Abstract
Mutations in the Drosophila gene pavarotti result in the formation of abnormally large cells in the embryonic nervous system. In mitotic cycle 16, cells of pav mutant embryos undergo normal anaphase but then develop an abnormal telophase spindle and fail to undertake cytokinesis. We show that the septin Peanut, actin, and the actin-associated protein Anillin, do not become correctly localized in pav mutants. pav encodes a kinesin-like protein, PAV–KLP, related to the mammalian MKLP-1. In cellularized embryos, the protein is localized to centrosomes early in mitosis, and to the midbody region of the spindle in late anaphase and telophase. We show that Polo kinase associates with PAV–KLP with which it shows an overlapping pattern of subcellular localization during the mitotic cycle and this distribution is disrupted in pav mutants. We suggest that PAV–KLP is required both to establish the structure of the telophase spindle to provide a framework for the assembly of the contractile ring, and to mobilize mitotic regulator proteins.
Keywords: Cytokinesis, kinesin-like protein, Polo kinase, Drosophila
Cytokinesis is the ultimate act of mitosis, whereby segregated daughter nuclei are partitioned into two separate cells. In eukaryotes, this is accomplished by an actin–myosin contractile ring that forms around the cell equator during mitosis and constricts inwards at telophase (Schroeder 1972). In higher eukaryotes the correct positioning and assembly of the contractile ring requires the mitotic spindle, although the mechanism by which this occurs is a matter of controversy (Swann and Mitchison 1956; Zhang and Nicklas 1996). The classical experiments of Rappaport (1961) suggest that the spindle poles are sufficient to stimulate cytokinesis. By generating horseshoe-shaped single cell echinoderm embryos containing two mitotic spindles, not only were cleavage furrows formed in the central region of the mitotic spindles, but an extra furrow was formed equidistant between two asters lacking any intervening chromosomes or spindle microtubules. Similar phenomena have been observed in cultured mammalian cells, and although this does occur infrequently it suggests that this mechanism may act universally (Eckley et al. 1997; Rieder et al. 1997). Two mechanisms have been put forward to demonstrate how the spindle poles might induce cleavage furrows. In the first model, the poles signal directly to the equatorial cortex of the cell, perhaps by the action of astral microtubules (Devore et al. 1989). Alternatively, the astral relaxation model (White and Borisy 1983) proposes that spindle poles induce a relaxation of the cell cortex nearest to the poles, leading to a tension differential between the poles and the equator that results in equatorial contraction. However, this model has been discredited recently by force measurements showing that tension increases at the equator without a concomitant decrease at the cell poles (Burton and Taylor 1997).
On the other hand, there is growing evidence that the central spindle plays an essential role in the positioning and assembly of the contractile ring. The central spindle is composed of a dense network of overlapping antiparallel microtubules that forms between the separating daughter nuclei during anaphase (Mastronarde et al. 1993). In cultured cells induced to develop multipolar spindles as result of treatment with low concentrations of the microtubule-destabilizing drug colcemid, the formation of a cleavage furrow absolutely depends on the presence of central spindle microtubules that are required throughout cytokinesis (Wheatley and Wang 1996). Creation of a barrier between the central spindle and the cell cortex prevents cleavage, indicating that a signal for cytokinesis may emanate from the central spindle (Cao and Wang 1996). Manipulation of grasshopper neuroblast spindles results in the formation of a cleavage furrow when the spindle is moved close to the cell cortex (Kawamura 1977).
In addition to the contribution made by the spindle, several proteins, known as the chromosomal passenger proteins, dissociate from chromosomes at the metaphase–anaphase transition to be deposited at the cell equator. The inner centromere proteins (INCENPs), for example, transfer to the central spindle and the cell cortex and are necessary for completion of cytokinesis (Eckley et al. 1997; Earnshaw and Cook 1991), whereas TD-60 forms a disc structure in the position of the metaphase plate anticipating the cleavage plane (Andreassen et al. 1991). This disc is proposed to link the central spindle to the cell cortex (Martineau et al. 1995).
