fused in frame to the preceding exon (Costanzo and Fox 1990; Pel and Grivell 1993; Grivell 1995). Splicing of these introns is catalyzed by the intron-encoded maturase and thus depends on its synthesis. Analyzing mtRNA from Δpim/SUP cells, no excised lariat structure was detectable with a probe specific for intron 1 of COXI (aI1), a 2.4-kb group II intron that encodes an mRNA maturase (Fig. 5). The deficiency in the splicing of this maturase-encoding intron is in agreement with the observed requirement of PIM1 protease for CoxI translation. Notably, a defect in the splicing of intron aI1 did not impair the processing of the downstream intron aI5γ, which does not encode a maturase (Fig. 5). Indeed, cotranscriptional splicing has been demonstrated for introns that do not encode a maturase (Lewin et al. 1995). The excised intron aI5γ, however, accumulated at a reduced level in Δpim1/SUP mitochondria when compared to wild type, most likely caused by rapid degradation of nonprocessed COXI pre-mRNA.
Discussion
Cells lacking PIM1 protease lose the integrity of mtDNA and thereby their respiratory competence (Suzuki et al. 1994; van Dyck et al. 1994). This phenotype has prevented a detailed characterization of the role of PIM1 protease in mitochondrial biogenesis. In the present manuscript, we took advantage of the identification of a multicopy suppressor that stabilizes mtDNA in the absence of PIM1. The analysis of pim1 mutants containing mtDNA revealed deficiencies in the synthesis of mitochondrially encoded CoxI and Cob and thereby in the assembly of respiratory chain complexes. The defect in the synthesis of CoxI and Cob provides an explanation for the observed respiratory deficiency of pim1 mutant cells containing intact mtDNA. These results establish essential proteolytic functions of the ATP-dependent PIM1 protease in mitochondria that control the biogenesis of the respiratory chain (summarized in Fig. 6).
Figure 6.
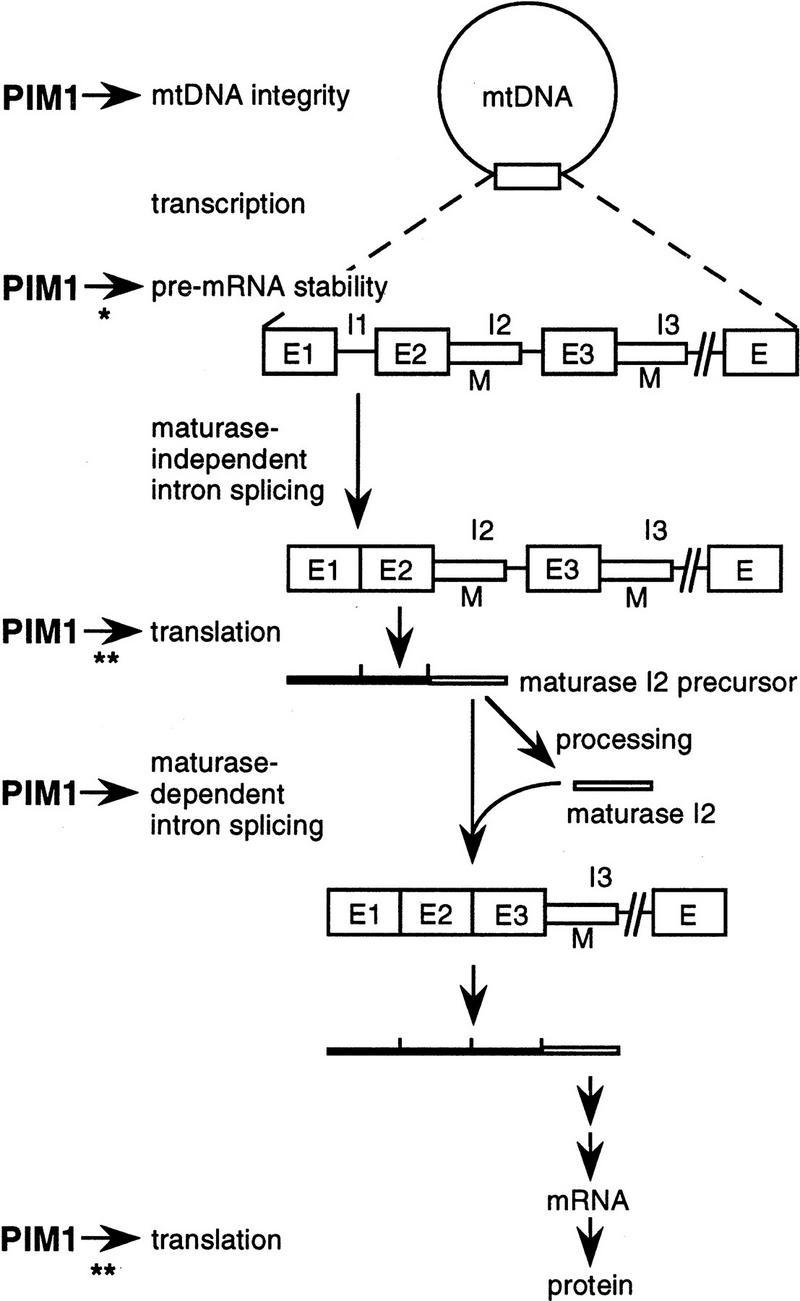
Roles of PIM1 protease in mitochondrial biogenesis. PIM1-mediated proteolysis is required for mtDNA integrity and the expression of the intron-containing COXI and COB genes in mitochondria (see text for details). (*) The instability of pre-RNA in the absence of PIM1 may be a secondary effect of pre-mRNA processing deficiencies. (**) PIM1 protease is only required for translation of COXI mRNA. (E, E1–3) exons; (I, I1–3) introns; (M) mRNA maturases.
