Abstract
The fission yeast Sty1 stress-activated MAP kinase is crucial for the cellular response to a variety of stress conditions. Accordingly, sty1− cells are defective in their response to nutrient limitation, lose viability in stationary phase, and are hypersensitive to osmotic stress, oxidative stress, and UV treatment. Some of these phenotypes are caused by Sty1-dependent regulation of the Atf1 transcription factor, which controls both meiosis-specific and osmotic stress-responsive genes. However, in this report we demonstrate that the cellular response to oxidative stress and to treatment with a variety of cytotoxic agents is the result of Sty1 regulation of the Pap1 transcription factor, a bZip protein with structural and DNA binding similarities to the mammalian c-Jun protein. We show that both Sty1 and Pap1 are required for the expression of a number of genes involved in the oxidative stress response and for the expression of two genes, hba2+/bfr1+ and pmd1+, which encode energy-dependent transport proteins involved in multidrug resistance. Furthermore, we demonstrate that Pap1 is regulated by stress-dependent changes in subcellular localization. On imposition of oxidative stress, the Pap1 protein relocalizes from the cytoplasm to the nucleus in a process that is dependent on the Sty1 kinase. This relocalization is the result of regulated protein export, rather than import, and involves the Crm1 (exportin) nuclear export factor and the dcd1+/pim1+ gene that encodes an Ran nucleotide exchange factor.
Keywords: MAP kinase, stress activation, oxidative stress, Pap1 transcription factor, regulated localization
A rapid molecular response is induced in all eukaryotic cells on exposure to adverse environmental conditions. The response invariably involves changes in the level of gene transcription resulting from modulation of the activity of particular transcription factors (Treisman 1996). The link between stress signals and the transcription machinery is provided by certain MAP kinase signaling pathways that are strongly activated by a variety of adverse stimuli including osmotic, oxidative, and heat shock, UV light, and inhibition of protein synthesis. In mammalian cells these pathways are also activated by agonists, such as proinflammatory cytokines, which are released following stress or infection (for review, see Waskiewicz and Cooper 1995; Kyriakis and Avruch 1996). There are two classes of stress-activated MAP kinase pathways in mammalian cells: the stress-activated protein kinases (SAPKs), also known as the c-Jun-amino-terminal kinases (JNKs), and the p38/RK/CSBP kinases (Marshall 1994; Kyriakis and Avruch 1996). These pathways have been implicated in a number of biological processes, many of which are related to the stress response; they include cell transformation (Rodrigues et al. 1997), the promotion of apoptosis, and the synthesis of certain cytokines (for reviews, see Kyriakis and Avruch 1996; Cohen 1997; Karin et al 1997).
An important feature of these kinase pathways is their high degree of conservation: Structurally similar stress-activated kinases have been identified in Drosophila (Riesgo-Escovar et al. 1996; Sluss et al. 1996) as well as in budding (Brewster et al. 1993) and fission yeast (Millar et al. 1995; Shiozaki and Russell 1995; Kato et al. 1996). In fission yeast, the stress activated kinase Sty1 (also known as Spc1 and Phh1) is necessary for the cell to survive a number of different stress conditions including osmotic stress, UV irradiation, and nutrient limitation (Warbrick and Fantes 1991; Millar et al. 1995; Shiozaki and Russell 1995; Degols et al. 1996; Kato et al. 1996; Shiozaki and Russell 1996; Degols and Russell 1997).
Similar transcription factors have been found to be substrates for the stress activated kinases in mammalian cells, Drosophila, and fission yeast. Two transcription factor targets for the mammalian stress activated kinases are c-Jun (Hibi et al. 1993; Derijard et al. 1994; Kyriakis et al. 1994) and ATF2 (Gupta et al. 1995; Livingstone et al. 1995; Raingeaud et al. 1995; van Dam et al. 1995), both of which contain bZip DNA binding/dimerization domains. In both cases, phosphorylation enhances the transcriptional activity of the factors, the important phosphorylation sites being within the functionally defined transcriptional activation domains. The factors also contain a high affinity binding site for the kinases that is distinct from the phosphoacceptor sites. These interaction domains are crucial for phosphorylation and provide a mechanism for determining kinase specificity. A conserved link between kinase and transcription factor is found in Drosophila in which genetic evidence suggests that Drosophila Jun (DJun) is a critical target for DJNK (for review, see Noselli 1998). In the case of fission yeast, both biochemical and genetic evidence has demonstrated that one target of the Sty1 kinase is the bZip containing transcription factor Atf1, which shares homology to the mammalian factor ATF2 (Shiozaki and Russell 1996; Wilkinson et al. 1996). In addition, Sty1 and Atf1 can form a stable complex.
Inactivation of the fission yeast Sty1 kinase, or upstream components of the signaling pathway, results in a pleiotropic phenotype. sty1− cells are sensitive to osmotic stress, heat stress, oxidative stress, and UV irradiation; additionally they are defective in their response to nutrient deprivation (Millar et al. 1995; Shiozaki and Russell 1995; Kato et al. 1996; Degols and Russell 1997). When starved for nutrients, fission yeast cells exit the cell cycle and enter a metabolically inert stationary phase or, if mating partners are available, undergo sexual development culminating in meiosis and spore formation. However, sty1− cells are profoundly sterile and rapidly lose viability on growth to saturation indicating a failure to enter stationary phase. These defects emphasize the pivotal role the Sty1 kinase plays in the stress response of fission yeast cells. Sty1 also has a role in the regulation of cell cycle progression. Cells lacking Sty1 are substantially longer at cell division and show synthetic lethality with loss of the Cdc25 tyrosine phosphatase, a known regulator of entry into mitosis (Warbrick and Fantes 1991; Millar et al. 1992, 1995; Shiozaki and Russell 1995). To date, the only known substrate for the Sty1 kinase is the transcription factor Atf1. Cells lacking Atf1 show some, but not all of the phenotypes demonstrated by sty1− cells: They are sterile and sensitive to osmotic stress (Kanoh et al. 1996; Shiozaki and Russell 1996; Wilkinson et al. 1996) but are not sensitive to oxidative stress or to UV irradiation (Degols and Russell 1997). Furthermore, atf1− cells show no obvious cell cycle defects (Kanoh et al. 1996; Shiozaki and Russell 1996; Wilkinson et al. 1996). These observations suggest that alternative Sty1 substrates exist including additional transcription factor(s) and a regulator of mitotic initiation.
In this report, we identify the transcription factor Pap1 as a downstream target of Sty1. Pap1 is a bZip containing protein that has homology and similar DNA binding specificity to the mammalian c-Jun protein (Toda et al. 1991). Previous studies have identified a role for Pap1 in multidrug resistance, its deletion resulting in sensitivity to a variety of toxic compounds (Toda et al. 1991; Kumada et al. 1996). We show that sty1− cells have a similar multidrug-sensitive phenotype and that two genes encoding members of the ABC transporter family are expressed in a Sty1- and Pap1-dependent manner. We also show that cells lacking Pap1 are sensitive to oxidative stress but not to osmotic stress or nutrient deprivation. This sensitivity is caused by the role that Pap1 plays in regulating the expression of multiple genes involved in the oxidative stress response. Thus pap1− cells and atf1− cells show differing and nonoverlapping subsets of the phenotypes observed in sty1− cells. Our data demonstrate that Pap1 is regulated at the level of nuclear localization in a stress- and Sty1-dependent manner. Furthermore, regulated localization appears to be mediated at the level of nuclear export rather than import, in a mechanism that is dependent on the nuclear export factor Crm1 (Exportin). Because Pap1 and Atf1 have homology to c-Jun and ATF2 respectively, their identification as transcription factor targets of Sty1 highlights a remarkable level of conservation in the stress response between fission yeast and mammalian cells.
Results
pap1− and sty1− cells have overlapping phenotypes
Because deletion of atf1+ results in only a subset of the phenotypes seen when the Sty1 pathway is inactivated, additional targets for Sty1 must exist. A second transcription factor that has a role in the Schizosaccharomyces pombe stress response is a bZip protein called Pap1. Pap1 is a homolog of the budding yeast Yap1 protein and was initially identified on the basis of its ability to confer a multidrug resistance phenotype in fission yeast (Toda et al. 1991, 1992). The DNA binding domains of both Pap1 and Yap1 show good similarity to the DNA binding domains of the mammalian AP-1 family of transcription factors and both Pap1 and Yap1 can bind to, and activate transcription from, an AP-1 binding site.