In Drosophila, mutational analysis has identified several genes encoding structural elements of the contractile ring essential for cytokinesis, such as myosin light chain, cofilin, profilin, and Peanut, a homolog of the yeast septins (Karess et al. 1991; Neufeld and Rubin 1994; Gunsalus et al. 1995; Giansanti et al. 1996). Recently, cloning of the gene defective in KLP3A mutants identified a kinesin-like protein (KLP) as having a role in cytokinesis in male meiosis (Williams et al. 1995). KLPs are microtubule motor proteins responsible for many of the dynamic aspects of mitosis such as centrosome separation, spindle assembly, and anaphase movement of chromosomes (Moore and Endow 1996).
In this paper we describe the phenotype of lethal mutations in the essential gene pavarotti (pav),which encodes a fly homolog of the mammalian mitotic kinesin-like protein-1 (MKLP-1). MKLP-1 has been suggested to play a role in anaphase B spindle pole elongation based on in vitro motility assays (Nislow et al. 1992). However, we show that in pav mutants anaphase B occurs normally but cytokinesis is defective, due to a disruption of the central spindle structure and subsequent failure to assemble a contractile ring. In mitosis, the KLP encoded by pav (PAV–KLP) first associates with the centrosomes and then becomes localized at the spindle equator at late anaphase. We also show that PAV–KLP associates with Polo kinase and is required for its localization to the centrosome and central spindle. The Polo-like kinases (plks) are known to be required for centrosome function: Mutations in the Drosophila gene polo and its fission yeast homolog plo1 lead to spindle defects including the formation of monopolar structures (Sunkel and Glover 1988; Lamazares et al. 1991; Ohkura et al. 1995). Injection of antibodies against human plk also result in the formation of monopolar structures in HeLa cells (Lane and Nigg 1996). An additional role for plks in cytokinesis is suggested from the late mitotic phenotype of fission yeast plo1 and its budding yeast counterpart cdc5 (Hartwell et al. 1973; Ohkura et al. 1995). Multiple septation has been shown to result from plo1+ overexpression in fission yeast, or the expression of mammalian Plk in budding yeast (Ohkura et al. 1995; Lee and Erikson 1997). In light of the association of Polo kinase with PAV–KLP, we discuss whether the plks might also have a role in cytokinesis in metazoan cells that has been obscured by the earlier requirement for mitosis.
Results
pav embryos show defects at cytokinesis
Preliminary characterization of the phenotype of pav mutants by staining embryos with the neuronal marker mAb 22C10 indicated the formation of fewer and larger neurons than usual in the embryonic development of the PNS, and an absence of support cells (Salzberg et al. 1994). To investigate whether the large cells seen in pav mutant embryos could arise from cell division defects, we looked for mitotic defects in pav mutants from cycle 14 (the first cycle under the control of the zygotic genome) onward through cycles 15 and 16 (Materials and Methods). We could detect no differences between wild-type and pav embryos in cycle 14 or 15, but in cycle 16 we observed that although mitosis appeared to be normal within the ventral epidermis, interphase nuclei were frequently closely associated in pairs in both P-element and EMS-induced alleles of pav (Fig. 1B). The boundaries of the cells of the embryo shown in this field are defined using an antibody against α-spectrin, a membrane-bound structural protein. It can be seen that the cell membrane does not invaginate during telophase in pav embryos as it does in wild type (Fig. 1A). Despite this failure of cytokinesis, nuclei appear to enter interphase 17 successfully, as nuclear laminae reform around both nuclei in binucleate cells (Fig. 1D). Such binucleate cells are never seen in wild type (Fig. 1C).
Figure 1.