PIM1 protease is required for the translation of mature COXI mRNA. Synthesis of CoxI was impaired in Δpim1 cells carrying intronless mtDNA, although mature mRNA accumulated in these cells. CoxI synthesis was not restored upon expression of a proteolytically inactive PIM1 mutant, demonstrating the requirement of PIM1-mediated proteolysis for CoxI translation. Membrane-bound translational activator proteins have been identified in mitochondria (Fox 1996; Rödel 1997). They physically interact with mitochondrial ribosomal subunits and the 5′ untranslated leader of their target and thereby allow the post-transcriptional control of gene expression (Haffter et al. 1991; Mulero and Fox 1993; Brown et al. 1994). Activator proteins are mRNA-specific and regulate the synthesis of mitochondrially encoded proteins in a gene-specific manner (Pel and Grivell 1994; Fox 1996). Similarly, PIM1 protease is required for the translation of COXI but not of other mitochondrially encoded proteins. Furthermore, PIM1 was found in association with the inner surface of the mitochondrial inner membrane after sonication of isolated mitochondria (L.van Dyck, I. Wagner, and T. Langer, unpubl.). It is conceivable that PIM1 exerts its function in CoxI synthesis by regulating the activity of other mitochondrial proteins. This might include the activation of proteins specifically involved in the translation of the COXI gene by PIM1-mediated processing. Alternatively, PIM1 protease might be required to degrade specific RNA-binding proteins that inhibit the translation of COXI transcripts.
Several introns of the COXI and COB genes contain an open reading frame that encodes an mRNA maturase fused in frame to the preceding exon (Costanzo and Fox 1990; Pel and Grivell 1993, 1995). Splicing of these introns is catalyzed by the intron-encoded maturase and thus depends on its translation. The deficiency in COXI pre-mRNA splicing in pim1 mutants can therefore be attributed to the impaired synthesis of intron-encoded mRNA maturases. Moreover, the failure to remove introns in the absence of PIM1 protease may result in the rapid degradation of COXI pre-mRNA transcripts, as a linkage between RNA processing and stability has been observed in mitochondria of various organisms (Grivell 1995). Thus, deficiencies in COXI pre-mRNA stability and splicing can be explained satisfactorily by the requirement of PIM1 protease for CoxI translation. The pleiotropic effect of pim1 mutants on the expression of the COXI gene is reminiscent of other proteins involved in mitochondrial gene expression (Groudinsky et al. 1993; Manthey and McEwen 1995). The product of the yeast nuclear gene PET309 is required for the translation of mature COXI and the stability of COXI pre-mRNA, as is PIM1 protease (Manthey and McEwen 1995). In contrast to pim1 cells, however, COB transcripts are not affected in pet309 mutants. Furthermore, SUV3, encoding a putative RNA helicase, is necessary for the stability of intron-containing COXI and COB transcripts, but not for the translation of mature COXI (Golik et al. 1995).
