Summary
The methylation of cytosines within cytosine–guanine (CG) dinucleotides is an epigenetic mark that can modify gene transcription. With the advent of high-throughput sequencing, it is possible to map methylomes, i.e. detect methylated CGs on a genome-wide scale. The methylomes sequenced to date reveal a divergence in prevalence and targeting of CG methylation between taxa, despite the conservation of the DNA methyltransferase enzymes that cause DNA methylation. Therefore, interspecific methylation usage is predicted to diverge. In various taxa, this tenet gains support from patterns of CG depletion that can be traced in DNA before methylomes are explicitly mapped. Depletion of CGs in methylated genomic regions is expected because methylated cytosines are subject to increased mutability caused by nucleotide deamination. However, the basis of diverging interspecific methylation usage is less clear. We use insights from the methylome of honeybees (Apis mellifera) to emphasize the possible importance of organismal life histories in explaining methylation usage and the accuracy of methylation prediction based on CG depletion. Interestingly, methylated genes in honeybees are more conserved across taxa than non-methylated genes despite the divergence in utilization of methylation and the increased mutability caused by deamination.
Key words: CG methylation, DNA sequencing, genome analysis, honeybee genome, epigenetics
Introduction
The advent of deep sequencing technology is responsible for most of thetab ∼1300 prokaryotic genome and ∼900 eukaryotic genome sequencing projects currently listed in the NCBI BioProject database (http://www.ncbi.nlm.nih.gov/bioproject). In addition, deep sequencing can assay layers of molecular regulation that act on genes, thereby providing information on gene regulatory networks with high sensitivity on a genome-wide scale. Examples of such assays include measuring the abundance of DNA binding proteins (chromatin immunoprecipitation sequencing), RNAs (whole transcriptome sequencing) and the intensity of DNA methylation (bisulfite sequencing), all at base-pair resolution (Lister et al., 2008; Park, 2009; Wang et al., 2009). Cross-taxa comparisons of these layers of control can provide evidence of evolutionary divergence as well as conservation in gene regulation, and allow us to understand the evolution of gene sequences in the context of how they are regulated.
Here, we focus on the mechanism of DNA methylation. We review how different taxa use methylation marks as regulatory devices and discuss the genomic signatures that these marks leave in DNA. We underline how an organism's life history can open the door to understanding variation in the DNA methylation layer of control, using the recently available, comprehensive, whole-genome methylation data for the honeybee Apis mellifera as a resource (Lyko et al., 2010). The life history of A. mellifera has been studied since Aristotle (3000 BC). The honeybee is now an economically important pollinator and a research tool in sociobiology, behavioral biology and neuroscience. The honeybee research community is a model for developing coordinated genomic resources (The Honeybee Genome Sequencing Consortium, 2006). This insect can provide insights into DNA methylation that are of broad interest and relevance to basic and applied science.
DNA methylation: mechanism and utilization
DNA methyltransferases (DNMTs) are a family of enzymes capable of methylating cytosines within a CHH nucleotide context (where H is an A, C, T or G nucleotide) (Law and Jacobsen, 2010). However, methylation is most prevalent in cytosine–phosphate–guanine (CpG) dinucleotides in plants and mammals (Lister et al., 2008; Lister et al., 2009; Law and Jacobsen, 2010). The DNMT enzymes have different functions: DNMT1 copies methylation onto the daughter strand during cell replication, ensuring that DNA methylation is transmitted across cell generations and inherited in offspring through imprinted germlines; DNMT2 methylates tRNA; and DNMT3 is the de novo DNA methyltransferase that methylates new CpGs in response to maturational, physiological, behavioral and environmental influences or trauma that changes the nuclear milieu of cells (Law and Jacobsen, 2010). Not all species contain a full complement of DNMTs (examples in Table 1), but those that do have homologous methyltransferases.
Table 1.
Species-specific methylome attributes
DNMTs have the potential to influence gene expression, and thus cell phenotypes, by methylating DNA nearby or within genes. DNA methylation is a stable epigenetic mark that acts in concert with methyl binding proteins and other histone modifiers to alter the local chromatin state and change the accessibility of the DNA to RNA polymerase II for transcription (Klose and Bird, 2006; Bogdanovic and Veenstra, 2009). Thereby, DNA methylation can influence or dictate the phenotype of a cell via stable modifications in transcription rates.
Phenotypic differences that arise in conjunction with changes in DNA methylation are evident between cells, e.g. in human embryonic stem cells and fibroblasts that can be differentiated by intensities of DNA methylation in regions of imprinting (Lister et al., 2009). At the level of entire organisms, functional utilization of DNA methylation is evident in species such as Apis mellifera, where females differentiate into castes of reproductive queens or essentially sterile workers. This bifurcation is socially induced through controlled feeding of larvae, and de novo DNA methylation is used to internalize a restricted diet as a step in the process of worker development (Kucharski et al., 2008). In addition to differential methylation between phenotypes at the level of cell type or the entire organism, variable methylation has been measured across biological samples of the same phenotype. In humans for example, differentially methylated regions are observed within the same tissue in different individuals (Feinberg and Irizarry, 2010). The ability to imprint variable DNA methylation within cells of the same type is likely an adaptive trait, and has been used to explain synaptic plasticity in memory and stress-induced behavior in vertebrates (LaPlant et al., 2010; Miller and Sweatt, 2007; Miller et al., 2010). Not all invertebrates may similarly depend on DNA methylation for brain function and behavior because several species lack a complete and functional DNA methylation system. However, a dynamic use of DNA methylation in brain tissue is supported by recent data from honeybees (Lockett et al., 2010). Thus, generally speaking, the genome-wide transcriptional regulation that can be achieved with DNA methylation is functionally manifested in various animal cells, tissues, individuals, castes and behaviors.
