Abstract
The U2 snRNP component SAP 155 contacts pre-mRNA on both sides of the branch site early in spliceosome assembly and is therefore positioned near or at the spliceosome catalytic center. We have isolated a cDNA encoding human SAP 155 and identified its highly related Saccharomyces cerevisiae homolog (50% identity). The carboxy-terminal two-thirds of SAP 155 shows the highest conservation and is remarkably similar to the regulatory subunit A of the phosphatase PP2A. Significantly, SAP 155 is phosphorylated concomitant with or just after catalytic step one, making this the first example of a protein modification tightly regulated with splicing catalysis.
Keywords: U2 snRNP, SAP 155 phosphorylation, spliceosome, splicing
Spliceosomal complexes assemble on pre-mRNA in the order E, A, B, and C, with the catalytic steps taking place in the C complex (for review, see Kramer 1996). Step 1 is a nucleophilic attack on the 5′ splice site (ss) by the branch-site adenosine, which is bulged from a duplex between U2 snRNA and the branchpoint sequence (BPS). This duplex is established in the A complex concomitant with binding of six U2 snRNP proteins [spliceosome-associated proteins (SAPs) 49, 61, 62, 114, 145, and 155] near the BPS. These proteins are components of two multiprotein complexes, SF3a and SF3b, which are required for A complex assembly (for review, see Kramer 1996; Reed 1996). SF3a consists of three subunits, SF3a60, SF3a66, SF3a120 (corresponding to SAPs 61, 62, and 114, respectively), and SF3b consists of at least four subunits, SF3b50, SF3b130, SF3b145, and SF3b155 (SAPs 49, 130, 145, and 155, respectively; we will use the SAP nomenclature). cDNAs encoding all of the SF3a and two of the SF3b subunits (SAPs 145 and 49) have been isolated and Saccharomyces cerevisiae homologs identified (Kramer 1996; Reed 1996). The SF3a subunits are all essential in yeast and required for A complex assembly. ySAP 145, originally identified by its similarity to human SAP 145 (Gozani et al. 1996), corresponds to the CUS1 gene, isolated as a suppressor of a U2 snRNA mutation (Wells et al. 1996). CUS1 is essential in yeast and is required for A complex assembly (Wells et al. 1996).
In the mammalian A complex, the SF3a/b subunits cross-link to a 25-nucleotide region in the pre-mRNA located immediately upstream of the BPS (Gozani et al. 1996). These RNA–protein interactions are thought to function in part to anchor U2 snRNP to the BPS (Gozani et al. 1996). SAP 155 cross-links to pre-mRNA on both sides of the branch site (O. Gozani, J. Potashkin, and R. Reed, in prep.) and is thus strategically positioned over the branch-site adenosine before this residue undergoes nucleophilic attack on the 5′ ss. This location and timing of the SAP 155–pre-mRNA interactions suggest that SAP 155 is a critical component of the spliceosome active site. Here we report the isolation of a cDNA encoding SAP 155 and find that it is the most highly conserved SF3 component. We also show that SAP 155 is phosphorylated concomitant with splicing catalysis. The functional importance of SAP 155 phosphorylation for splicing is indicated by the observations that phosphorylation is detected only in functional spliceosomes, is regulated with catalysis and occurs generally with different pre-mRNAs, and that phosphorylated SAP 155 contacts both sides of the branch site during catalysis.
Results
Isolation of SAP 155 cDNA
To isolate a human SAP 155 cDNA, spliceosomal complex A was purified in large scale and fractionated by SDS-PAGE, and peptide sequences were obtained from tryptic digestion products of SAP 155 (Bennett and Reed 1993). One of the peptide sequences matched an EST in GenBank, and this EST was used to screen libraries to obtain the SAP 155 cDNA. This cDNA is predicted to encode a 1304-amino-acid protein with a molecular mass of 146 kD (Fig. 1A). SAP 155 is most similar to the regulatory subunit A of the phosphatase PP2A (Ruediger et al. 1994). PP2A-A is organized in 15 nonidentical repeats, 38–43 amino acids in length; the consensus is shown below the SAP 155 sequence (Ruediger et al. 1994; Fig. 1A). These repeats are thought to fold PP2A-A into a rod-like structure that interacts directly with the regulatory B and catalytic C subunits (Ruediger et al. 1994). The carboxy-terminal two-thirds of SAP 155 can be aligned into 22 PP2A-like repeats. SAP 155 and PP2A-A contain few exact matches at many positions in the PP2A-A consensus; rather, the main characteristic of these repeats is a specific distribution of hydrophobic and charged residues. In PP2A-A, the amino acids at positions 5–7 in the consensus are LLP (Fig. 1A). Only two repeats have this exact sequence; the others are composed of related residues (e.g., IIP, LVP). Similarly, in SAP 155, there are two LLPs at positions 5–7 of the repeats and several related sequences. Like PP2A-A, SAP 155 is enriched in serines or threonines at position 1, pairs of acidic residues at positions 15 and 16, hydrophobic residues at positions 24 and 31, and glycines at position 36. The similar organization of SAP 155 and PP2A-A suggests that these proteins fold into a similar structure and that the PP2A-A repeats in SAP 155 play a role in as yet unidentified protein–protein interactions.