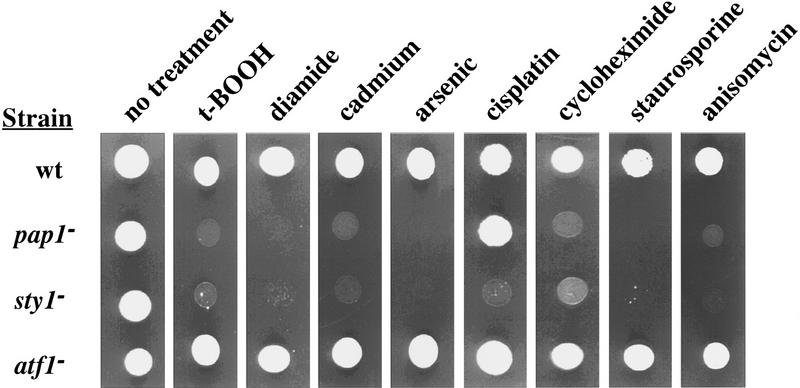
As a first step in determining whether Pap1 is regulated by the Sty1 pathway, we investigated whether or not pap1− and sty1− cells share overlapping phenotypes. As shown in Figure 1 inactivation of either pap1+ or sty1+ results in cells sensitive to oxidative stress elicited by the prooxidants diamide and t-butyl hydroperoxide (t-BOOH). In addition, both strains were found to be hypersensitive to treatment with H2O2 (data not shown) (Degols et al. 1996). Furthermore, both sty1− and pap1− cells were hypersensitive to treatment with a variety of structurally unrelated drugs including staurosporine, cycloheximide, and anisomycin as well as to heavy metal toxicity elicited by cadmium (Cd2+) or arsenite (As3+). In contrast, deletion of atf1+ did not result in increased sensitivity to any of these treatments.
Figure 1.
Phenotypes of strains carrying mutations in pap1+, sty1+, or atf1+. pap1− and sty1− cells were hypersensitive to a variety of cytotoxic compounds. Approximately 103 cells from exponentially growing cultures of each strain were spotted onto YE5S plates containing the indicated compounds: t-BOOH (0.5 mm); diamide (2 mm); cadmium sulfate (0.1 mm); sodium arsenite (0.5 mm); cisplatin (0.5 mm); cycloheximide (15 μg/ml); staurosporine (0.5 μg/ml); anisomycin (7.5 μg/ml). Plates were incubated at 30°C for 2–3 days.
Cisplatin (cis-diamminedichloroplatinum) is a platinum-containing compound that forms DNA–intrastrand cross-links and which is used in cancer chemotherapy. Studies performed in mammalian cells have shown a correlation between cisplatin resistance and resistance to heavy metals such as Cd2+ and As3+ (Naredi et al. 1995). Therefore, we tested the sensitivities of the sty1−, pap1−, or atf1− strains to cisplatin. sty1− cells were found to be hypersensitive to cisplatin treatment, whereas pap1− and atf1− cells were not (Fig. 1). However, a strain with both pap1+ and atf1+ inactivated showed a similar hypersensitive phenotype to cisplatin treatment as the sty1− strain (data not shown); this suggests that in the DNA damage response pathway, Pap1 and Atf1 may be functionally redundant.
Thus, sty1− cells share a number of stress-sensitive phenotypes with pap1− cells. These phenotypes included sensitivity to oxidative stress, heavy metal toxicity, and to treatment with a range of unrelated drugs. None of these phenotypes were seen in atf1− cells. These results suggest that Pap1, like Atf1, may be a downstream target of the Sty1 kinase, controlling the expression of specific Sty1-responsive genes.
Pap1 and Sty1 are required for stress-dependent gene expression
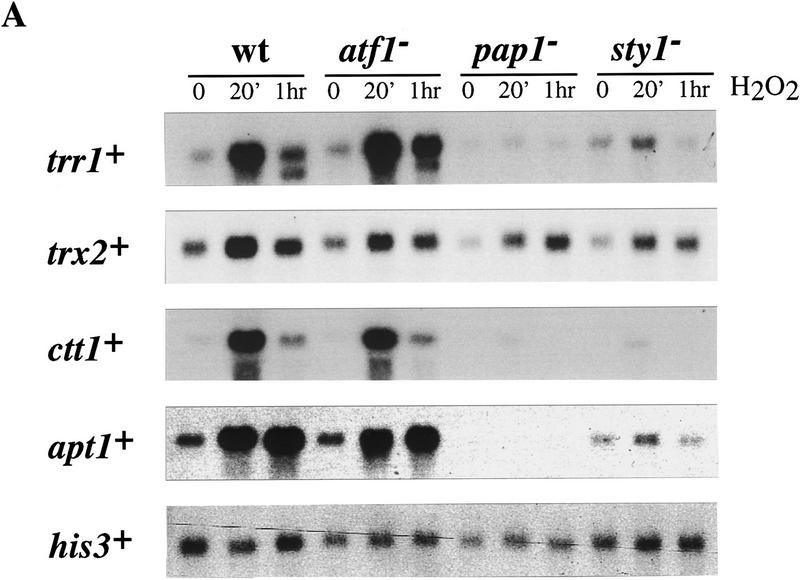
A number of genes in S. pombe that have functional roles in either the oxidative stress response, heavy metal detoxification, or multidrug resistance have been identified. Given their involvement in these responses, we tested whether or not they are transcriptionally induced following a specific stress or treatment and if so, whether this induction is dependent on Pap1, Sty1, or both. Thioredoxin is a small protein with a redox active disulfide bond. In Saccharomyces cerevisiae, TRX2, and TRR1 encoding thioredoxin and thioredoxin reductase respectively, are induced by oxidative stress in a Yap1-dependent manner (Kuge et al. 1994; Morgan et al. 1997). The S. pombe homolog of TRR1, trr1+, has been cloned recently (Casso and Beach 1996), and we have identified a homolog of TRX2, called trx2+ (B. Morgan unpubl.). trr1+ transcription was induced following oxidative stress in wild-type cells but was greatly reduced in pap1− and sty1− cells (Fig. 2A). In contrast, the response in cells deleted for the atf1+ gene was indistinguishable from that of wild-type cells. trx2+ expression was quantitatively similar; its expression was induced by oxidative stress in wild-type cells and this induction was partially dependent on Sty1 and Pap1. However, the basal level of expression of trx2+ was higher than that of trr1+ and there remained significant residual stress-dependent induction of trx2+ in both pap1− and sty1− cells. Thus, it is likely that additional factors are involved in trx2+ regulation.
Figure 2.
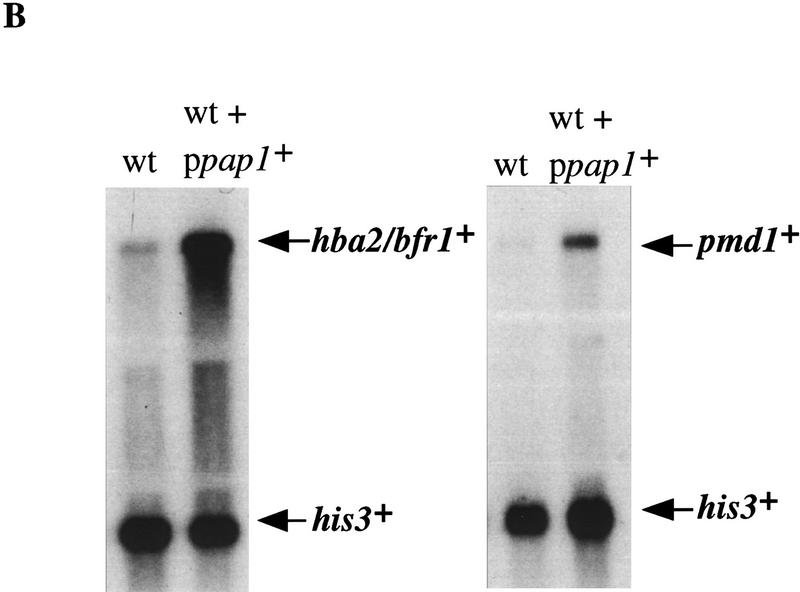
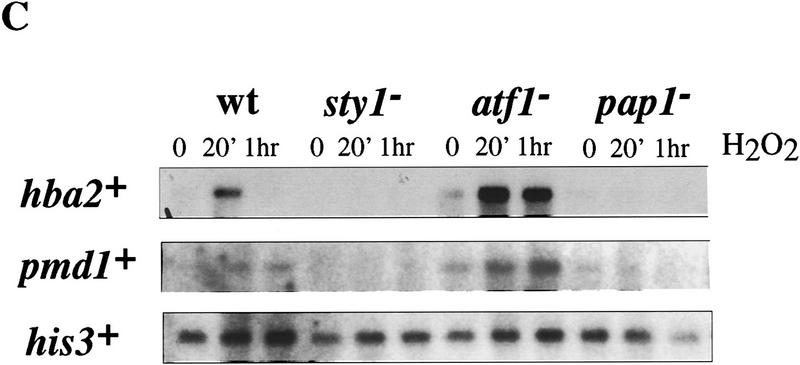
Northern analysis of stress-responsive genes in wild-type (wt), pap1−, sty1−, and atf1− strains following H2O2 treatment. (A) Total RNA was isolated from each of the indicated strains at 0, 20, and 60 min, following addition of H2O2 to a final concentration of 0.2 mm, electrophoresed, transferred to a nylon membrane, and probed with a 32P-labeled DNA specific to each of the genes denoted. The invariant his3+ mRNA was included as a loading control. (B) Total RNA was isolated from wild type (HM123) and HM123 carrying pRep1pap1+ Five micrograms of RNA was electrophoresed, transferred to a nylon membrane, and hybridized with 32P-labeled probes specific to either hba2+/bfr1+, pmd1+, or his3+. (C) Total RNA was isolated from the indicated strains before and after H2O2 treatment as described above, transfered to a nylon membrane, and probed with 32P-labeled probes specific to either hba2+/bfr1+, pmd1, or his3+.