Genomic DNA signatures of CpG depletion
DNA methylation is unique from other epigenetic marks, such as the modification of histone tails, because its heritability results in CpG depletion. In humans, a C to T mutation at methylated cytosines (mCs) occurs at a rate 10- to 50-fold higher than any other mutation in part because mCs are subject to spontaneous deamination (Duncan and Miller, 1980; Bulmer, 1986; Britten et al., 1988; Sved and Bird, 1990). Deamination turns the mC to T, eliminating the CpG dinucleotide following DNA mismatch repair (Duncan and Miller, 1980). Thus, CpG depletion occurs in genomic regions that are targeted for consistent methylation over several consecutive generations when there is deamination in the germline.
Genomic regions that have signatures of CpG depletion are presumed to have been methylated over evolutionary time. Such signatures can be generally informative about the prevalence and targeting of DNA methylation in a genome prior to the determination of the methylome. For instance, approximately half of all honeybee genes have less CpGs than expected, indicating that only half of the genes are targeted for DNA methylation (Elango et al., 2009). The ability to predict methylated DNA regions in non-germline cells by measuring CpG depletion from the genome is limited because: (1) mutations caused by DNA methylation can only be passed through the germline and (2) DNA methylation changes with cell differentiation.
Comparing CpG depletion within the genomes of various species reveals the evolutionary divergence of DNA methylation targeting systems. In other words, the genome-wide patterns of CpG depletion reveal the methylation marks that were laid down in the past, and from these interspecific signatures we can infer how functional regions (e.g. genes, exons, introns, promoters, transposons, repeats and intergenic regions) have been differentially targeted. Currently, there is no complete theory that connects the prevalence of DNA methylation in the genome to signatures of CpG depletion. In the following, we argue that a complete formulation of such a theory must incorporate the biological usage of DNA methylation, instead of focusing solely on the genome.
Methylomes
Deep sequencing technology can be modified to detect DNA methylation at base-pair resolution. Deep sequencing generates billions of short (typically between 35 and 200 nucleotides long) sequences, called ‘reads’, from a given input sample. These reads are then used to construct an entire genome from scratch or to detect differences between the input sample and a preassembled genome by allowing mismatches during the realignment of the reads to the genome. The process of using sequencing to determine mCs involves the experimental conversion of C, but not mC, to U by treating the DNA with sodium bisulfite, then U to T during the sequencing process. In this way, the number of contexts (i.e. sequencing reads) in which a specific C is found to be methylated or unmethylated is tallied to create a genome-wide methylation intensity profile: the methylome (Lister et al., 2008). Thus far, the methylomes of 19 organisms have been sequenced, elucidating the divergence of DNA methylation targeting in a wide range of taxa (Lister et al., 2008; Lister et al., 2009; Laurent et al., 2010; Lyko et al., 2010; Xiang et al., 2010; Zemach et al., 2010).
The honeybee methylome
Recently, bisulfite sequencing was used to delineate the methylome of the brains of honeybee queens and workers (Lyko et al., 2010). The samples used to generate the methylome data were pooled from 50 individuals per caste, where the queens and workers were first age-matched as 10-day-olds and later as 2.5-week-olds. This methylome analysis revealed that >75% of methylated CpGs in the honeybee are localized to exons, and that bees have negligible DNA methylation outside of CpG dinucleotides (Lyko et al., 2010). Only a small fraction of data was classified as intron or promoter methylation.
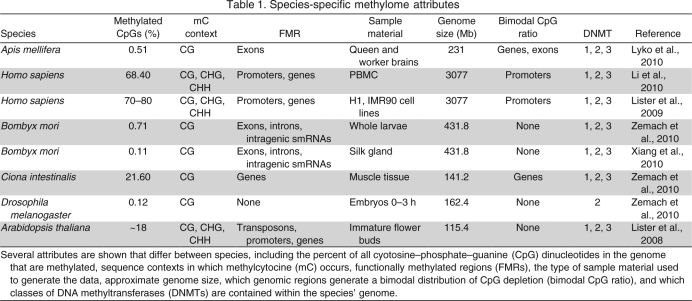
The exons that are targeted for methylation in honeybees lie in approximately half of the annotated genes of the genome. The remaining half of the unmethylated genes has methylation intensity below the background rate of adjacent intergenic regions, indicating that their lack of methylation is actively maintained (Fig. 1, top panel). Unmethylated exons also have methylation intensities below the adjacent introns that are not targeted for methylation (Fig. 1, bottom panel). We can conclude that the methylation targeting system in the honeybee is specific, to the extent that it recognizes intron–exon boundaries and that non-targeted genes and exons are not spuriously methylated.
Fig. 1.