Figure 1.


(A) The amino acid sequence predicted from the human SAP 155 cDNA sequence. The RWDETP repeats at the amino terminus are aligned for convenience. These repeats and the TPGX repeats are indicated in boldface type, and the TPGH repeats are also underlined. The PP2A-like repeats are aligned according to the consensus for PP2A. This consensus is shown below the SAP 155 sequence. The underlined amino acids indicate those that match the consensus or are similar in type (hydrophobic or charged) to the consensus residue. The brackets indicate the 82-amino-acid region that is missing from the ΔSAP 155 cDNA (see Materials and Methods). (B) SAP 155 homologs are shown aligned with human SAP 155. Identities are indicated in white on black. ceSAP 155, spSAP 155, and scSAP 155 correspond to the C. elegans, S. pombe, and S. cerevisiae proteins, respectively.
The amino terminus of SAP 155 is hydrophilic and contains two types of repeats (Fig. 1A). There are three perfect and several degenerate repeats of the sequence RWDETP. There are also five repeats of the sequence TPGH and several derivatives in which the last amino acid is either basic (K, R) or small (A, S). These repeats closely resemble phosphorylation sites for the Cdk serine–threonine kinases (S/T-P-X-R/K) (Moreno and Nurse 1990).
SAP 155 is the most highly conserved SF3 subunit
Probable S. cerevisiae, Schizosaccharomyces pombe, and Caenorhabditis elegans homologs of SAP 155 were identified in GenBank (Fig. 1B). Both the size and sequence of these homologs are strikingly conserved: scSAP 155 is 50% identical to human SAP 155; ceSAP 155 and spSAP 155 are ∼66% and 56% identical to SAP 155, respectively. The highest conservation lies in the carboxy-terminal two-thirds of the protein, which contains the PP2A-like repeats. Over this region, ceSAP 155 and spSAP 155 are 86% and 75% identical to SAP 155, respectively. Virtually the entire scSAP 155 protein consists of these repeats, and they are 55% identical to SAP 155.
SAP 155 is phosphorylated late in the splicing pathway
To investigate the role of SAP 155 in splicing, we raised antibodies to two peptides. These antibodies specifically immunoprecipitate in vitro-translated SAP 155, detect SAP 155 on Western blots of the A complex or nuclear extract (NE), and immunoprecipitate a small fraction of U2 snRNP from NE (see below; data not shown). The antibodies do not inhibit either A complex assembly or the splicing reaction, possibly because the epitope is mostly masked in extracts (data not shown).
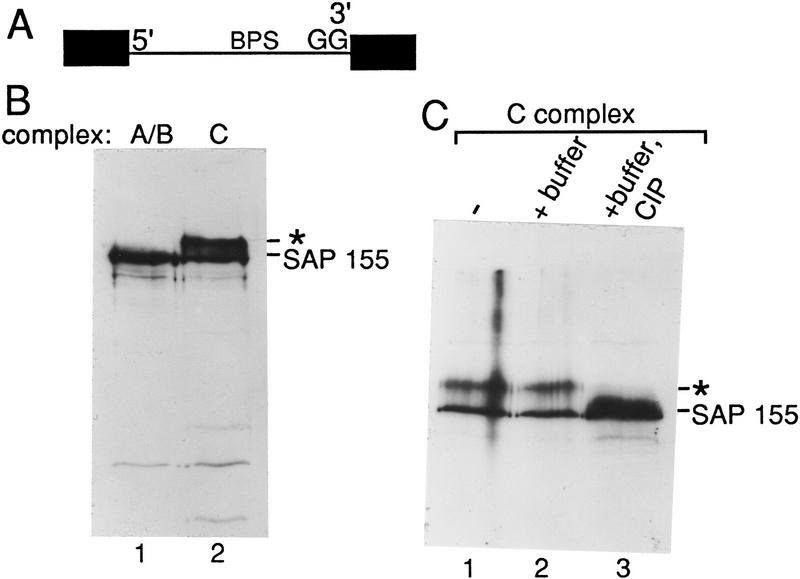
The antibody was used for Western analysis of spliceosomal complexes at different steps of the splicing pathway. Initially, complexes were assembled on ΔAG pre-mRNA, which contains a GG → AG substitution at the 3′ ss (Gozani et al. 1994) (Fig. 2A). This mutation does not affect assembly of the A, B, and C complexes but blocks step 2 of splicing. After a long incubation under splicing conditions, the C complex containing high levels of step 1 products accumulates on ΔAG pre-mRNA (Gozani et al. 1996). Spliceosomal complexes A/B or C were isolated and used for Western analysis. SAP 155 was detected in the A/B complex (Fig. 2B, lane 1). Surprisingly, another major band was detected in the C complex (asterisk; Fig. 2B, lane 2). As SAP 155 contains multiple potential phosphorylation sites (see Fig. 1A), we tested whether this additional band is a phosphorylated form of SAP 155. Incubation of affinity-purified C complex with calf intestinal alkaline phosphatase (CIP) results in the loss of the slower-mobility band and an increase in the levels of the SAP 155 band (Fig. 2C). Addition of the phosphatase inhibitor vanadate eliminates the effect of CIP treatment (data not shown). These data indicate that SAP 155 is modified by phosphorylation.