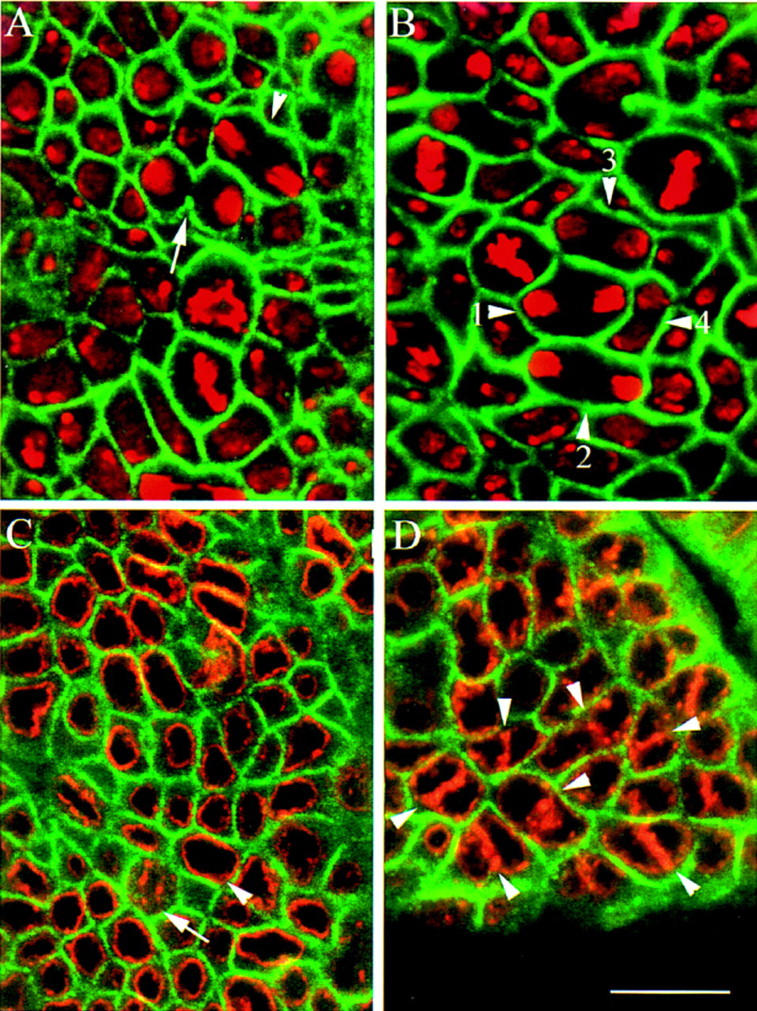
pav embryos show cytokinesis defects at cycle 16. (A) A wild-type embryo at cycle 16 stained to reveal α-spectrin (green) and DNA (red). The contraction of the cleavage furrow can be seen in cells in late anaphase (arrowhead) and telophase (arrow). (B) Cells in homozygous pavB200 embryos do not develop this constriction in cycle 16. Arrowheads 1–4 show the development of binucleate cells at progressively later stages from anaphase through telophase. (C) A wild-type embryo at cycle 16 stained to reveal lamin A (red) and α-spectrin (green). There is only one nuclear lamina per cell (arrowhead), which is dismantled during mitosis (arrow). (D) Homozygous pavB200 embryo at cycle 16, with many cells containing two nuclear laminae (arrowheads). Scale bar, 10 μm.
In addition to the cytokinesis defects that can be observed in essentially all cells at cycle 16, we also observed other mitotic defects at a very low frequency in pavB200 and pavM187 embryos but not in the P-element alleles. The affected cells show multipolar spindles and multiple centrosomes that are always associated with what appear to be tetraploid masses of chromatin (data not shown). This would suggest that in the pavM187 and pavB2004 alleles, binucleate cells are formed that can undergo another round of mitosis with double the usual number of centrosomes. This could occur if the maternal supply of pav gene product were to become exhausted in some cells during cycle 15. Such cells would fail to undertake cytokinesis but then proceed to attempt a sixteenth division, the last cell cycle for the majority of epidermal cells.
pav cells show defects in the central spindle at telophase, and septin and actin rings fail to form
To address how pav mutations might affect cytokinesis, we examined the structure of the mitotic spindle in mutant embryos. The morphology of the spindle was indistinguishable from wild type in the mitotic stages up until late anaphase. We observe no differences between wild-type and pav mutants in the degree of elongation of the spindle as cells progress from metaphase to late anaphase (see legend to Fig. 2). However, in pav mutants the distinctive morphology of the central spindle is substantially changed at telophase so that it appears to contain fewer bundles of microtubules (Fig. 2). In addition, whereas peripheral spindle microtubules (i.e., those microtubules emanating from the centrosome that contact the cell cortex) are seen to become constricted in wild type, this appears not to occur in the pav mutant.