The level of absolute methylation targeted to genes and exons in honeybee queens and workers. (Top) Absolute methylation (total intensity of CpG methylation divided by sequence length) was calculated by dividing the region ±1 kbp of all genes into 20 equal intervals. This calculation is similar for the percentage of gene length. (Bottom) Similar methods were used to calculate absolute methylation over the length of all exons and the regions ±1 kbp of all exons. There is a clear recognition of intron–exon boundaries and unmethylated exons have methylation intensity below the background methylation in adjacent introns. Methylation data were obtained from bisulfite sequencing (Lyko et al., 2010).
Human versus honeybee methylomes
Although honeybee introns are sparsely methylated compared with exon regions, the methylation intensities of the human genome increase in introns that are adjacent to methylated exons (Lister et al., 2009). This difference is reflected in an opposing pattern of exon–intron CpG depletion that we describe in more detail below (see The normalized CpG ratio and methylation targeting). When further contrasting patterns of methylation in the two genomes, an obvious difference is that >70% of CpGs are targeted for methylation in humans compared with <3% in honeybees. With only a small fraction of methylation detected in the promoter regions of genes, the major functional role of methylation in honeybees may be the regulation of splice variant diversity rather than to silence gene transcription (Lyko et al., 2010). In contrast, promoter methylation is a widely used mechanism for transcriptional regulation in humans (Saxonov et al., 2006). Humans, moreover, methylate in the CA dinucleotide context and this type of non-CpG methylation may help to maintain the pluripotency of the stem cells, as it is not observed in differentiated cells (Lister et al., 2009). Similar interspecific differences are apparent from the methylome data summarized in Table 1.
Broader comparative aspects
These differences (Table 1) point to a divergence in the functional utilization of DNA methylation between species. A conserved role for gene body methylation has been proposed that associates DNA methylation with regulation of gene expression. Yet, the current available overlays of methylomes and transcriptomes do not resolve whether increased methylation causes a consistent pattern of upregulation or downregulation of genes. In rice, sea squirts, silkworms, honeybees, anemones and puffer fish, a parabolic relationship with transcription is observed, i.e. intermediately transcribed genes are more likely to be methylated than low or highly expressed genes (Zemach et al., 2010). The anemone and silkworm are the only organisms reported, thus far, to show a direct positive correlation between gene body methylation and gene transcription (Xiang et al., 2010; Zemach et al., 2010). In the honeybee, the highest decile of expressed genes is the least methylated, whereas all other deciles have approximately the same intermediate level of gene body methylation (Zemach et al., 2010).
It may be difficult to detect a direct correlation between DNA methylation and transcript abundance in honeybees, and in other species, if methylation is targeted to a specific fraction of the entire gene while expression is measured at a different segment or over the whole gene body. An example of the former is the genome-wide oligonucleotide microarray for honeybees that, currently, contains an average of two probes per gene. Whenever DNA methylation influences exons that are not printed on the array, associations pass unnoticed. As exon methylation in honeybees may control the abundance of splice variants by directly increasing the rate at which the methylated exon is included in transcription, a stronger genome-wide correlation may be discerned in future work by using the intragenic regions targeted for methylation, i.e. by comparing exon expression with exon methylation. Similar scenarios may arise in other species.
Methylation and organismal life history
Biological scenarios may help explain distinct implementations of transcriptional regulation by DNA methylation between taxa. In the honeybee, eusociality provides a reasonable justification for a low prevalence of DNA methylation. Dependence on a multi-caste social system necessitates phenotypic accommodation in one caste for beneficial phenotypic innovations in another. In honeybees, the whole-body amount of DNA methylation in queens is lower than in workers and inhibition of de novo methylation during the development of larvae-fed workers results in queen-like individuals (Kucharski et al., 2008). During the adult stage, a structure of lower genome-wide methylation in queens may accommodate a wider use of dynamic DNA methylation in worker tissues, including brain and fat body (functionally homologous to white adipose tissue), that are central to worker behavioral expression and regulation (Amdam, 2011). Honeybee worker phenotypes are plastic and diverse, but correlations between specific suites of physiological and behavioral expression are usually fitted into a predictable and temporal work schedule that may be governed, in part, by the use of DNA methylation.
We speculate that this governing of temporal worker phenotypes can result in a higher degree of inter-individual variability in DNA methylation between worker tissues than between queen tissues. It remains to be tested whether the resulting heterogeneity of variances can preclude detection of differential methylation between castes. However, it is encouraging that other authors have found considerable differences between the brains of mated 2.5-week-old queens and 8-day-old workers (Lyko et al., 2010). Future studies on honeybees can also take questions of variability and heterogeneity more directly into account by using individual rather than pooled samples in analyses of DNA methylation. These results can address questions of general interest, such as whether or when inter-individual variability in DNA methylation compresses or expands during processes of development, maturation, behavioral change and aging.
In addition to accommodating caste differences in adult brain function and behavioral plasticity, variable DNA methylation could also result in adaptive phenotypic variations between newly emerged (‘newborn’) worker bees. Such early phenotypic heterogeneity within the worker caste is proposed to be fundamental to the initial seeding division of labor, a hallmark trait of insect sociality (Oldroyd and Fewell, 2007; Waibel et al., 2006). Methylation-mediated heterogeneity could arise during early development, when honeybee larvae are isolated in separate wax cells and exposed to similar yet unique microenvironments. DNA methylation could internalize this variability by potentially influencing the expression of thousands of genes and hence imprint cumulative developmental experiences. The life history of the honeybee, therefore, can make it a unique model organism to study the benefits and roles of epigenetic variability at the levels of individual and society. Analogies may extend to other taxa.