The ctt1+ gene encoding catalase has been shown previously to be transcriptionally induced following exposure to a wide variety of stresses, including many of the stresses that activate the Sty1 pathway (Nakagawa et al. 1995). Furthermore, previous work has shown that induction of ctt1+ following osmotic stress is dependent on both Sty1 and the Atf1 transcription factor (Wilkinson et al. 1996). Surprisingly, following oxidative stress, induction of ctt1+ transcription was no longer dependent on Atf1 but rather was dependent on the Pap1 transcription factor (Fig. 2A). Moreover, this induction required the Sty1 kinase. Thus, expression of trx2+, trr1+, and ctt1+ is induced by oxidative stress in a manner that is dependent on both the Sty1 kinase as well as the Pap1 transcription factor.
Another gene, called apt1+ for AP-1 target gene, which is a known target of Pap1, encodes a protein called p25 (Toda et al. 1992). The function of p25 is unknown although it has homology to flavodoxins and, therefore, might have a role in the oxidative stress response. As shown in Figure 2A, apt1+ was strongly induced following oxidative stress and, again, this induction was dependent on the Sty1 kinase and the Pap1 transcription factor.
Several genes in S. pombe have been cloned on the basis of their ability to confer a multidrug resistant phenotype when overexpressed. These genes include: pap1+ (Toda et al. 1991); pad1+, a gene encoding both an apparent coactivator of Pap1 and a component of the 26S proteasome (Shimanuki et al. 1995; Spataro et al. 1997); hba1+, encoding a protein homologous to RanBPs (Turi et al 1996); and two genes encoding members of the ABC transporter family called hba2+/bfr1+ and pmd1+ (Turi and Rose 1995; Nagao et al. 1995; Nishi et al. 1992). Overexpression of pap1+ from the full strength nmt1+ promoter resulted in a multidrug resistant phenotype. This level of pap1+ expression also resulted in the induction of both hba2+/bfr1+ and pmd1+ (Fig. 2B); (expression of pad1+ and hba1+ was not affected by pap1+ overexpression; data not shown). Furthermore, both hba2+/bfr1+ and pmd1+ were induced following oxidative stress and this induction was dependent on both Pap1 and Sty1 (Fig. 2C). These results suggest that the hypersensitivity of both sty1− and pap1− cells to a variety of cytotoxic drugs is caused by the lack of hba2+/bfr1+ and pmd1+ expression.
With the exception of ctt1+, all genes described above, which appear to be direct targets of the Pap1 transcription factor, are not induced by osmotic stress even though such stress is a perfectly good activator of the Sty1 kinase pathway (Millar et al. 1995; Shiozaki and Russell 1995). Thus, activation of the Sty1 kinase is necessary, but not sufficient, for their induced expression, suggesting that a second signal, specific to oxidative stress, is required.
Pap1 is regulated at the level of nuclear localization
The observations described above suggest that Pap1 and Sty1 participate in the same pathway. Recent work in budding yeast has shown that the Pap1 homolog, Yap1, is regulated at the level of nuclear localization (Kuge et al. 1997). Under normal growth conditions Yap1 is predominantly cytoplasmic but following oxidative stress, it translocates to the nucleus. The cytoplasmic localization of Yap1 is dependent on a cysteine-rich domain (CRD) in its extreme carboxyl terminus. Thus, deletion of the Yap1 CRD results in constitutive nuclear localization of Yap1, and fusion of the CRD to a heterologous transcription factor imparts regulated nuclear localisation to that fusion protein (Kuge et al. 1997). Because the S. pombe Pap1 protein contains a region homologous to the CRD in its carboxyl terminus (Toda et al. 1991), we suspected that Pap1 is also regulated, at least in part, at the level of nuclear localization.
To test this hypothesis, we constructed a fusion of green fluorescent protein (GFP) to the amino-terminus of full-length Pap1. Expression of the fusion protein was placed under control of the intermediate strength thiamine repressible nmt1+ promoter (pRep41) (Fig. 3A). The GFP–Pap1 fusion protein was able to suppress the oxidative stress-sensitive phenotype of pap1− cells indicating that it retained Pap1 activity (data not shown). As shown in Figure 3B, in nonstressed cells the fusion protein was found to be cytoplasmic and visibly excluded from the nucleus. However, following treatment of the cells with H2O2 for 30 min, the GFP–Pap1 protein concentrated in a single spot that colocalized with DAPI-stained nuclei (Fig. 3C). Thus, like Yap1 in budding yeast, the Pap1 protein was regulated by stress-dependent nuclear localization. Intriguingly, induction of osmotic stress with 1.5 m sorbitol did not result in nuclear localization of GFP–Pap1 (data not shown). Therefore, Pap1 relocalization, like Pap1-dependent gene expression, appeared to be controlled specifically by oxidative stress.
Figure 3.
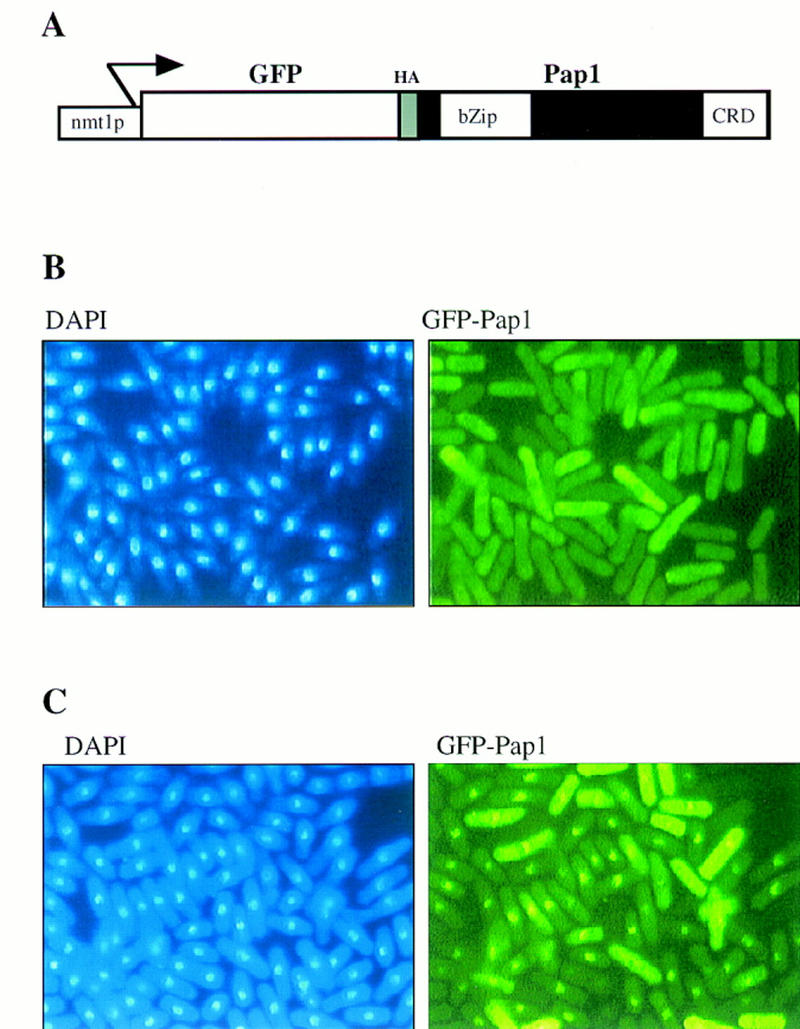
GFP–Pap1 localizes to the nucleus following oxidative stress. (A) A schematic of the GFP–pap1+ fusion construct expressed under the control of the intermediate strength nmt1+ promoter (pRep41). (B) Fluorescence microscopic analysis of untreated wild-type cells containing Rep41–GFP–Pap1, stained with DAPI to visualize the nucleus at 365 nm or at 390 nm to visualize the cellular distribution of GFP-tagged Pap1. (C) Fluorescence analysis of the same cells treated for 30 min with 0.2 mm H2O2.