Figure 2.
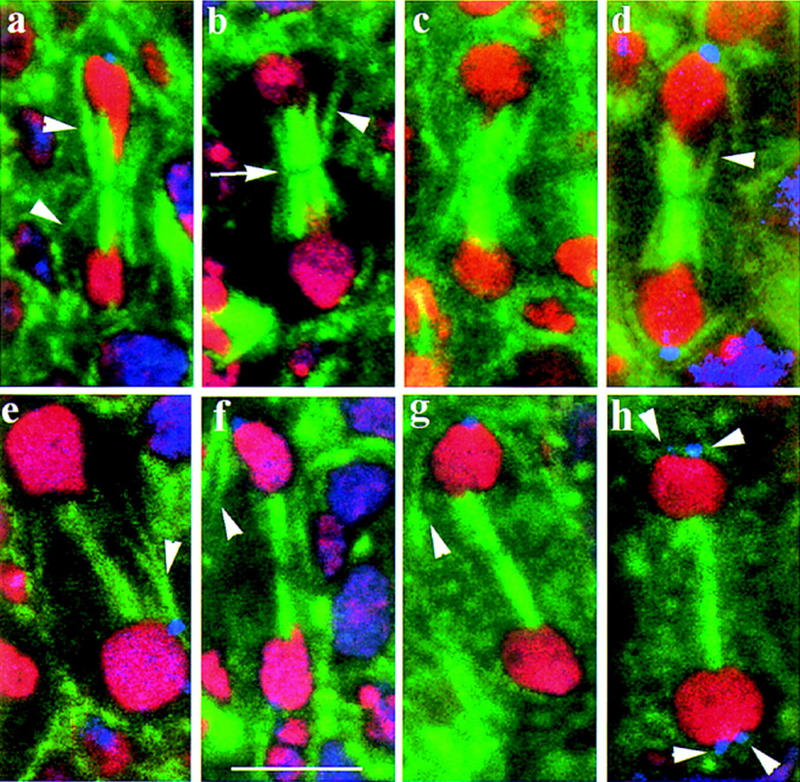
pav embryos show abnormal central spindles. A selection of confocal images showing telophase spindles from cycle 16 wild type (a–d) and pav (e–h) embryos stained to reveal β-tubulin (green), DNA (red), and the centrosomal antigen CP190 (blue). The wild-type spindle at telophase comprises a dense network of microtubules extending between the separated nuclei. Peripheral microtubules taper inward at the spindle equator due to the action of the contractile ring (arrowheads in a–d). A clear gap between the two halves of the spindle is also apparent (arrow in b). In contrast, the peripheral microtubules in pav telophases fail to taper inward (arrowheads in e–g) and the microtubules of the central spindle are generally more compact (a–h). (h) A rare telophase cell with four centrosomes and large nuclei. Measurements of centrosome–centrosome distances at metaphase and anaphase revealed no differences between wild type and pav mutants. Spindle lengths were 5.46 ± 0.46 μm (n = 32) wild-type metaphase; 9.41 ± 1.00 μm (n = 43) wild-type anaphase; 5.43 ± 0.41 μm (n = 10) pav metaphase; 10.22 ± 0.96 μm (n = 17) pav anaphase. Scale bar, 10 μm.
We also examined the localization of Peanut, Actin, and Anillin, proteins known to be required for cytokinesis. We first looked at the formation of the septin ring using antibodies to the Drosophila septin, Peanut. The septin family of proteins was originally identified in Saccharomyces cerevisiae as required for bud neck formation and subsequently shown to be present in most eukaryotes (Longtine et al. 1996). Although peanut has been shown to be required for cytokinesis in Drosophila, its exact role is unknown (Neufeld and Rubin 1994). Recent results suggest that it may mediate attachment between the actin network and the spindle (Glotzer and Hyman 1996). We observe that in wild-type cellularized embryos, Peanut protein is confined to the cell surfaces, with punctate spots of the protein at the site of the intercellular bridges remaining from the previous division (Fig. 3A). At late anaphase, Peanut accumulates in the equatorial region of the cell and appears at the contractile ring at telophase. In pav embryos, Peanut staining is generally weaker and more delocalized than in wild type, although some cells appear to have a near normal surface distribution of the protein. However, although areas of punctate Peanut staining can sometimes be seen in apparent association with the plasma membrane midway between the telophase nuclei, Peanut never appears at the site of the contractile ring (Fig. 3D).