Differential and variable methylation
To date, individual heterogeneity has not been much discussed in the context of methylome sequencing, which can assess whether DNA methylation acts genome-wide as a mechanism to stochastically generate variable phenotypes from similar precursors. Variable methylation within the same phenotype may be of significance in biological settings in development and disease, during which epigenetic variation can be adaptive and selectable. DNA methylation during development has traditionally been characterized by its ability to aid in cell differentiation by imprinting stable (temporally independent) and heritable modifications in transcription (Li, 2002; Weaver et al., 2009). However, a mechanism to produce random DNA methylation that results in the overproduction of variants may be involved in several developmental processes. In such processes, these variants are then selected upon, leaving behind the phenotypes that are most fit with respect to a developmental cue. For example, neural growth involves the overproliferation of neurons followed by the selection of groups with the strongest response to a given input; it is estimated that 70% of neurons are eliminated in some regions during vertebrate nervous system development (Edelman, 1988). In cancer, initiation and progression is caused by somatic selection of the malignant phenotype from a heterogeneous population of cells. Indeed, the production of aberrant epi-genotypes, either marked or induced by variable DNA methylation, may be required to produce the large degree of cellular heterogeneity needed for malignant progression (Feinberg, 2007).
Deep sequencing and the detection of variably methylated regions
Sequencing can be used to measure the state of genome-wide DNA methylation during the processes of disease and development (Lister et al., 2009; Laurent et al., 2010). By measuring genome-wide methylation with high resolution, one can distinguish processes that require global instability of methylation, marked by genome-wide variation, from processes that are a consequence of programmed sensitivity of methylation in specific genomic regions with respect to variable environmental signals.
Deep sequencing has been used for the detection of differentially methylated regions (DMRs); however, it has been underutilized in the detection of variable methylated regions (VMRs) in any species. VMRs are found in the human genome (Feinberg and Irizarry, 2010) and the high resolution of deep sequencing enables us to search for VMRs that cannot be detected with lower resolution technology. The potential of deep sequencing for VMR detection has already been exhibited; >2 and >4% of methylated CpGs were found to be unique amongst biological replicates of human embryonic stem cells and fetal lung fibroblasts, respectively (Lister et al., 2009).
Deep sequencing must be coupled with species-independent statistical methods for detecting VMRs to quantify the conservation of variability in methylation across species. Thus far, methods developed for DMR detection have been largely species specific and focused on densely methylated genomes. This focus can misrepresent the ability of deep sequencing to detect small yet statistically significant differences. Extending these DMR methods for VMR detection, thereby, may prove to be inappropriate for sparsely methylated genomes. For instance, the DNA methylation density thresholds used for human DMR detection are seldom attained in the genome of the honeybee (Lister et al., 2009; Laurent et al., 2010; Li et al., 2010). Along these lines, the recently reported honeybee methylome used a species-independent method to exemplify that DMRs can be detected in a genome with a low density of methylation; a generalized linear model of the binomial family was used to detect 650 differentially methylated genes (DMGs) between queens and workers in a genome with less than 70,000 mCs (Lyko et al., 2010).
The sparsity of the honeybee methylome, the confinement of methylation targeting to exons, and the aforementioned benefits of epigenetic heterogeneity make the honeybee an ideal organism in which to use deep sequencing to explore the variability of DNA methylation between similar individuals, e.g. within the worker caste of a single colony. By using an artificial mating scheme, one can control the genotype of the haploid drone fathers and create full sister workers that are largely homogeneous genetically. Such schemes reduce the genetic contribution to phenotypic variability within the worker caste. Between castes, deep sequencing can be leveraged to test whether mechanisms that can internalize developmental cues into adaptive heterogeneity in DNA methylation are desensitized in queens relative to workers (see above). Specifically, deep sequencing could detect whether the lower amount of DNA methylation in honeybee queens co-occurs with a lower inter-individual variability in DNA methylation.
The normalized CpG ratio and methylation targeting
The scenario in which non-reproductive individuals use methylation more than reproductive individuals presents an obstacle for interpreting CpG depletion by deamination because deamination in a region requires consistent methylation to be passed through several consecutive generations via the germline. Because workers bees do not normally reproduce, it is possible that the current honeybee methylation targeting system evolved largely without the effects of deamination in genes used to regulate worker phenotypes.
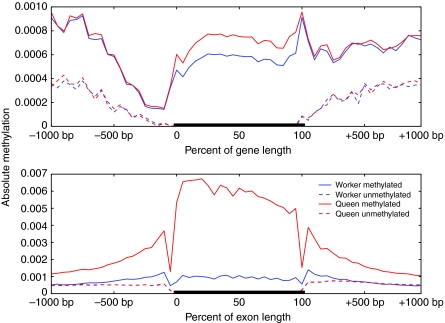
However, previous studies have use the normalized CpG ratio of a genomic region {the observed CpG frequency normalized by the expected CpG frequency, where the expected CpG frequency = [(C+G)/2]2} to predict the methylation status of genes in the honeybee genome as well as the genomes of several other species (Yi and Goodisman, 2009). Many species whose genomes contain DNMT1 and DNMT3, including honeybees, generate a bimodal distribution when the CpG ratio is computed over all genes (Fig. 2, left panel). In genomes with a bimodal distribution, the lower mode is assumed to result from CpG depletion by deamination and the higher mode contains unmethylated genes that are enriched with CpGs, whereas no such distinction can be made in genomes with a unimodal distribution (Fig. 2, right panel). We propose that this assumption can be an inaccurate tool for understanding DNA methylation in honeybees, particularly in workers, as genes that are methylated in workers may not be methylated in queens, and hence they will not be subject to the effects of deamination.