Stress-dependent nuclear accumulation of Pap1 is dependent on an active Sty1 pathway
Next, we examined how the GFP–Pap1 fusion protein responded to stress in sty1− cells. The GFP–Pap1 protein was found to be cytoplasmic in nonstressed sty1− cells. However, following oxidative stress GFP–Pap1 localized to the nucleus in only a small minority (<5%) of cells relative to the wild-type control (Fig. 4). Thus, the Sty1 kinase was required for the regulated accumulation of Pap1 in the nucleus.
Figure 4.
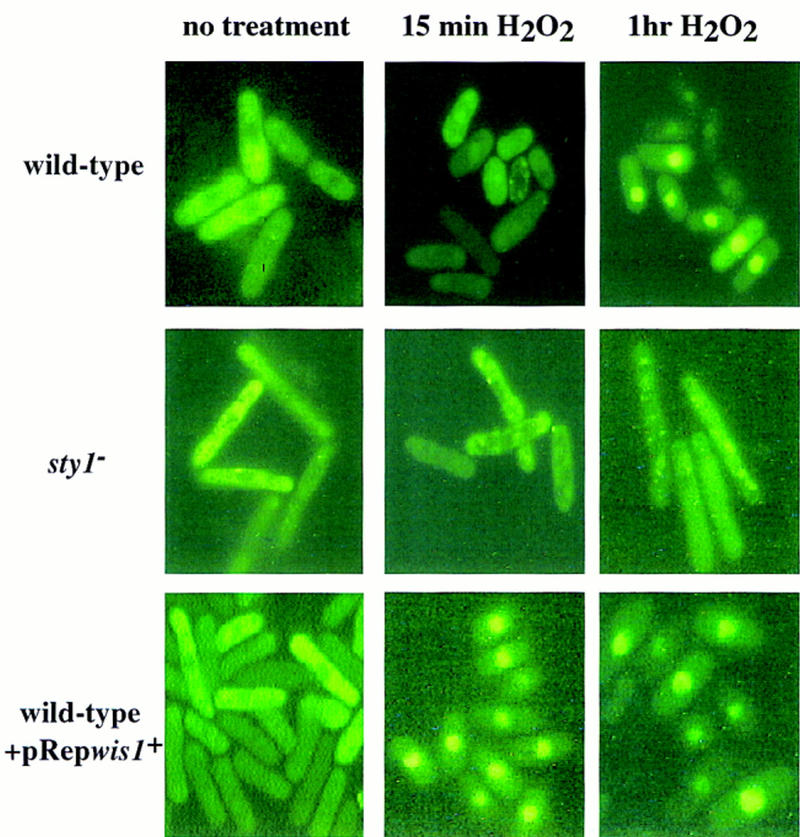
Stress-dependent relocalization of GFP–Pap1 to the nucleus involves the Sty1 pathway. Wild-type cells (TP114-2A + pRep41–GFP–pap1+ + pRep42), sty1− cells (NT224 + pRep41–GFP–pap1+ + pRep42), or wild-type cells overexpressing the Wis1 kinase (TP114-2A + pRep41–GFP–pap1+ + pRep42–wis1+) were exposed to 0.2 mm H2O2 for the indicated times and subjected to fluorescence microscopy to visualize the GFP-tagged Pap1 fusion protein.
Previous results have shown that overexpression of the MAP kinase, Wis1, can activate the Sty1 kinase (J. Millar, pers. comm.; Millar et al. 1995; Shiozaki and Russell 1995). By use of an nmt1+ promoter of intermediate strength (pRep41), we examined the effect of moderate overexpression of wis1+ on the cellular localization of GFP–Pap1. Interestingly, overexpression of wis1+ on its own had no obvious effect on GFP–Pap1 localization. However, overexpressing wis1+, in combination with oxidative stress, had an enhancing effect on the kinetics of GFP–Pap1 localization to the nucleus. Thus, GFP–Pap1 was seen to be concentrated in the nucleus ∼5–10 min after treatment with H2O2 in Rep41–wis1+-treated cells compared with wild-type cells that took at least 30 min before a comparable number of cells showed GFP–Pap1 nuclear localization (Fig. 4).
Pap1 nuclear localization is controlled by regulated nuclear export
Genetic studies have implicated a number of gene products in the control of multidrug resistance in fission yeast. Most of these proteins are predicted either to activate Pap1, or, as shown here for pmd1+ and hba2+/bfr1+, are targets for Pap1-dependent transcription. However, one gene product, Crm1, appears to function as a negative regulator of Pap1 activity because mutant alleles of crm1+ show a pap1+-dependent multidrug resistant phenotype (Toda et al. 1992; Kumada et al. 1996). crm1+ is an essential gene that encodes an evolutionarily conserved protein related to β-importin-like nuclear transport factors (Adachi and Yanagida 1989; Gorlich et al. 1997). Recently it has been shown that in S. cerevisiae, Xenopus, and mammalian cells, Crm1 is a nuclear export factor (Fornerod et al. 1997; Fukuda et al. 1997; Neville et al. 1997; Ossareh-Nazari et al. 1997, Stade et al. 1997). We analyzed the expression of Pap1-dependent genes in wild-type cells and cells carrying conditional mutations in crm1+ (Fig. 5A). Even at the permissive temperature, crm1-809 and crm1-119 cells showed increased expression of apt1+ and hba2+/bfr1+. The increased expression of hba2+/bfr1+ and potentially other genes encoding ABC transporters, such as pmd1+, could explain the multidrug resistant phenotype associated with crm1 mutants.
Figure 5.
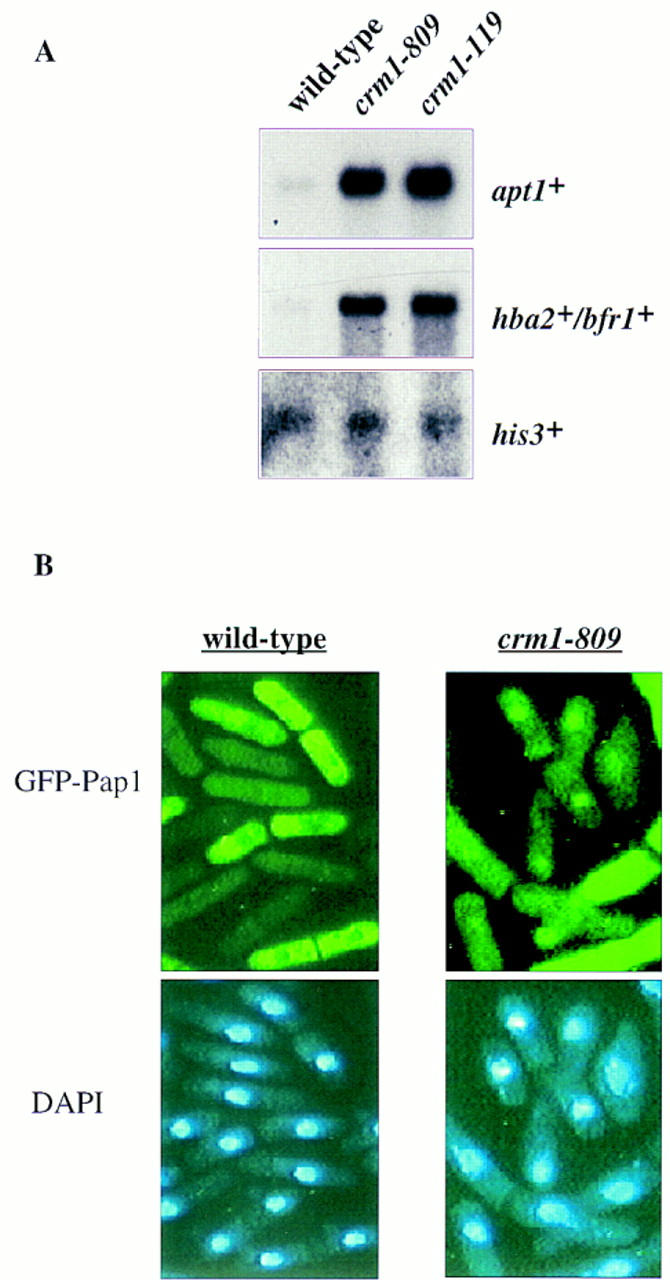
crm1 mutants show increased nuclear accumulation of GFP–Pap1. (A) Total RNA was isolated from exponentially growing cultures (30°C) of wild-type, crm1-809, or crm1-119 cells. Five micrograms of RNA was electrophoresed, transfered to a nylon (Genescreen) membrane and probed with a 32P-labeled probe specific to hba2+/bfr1+, apt1+, or his3+. (B) Exponentially growing wild-type cells, or crm1-809 cells, grown at 30°C, were subjected to fluoresence microscopy to determine the intracellular location of the GFP–Pap1 fusion protein.