Figure 3.
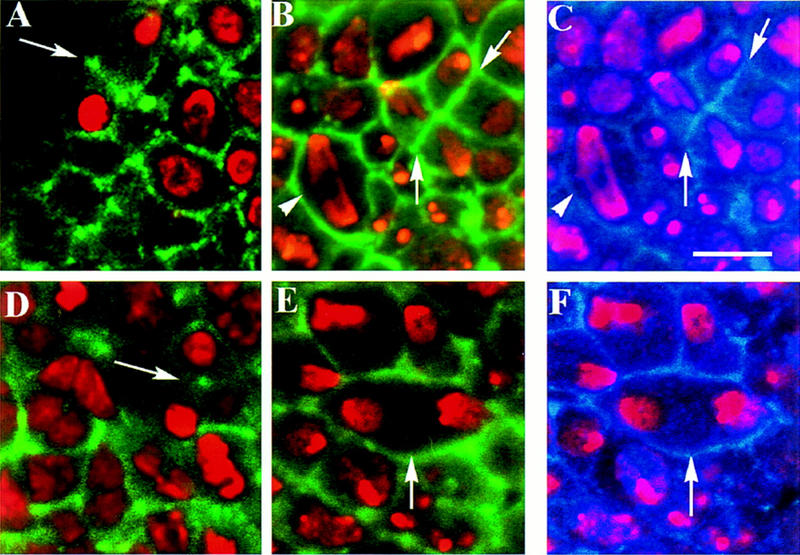
Neither Peanut protein, Actin, or Anillin becomes correctly localized at the end of mitosis in pav embryos. Wild-type (A-C) and pav (D–F) cycle 16 embryos were stained to reveal DNA (red) and either Peanut (A and D, green), Actin (B and E, green), or Anillin (C and F, blue). In wild type (A), Peanut is distributed beneath the cell surface, and at anaphase it accumulates near the nascent contractile ring (arrow) and remains associated with this region through telophase (arrow). In pav embryos (D) Peanut protein is variable in distribution and more diffuse than in wild type. Peanut is notably absent from the equatorial region of the cell during telophase (arrow). Dividing cells from wild-type embryos (B and C) accumulate actin (B) in the equatorial region at late anaphase (short arrow) through telophase (long arrow). The arrowhead shows a mid-anaphase figure, before this accumulation has occurred. The distribution of Anillin (C) at the end of mitosis is very similar to that of Actin. In addition, anillin shows faint nuclear staining during interphase. In pav embryos, neither Actin nor Anillin localize to the equatorial region during telophase (arrows in E and F, respectively). Scale bar, 10 μm.
We also examined pav mutants to determine whether Actin or Anillin localized at the contractile ring and if cleavage furrows would form. In wild-type cellularized embryos, Actin is present around the surface of the cell throughout the cell cycle and accumulates in the equatorial region of the cell during telophase before forming the contractile ring (Fig. 3B). The Actin binding protein Anillin is thought to be involved in regulating or organizing the contractile domains of the Actin cytoskeleton (Field and Alberts 1995). It is found in the nucleus of wild-type cellularized embryos during interphase but is relocalized to the cortex during mitosis and to the cleavage furrow at cytokinesis (Fig. 3C). It appears at the cleavage furrow slightly preceding Actin. In pav mutant embryos, there is no indication of the accumulation of either Actin or Anillin at the position that the contractile ring and cleavage furrow normally form. However, the distribution of these proteins at other stages of the cell cycle appears to be unaffected (Fig. 3E,F).