The analysis of COB gene expression in pim1 mutant cells, however, points to additional functions of PIM1 protease in mitochondrial gene expression. In contrast to COXI, translation of mature COB mRNA does not depend on the presence of PIM1 in mitochondria as demonstrated by the efficient synthesis of Cob in Δpim1/SUP cells carrying an intronless mitochondrial genome and in Δpim1/LON cells. Indirect effects on the splicing of COB transcripts because of impaired synthesis of intron-encoded mRNA maturases can therefore be excluded. Still, Northern blot analysis of Δpim1/LON cells harboring a Lon-like protease with reduced enzymatic activity revealed deficiencies in the processing of COB transcripts, indicating a role of PIM1 protease for the splicing of COB pre-mRNAs. Notably, the maturase encoded by the intron bI4 of the COB gene is required for the splicing of both intron bI4 itself and intron aI4 of the COXI gene (Dhawale et al. 1981; Banroques et al. 1987). Defects in the processing of COB pre-mRNAs result therefore in an impaired splicing of COXI transcripts.
How may PIM1 affect the splicing of mitochondrial transcripts? PIM1 protease could regulate the activity or stability of a protein directly involved in the splicing process. It is, for instance, an attractive possibility that PIM1 mediates the proteolytic processing of some mRNA maturases that are synthesized as fusion proteins with the peptide products of preceding exons (Costanzo and Fox 1990; Pel and Grivell 1993; Grivell 1995). Indeed, an energy-dependent step in the splicing of intron bI4 of the COB gene has been proposed (Muroff and Tzagoloff 1990). Further studies, however, are necessary to substantiate this hypothesis.
The stability of COXI and COB mRNAs containing multiple introns was impaired in Δpim1 cells lacking a Lon-like protease but containing mtDNA. Impaired splicing may result in the degradation of the intron-containing transcripts. Otherwise a direct role of PIM1 protease for pre-mRNA stability has to be envisioned. It should be noted in this context that expression of Lon protease did result in the stabilization of COXI and COB pre-mRNAs in Δpim1 cells, although splicing was impaired in these cells.
Although our results assign crucial functions to PIM1 for COXI and COB pre-mRNA stability and COXI translation, the protease is not required for transcription of these genes. The Northern blot analysis of Δpim1 cells harboring intact mtDNA with intron-specific probes revealed normal transcription of COXI and COB genes in these cells. Thus, the lack of transcripts in the absence of PIM1 is caused by degradation of COXI and COB pre-mRNAs. As most mitochondrially encoded genes, COXI and COB are initially transcribed into polycistronic RNAs followed by the processing of the primary transcript (Grivell 1989). The COXI gene is cotranscribed with the genes encoding ATP synthase subunits 6 and 8, synthesis of which occurred at wild-type levels in mitochondria lacking PIM1 protease. Similarly, the COB gene is transcribed into a precursor also containing tRNAGlu, which is essential for the synthesis of all mitochondrially encoded proteins. Thus, polycistronic precursor processing does not depend on the presence of PIM1 protease in mitochondria.
PIM1 protease has recently been proposed to serve as a chaperone in the assembly of respiratory complexes independent of its proteolytic activity (Rep et al. 1996b). Our results do not exclude chaperone-like properties of PIM1, however, they explain the respiratory deficiency of pim1 mutant cells by the lack of essential proteolytic functions of PIM1. PIM1-mediated proteolysis is required for mtDNA integrity (Wagner et al. 1997) and for the synthesis of respiratory chain subunits. Thus, impaired respiration in the absence of PIM1 protease is not caused by deficiencies in the assembly process per se or misfolded polypeptides accumulating within mitochondria, but reflects specific requirements of PIM1-mediated proteolysis for the biogenesis of the respiratory chain. The homologous Lon protease from E. coli can partially substitute for PIM1 in these processes, suggesting a conserved mode of action.