Next, we examined the cellular localization of the GFP–Pap1 fusion protein in crm1-809 cells. Derepression of the nmt1+ controlled GFP–Pap1 gene followed by flouresence microscopy 24 hr later revealed that the GFP-Pap1 protein was strongly localized to the nucleus in ∼10% of cells (Fig. 5B). This result was significant because in wild-type cells the fusion protein was never seen in the nucleus of nonstressed cells. In the remaining 90% of crm1-809 (pRepGFP–Pap1) cells the nuclear exclusion of the fusion protein that was seen in wild-type cells was not apparent, suggesting that there was equal partitioning of the GFP–Pap1 protein between the nucleus and the cytoplasm.
RCC1 (nucleotide exchange factor) is required for nuclear export of Pap1
The small GTP-binding protein, Ran, is critical for nucleocytoplasmic trafficking (Nigg 1997). Recent experiments have shown that nuclear export is dependent on RanGTP, the high nuclear levels of which are maintained by the action of the RCC1 nucleotide exchange factor (which regenerates Ran–GTP from Ran–GDP). In S. pombe, the RCC1 homolog is encoded by the pim1+/dcd1+ gene (Matsumoto and Beach 1991; Sazer and Nurse 1994). Because dcd1+ is an essential gene, we used a dcd1ts strain to observe what effect inactivating this gene had on Pap1 activity and on GFP–Pap1 localization. As seen in Figure 6A, Pap1-dependent transcripts were high in the dcd1ts strain in unstressed conditions and at the permissive temperature. Even higher levels of expression were obtained when the cells were shifted to the nonpermissive temperature (36°C). Additionally, GFP–Pap1 was found to be predominantly nuclear in ∼10% of nonstressed dcd1ts cells at the permissive temperature and this nuclear localization increased dramatically (to ∼50%) following a shift to 36°C for 2–4 hr (Fig. 6B).
Figure 6.
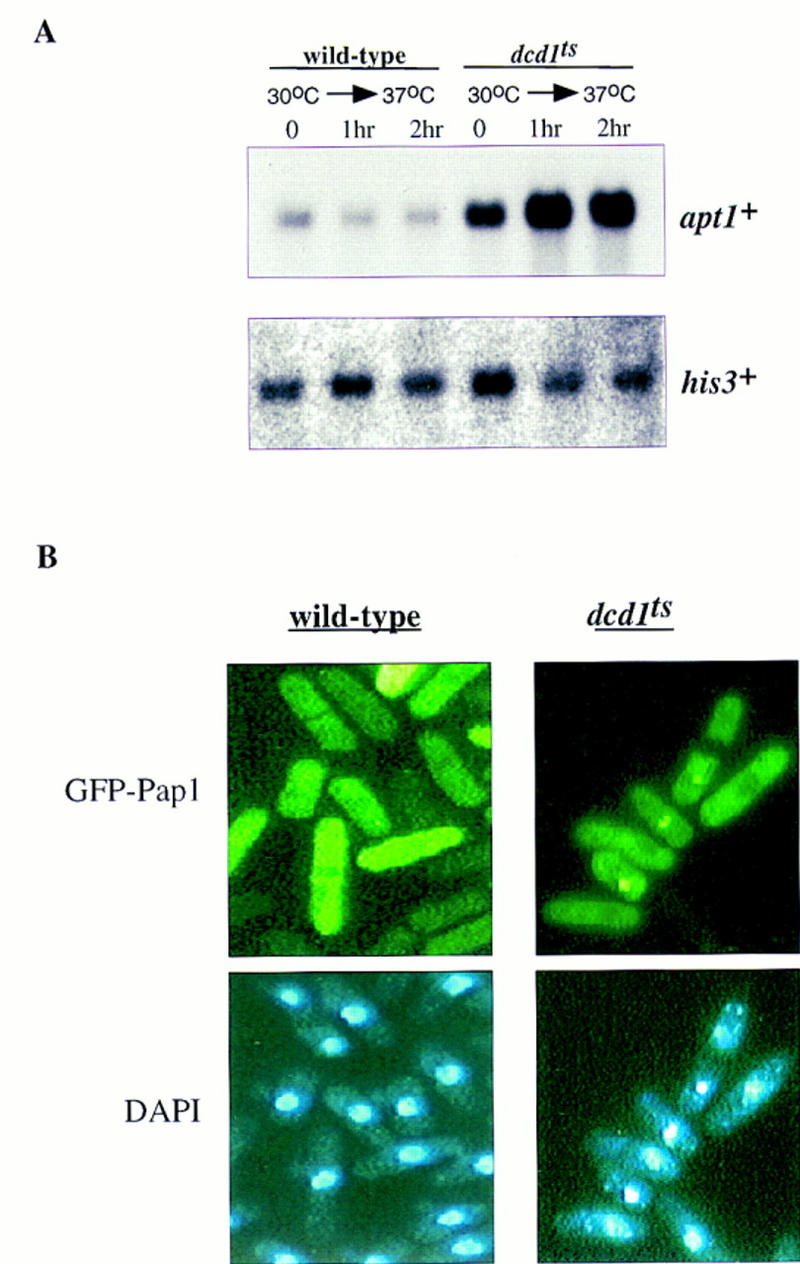
RCC1 mutants show increased nuclear accumulation of GFP–Pap1. (A) Total RNA was isolated from exponentially growing cultures of wild-type cells and dcd1ts cells grown at 30°C or cells shifted to the nonpermissive temperature (36°C) for 1 or 2 hr. Five micrograms of RNA was electrophoresed, transferred to a nylon (Genescreen) membrane, and probed with a 32P-labeled probe specific to apt1+ and his3+ as a loading control. (B) Exponentially growing wild-type cells or dcd1ts cells were subjected to fluoresence microscopy to determine the intracellular location of the GFP–Pap1 fusion protein after a shift of cells from the permissive (30°C) temperature to 36°C for 2 hr.
Discussion
Pap1 and Atf1 mediate different aspects of the Sty1-dependent stress response
In this report, we demonstrate that the activity of the Pap1 transcription factor is regulated by the Sty1 stress activated kinase. Accordingly, pap1− cells display a subset of the pleiotropic phenotypes that result from inactivation of sty1+ or components upstream in the Sty1 pathway. Both sty1− and pap1− cells are hypersensitive to treatment with a variety of drugs, to heavy metal toxicity, and to oxidative stress. This sensitivity derives from the role that Pap1 plays in regulating the expression of a number of genes involved in the oxidative stress response and in drug resistance.
Pap1 represents a second example of a transciption factor regulated by Sty1, the other being Atf1. Atf1 and Sty1 are critical for sexual differentiation and for surviving osmotic stress and entry into stationary phase (Takeda et al. 1995; Kato et al. 1996; Shiozaki and Russell 1996; Wilkinson et al. 1996). Therefore, apart from the cell cycle defect, all the phenotypes associated with deletion of the sty1+ gene can be rationalized through the role of this kinase in regulating the expression of Atf1- and Pap1-dependent genes (Fig. 7). It is clearly significant that these two transcription factors share considerable similarity both in structure and DNA-binding activity to the mammalian factors ATF2 and c-Jun, which are also regulated by stress-activated MAP kinase signaling. These findings further highlight the considerable conservation between mammalian cells and S. pombe in the transcriptional activities controlled by such signaling pathways.
Figure 7.