Figure 2.
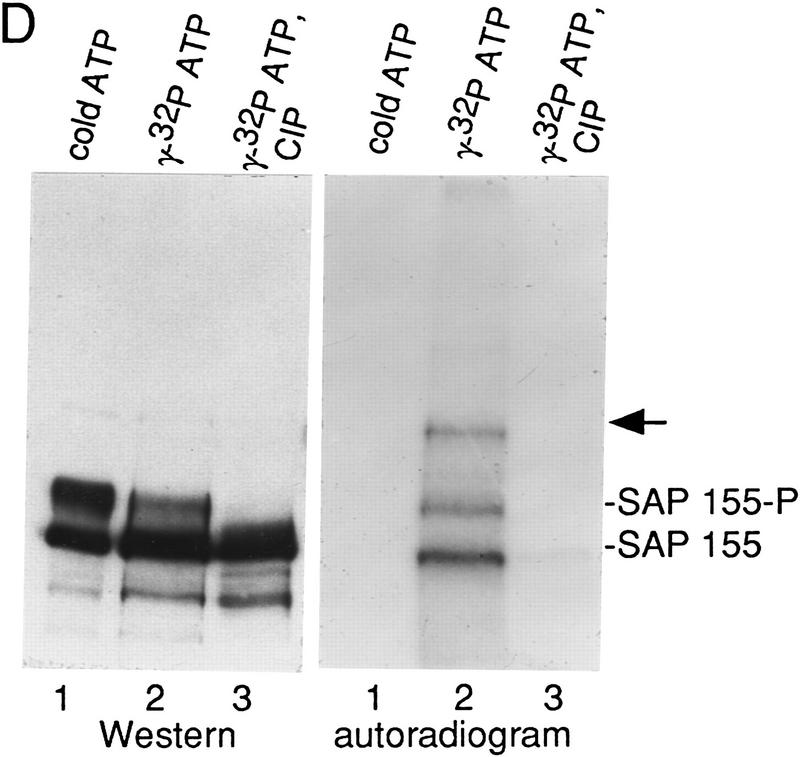
SAP 155 is phosphorylated in the C complex assembled on ΔAG pre-mRNA. (A) Schematic of ΔAG AdML pre-mRNA; (B) Western blot of A/B (lane 1) and C (lane 2) complexes probed with SAP 155 antibodies; (C) Western blot of the C complex alone (lane 1), incubated in buffer (lane 2), or incubated in buffer and phosphatase (CIP) (lane 3). The asterisk indicates the novel band detected in the C complex. (D) The C complex was assembled under standard splicing conditions (lane 1) or in a reaction containing [γ-32P]ATP (lane 2). An aliquot of the radiolabeled C complex was treated with CIP (lane 3). The autoradiograph (right) and Western blot (left) probed with SAP 155 antibodies are shown. The arrow indicates the 200-kD protein that is phosphorylated and also weakly detected by the antisera.
To obtain further evidence that the CIP-sensitive form of SAP 155 is phosphorylated, the C complex was assembled using [γ-32P]ATP fractionated by SDS-PAGE and immobilized on a membrane. 32P-Labeled proteins were detected by autoradiography and compared with Western analysis of the same blot using SAP 155 antisera (Fig. 2D). As shown in lane 2, three major high-molecular- mass bands were detected in the C complex on the autoradiograph (right). Significantly, two of these comigrate with the major bands detected by the SAP 155 antibody (Fig. 2D, lane 2, cf. Western and autoradiograph). A 200-kD protein detected on the autoradiograph is weakly detected by the SAP 155 antibody (arrow). The 32P-labeled bands are largely abolished by treatment with CIP, but the fastest migrating form of SAP 155 is still detected by Western blotting (Fig. 2D, lane 3). Together, these data indicate that SAP 155 is phosphorylated in the C complex. We have designated the major phosphorylated form of this protein SAP 155-P.
The fast migrating form of SAP 155 is detected on the autoradiograph (Fig. 2D, right, lane 2), indicating that it is phosphorylated on at least one site. Thus, the large mobility shift of SAP 155-P is most likely a result of multiple phosphorylation sites. The 200-kD protein that is detected weakly on the Western blot (Fig. 2D, arrow) could either be a hyperphosphorylated form of SAP 155 or a protein that weakly cross-reacts with SAP 155 antisera [suggested because a 200-kD protein is faintly detected by the SAP 155 antisera in complexes isolated at all stages of spliceosome assembly (see below)].