Materials and methods
Yeast strains and genetic analysis
Yeast strains used in this study are described in Table 1. The wild-type strain YPH500 contains long gene variants of COXI and COB. Cells were grown on YEP medium (1% yeast extract, 2% peptone) or on minimal medium (0.7% yeast nitrogen base containing ammonium sulfate) that was supplemented with the auxotrophic requirements and contained glucose (2%), galactose (2%), or glycerol (3%) as the sole carbon source.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
| YPH500 | MATα ura3-52 lys2-801amberade101ochretrp1-Δ63 his3-200 leu2-Δ1 (ρ+) | Sikorski and Hieter (1989) |
| Δpim1/LON* | MATα pim::HIS3 (ρ+) pVT100-U::ADH1–Su9(69)LonK362A: 2μ, URA3 | Teichmann et al. (1996) |
| Δpim1/LON/SUP* | MATα pim1::HIS3 (ρ+) pVT100-U::ADH1–Su9(69)LonK362A: 2μ, URA3, YEp13–SUP: 2μ, LEU2 | this study |
| Δpim1/SUP* | MATα pim1::HIS3 (ρ+) YEp13–SUP: 2μ LEU2 | this study |
| kar167-1 | MATa kar1-1 trp5 (ρ+ intronless) | Seraphin et al. (1987) |
Strains labeled with an asterisk are isogenic to YPH500.
The genetic analysis of yeast mutants was carried out according to published procedures (Sherman 1991). ρ0 derivative strains were prepared by ethidium bromide treatment (Fox et al. 1991). Cytoduction was performed essentially as described (Conde and Fink 1976; Berlin et al. 1991): ρ0 derivatives were transformed with plasmids expressing the suppressor gene and crossed with kar1 strains bearing the mitochondrial genome of interest. Cytoductants were selected both for their ability to grow on YEP glycerol and the presence of auxotrophic markers of the ρ0 parental strain. Mutant cytoductants were generated by disruption of the PIM1 gene using a pim1::HIS3 disruption cassette (Wagner et al. 1997).
Nucleic acid procedures
Standard DNA manipulations were carried out as previously described (Sambrook et al. 1989; Ausubel et al. 1992). Double-stranded DNA templates were sequenced using Sequenase (USB Corp.) according to the manufacturer’s guidelines. mtRNA was extracted essentially as described (Schmitt et al. 1990). Mitochondria were isolated according to published procedures (Herrmann et al. 1994; Zinser and Daum 1995) and lysed at a concentration of 10 mg/ml in 50 mm Tris-HCl (pH 7.4), 10 mm EDTA, 1% (wt/vol) SDS in the presence of proteinase K (100 μg/ml). After addition of NaCl to a concentration of 150 mm, mtRNA was phenol-extracted. Northern blotting of mtRNA (3 μg) was performed with Hybond-N nylon membrane (Amersham Corp.) using the protocol of the vendor. Probes were labeled with [α-32P]dATP using the Random Prime DNA labeling kit (Boehringer Mannheim). Hybridization was carried out for 15 hr at 42°C in 5× SSC, 0.5% (wt/vol) SDS, 40% formamide, 5× Denhardt’s reagent, and 20 mg/ml denatured salmon sperm DNA. Membranes were washed three times with 2× SSC, 0.5% (wt/vol) SDS, at room temperature for 5 min and twice in 1× SSC, 0.5% SDS, at 50°C for 30 min. The indicated sizes of the transcripts were estimated using the RNA molecular weight marker II (Boehringer Mannheim).
The following DNA fragments were used as probes for the Northern blot analysis: COB exon probe, pA12/Mb2 (Nobrega and Tzagoloff 1980); COXI exon probe, pCOX1/A4-I corresponding to a DNA fragment from COXI containing exon A4 and part of intron aI4 (kindly provided by A. Tzagoloff, Columbia University, New York, NY); COXII probe, PCR-amplified 689-bp internal DNA fragment of COXII. COB intron probes: bI1, pYJL12; bI2, pYJL5 (Lazowska et al. 1989); COXI intron probes: aI1, 766 bp HinDII–MboI fragment in pUC13; aI5γ, pYJL14, 533-bp TaqI–RsaI fragment in pUC13 kindly provided by J. Lazowska (CNRS, Gif-Sur-Yvette, France).