pav encodes a KLP with homology to MKLP1
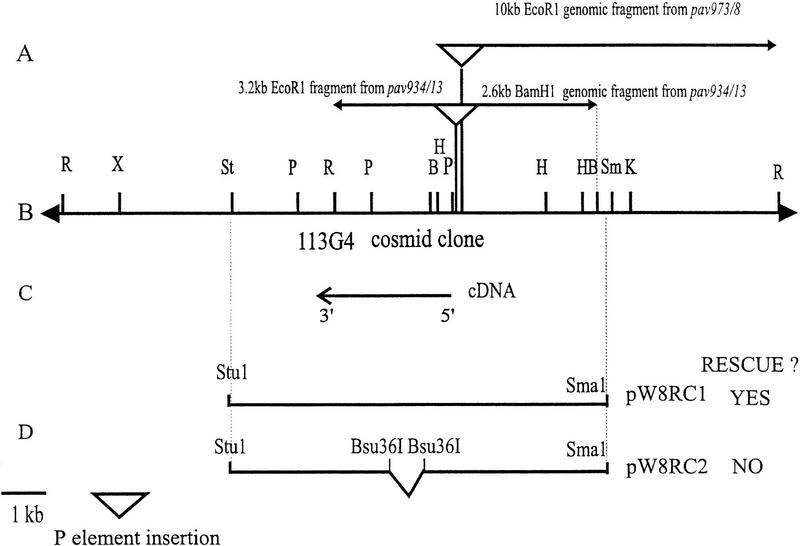
A route toward the cloning of the pav gene was provided by the finding of P-element-induced alleles pav934/13 and pav973/8 in a collection of third chromosome P-element insertion mutants (Deak et al. 1997; Salzberg et al. 1997). One of these mutants, pav934/13, reverted to wild type following mobilization of the P element under dysgenic conditions, indicating that the P element is likely to be responsible for the mutation (Salzberg et al. 1997; Y. Inoue, pers. comm.). We rescued sequences flanking the P insertion at 64B 2-10 that detected 3.2-kb cDNA clones in 0- to 4-hr and 0- to 8-hr embryonic cDNA libraries (Brown and Kafatos 1988). DNA sequencing revealed that the P element lay 191 bp immediately 5′ to the first ATG of the ORF within the cDNA. To test whether this candidate gene would rescue pav mutations, we introduced a 10.5-kb segment of genomic DNA containing 4 kb of DNA upstream and 3 kb downstream of the transcription unit into the transformation vector pW8 (Klemenz et al. 1987) termed pRC1 in Figure 4. A transformant containing this 10.5-kb fragment is able to rescue two different alleles of pav that were tested (see Materials and Methods). To demonstrate that pav was rescued by the candidate gene immediately downstream of the P-element insertion, and not by some other gene present in the rescue construct, we made a second construct, pRC2, containing an internal 1.3-kb deletion within the coding region of the gene. Two independent transformants carrying this construct were unable to rescue pav, confirming the correct identity of the candidate gene.
Figure 4.
Physical map of the pav region. (A) Genomic fragments obtained by plasmid rescue of P-element mutants. (B) Restriction endonuclease cleavage map of a 20.5-kb segment of genomic DNA. Cleavage sites are: EcoRI (R); XbaI (X); StuI (St); PstI (P); HindIII (H); SmaI (Sm). (C) Location and polarity of 3.2-kb pNBpav cDNA. (D) Transformation constructs to test rescue of pav phenotype.
The sequence of a 3.2-kb pav cDNA reveals an ORF of 2658 bases coding for an 886-amino-acid protein of 100 kD predicted molecular mass with homology to the KLP family of microtubule motor proteins (Fig. 5). It contains a predicted microtubule motor domain of ∼350 amino acids that includes a nuclear localization signal, an ATP binding site, and several motifs common to plus end-directed KLPs. A predicted coiled–coil region lies between residues 500 and 700, whereas residues 700–886 show homology only with the MKLP-1 subfamily of KLPs. PAV–KLP is most similar to this subfamily throughout its entire sequence, showing 34% identity with MKLP-1 as compared to 13% identity to other mitotic KLPs. The highest degree of identity (48%) lies in the motor and hinge domains (residues 1–450), although patches of identity extend throughout the entire carboxy-terminal region of the protein (residues 700–886), a segment of KLPs known to be highly diverged. The MKLP-1 subfamily of KLPs comprises human MKLP-1 and its Chinese hamster counterpart CHO-1, first identified as a mitotic spindle antigen (Sellitto and Kuriyama 1988; Nislow et al. 1992.)