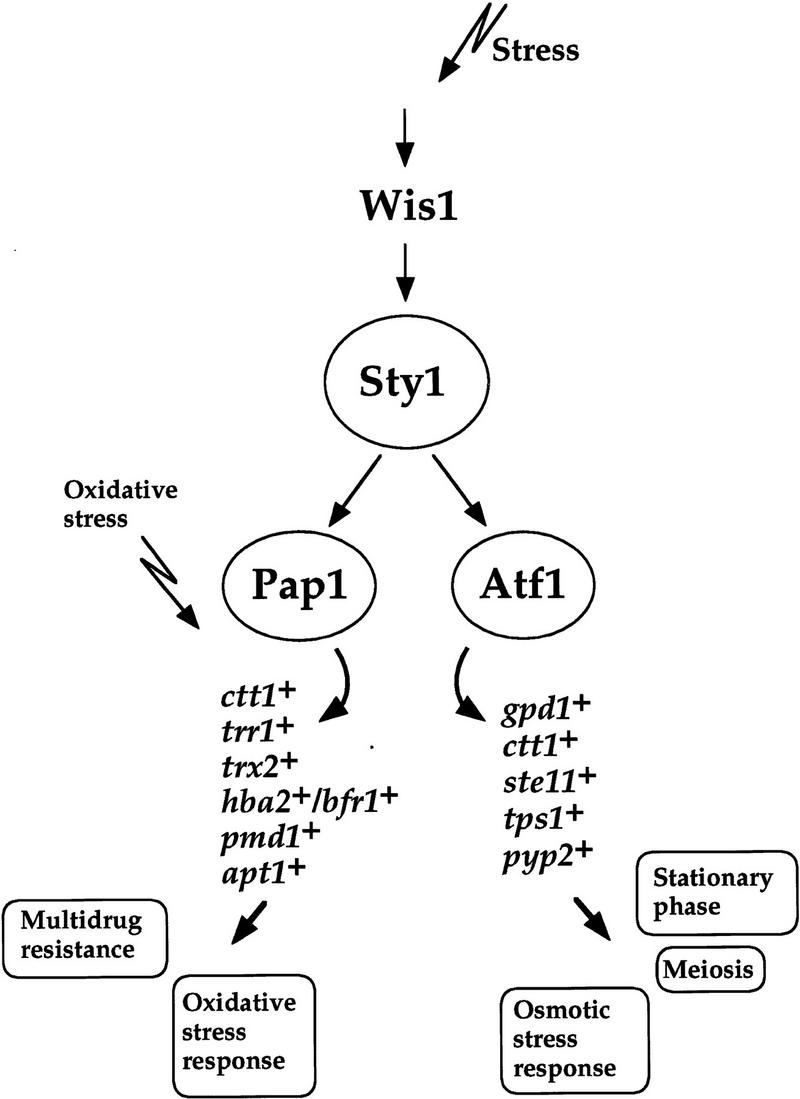
Model depicting the role of the Sty1 pathway in the fission yeast stress response. The Sty1 signaling pathway is involved in the response of cells to a variety of different stress conditions. The role of Sty1 is to regulate the activity of the two transcription factors Atf1 (Shiozaki and Russell 1996; Wilkinson et al. 1996) and Pap1 (this study), which in turn regulate the expression of a number of genes encoding products that mediate different stress responses. The physiological events controlled by each factor are indicated.
Pap1-mediated gene expression is essential for the oxidative stress response
We have identified a number of genes involved in the oxidative stress response that require both Pap1 and Sty1 for their expression. Three such genes are trr1+, trx2+, and ctt1+. Thioredoxin, which is ubiquitously expressed in both prokaryotes and eukaryotes, has been credited with a number of important properties. It plays a role in the reduction of reactive oxygen species and in the regeneration of damaged proteins by means of its protein oxidoreductase activity (Fernando et al. 1992; Mitsui et al. 1992). In this study we show that trx2+ and trr1+, encoding thioredoxin and thioredoxin reductase respectively, are induced following oxidative stress in a Pap1- and Sty1-dependent manner. Pap1 binding sites, but not Atf1 binding sites, could be found in both trx2+ and trr1+ promoters (W.M. Toone and N. Jones, unpubl.) consistent with the observation that neither gene was responsive to osmotic stress.
Catalase, encoded by ctt1+, is involved in the decomposition of H2O2. However, unlike trx2+ and trr1+, ctt1+ is transcriptionally induced following exposure to a variety of stresses including UV irradiation, osmotic stress, and oxidative stress (Nakagawa 1995). ctt1+ induction following osmotic stress is dependent on the Atf1 transcription factor as well as the Sty1 kinase (Wilkinson et al. 1996). However, in response to oxidative stress, induction of ctt1+, like that of trr1+ and trx2+, was found to be wholly dependent on the Pap1 transcription factor. Consistent with these observations, the ctt1+ promoter contains potential binding sites for both Atf1 and Pap1 (W.M. Toone and N. Jones, unpubl.). ctt1+ regulation in response to different stresses is, therefore, complex. Under osmotic stress conditions, Pap1 is not activated and, hence, expression of ctt1+ is dependent solely on Atf1. However, under oxidative stress conditions both factors are activated and yet ctt1+ expression is dependent on Pap1. This raises the question of why Atf1 cannot compensate for the loss of Pap1 under these conditions. This question is currently under investigation.
Pap1 and Sty1 are required for multidrug resistance
Previous studies in S. pombe have identified two genes, hba2+/bfr1+ and pmd1+, which encode ATP-binding cassette-type (ABC) transporter proteins. Overexpression of either gene confers a drug resistant phenotype (Nishi et al. 1992; Nagao et al. 1995; Turi and Rose 1995). We show that these genes are downstream targets of the Pap1 transcription factor and that overexpression of pap1+ results in the concomitant overexpression of hba2+/bfr1+ and pmd1+; this likely explains the multidrug resistant phenotype of pap1+ overexpressing cells. Moreover, both hba2+/bfr1+ and pmd1+ were found to be transcriptionally induced by oxidative stress in a Pap1- and Sty1-dependent manner. Both transporters are homologs of the human P-glycoprotein, encoded by the MDR1 gene, which is involved in the energy-dependent efflux of drugs used in cancer chemotherapy (for review, see Gottesman and Pastan 1993). Interestingly, MDR1 is known to be transcriptionally induced following heat stress, exposure to heavy metals such as arsenite and cadmium, UV irradiation, and drug treatment (Chin et al. 1990; Uchiumi et al. 1993). These are, in fact, many of the same treatments that activate the SAPK pathways in mammalian cells. Furthermore, multidrug resistant cell lines that overexpress MDR1 have been found to contain an activated form of the JNK (SAPK), which may in turn facilitate MDR1 expression through a noncanonical AP-1 consensus element in the human MDR1 promoter (Ueda et al. 1987).
The cancer therapeutic agent cisplatin induces DNA damage by the formation of 1,2-intrastrand crosslinks. In human cells, response to such damage appears to involve stress-activated signaling because the JNK/SAPK pathway is activated by cisplatin treatment and expression of a dominant negative Jun, which inhibits c-Jun-dependent transcription, results in cisplatin hypersensitivity (Potapova et al. 1997). Likewise, in fission yeast, sty1− cells are hypersensitive to cisplatin treatment. However, the involvement of Pap1 or Atf1 in this response is complex; neither pap1− nor atf1− cells are hypersensitive to cisplatin, whereas a pap1−atf1− double mutant is sensitive. This would suggest that the response could be mediated by either transcription factor. Clearly, to resolve this question, genes involved in cisplatin resistance would need to be identified and examined for their dependence on Pap1 or Atf1. Human cells deficient in nucleotide excision repair are unable to repair lesions resulting from either cisplatin or UV treatment (Zamble and Lippard 1995) suggesting a common response to both DNA damaging agents. Interestingly, the sensitivity of various mutant cells to UV irradiation is similar to that seen with cisplatin treatment; thus, neither pap1− nor atf1− cells are sensitive to UV irradiation whereas pap1−atf1− and sty1− cells are sensitive (W.M. Toone, unpubl.).
Pap1 is regulated by stress-dependent changes in subcellular localization
A growing number of transcription factors translocate from the cytoplasm to the nucleus in response to specific stimuli. Previous work has concentrated on mechanisms governing the regulated nuclear entry of these proteins (Nigg 1997). Thus, for example, the nuclear localization of the S. cerevisiae transcription factor Swi5, is regulated in a cell cycle-dependent manner through direct phophorylation by a cyclin-dependent kinase (Moll et al. 1991); nuclear entry of the mammalian transcription factor NF-κB is regulated by various cytokines and stress-induced signals (Baeuerle and Henkel 1994); whereas nuclear localization of the S. cerevisiae transcription factor Pho4 is negatively regulated by the nutrient-sensitive Pho80–Pho85 kinase (O’Neill et al. 1996).
Evidence presented here indicates that the fission yeast transcription factor, Pap1 is regulated by a mechanism acting primarily on the nuclear export machinery. Treatment of cells containing a GFP–Pap1 fusion protein with H2O2 resulted in the rapid accumulation of the protein in the nucleus. In agreement with the postulated role for Crm1 as a nuclear export factor and the genetic evidence implicating crm1+ in Pap1 regulation, GFP–Pap1 also accumulated in the nuclei of unstressed cells carrying a conditional allele of crm1+. Concurrent studies in budding yeast show that Yap1 activation is mediated in a similar Crm1-dependent manner (S. Kuge, T. Toda, N. Jones, N. Iizuka, and A. Nomoto, in prep.). Furthermore, the carboxy-terminal CRD of Yap1, which is required for the cytoplasmic localization of Yap1 in unstressed cells, has been shown to be a specialized nuclear export sequence.
A critical component involved in both nuclear import and export is the small GTPase, Ran. There is a gradient of Ran–GTP across the nuclear membrane with high concentrations of Ran–GTP inside the nucleus and low Ran–GTP in the cytoplasm. This gradient of Ran–GTP is thought to impart directionality to nucleocytoplasmic transport (Nigg 1997). High nuclear levels of Ran–GTP are maintained by the RCC1 nucleotide exchange factor, and recent results have shown that RCC1 is crucial for the nuclear export of importin-α (Koepp et al. 1996). Our results show that the RCC1 protein also drives Crm1-mediated export in fission yeast because GFP-Pap1 accumulates in the nucleus in pim1ts/dcd1ts cells.