SAP 155-P is bound on both sides of the branch site in the C complex
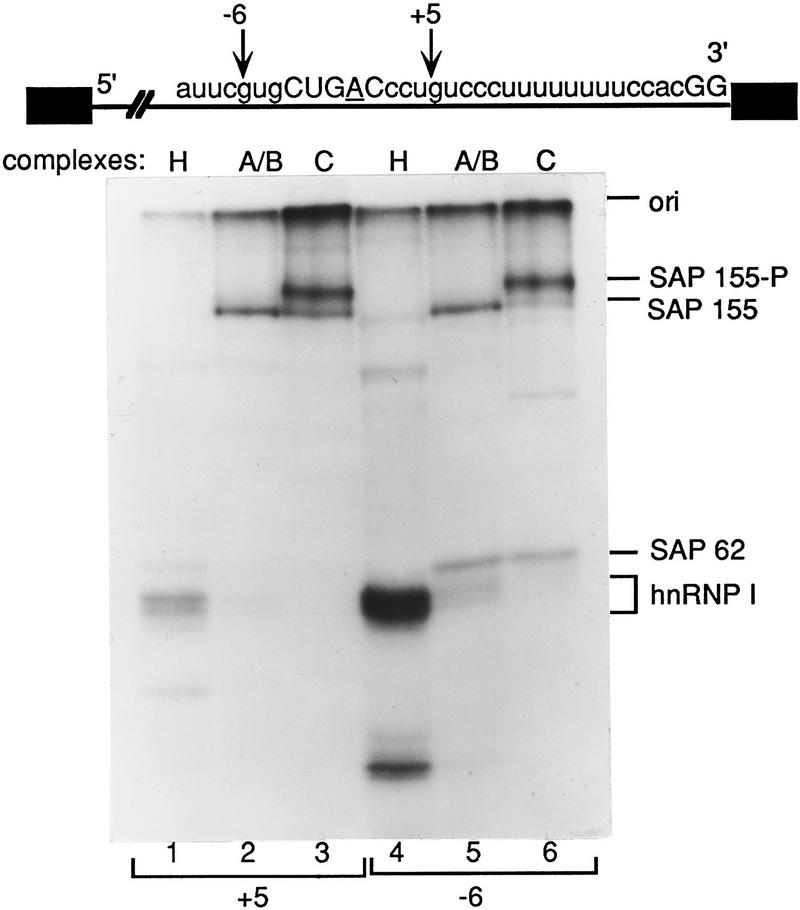
Prior to catalytic step 1, SAP 155 cross-links to pre-mRNA immediately upstream and downstream, but not directly at, the branch site (O. Gozani, J. Potashkin, and R. Reed, in prep.). Because phosphorylation can trigger significant conformational changes in proteins, we asked whether phosphorylation of SAP 155 affects its interactions with pre-mRNA. ΔAG pre-mRNA that contained a single 32P-labeled guanosine, either 6 nucleotides upstream (−6) or 5 nucleotides downstream (+5) from the branch site (Fig. 3), was synthesized and assembled into the hnRNP complex H, or into the A/B or C spliceosomal complexes, isolated by gel filtration, and UV irradiated. After RNase A digestion, proteins were fractionated by SDS-PAGE. As observed (O. Gozani, J. Potashkin, and R. Reed, in prep.), SAP 155 specifically cross-links to pre-mRNA at +5 and −6 sites in the A/B complex but not in the H complex (Fig. 3, lanes 1,2,4,5); the SF3a component SAP 62 is also detected at the −6 site in the A/B complex (lane 5) (Gozani et al. 1996; and in prep.). Strikingly, in the C complex, SAP 155 at both −6 and +5 is largely replaced by the cross-linking of a more slowly migrating protein, which comigrates with SAP 155-P (data not shown). Together, these observations indicate that SAP 155 remains bound on both sides of the branch site in the C complex and is phosphorylated near or at this stage in the splicing pathway. The ratio of SAP 155-P to SAP 155 is greater in some C complex preparations than in others (see Figs. 2 and 3; data not shown); this ratio correlates with efficiency of conversion to the C complex, which varies between preparations. The observation that SAP 155-P is bound on both sides of the BPS on the lariat exon, rather than, for example, dissociating from the pre-mRNA, is consistent with the possibility that SAP 155 phosphorylation is functionally important in the splicing pathway.
Figure 3.