Isolation of the multicopy supressor gene
A YEp13 yeast genomic library was used to transform Δpim1/LON cells to leucine prototrophy (Broach et al. 1979). Ura3+ Leu2+ transformants were replica plated on YEP glycerol and incubated at 36°C for 7 days. YEp13–SUP was selected for its ability to rescue the thermosensitive growth defect of Δpim1/LON cells. Plasmid linkage of the suppression was confirmed by retransformation. The insert extremities of the rescuing plasmid YEp13–SUP were sequenced using primers YEP13a (5′-GCTTCGCTACTTGGAG-3′) and YEP13b (5′-ATCGGTGATGTCGGCG-3′). A search for homology using the BLAST program led to the identification of a 5.4-kb DNA fragment on the right arm of chromosome IV encoding SSS1, YDR087c, and SLU7. The suppressive effect of Sss1p was demonstrated by transforming the multicopy plasmid pTX64 harboring SSS1 (kindly provided by T. Sommer, Max-Delbrück-Center, Berlin, Germany) in the wild-type strain YPH500 and subsequent disruption of the PIM1 gene.
Labeling of mitochondrial translation products in vivo
Mitochondrial translation products were labeled in vivo essentially as described (Douglas et al. 1979; McKee and Poyton 1984; Langer et al. 1995). Cells were grown in minimal medium galactose lacking methionine. For each time point, cells (0.5 OD578 units) were harvested in midexponential phase by 15-sec centrifugation in a bench centrifuge, washed and resuspended in 500 μl of labeling buffer (40 mm K2HPO4 at pH 6, 2% galactose). Cells were incubated for 10 min at 30°C and cycloheximide was added to a final concentration of 150 μg/ml to inhibit the cytosolic protein synthesis. After a further incubation for 2 min, labeling of translation products with [35S]methionine (40 μCi; 1000 Ci/mmole) was performed for the times indicated and stopped by the addition of unlabeled methionine (10 mm). Cells were isolated by 15-sec centrifugation and washed with 10 mm methionine. Total cell proteins were extracted by alkaline lysis (Yaffe and Schatz 1984) and solubilized by shaking for 30 min at 4°C in LiDS sample buffer (2% lithium dodecylsulfate, 10% glycerol, 2.5% β-mercaptoethanol, 0.02% bromphenol blue, 60 mm Tris/Cl at pH 6.8). Proteins were separated by SDS-PAGE and visualized by autoradiography.
For pulse chase experiments, cells (3 OD578 units) were resuspended in labeling buffer (1.5 ml). After addition of cycloheximide, labeling was performed for 30 min at 30°C with [35S]methionine (100 μCi, 1000 Ci/mmole). After addition of methionine (10 mm), reisolation and washing, cells were resuspended in labeling medium (600 μl) containing methionine (10 mm) and further incubated at 30°C. At the time points indicated, aliquots of the cells were harvested and analyzed.
Acknowledgments
We thank J. Lazowska, T. Sommer, A. Tzagoloff, and C. Jacq for plasmids and yeast strains, and we are grateful to J. Lazowska for stimulating discussions. The excellent technical assistance of Gabi Ludwig, Petra Robisch, and Alexandra Weinzierl is gratefully acknowledged. L.v.D. was a recipient of a Senior Research Fellowship of the European Union (DGXII; biotechnology). The work was supported by grants from the Deutsche Forschungsgemeinschaft (La918/1-2; SFB184, B21) to T.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL: Langer@bio.med.uni-muenchen.de; FAX 49 89 5996 270.
References
- Adam Z. Protein stability and degradation in chloroplasts. Plant Mol Biol. 1996;32:773–783. doi: 10.1007/BF00020476. [DOI] [PubMed] [Google Scholar]
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12-complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- Ausubel FJ, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: Greene Publishing Associates and Wiley-Interscience; 1992. [Google Scholar]
- Banroques J, Perea J, Jacq C. Efficient splicing of two yeast mitochondrial introns controlled by a nuclear-encoded maturase. EMBO J. 1987;6:1085–1091. doi: 10.1002/j.1460-2075.1987.tb04862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Lupas A. The proteasome. Curr Opin Struct Biol. 1997;7:273–282. doi: 10.1016/s0959-440x(97)80036-x. [DOI] [PubMed] [Google Scholar]