Fig. 2.
CpG ratio distributions for several species. (Left) Bimodal CpG ratio distributions are present in Apis mellifera (honeybee) genes, Homo sapiens (human) promoters, Ciona intestinalis (sea squirt) genes and Acyrthosiphon pisum (pea aphid) genes. (Right) Unimodal CpG ratio distributions are present in H. sapiens, Drosophila melanogaster (fly) and Bombyx mori (silkworm) genes. Bimodal distributions are indicative of methylated and unmethylated classes of genes within each genome. Genes in the lower mode are assumed to be methylated, whereas genes in the higher mode are unmethylated. The promoter regions used for human genes were 1 kbp directly upstream of the transcription start sites. All genome annotation and sequences were taken from publicly available databases: for silkworms, the Silkworm Genome Database (http://silkworm.genomics.org.cn/); for all other species, the NCBI FTP site (http://www.ncbi.nlm.nih.gov/Ftp/).
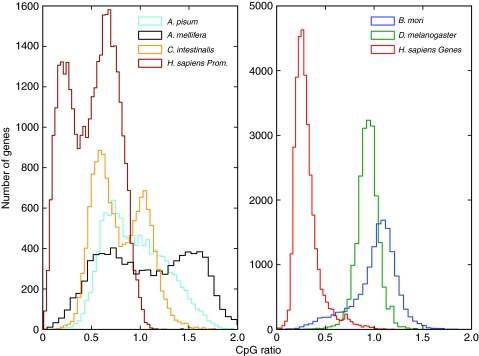
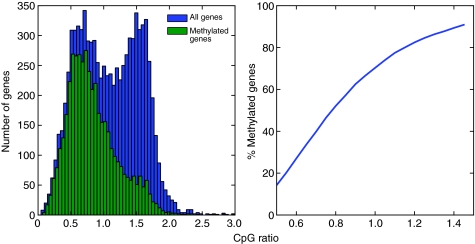
Using the CpG ratio to predict whether a gene is methylated can also produce false positives in the honeybee because of the underlying assumption that an entire gene is evenly targeted for methylation. For instance, honeybee heparan sulfate sulfotransferase has a CpG ratio of 1.03, which suggests a lack of depletion and, therefore, that the gene is not methylated. However, the gene's second and third exons are clearly methylated with no significant variation in queens and workers. The neutral CpG ratio can be attributed to the uneven distribution of CpGs and methylation targeting over the entire gene (Fig. 3). Previous studies that relied on the CpG ratio as proxy for methylation status may therefore need to be confirmed with new methylome data (e.g. Hunt et al., 2010; Elango et al., 2009). Explicitly, a CpG ratio of approximately 1 does not accurately distinguish between methylated and unmethylated genes in the honeybee brain because 28.5% of methylated genes have a CpG ratio greater than 1 (Fig. 4).
Fig. 3.
Methylation and CpG depletion in honeybee heparan sulfate 6-0-sulfotransferase. (Top) Bisulfite sequencing data for the gene length. Red lines above the x-axis indicate queen methylation intensity; blue lines below the x-axis indicate worker methylation intensity. The four exons (1–4 from left to right) for this gene are shown on the x-axis as boxes. The x-axis is labelled as the distance from the translation start site (TSS) with negative coordinates because this gene is transcribed from the antisense strand. Red and blue plus signs on the x-axis indicate CpG coverage from bisulfite sequencing data in queens and workers, respectively. Exons 2 and 3 are methylated in both queens and workers. (Bottom) The CpG ratio is calculated with a 200 bp sliding window along the gene length. There is CpG enrichment in unmethylated exons (exons 1 and 4) and CpG depletion in methylated exons (exons 2 and 3), leaving an average CpG ratio of 1.03 over the entire gene. Methylation data were obtained from bisulfite sequencing (Lyko et al., 2010).
Fig. 4.
Location of methylated honeybee genes in the bimodal CpG ratio distribution. (Left) The CpG ratio distribution for honeybee genes (blue) is shown together with the distribution limited to just methylated genes (green). (Right) The cumulative distribution function for methylated genes is shown as a function of the CpG ratio. Although the majority of methylated genes lie in the lower mode of the bimodal distribution, there are a significant number of methylated genes that lie in the higher mode. The CpG ratio can misrepresent the methylation status; for instance, over 28% of methylated genes have a CpG ratio greater than 1. Methylation data were obtained from bisulfite sequencing (Lyko et al., 2010).