SAP 155-P cross-links on both sides of the branch site. A schematic of ΔAG AdML pre-mRNA showing the sequence of the 3′ portion of the intron. (+5 and −6) The guanosine residues that were 32P-labeled. The GG at the 3′ ss and BPS are shown in uppercase letters; the branch-site adenosine is underlined. Proteins were UV cross-linked in the H (lanes 1,4), A/B (lanes 2,5), and C (lanes 3,6) complexes assembled on pre-mRNAs labeled at the +5 (lanes 1–3) or −6 (lanes 4–6) sites. (ori) The gel origin. SAP 155 and SAP 62 were identified on two-dimensional gels (data not shown).
Phosphorylation of SAP 155 accompanies splicing catalysis
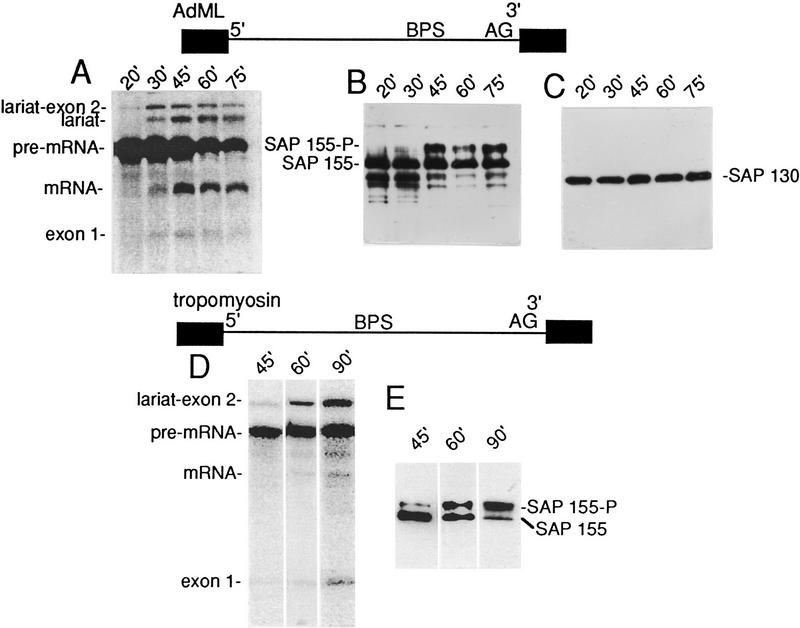
As described above, catalytic step 2 of splicing is blocked on ΔAG pre-mRNA, and the C complex containing the products of step 1 accumulates. To determine whether SAP 155 phosphorylation occurs during the normal splicing pathway and to investigate its kinetics, complexes were assembled on wild-type AdML pre-mRNA for varying lengths of time and then purified (Fig. 4). As shown in Figure 4B, SAP 155 is phosphorylated during the normal splicing pathway. In contrast, there is no change in another SF3b component, SAP 130 (Fig. 4C). The highest levels of step 1 and 2 splicing products are detected at the 45-, 60-, and 75-min time points (Fig. 4A). Similarly, SAP 155-P is present at the highest levels at these time points (Fig. 4B). These data indicate that phosphorylation of SAP 155 occurs concomitant with the catalytic steps of splicing. Because the kinetics of steps 1 and 2 are closely coupled on wild-type pre-mRNA, it is not possible to determine whether phosphorylation accompanies catalytic step 1 or step 2. However, the observation that SAP 155-P accumulates on ΔAG pre-mRNA (Fig. 2), which is unable to undergo step 2, indicates that the phosphorylation normally precedes step 2. The high levels of SAP 155-P at the 45- to 75-min time points versus the 30-min time point suggests that SAP 155-P is bound to the lariat–intron as well as the lariat–exon intermediate.
Figure 4.
Phosphorylation of SAP 155 occurs concomitant with splicing catalysis. Wild-type AdML (A–C) or tropomyosin (D,E) pre-mRNA was incubated under splicing conditions for the times indicated. Total RNA (A,D) or protein from spliceosomal complexes (B,C,E) was analyzed. Western blots were probed with SAP 155 (B,E) or SAP 130 (C) antisera. The bands below SAP 155 may be breakdown products.
Coupling of SAP 155 phosphorylation to catalysis occurs generally
The observation that SAP 155 phosphorylation accompanies splicing catalysis of wild-type AdML pre-mRNA suggests that this phosphorylation is functionally important for splicing. At present, it is not technically feasible to assemble SF3b or U2 snRNP using recombinant SAP 155. As SAP 155 is an integral component of these complexes, the effects of mutations in the SAP 155 phosphorylation sites cannot be tested directly in splicing. Thus, we asked whether the phosphorylation occurs on a pre-mRNA unrelated to AdML. As shown in Figure 4, D and E, SAP 155 phosphorylation does accompany catalysis on α-tropomyosin pre-mRNA. Step 1 occurs with slower kinetics on α-tropomyosin than on AdML, and step 2 is much less efficient (Fig. 4, cf. A and D) (Chiara et al. 1997). Significantly, the kinetics of appearance of SAP 155-P parallel the kinetics of catalytic step 1 (Fig. 4D,E). The observation that SAP 155 phosphorylation accompanies catalysis on two pre-mRNAs (AdML and tropomyosin) with different step 1 kinetics suggests that step 1 and SAP 155 phosphorylation are functionally coupled and therefore that SAP 155 phosphorylation is likely to be a general step in the splicing reaction.