- Berlin V, Brill JA, Trueheart J, Boeke JD, Fink GR. Genetic screens and selections for cell and nuclear fusion mutants. Methods Enzymol. 1991;194:774–792. doi: 10.1016/0076-6879(91)94058-k. [DOI] [PubMed] [Google Scholar]
- Broach JR, Strathern JN, Hicks JB. Transformation in yeast: Development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979;8:121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Brown NG, Costanzo MC, Fox TD. Interactions among three proteins that specifically activate translation of the mitochondrial COX3 mRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:1045–1053. doi: 10.1128/mcb.14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow RA, Perlman PS, Grossman LI. The unusual var1 gene of yeast mitochondrial DNA. Science. 1985;228:1496–1501. doi: 10.1126/science.2990030. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Tanaka N, White KH, Thorsness PE. Mitochondrial morphological and functional defects in yeast caused by yme1 are suppressed by mutation of a 26S protease subunit homologue. Mol Biol Cell. 1994;5:899–905. doi: 10.1091/mbc.5.8.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde J, Fink GR. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confalonieri F, Duguet M. A 200-amino acid ATPase module in search of a basic function. BioEssays. 1995;17:639–650. doi: 10.1002/bies.950170710. [DOI] [PubMed] [Google Scholar]
- Costanzo MC, Fox TD. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Dhawahle S, Hanson DK, Alexander NJ, Perlman PS, Mahler HR. Regulatory interactions between mitochondrial genes: Interactions between two mosaic genes. Proc Natl Acad Sci. 1981;78:1778–1782. doi: 10.1073/pnas.78.3.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas M, Finkelstein D, Butow RA. Analysis of products of mitochondrial protein synthesis in yeast: Genetic and biochemical aspects. Methods Enzymol. 1979;56:58–66. doi: 10.1016/0076-6879(79)56009-1. [DOI] [PubMed] [Google Scholar]
- Esnault Y, Blondel MO, Deshaies RJ, Schekman R, Kepes F. The yeast SSS1 gene is essential for secretory protein translocation and encodes a conserved protein of the endoplasmic reticulum. EMBO J. 1993;12:4083–4093. doi: 10.1002/j.1460-2075.1993.tb06092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault Y, Feldheim D, Blondel MO, Schekman R, Képès F. SSS1 encodes a stabilizing component of the Sec61 subcomplex of the yeast protein translocation apparatus. J Biol Chem. 1994;269:27478–27485. [PubMed] [Google Scholar]
- Finke K, Plath K, Panzner S, Prehn S. A second trimeric complex containing homologs of the Sec61p complex functions in protein transprot across the ER membrane of S. cerevisiae. EMBO J. 1996;15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- Fox TD. Translational control of endogenous and recoded nuclear genes in yeast mitochondria: Regulation and membrane targeting. Experientia. 1996;52:1130–1135. doi: 10.1007/BF01952112. [DOI] [PubMed] [Google Scholar]
- Fox TD, Folley LS, Mulero JJ, McMullin TW, Thorsness PE, Hedin LO, Costanzo MC. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- Frank D, Guthrie C. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes & Dev. 1992;6:2112–2124. doi: 10.1101/gad.6.11.2112. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur J Biochem. 1992;203:9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- Golik P, Szczepanek T, Bartnik E, Stepien PP, Lazowska J. The S. cerevisiae nuclear gene SUV3 encoding a putative RNA helicase is necessary for the stability of mitochondrial transcripts containing multiple introns. Curr Genet. 1995;28:217–224. doi: 10.1007/BF00309780. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Maurizi MR. Regulation by proteolysis: Energy-dependent proteases and their targets. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivell LA. Nucleo-mitochondrial interactions in yeast mitochondrial biogenesis. Eur J Biochem. 1989;182:477–493. doi: 10.1111/j.1432-1033.1989.tb14854.x. [DOI] [PubMed] [Google Scholar]
- ————— Nucleo-mitochondrial interactions in mitochondrial gene expression. Crit Rev Biochem Mol Biol. 1995;30:121–164. doi: 10.3109/10409239509085141. [DOI] [PubMed] [Google Scholar]
- Grivell LA, Schweyen RJ. RNA splicing in yeast mitochondria: Taking out the twists. Trends Genet. 1989;5:39–41. doi: 10.1016/0168-9525(89)90018-8. [DOI] [PubMed] [Google Scholar]