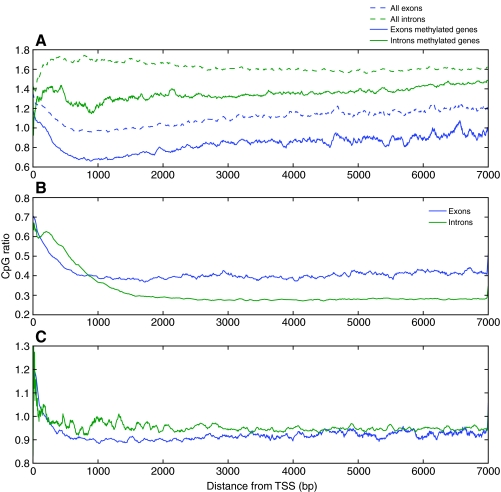
Similar aspects of DNA methylation targeting that preclude computational interpretation of these marks can be broadly present in animals and plants. If such aspects are better understood, we may also more fully comprehend the disparities that are apparent from cross-species computational comparisons of methylation. For example, the vast divergence in intragenic methylation targeting between humans and honeybees can be discerned by using the CpG ratio to calculate the CpG depletion in exons and introns. Human exons have a uniformly higher CpG ratio than introns over the gene length, whereas this pattern is reversed in honeybees independent of gene methylation status (Fig. 5). This reversal may be a consequence of the divergent roles of intragenic methylation marks in humans and honeybees. To date, the only proposed effect of intragenic methylation in honeybees is the regulation of splice variants through methylation of exons (Lyko et al., 2010). In humans, it has recently been shown that the regulation of alternative promoters can be achieved with intragenic methylation through the methylation of introns by shifting the transcription start site to inside the gene body (Maunakea et al., 2010). Confirming these intragenic methylation regulatory mechanisms in honeybees and humans on a genome-wide scale could explain opposing pattern of exon and intron CpG depletion seen in these species.
Fig. 5.
Intragenic CpG depletion and enrichment in (A) Apis mellifera, (B) Homo sapiens and (C) Drosophila melanogaster. The CpG ratio is calculated within introns and exons from 0 to 7 kbp from the translation start site (TSS), using a sliding window of 100 bp along the gene lengths. Exons are more CpG depleted than introns in honeybees regardless of gene methylation and this pattern is reversed in humans. Flies show no CpG depletion because there is no DNA methylation present in the fly genome. Methylation data were obtained from bisulfite sequencing (Lyko et al., 2010).
The association of methylation and gene conservation
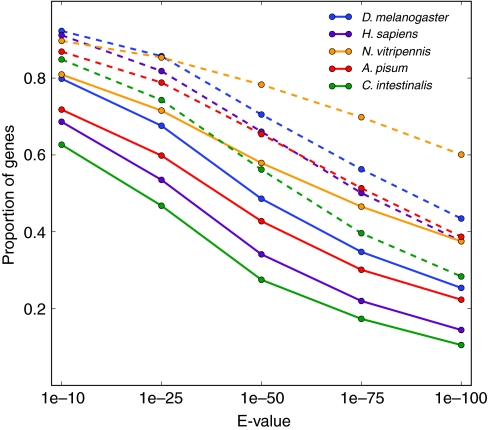
Despite differences in targeting and prevalence of methylation between taxa, we find that there is a higher cross-species conservation in methylated A. mellifera genes than in unmethylated genes (Fig. 6). There are two mechanisms that can contribute to this conservation. First, maintaining DNA methylation in a gene requires preservation of the target sequences used to guide DNA methylation to the region (e.g. by RNA-directed DNA methylation). Maintenance of target sequences confines the evolutionary landscape of a gene by requiring that each target sequence (∼30 bp each) remains unaltered. Second, if reproductive success is increased at the advent of DNA methylation in a gene region, the region could undergo purifying selection at all non-synonymous methylated cytosines because of deamination. Deamination at methylated Cs in a gene would produce all tolerable C to T transitions at a higher rate than other mutations. This purifying selection would increase the lethality of other non-CpG replacement mutations, thereby also confining the future evolution of the gene. An expected outcome of this restriction is that genes that are currently methylated in the honeybee are more highly conserved compared with other species, even those species that have lost a functional DNA methylation system such as Drosophila melanogaster (Fig. 6).
Fig. 6.
Cross-species conservation of methylated and unmethylated honeybee genes. Conservation of a honeybee gene to another species is calculated by using protein BLAST (http://blast.ncbi.blm.bih.gov/) to calculate and E-value (lower E-values correspond to higher protein similarity). The proportion of all methylated and unmethylated honeybee genes with E-values calculated from Drosophila melanogaster (fly), Homo sapiens (human), Nasonia vitripennis (jewel wasp), Acyrthosiphon pisum (pea aphid) and Ciona intestinalis (sea squirt). The proportion of methylated genes (data points are solid circles connected by dashed lines) is significantly higher than the proportion of unmethylated genes (data points are solid circles connected by solid lines) for all E-values plotted (P<0.05, Fisher's exact test), indicating that methylated honeybee genes are more highly conserved than unmethylated genes across all species shown. Methylation data were obtained from bisulfite sequencing (Lyko et al., 2010).