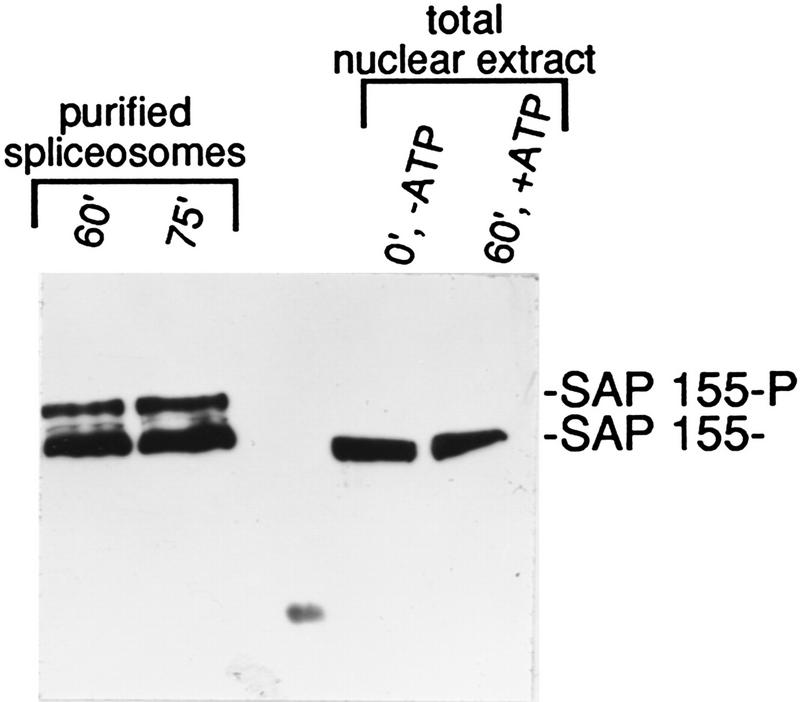
Other splicing factors, including arginine–serine repeat (SR) proteins, U1–70K and U2AF, are phosphorylated when NEs are incubated in the presence of ATP–Mg2+ (Tazi et al. 1993; Woppmann et al. 1993; Xiao and Manley 1997; data not shown). To determine whether SAP 155-P can be detected in NEs, we incubated the extract under spliceosomal assembly conditions in the absence of pre-mRNA (Fig. 5). A Western blot was then performed on total protein from the NE or purified from 60- or 75-min spliceosomal complexes. SAP 155-P is not detected in total NE incubated for 60 min (Fig. 5) or at any other time point between 0 and 75 min (data not shown). We conclude that SAP 155 phosphorylation is tightly regulated and is detected only in spliceosomal complexes and only at a specific time in the splicing pathway, concomitant with catalysis. Together, these data indicate that SAP 155 phosphorylation is functionally important for the splicing reaction.
Figure 5.
SAP 155-P is not detected in nuclear extracts incubated under splicing conditions. Shown is Western analysis of purified spliceosomal complexes assembled on AdML pre-mRNA (60- and 75-min time points) or total NE incubated as indicated. SAP 155 antibodies were used as a probe.
Discussion
We have isolated a cDNA encoding human SAP 155, a subunit of the splicing factor SF3b, a U2 snRNP component, and a component of the A, B, and C spliceosomal complexes. UV cross-linking studies revealed that SAP 155 binds on both sides of the branch site prior to catalytic step 1 of the splicing reaction (O. Gozani, J. Potashkin, and R. Reed, in prep.). SAP 155 is subsequently phosphorylated and remains bound on both sides of the branch site prior to catalytic step 2 (this study). As the branch-site adenosine functions as the nucleophile for step 1, and the branch structure is required for step 2, the strategic positioning of SAP 155 over the branch site suggests that this protein is a critical component of the spliceosome active site. We have identified the apparent S. pombe, C. elegans, and S. cerevisiae homologs of SAP 155 in GenBank. scSAP 155 is 50% identical to human SAP 155, whereas the other SF3 subunits are ∼25% identical between yeast and human. The SAP 155 carboxyl terminus is organized into 22 nonidentical, tandem repeats, which are similar to those found in the regulatory subunit A of the phosphatase PP2A. We have not detected any phosphatase activity of SAP 155 nor an interaction with the catalytic subunit of PP2A. Nevertheless, the repeats must have an important function because scSAP 155 consists almost entirely of them and the scSAP 155 gene is essential for viability in yeast (O. Gozani and R. Reed, unpubl.). The SAP 155 amino terminus is less well conserved. It contains multiple TPGH motifs and several RWDETP motifs, neither of which are present in scSAP 155.