- Groudinsky O, Bousquet I, Wallis MG, Slonimski PP, Dujardin G. The NAM1/MTF2 nuclear gene product is selectively required for the stability and/or processing of mitochondrial transcripts of the atp6 and of the mosaic, cox1 and cytb genes in Saccharomyces cerevisiae. Mol & Gen Genet. 1993;240:419–427. doi: 10.1007/BF00280396. [DOI] [PubMed] [Google Scholar]
- Guélin E, Rep M, Grivell LA. Sequence of the AFG3 gene encoding a new member of the FtsH/Yme1/Tma subfamily of the AAA-protein family. Yeast. 1994;10:1389–1394. doi: 10.1002/yea.320101016. [DOI] [PubMed] [Google Scholar]
- Guélin E, Rep M, Grivell LA. Afg3p, a mitochondrial ATP-dependent metalloprotease, is involved in the degradation of mitochondrially-encoded Cox1, Cox3, Cob, Su6, Su8 and Su9 subunits of the inner membrane complexes III, IV and V. FEBS Lett. 1996;381:42–46. doi: 10.1016/0014-5793(96)00074-9. [DOI] [PubMed] [Google Scholar]
- Haffter P, McMullin TW, Fox TD. Functional interactions among two yeast mitochondrial ribosomal proteins and an mRNA-specific translational activator. Genetics. 1991;127:319–326. doi: 10.1093/genetics/127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Fölsch H, Neupert W, Stuart RA. Isolation of yeast mitochondria and study of mitochondrial protein translation. In: Celis DE, editor. Cell biology: A laboratory handbook. San Diego, CA: Academic Press; 1994. pp. 538–544. [Google Scholar]
- Hilt W, Wolf D. Proteasomes: Destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- Kunau WH, Beyer A, Franken T, Gotte K, Marzioch M, Saidowsky J, Skaletz-Rorowski A, Wiebel FF. Two complementary approaches to study peroxisome biogenesis in Saccharomyces cerevisiae: Forward and reversed genetics. Biochimie. 1993;75:209–224. doi: 10.1016/0300-9084(93)90079-8. [DOI] [PubMed] [Google Scholar]
- Langer T, Neupert W. Regulated protein degradation in mitochondria. Experientia. 1996;52:1069–1076. doi: 10.1007/BF01952104. [DOI] [PubMed] [Google Scholar]
- Langer T, Pajic A, Wagner I, Neupert W. Proteolytic breakdown of membrane-associated polypeptides in mitochondria of Saccharomyces cerevisiae. Methods Enzymol. 1995;260:495–503. doi: 10.1016/0076-6879(95)60161-9. [DOI] [PubMed] [Google Scholar]
- Lazowska J, Claisse M, Gargouri A, Kotylak Z, Spyridakis A, Slonimski PP. Protein encoded by the third intron of cytochrome b gene in Saccharomyces cerevisiae is an mRNA maturase. Analysis of mitochondrial mutants, RNA transcripts, proteins, and evolutionary relationships. J Mol Biol. 1989;205:275–289. doi: 10.1016/0022-2836(89)90341-0. [DOI] [PubMed] [Google Scholar]
- Leonhard K, Herrmann JM, Stuart RA, Mannhaupt G, Neupert W, Langer T. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 1996;15:4218–4229. [PMC free article] [PubMed] [Google Scholar]
- Lewin AS, Thomas Jr J, Tirupati HK. Cotranscriptional splicing of a group I intron is facilitated by the Cbp2 protein. Mol Cell Biol. 1995;15:6971–6978. doi: 10.1128/mcb.15.12.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey GM, McEwen JE. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizi MR. Proteases and protein degradation in Escherichia coli. Experientia. 1992;48:178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- McEwen JE, Ko C, Kloeckner-Gruissem B, Poyton RO. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae. Characterization of mutants of 34 complementation groups. J Biol Chem. 1986;261:11872–11879. [PubMed] [Google Scholar]
- McKee EE, Poyton P. Mitochondrial gene expression in Saccharomyces cerevisiae. Optimal conditions for protein synthesis in isolated mitochondria. J Biol Chem. 1984;259:9320–9331. [PubMed] [Google Scholar]
- Mulero JJ, Fox TD. PET111 acts in the 5′-leader of Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics. 1993;133:509–516. doi: 10.1093/genetics/133.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroff I, Tzagoloff A. CBP7 codes for a co-factor required in conjunction with a mitochondrial maturase for splicing of its cognate intervening sequence. EMBO J. 1990;9:2765–2773. doi: 10.1002/j.1460-2075.1990.tb07464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega FG, Tzagoloff A. Assembly of the mitochondrial membrane system. DNA sequence and organization of the cytochrome b gene in Saccharomyces cerevisiae. J Biol Chem. 1980;255:9828–9837. [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport T. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Pel HJ, Grivell LA. The biology of yeast mitochondrial introns. Mol Biol Rep. 1993;18:1–13. doi: 10.1007/BF01006890. [DOI] [PubMed] [Google Scholar]