Concluding remarks
Variable methylation in the honeybee caused by the sensitivity of DNA methylation to developmental signals could provide phenotypic heterogeneity without genetic heterogeneity. If stochasticity was intrinsic to an ancestral DNA methylation system, the prevalence of this source of variability remains to be measured within species. We speculate that the mechanism may be conserved because of the beneficial impact on developmental plasticity and population heterogeneity. For instance, DNA methylation can increase bet-hedging by internalizing environmental variation encountered during development so that cohorts of offspring can display a diverse spectrum of phenotypes under stressful environments. As costs of deep sequencing decrease, it will be possible to test these general predictions in honeybees through better access to individual sequence data.
In broad terms, the mechanism of DNA methylation emphasizes the importance of integrating multiple types of data from high-throughput sequencing to create a multi-scale perspective of genetic and epigenetic regulation. In future work, the gap in understanding how species with homologous DNA methyltransferases can exhibit epigenetic divergence can be filled by probing the sequences that encode the methylation targeting system. It is widely maintained that PIWI-interacting RNAs (piRNAs), a class of RNAs 24–26 nt in length transcribed from non-coding regions, are used to guide methylation to target sequences (Lister et al., 2008; Bird, 1999); chromatin immunoprecipitation sequencing and RNA immunoprecipitation sequencing can be used to locate and understand how the sequences encoding these piRNAs have evolved in different taxa. The mouse genome is estimated to have approximately 200,000 piRNAs, showing that there is an entire scale of regulatory complexity that is yet to be investigated (Betel et al., 2007). By connecting DNA methylation to its targeting regulators, the functional RNA world will be drawn into systems biology to create a more complete picture of gene network regulation. The determination of sequences encoding piRNAs from several species will allow calculations of the evolutionary divergence of species, currently based on gene sequences alone, to expand and include regions of DNA that were once described as ‘junk’.
Acknowledgements
We thank Y. Wang, K. Traynor, M. G. Forero and the anonymous referees for helpful comments on the manuscript, and F. Wolschin, S. Kumar and G. Mendez for valuable discussions.
Glossary
- Differentially methylated region (DMR)
A contiguous region of DNA that is differentially methylated between two different samples.
- DNA methylation
The methylation of DNA involves the addition of a methyl group onto the 5 position of the pyrimidine ring on cytosines. Here, we focus on methylated cytosines, but methylated adenine is known to occur in Escherichia coli.
- Epigenetic divergence
Differences in the prevalence or targeting of epigenetic features that occur between species. For instance, gene body methylation is basal to plants and animals, yet many invertebrates lack the promoter and transposon methylation that is ubiquitous among vertebrates. Thus, the additional targets (e.g. promoters) of DNA methylation in vertebrates that are not present in invertebrates indicate an epigenetic divergence.
- Epigenetic heterogeneity
Variability in epigenetic features that is observed within a genetically uniform population of cells or individuals.
- Epigenome
The set of all epigenetic features that belong to a specific phenotype of a cell, tissue or organism. An example is the genome-wide location and intensity of DNA methylation.
- Eusociality
Life within a colony of individuals that contains sterile individuals that help raise the offspring of reproductive individuals within that same colony.
- Exon
Any contiguous nucleotide sequence within a gene that is present in the mature form of an mRNA produced after splicing.
- Gene body methylation
DNA methylation that occurs within a gene sequence.
- Intergenic region
A contiguous nucleotide sequence located outside of genomic regions that have been annotated as genes. Intergenic regions can be methylated (intergenic methylation), although the precise functional role of this methylation is largely unknown for the honeybee.
- Intron
Any contiguous nucleotide sequence within a gene that is removed by RNA splicing to produce the mature mRNA product of a gene.
- Mature messenger RNA (mRNA)
An RNA transcript that has been spliced and is ready for translation into a protein.
- Phenotypic heterogeneity
Variability in phenotype that is observed within a genetically uniform population of cells or individuals.
- PIWI-interacting RNAs (piRNAs)
A class of RNAs 24–26 nt in length transcribed from non-coding regions that form RNA–protein complexes through interactions with PIWI proteins. It is hypothesized that piRNA–PIWI complexes are used to guide methyltransferases to specific DNA target sequences that are complementary to the piRNA sequence.
- Promoter
A contiguous region of DNA that regulates the transcription of a specific gene. Promoters are typically located within 2 kbp upstream of the transcription start site.
- Splice variant
Exons contained in post-transcriptional RNA can occur in different arrangements within the final mature mRNA product of a gene as a result of RNA splicing. Exons can be skipped during transcription, reducing the total number of possible arrangements of exons within the mature mRNA. Similarly, introns may be included in the mature mRNA despite RNA splicing. Any specific mature mRNA produced after RNA splicing is called a splice variant. The set of all possible splice variants produced by a genome is the splice variant diversity.
- Variably methylated region (VMR)
A contiguous region of DNA that is variably methylated between two or more different samples.
Footnotes
G.V.A. was supported by the Research Council of Norway the National Institute on Aging (NIA P01 AG22500) and the PEW Charitable Trust. Deposited in PMC for release after 12 months.