Remodeling at the 3′ ss for catalytic step 2
Prior to catalytic step 2 of the splicing reaction, a major change occurs in the proteins that interact at the pyrimidine tract and 3′ ss. The splicing factor U2AF is replaced by three components of U5 snRNP (U5220, U5110, and U5116), and AG100 bound near or at the AG is replaced by AG75 (Chiara et al. 1996, 1997). Here, we have shown that SAP 155 remains bound on both sides of the branch site after step 1 but is phosphorylated. SAP 155 phosphorylation is likely to be important in the basic splicing mechanism as it is detected only in functional spliceosomes and is coupled with catalysis on unrelated pre-mRNAs having different step 1 kinetics. One of these pre-mRNAs, α-tropomyosin, undergoes step 2 inefficiently and thus accumulates high levels of step 1 products. SAP 155-P also accumulates to high levels in C complexes assembled on this pre-mRNA. These data suggest that SAP 155 phosphorylation occurs during or just after step 1, but before step 2. SAP 155-P also accumulates on a pre-mRNA containing an AG → GG mutation at the 3′ ss, a mutation that blocks step 2. Thus, phosphorylation of SAP 155 does not require, and likely precedes, recognition of the AG for step 2.
One possible role for SAP 155 phosphorylation is to trigger the remodeling of the 3′ ss for catalytic step 2. The amino terminus of SAP 155 contains multiple TPGH motifs that are similar to phosphorylation sites for the Cdks. The same region of SAP 155 also contains a domain that interacts with U2AF (O. Gozani, J. Potashkin, and R. Reed, in prep.). Thus, it is possible that SAP 155–U2AF interactions established early in spliceosome assembly are disrupted when SAP 155 is phosphorylated. This phosphorylation may in turn allow the replacement of U2AF by U5 snRNP on the 3′ ss. In S. cerevisiae, the U2AF binding site (pyrimidine tract at the 3′ ss) is not well-conserved, and the U2AF homolog MUD2 is not essential (Abovich et al. 1994). Thus, if scSAP 155–MUD2 interactions occur at all in yeast, they almost certainly do not play as important a role as in mammals, which may explain why the scSAP 155 amino terminus is not well conserved.
Several splicing factors are known to be phosphorylated, including the SR protein family (Roth et al. 1991; Gui et al. 1994; Colwill et al. 1996; Xiao and Manley 1997), the U1 snRNP component U1–70K (Tazi et al. 1993; Woppmann et al. 1993), and a 27-kD protein in the U4/U5/U6 tri-snRNP (Fetzer et al. 1997). These proteins share the presence of a domain rich in SR repeats that contain the known or presumed phosphorylation sites (Roth et al. 1991; Woppmann et al. 1993; Gui et al. 1994; Colwill et al. 1996). Several kinases that phosphorylate SR domains have also been reported (Woppmann et al. 1993; Giu et al. 1994; Colwill et al. 1996), and several studies indicate that phosphorylation/ dephosphorylation of SR domains play a role in splicing (Mermoud et al. 1992, 1994; Tazi et al. 1993; Xiao and Manley, 1997) and in regulating the intranuclear distribution of SR proteins (Gui et al. 1994; Colwill et al. 1996).
SAP 155 is the first reported example of a phosphorylated splicing factor that lacks an SR domain. Moreover, unlike the SR domain-containing proteins, SAP 155 phosphorylation cannot be detected by incubating nuclear extract with ATP–Mg2+. Rather, SAP 155 phosphorylation can only be detected in functional spliceosomes. This is the first example of a phosphorylation event that occurs exclusively at the time of the catalytic steps of the splicing reaction. These observations make it highly likely that SAP 155 phosphorylation has a function in splicing. A phosphatase activity required for SAP 155 dephosphorylation must also exist and would be expected to play an important role in splicing.
Materials and methods
SAP 155 cDNA and antibody
SAP 155 was isolated from an SDS gel containing spliceosomal complex A3′ and used for peptide sequencing. One peptide (TPIGTPAMNMAT) matched a partial cDNA (17F6, gift from Dr. J.-M. Frigerio, INSERM, Paris, France) in GenBank. Sequencing of 17F6 revealed that it encoded another SAP 155 peptide (VNDQPSGNLPFLKP). Additional cDNAs were obtained by screening human cDNA libraries using the 17F6 probe and were used to construct a clone that encodes most of SAP 155. This clone, designated ΔSAP 155, lacks a region of 246 nucleotides in the 3′ portion of the SAP 155 cDNA (nucleotides 2797–3042). ΔSAP 155 is in-frame, but lacks amino acids 933–1014 (missing amino acids are bracketed in Fig. 1A). The 82-amino-acid region lacking from ΔSAP 155 is toxic to Escherichia coli, as we were unable to insert this region into SAP 155 without obtaining deletions or other mutations. The sequence of the 246-nucleotide region was therefore obtained from a PCR product from a HeLa cDNA library. Rabbit antisera were raised to two SAP 155 peptides (GASKRKSRWDETPAS and DRTMKSVNDQPSGNL) (BabCo). For Western blots, 6% or 9% SDS-PAGE was carried out, and proteins were immobilized on PVDF. Membranes were blocked in 5% nonfat dry milk in PBS containing 0.1% Tween 20. The SAP 155 antibody was used at a 1/1000 dilution, and HRP-conjugated goat anti-rabbit secondary antibody was used at 1/5000. Bands were detected with ECL (Amersham).