- ————— Protein synthesis in mitochondria. Mol Biol Rep. 1994;19:183–194. doi: 10.1007/BF00986960. [DOI] [PubMed] [Google Scholar]
- Perlman PS. Genetic analysis of RNA splicing in yeast mitochondria. Methods Enzymol. 1990;181:539–558. doi: 10.1016/0076-6879(90)81150-s. [DOI] [PubMed] [Google Scholar]
- Rep M, Grivell LA. The role of protein degradation in mitochondrial function and biogenesis. Curr Genet. 1996;30:367–380. doi: 10.1007/s002940050145. [DOI] [PubMed] [Google Scholar]
- Rep M, Nooy J, Guélin E, Grivell LA. Three genes for mitochondrial proteins suppress null-mutations in both AFG3 and RCA1 when overexpressed. Curr Genet. 1996a;30:206–211. doi: 10.1007/s002940050122. [DOI] [PubMed] [Google Scholar]
- Rep M, van Dijl M, Suda K, Schatz G, Grivell LA, Suzuki CK. Promotion of mitochondrial membrane complex assembly by a proteolytically inactive yeast Lon. Science. 1996b;274:103–106. doi: 10.1126/science.274.5284.103. [DOI] [PubMed] [Google Scholar]
- Rödel G. Translational activator proteins required for cytochrome b synthesis in Saccharomyces cerevisiae. Curr Genet. 1997;31:375–379. doi: 10.1007/s002940050219. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B, Boulet A, Simon M, Faye G. Construction of a yeast strain devoid of mitochondrial introns and its use to screen nuclear genes involved in mitochondrial splicing. Proc Natl Acad Sci. 1987;84:6810–6814. doi: 10.1073/pnas.84.19.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling CJ, Hewitt EW. The S. cerevisiae SEC65 gene encodes a component of yeast signal recognition particle with homology to human SRP19. Nature. 1992;356:534–537. doi: 10.1038/356534a0. [DOI] [PubMed] [Google Scholar]
- Suzuki CK, Suda K, Wang N, Schatz G. Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science. 1994;264:273–276. doi: 10.1126/science.8146662. [DOI] [PubMed] [Google Scholar]
- Suzuki CK, Rep M, van Dijl JM, Suda K, Grivell LA, Schatz G. ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem Sci. 1997;22:118–123. doi: 10.1016/s0968-0004(97)01020-7. [DOI] [PubMed] [Google Scholar]
- Tauer R, Mannhaupt G, Schnall R, Pajic A, Langer T, Feldmann H. Yta10p, a member of a novel ATPase family in yeast, is essential for mitochondrial function. FEBS Lett. 1994;353:197–200. doi: 10.1016/0014-5793(94)01045-5. [DOI] [PubMed] [Google Scholar]
- Teichmann U, van Dyck L, Guiard B, Fischer H, Glockshuber R, Neupert W, Langer T. Substitution of PIM1 protease in mitochondria by Escherichia coli Lon protease. J Biol Chem. 1996;271:10137–10142. doi: 10.1074/jbc.271.17.10137. [DOI] [PubMed] [Google Scholar]
- Thorsness PE, White KH, Fox TD. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5418–5426. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Myers AM. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A, Yue J, Jang J, Paul MF. A new member of a family of ATPases is essential for assembly of mitochondrial respiratory chain and ATP synthetase complexes in Saccharomyces cerevisiae. J Biol Chem. 1994;269:26144–26151. [PubMed] [Google Scholar]
- van Dyck L, Pearce DA, Sherman F. PIM1 encodes a mitochondrial ATP-dependent protease that is required for mitochondrial function in the yeast Saccharomyces cerevisiae. J Biol Chem. 1994;269:238–242. [PubMed] [Google Scholar]
- Wagner I, Arlt H, van Dyck L, Langer T, Neupert W. Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO J. 1994;13:5135–5145. doi: 10.1002/j.1460-2075.1994.tb06843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner I, van Dyck L, Savel’ev A, Neupert W, Langer T. Autocatalytic processing of the ATP-dependent PIM1 protease: Crucial function of a pro-region for sorting to mitochondria. EMBO J. 1997;16:7317–7325. doi: 10.1093/emboj/16.24.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]