How is Pap1 localization regulated by oxidative stress? One consequence of stress is the activation of the Sty1 pathway, and Sty1 is clearly required for GFP–Pap1 nuclear accumulation. However, activation of Sty is not sufficient because its activation by osmotic stress or by overexpression of Wis1 does not lead to increased concentration of Pap1 in the nucleus. Thus, an additional event specific to oxidative stress is required in combination with an active Sty1 kinase. A possible clue to the nature of this additional event comes from studies involving the Yap1 transcription factor in S. cerevisiae. The carboxy-terminal CRD of Yap1 contains a functional nuclear export sequence and the conserved cysteines residues within the CRD are critical for this NES function (Kuge et al. 1997; S. Kuge, T. Toda, N. Jones, N. Iizuka, and A. Nomoto, in prep.). Furthermore, recent evidence indicates that, in S. cerevisiae, the CRD and Crm1 interact (S. Kuge, T. Toda, N. Jones, N. Iizuka, and A. Nomoto, in prep.). A reasonable scenario, therefore, would be that the Crm1–CRD interaction is redox-sensitive and is disrupted under conditions of oxidative stress. Because Pap1 contains a homologous CRD in its carboxyl terminus, it is likely that it is regulated in a similar manner. This leaves open the question of the involvement of Sty1 in the Pap1 relocalization process. One possibility is that Sty1 affects the export machinery in a general way such that when Sty1 is activated, export is reduced. Alternatively, and perhaps more likely, Sty1 may be specifically involved in regulating Pap1 localization. Evidence from two-hybrid and coimmunoprecipitation analysis indicates that Sty1 and Pap1 exist as a complex although, surprisingly, there is no evidence that Pap1 is a substrate for the Sty1 kinase (W.M. Toone and M. Samuels, unpubl.). Thus, one possibility is that Sty1 is responsible for transporting Pap1 into the nucleus; in the absence of oxidative stress Pap1 would then normally be exported by Crm1 as a default mechanism. This possibility is strengthened by recent evidence that Sty1 relocalizes from the cytoplsm to the nucleus following its activation by stress (Gaits et al. 1998). Future experiments will be aimed toward clarifying the role of Sty1 in Pap1 localization.
Materials and methods
Yeast strains and media
The yeast strains used in this study were as follows: HM123 (h− leu1), TP114-2A (h90 leu1 ura4), TP108-3C (h− leu1 his2 ura4 pap1::ura4+), NT224 (h− leu1 ura4 sty1-1), NT147 (h90 leu1 ura4 atf1::ura4+), TP113-6B (h leu1 ura4 crm1-809), AT3 (h− leu1 ura4 crm1-119). Cells were grown in rich medium (YE5S) or in synthetic minimal medium (EMM2) as described (Moreno et al. 1991; Alfa et al. 1993).
Drug sensitivity assay
S. pombe strains to be assayed for sensitivity to various toxic compounds were grown to an OD595 of ∼1.0. Cells were then diluted in YE5S liquid media, and 103 cells in 5 μl were spotted onto YE5S agar media containing the specific compound at the concentration indicated. The spots were allowed to dry and the plates were incubated at 30°C for 2–3 days.
Yeast transformations
S. pombe strains were grown to 0.2–0.5 OD595, pelleted, and washed three times with 10 ml of ice-cold 1 m sorbitol. Cells were resuspended in 1 m sorbitol at a final concentration of ∼109 cells/ml. Transformations were performed on 200 μl of cells plus plasmid DNA by electroporation with a Bio-Rad Gene Pulser II set at 200 Ω, 25 μF, and 2.25 kV with a resulting time constant of ∼4.5 msec. Four hundred microliters of cold 1 m sorbitol was added and the cells plated immediately on selective EMM agar medium.
RNA analysis
Total RNA was extracted from cells grown under the conditions described with a hot phenol protocol (White et al. 1986). A 5-μg sample of total RNA was denatured with glyoxal, separated on a 1.2% agarose gel, and transferred to a GeneScreen hybridization membrane (Dupont NEN Research Products, Boston, MA). Probes for RNA–DNA hybridization were either restriction fragments or PCR-generated fragments internal to the gene concerned and labeled with 32P by use of a DNA Megaprime labeling kit (Amersham). The invariant his3+ transcript was used as a loading control.
Construction of GFP-tagged pap1 gene
The pRep41–GFPpap1+ plasmid consists of the S65T mutant form of GFP fused to the amino terminus of the pap1+ ORF with an intervening in-frame HA tag. A PCR fragment containing the HA epitope (underlined) CACCTGCCATGTACCCATACGATGTTCCAGATTACGCT and the 5′ end of the pap1+ coding sequence from the second codon to the BglII site at position +119 was ligated to a restriction fragment containing the rest of the pap1+ coding sequence as well as a portion of the 3′ non-coding sequence (BglII–VspI). This HA-pap1+ fragment was inserted into the pRS cp-GFP-HA–YAP1 plasmid (Kuge et al 1997) digested with PvuII (at the 3′ end of GFP) and NdeI (in the 3′ end of YAP) to release the YAP1 ORF. A restriction fragment from the resulting plasmid (SmaI–SalI) containing GFP–HA–pap1+ was ligated into the pRep41 vector (behind the nmt1+ promoter; Maundrell 1993) digested with NdeI (end-filled)–SalI.
Fluorescence microscopy
Samples from cells grown under the conditions described were centrifuged and fixed by resuspending in −20°C methanol for at least 10 min. The fixed cells were gently centrifuged and resuspended in 1 ml dH2O. The cells were centrifuged and all but the last few microlitres of H2O were removed. Cells were resuspended in the remaining H2O, and 3 μl of this suspension was spread on a poly-l-lysine-coated coverslip and allowed to dry. Three microliters of DAPI (1 μg/ml) was added, and the coverslip was placed on a microscope slide. Fluorescence microscopy was carried out on a Nikon Microphot FX microscope at 100× magnification. GFP fluorescence was observed in fixed cells with illumination at 390 nm. DAPI fluorescence was observed with illumination at 365 nm. Images were captured by use of a Hammatsu C5310 CCD camera. Images were converted to Photoshop version 3.0 (Adobe Systems, Mountain View, CA) and collated by use of Clarisdraw.
Acknowledgments
We thank Dr. J. Millar and members of the Gene Regulation Laboratory for helpful advice and discussions, P. Nurse and S. Sazar for the dcd1ts strain, and M. Yanagida for the crm1cs strain. This work was supported by the ICRF (N.J. and T.T.), the Human Frontier Science Program Organization (N.J.), and the Training and Mobility Program of the European Commission (N.J.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jonesn@icrf.icnet.uk; FAX 44171-269-3581.