Analysis of spliceosomal complexes
Spliceosomal complexes were assembled on biotinylated pre-mRNAs, isolated by gel filtration and then purified by binding to avidin–agarose (Reed 1990). ΔAG AdML pre-mRNA was incubated for 10 or 60 min to assemble the A/B or C complexes, respectively (Gozani et al. 1994). A/B is a mixture of the A and B complexes that cannot be separated from each other by gel filtration. Phosphatase treatment of the C complex was carried out by washing a 30-μl aliquot of the complex bound on avidin–agarose beads with 1 ml of buffer (20 mm Tris-HCl at pH 7.9, 10 mm MgCl2, 50 mm NaCl, 1 mm DTT). The buffer was removed, and 30 μl of the same buffer was added. CIP (2 μl) was added, and the reaction mixture was incubated at 37°C for 1 hr. An additional 2 μl of CIP was added, and the incubation was continued for 1 hr. To phosphorylate SAP 155 with 32P, 500 ng of ΔAG pre-mRNA was incubated in a 500-μl splicing reaction containing 5 μl of [γ-32P]ATP (6000 Ci/mmole) and 375 μm cold ATP (500 μm cold ATP is present in the standard splicing reactions). Phosphatase treatment of the 32P-labeled complex was carried out as described above. For UV cross-linking experiments, ΔAG pre-mRNA was 32P site-specifically labeled (Moore and Sharp 1992) at guanosine residues located at −6 and +5 nucleotides from the branch site. Spliceosomal complexes H, A/B, and C were assembled, isolated by gel filtration, and UV cross-linked as described (Gozani et al. 1996). After digestion with RNase A, total protein was fractionated on a 9% SDS–polyacrylamide gel.
Acknowledgments
We are grateful to Stephen Lee for excellent technical assistance and to Z. Zhou for useful discussions and comments on the manuscript. This work was supported by National Institutes of Health (NIH) and Tobacco Research Council grants to R.R. and an NIH postdoctoral fellowship to C.Y.W. DNAX Research Institute is supported by Schering-Plough Corporation. The GenBank accession no. for SAP 155 is AF054284.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rreed@warren.med.harvard.edu; FAX (617) 432-3091.
References
- Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: An interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes & Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- Bennett M, Reed R. Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science. 1993;262:105–108. doi: 10.1126/science.8211113. [DOI] [PubMed] [Google Scholar]
- Chiara M, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara MD, Palandjian L, Kramer RF, Reed R. Evidence that U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. EMBO J. 1997;16:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Fetzer S, Lauber J, Will CL, Lührmann R. The [U4/U6.U5] tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA. 1997;3:344–355. [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Patton JG, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes & Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- Gui JF, Tronchere H, Chandler SD, Fu XD. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci. 1994;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Mermoud JE, Cohen PT, Lamond AI. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 1992;20:5263–5269. doi: 10.1093/nar/20.20.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA. Site-specific modification of pre-mRNA: The 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- Moreno S, Nurse P. Substrates for p34cdc2: In vivo veritas? Cell. 1990;61:549–551. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- Reed R. Protein composition of mammalian spliceosomes assembled in vitro. Proc Natl Acad Sci. 1990;87:8031–8035. doi: 10.1073/pnas.87.20.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruediger R, Hentz M, Fait J, Mumby M, Walter G. Molecular model of the A subunit of protein phosphatase 2A: interaction with other subunits and tumor antigens. J Virol. 1994;68:123–129. doi: 10.1128/jvi.68.1.123-129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J, Kornstadt U, Rossi F, Jeanteur P, Cathala G, Brunel C, Lührmann R. Thiophosphorylation of U1-70K protein inhibits pre-mRNA splicing. Nature. 1993;363:283–286. doi: 10.1038/363283a0. [DOI] [PubMed] [Google Scholar]
- Wells SE, Neville M, Haynes M, Wang J, Igel H, Ares M., Jr CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes & Dev. 1996;10:20–32. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- Woppmann A, Will CL, Kornstadt U, Zuo P, Manley JL, Lührmann R. Identification of an snRNP-associated kinase activity that phosphorylates arginine/serine rich domains typical of splicing factors. Nucleic Acids Res. 1993;21:2815–2822. doi: 10.1093/nar/21.12.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein-RNA interactions and is necessary for splicing. Genes & Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]