References
- Amdam G. (2011). Social context, stress, and plasticity of aging. Aging Cell 10, 18-27 [DOI] [PubMed] [Google Scholar]
- Betel D., Sheridan R., Marks D. S., Sander C. (2007). Computational analysis of mouse piRNA sequence and biogenesis. PLoS Comput. Biol. 3, e222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. (1999). DNA methylation de novo. Science 286, 2287-2288 [DOI] [PubMed] [Google Scholar]
- Bogdanovic O., Veenstra G. J. C. (2009). DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma 118, 549-565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Baron W. F., Stout D. B., Davidson E. H. (1988). Sources and evolution of human Alu repeated sequences. Proc. Natl. Acad. Sci. USA 85, 4770-4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. (1986). Neighboring base effects on substitution rates in pseudogenes. Mol. Biol. Evol. 3, 322-329 [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Miller J. H. (1980). Mutagenic deamination of cytosine residues in DNA. Nature 287, 560-561 [DOI] [PubMed] [Google Scholar]
- Edelman G. (1988). Topobiology. New York: Basic Books; [Google Scholar]
- Elango N., Hunt B. K., Goodisman M. A. D., Yi S. V. (2009). DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc. Natl. Acad. Sci. USA 106, 11206-11211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P. (2007). Phenotypic plasticity and the epigenetics of human disease. Nature 447, 433-440 [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Irizarry R. A. (2010). Evolution in health and medicine Sackler colloquium: stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc. Natl. Acad. Sci. USA 107 Suppl. 1, 1757-1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt B. G., Brisson J. A., Yi S. V., Goodisman M. A. D. (2010). Functional conservation of DNA methylation in the pea aphid and the honeybee. Genome Biol. Evol. 2, 219-728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R. J., Bird A. P. (2006). Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89-97 [DOI] [PubMed] [Google Scholar]
- Kucharski R., Maleszka J., Foret S., Maleszka R. (2008). Nutritional control of reproductive status in honeybees via DNA methylation. Science 319, 1827-1830 [DOI] [PubMed] [Google Scholar]
- LaPlant Q., Vialou V., Covington H. E., Dumitriu D., Feng J., Warren B. L., Maze I., Dietz D. M., Watts E. L., Iñiguez S. D., et al. (2010). Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 13, 1137-1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L., Wong E., Li G., Huynh T., Tsirigos A., Ong C. T., Low H. M., Kin Sung K. W., Rigoutsos I., Loring J., et al. (2010). Dynamic changes in the human methylome during differentiation. Genome Res. 20, 320-331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J. A., Jacobsen S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E. (2002). Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3, 662-673 [DOI] [PubMed] [Google Scholar]
- Li Y., Zhu J., Tian G., Li N., Li Q., Ye M., Zheng H., Yu J., Wu H., Sun J., et al. (2010). The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 8, e1000533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., O’Malley R. C., Tonti-Filippini J., Gregory B. D., Berry C. C., Millar A. H., Ecker J. R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523-536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Pelizzola M., Dowen R. H., Hawkins R. D., Hon G., Tonti-Filippini J., Nery J. R., Lee L., Ye Z., Ngo Q., et al. (2009). Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockett G. A., Helliwell P., Maleszka R. (2010). Involvement of DNA methylation in memory processing in the honey bee. Neuroreport 21, 812-816 [DOI] [PubMed] [Google Scholar]
- Lyko F., Foret S., Kucharski R., Wolf S., Falckenhayn C., Maleszka R. (2010). The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8, e1000506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea A. K., Nagarajan R. P., Bilenky M., Ballinger T. J., D’Souza C., Fouse S. D., Johnson B. E., Hong C., Nielsen C., Zhao Y., et al. (2010). Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. A., Sweatt J. D. (2007). Covalent modification of DNA regulates memory formation. Neuron 53, 857-869 [DOI] [PubMed] [Google Scholar]
- Miller C. A., Gavin C. F., White J. A., Parrish R. R., Honasoge A., Yancey C. R., Rivera I. M., Rubio M. D., Rumbaugh G., Sweatt J. D. (2010). Cortical DNA methylation maintains remote memory. Nat. Neurosci. 13, 664-666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd B. P., Fewell J. H. (2007). Genetic diversity promotes homeostasis in insect colonies. Trends Ecol. Evol. 22, 408-413 [DOI] [PubMed] [Google Scholar]
- Park P. J. (2009). ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 10, 669-680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S., Berg P., Brutlag D. L. (2006). A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 103, 1412-1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J., Bird A. (1990). The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl. Acad. Sci. USA 87, 4692-4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Honeybee Genome Sequencing Consortium (2006). Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931-949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waibel M., Floreano D., Magnenat S., Keller L. (2006). Division of labour and colony efficiency in social insects: effects of interactions between genetic architecture, colony kin structure and rate of perturbations. Proc. Biol. Sci. 273, 1815-1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver J. R., Susiarjo M., Bartolomei M. S. (2009). Imprinting and epigenetic changes in the early embryo. Mamm. Genome 20, 532-543 [DOI] [PubMed] [Google Scholar]
- Xiang H., Zhu J., Chen Q., Dai F., Li X., Li M., Zhang H., Zhang G., Li D., Dong Y., et al. (2010). Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat. Biotechnol. 28, 516-520 [DOI] [PubMed] [Google Scholar]
- Yi S. V., Goodisman M. A. D. (2009). Computational approaches for understanding the evolution of DNA methylation in animals. Epigenetics 4, 551-556 [DOI] [PubMed] [Google Scholar]
- Zemach A., McDaniel I. E., Silva P., Zilberman D. (2010). Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916-919 [DOI] [PubMed] [Google Scholar]