References
- Adachi Y, Yanagida M. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene, crm1+, which encodes a 115-kDa protein preferentially localized to the nucleus and its periphery. J Cell Biol. 1989;108:1195–1207. doi: 10.1083/jcb.108.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with fission yeast. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Baeuerle PA, Henkel T. Function and activation of NFκB in the immune system. Annu Rev Immunol. 1994;12:540–546. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- Casso D, Beach D. A mutation in a thioredoxin reductase homolog suppresses p53-induced growth-inhibition in the fission yeast Schizosaccharomyces pombe. Mol & Gen Genet. 1996;252:518–529. doi: 10.1007/BF02172398. [DOI] [PubMed] [Google Scholar]
- Chin K-V, Tanaka S, Darlington G, Pastan I, Gottesman MM. Heat shock and arsenite increase expression of the multidrug resistance (MDR1) gene in human renal carcinoma cells. J Biol Chem. 1990;265:221–226. [PubMed] [Google Scholar]
- Cohen P. The search for physiological substrates of MAP and SAP kinases in mammalian cells. Trends Cell Biol. 1997;7:353–361. doi: 10.1016/S0962-8924(97)01105-7. [DOI] [PubMed] [Google Scholar]
- Degols G, Russell P. Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:3356–3363. doi: 10.1128/mcb.17.6.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degols G, Shiozaki K, Russell P. Activation and regulation of the Spc1 stress- activated protein kinase in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:2870–2877. doi: 10.1128/mcb.16.6.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davies RJ. JNK1: A protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- Fernando MR, Nanri H, Yoshitake S, Nagata-Kuno K, Minakami S. Thioredoxin regenerates proteins inactivated by oxidative stress in endothelial cells. Eur J Biochem. 1992;209:917–922. doi: 10.1111/j.1432-1033.1992.tb17363.x. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Gaits, F.,G. Degols, K. Shiozaki, and P. Russell. 1998. Phosphorylation and association with the transcription factor Atf1 regulate localization of the Sty1/Spc1 stress-activated kinase in fission yeast. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Gorlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- Gupta S, Campbell D, Derijard B, Davies RJ. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes & Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- Kanoh J, Watanabe Y, Ohsugi M, Iino Y, Yamamoto M. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells. 1996;1:391–408. doi: 10.1046/j.1365-2443.1996.d01-247.x. [DOI] [PubMed] [Google Scholar]
- Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- Kato T, Okazaki K, Murakami H, Stettler S, Fantes P, Okayama H. Stress signal, mediated by a HOG1-like MAP kinase, controls sexual development in fission yeast. FEBS Lett. 1996;378:207–212. doi: 10.1016/0014-5793(95)01442-x. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Wong DH, Corbett AH, Silver PA. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumada K, Yanagida M, Toda T. Caffeine-resistance in fission yeast is caused by mutations in a single essential gene, crm1+ Mol Gen Genet. 1996;250:59–68. doi: 10.1007/BF02191825. [DOI] [PubMed] [Google Scholar]
- Kyriakis JM, Avruch J. Protein kinase cascades activated by stress and inflammatory cytokines. BioEssays. 1996;18:567–577. doi: 10.1002/bies.950180708. [DOI] [PubMed] [Google Scholar]
- Kyriakis JM, Banerjee P, Nikolakaki E, Dai T, Rubie EA, Ahmad MF, Avruch J, Woodgett JR. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ. MAP kinase kinase kinase, MAP kinase kinase, and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Beach D. Premature initiation of mitosis in yeast lacking RCC1 or an interacting GTPase. Cell. 1991;66:347–360. doi: 10.1016/0092-8674(91)90624-8. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Millar JBA, Russell P, Dixon JE, Guan KL. Negative regulation of mitosis by two functionally overlapping PTPases in fission yeast. EMBO J. 1992;11:4943–4952. doi: 10.1002/j.1460-2075.1992.tb05601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JBA, Buck V, Wilkinson MG. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes & Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- Mitsui A, Hirakawa T, Yodoi J. Reactive oxygen reducing and protein refolding activities of adult T cell leukemia-derived factor/human thioredoxin. Biochem Biophys Res Comm. 1992;186:1220–1226. doi: 10.1016/s0006-291x(05)81536-0. [DOI] [PubMed] [Google Scholar]
- Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morgan BA, Banks GR, Toone WM, Raitt D, Kuge S, Johnston LH. The Skn7 response regulator controls gene expression in the oxidative stress-response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997;16:1035–1044. doi: 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao K, Taguchi Y, Arioka M, Kadokura H, Takatsuki A, Yoda K, Yamasaki M. bfr1+, a novel gene of Schizosaccharomyces pombe which confers brefeldin-a resistance, is structurally related to the ATP-binding cassette superfamily. J Bacteriol. 1995;177:1536–1543. doi: 10.1128/jb.177.6.1536-1543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa CW, Mutoh N, Hayashi Y. Transcriptional regulation of the catalase gene in the fission yeast Schizosaccharomyces pombe: Molecular cloning of the catalase gene and northern blot of the transcript. J Biochem. 1995;118:109–116. doi: 10.1093/oxfordjournals.jbchem.a124864. [DOI] [PubMed] [Google Scholar]
- Naredi P, Heath DD, Enns RE, Howell SB. Cross-resistance between cisplatin, antimony potassium tartrate, and arsenite in human tumor cells. J Clin Invest. 1995;95:1193–1198. doi: 10.1172/JCI117768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: Signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Nishi K, Yoshida M, Nishimura M, Nishikama MM, Nishiama M, Horinouchi S, Beppo T. A leptomycin B resistance gene of Schizosaccharomyces pombe encodes a protein similar to the mammalian P-glycoproteins. Mol Microbiol. 1992;6:761–769. doi: 10.1111/j.1365-2958.1992.tb01526.x. [DOI] [PubMed] [Google Scholar]
- Noselli S. JNK signalling and morphogenesis in Drosophila. Trends Genet. 1998;14:33–38. doi: 10.1016/S0168-9525(97)01320-6. [DOI] [PubMed] [Google Scholar]
- O’Neill EM, Kaffman A, Jolly ER, O’Shea EK. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for the role of Crm1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Potapova O, Haghighi A, Bost F, Liu CT, Birrer MJ, Gjerset R, Mercola D. The jun kinase stress-activated protein-kinase pathway functions to regulate DNA-repair and inhibition of the pathway sensitizes tumor-cells to cisplatin. J Biol Chem. 1997;272:14041–14044. doi: 10.1074/jbc.272.22.14041. [DOI] [PubMed] [Google Scholar]
- Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davies RJ. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- Riesgo-Escovar JR, Hafen E. Drosophila Jun kinase regulates expression of decapentaplegic via the ETS-domain protein Aop and the AP-1 transcription factor DJun during dorsal closure. Genes & Dev. 1997;11:1717–1727. doi: 10.1101/gad.11.13.1717. [DOI] [PubMed] [Google Scholar]
- Rodrigues GA, Park M, Schlessinger J. Activation of the JNK pathway is essential for transformation by the MET oncogene. EMBO J. 1997;16:2634–2645. doi: 10.1093/emboj/16.10.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S, Nurse P. A fission yeast RCC1-related protein is required for the mitosis to interphase transition. EMBO J. 1994;13:606–615. doi: 10.1002/j.1460-2075.1994.tb06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Cell cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- ————— Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes & Dev. 1996;10:2276–2301. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- Shimanuki M, Saka Y, Yanagida M, Toda T. A novel essential fission yeast gene pad1+ positively regulates pap1+-dependent transcription and is implicated in the maintenance of chromosome structure. J Cell Sci. 1995;108:569–579. doi: 10.1242/jcs.108.2.569. [DOI] [PubMed] [Google Scholar]
- Sluss HK, Han ZQ, Barrett T, Davies RJ, Ip YT. A JNK signal-transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes & Dev. 1996;10:2745–2758. doi: 10.1101/gad.10.21.2745. [DOI] [PubMed] [Google Scholar]
- Spataro V, Toda T, Craig R, Seeger M, Dubiel W, Harris A, Norbury C. Resistance to diverse drugs and ultraviolet light conferred by overexpression of a novel human 26S proteasome subunit. J Biol Chem. 1997;272:30470–30475. doi: 10.1074/jbc.272.48.30470. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, Jones N. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–6208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Shimanuki M, Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP1 and budding yeast FUS3 and KSS1 kinases. Genes & Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- Toda T, Shimanuki M, Saka Y, Yamano H, Adachi Y, Shirakawa M, Kyogoku Y, Yanagida M. Fission yeast pap1-dependent transcription is negatively regulated by an essential nuclear protein, Crm1. Mol Cell Biol. 1992;12:5474–5484. doi: 10.1128/mcb.12.12.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- Turi TG, Rose JK. Characterization of a novel Schizosaccharomyces pombe multidrug-resistance transporter conferring brefeldin-a resistance. Biochem Biophys Res Com. 1995;213:410–418. doi: 10.1006/bbrc.1995.2147. [DOI] [PubMed] [Google Scholar]
- Turi TG, Mueller UW, Sazer S, Rose JK. Characterization of a nuclear-protein conferring brefeldin-a resistance in Schizosaccharomyces pombe. J Biol Chem. 1996;271:9166–9171. doi: 10.1074/jbc.271.15.9166. [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Kohno K, Tanimura H, Matsuo K, Sato S, Uchida Y, Kuwano M. Enhanced expression of the human multidrug resistance 1 gene in response to UV light irradiation. Cell Growth & Differ. 1993;4:147–157. [PubMed] [Google Scholar]
- Ueda K, Pastan I, Gottesman M M. Isolation and sequence of the promoter region of the human multidrug resistance (P-glycoprotein) gene. J Biol Chem. 1987;262:17432–17436. [PubMed] [Google Scholar]
- van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warbrick E, Fantes P. The Wis1 protein kinase is a dose-dependent regulator of mitosis in Schizosaccharomyces pombe. EMBO J. 1991;10:4291–4299. doi: 10.1002/j.1460-2075.1991.tb05007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasiewicz AJ, Cooper JA. Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- White JH, Barker DG, Nurse P, Johnston LH. Periodic transcription as a means of regulating gene expression during the cell cycle: Contrasting modes of expression of DNA ligase genes in budding and fission yeast. EMBO J. 1986;5:1705–1709. doi: 10.1002/j.1460-2075.1986.tb04414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MG, Samuels M, Takeda T, Toone WM, Shieh J-C, Toda T, Millar JBA, Jones N. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes & Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- Zamble DB, Lippard SJ. Cisplatin and DNA repair in cancer chemotherapy. Trends Biochem Sci. 1995;20:435–439. doi: 10.1016/s0968-0004(00)89095-7. [DOI] [PubMed] [Google Scholar]