Figure 5.
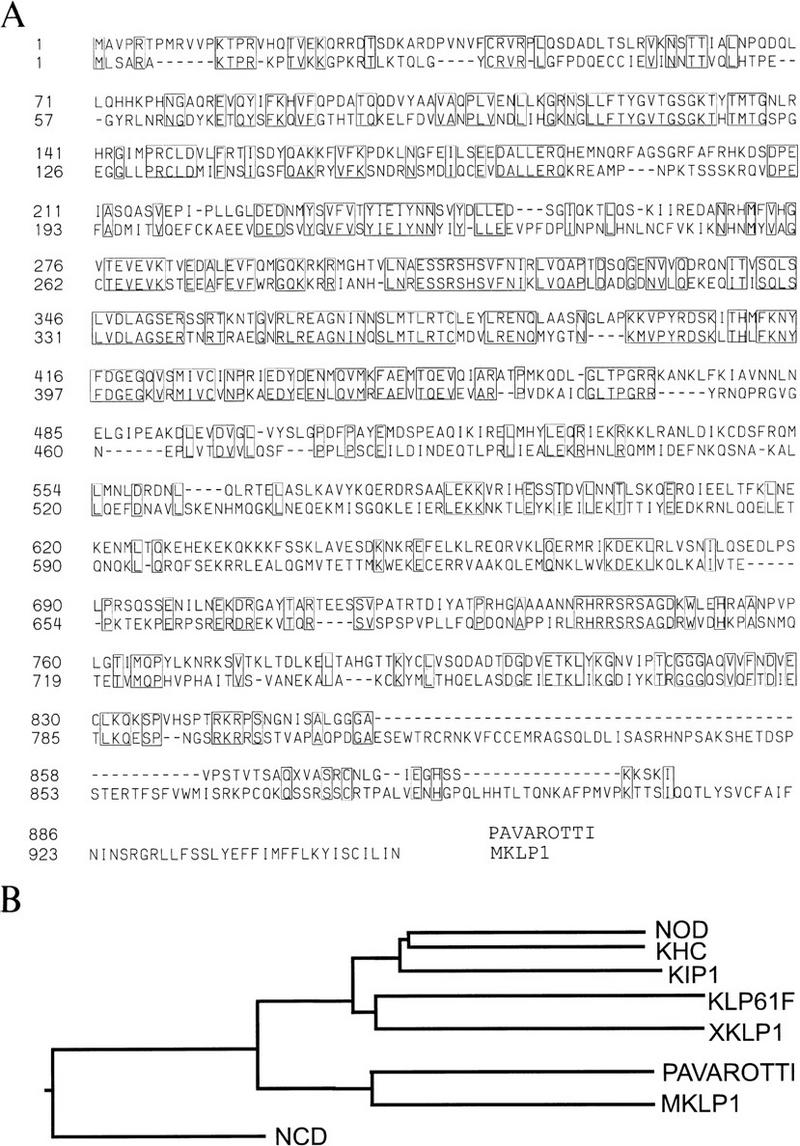
Comparison of the PAV–KLP with MKLP-1. (a) The full nucleotide sequence of the pNBpav cDNA and corresponding genomic region have been deposited in the EMBL databank. There are five putative initiation ATGs between the site of the P-element insertion and the longest ORF present in pNBpav. This ORF, located 191 bp, 3′ to the site of the P-element insertion identifies a protein of 886 amino acids with extensive homology to MKLP-1 and a predicted molecular mass of 100 kD. PAV–KLP contains a consensus nuclear localization motif (KTPR) at amino acids 13–17, an ATP binding site (GSGKT) at amino acids 132–137, and two motifs common to all KLPs (SSRSHS and LAGSE) at residues 308–313 and 349–352, respectively. Analysis of the sequence using the Lupas algorithm (Lupas et al. 1991) predicts the formation of coiled–coil regions in the central region of the protein (residues 500–700; data not shown). The comparison of the amino acid sequences of PAV–KLP (top row) and MKLP-1 (bottom row) was made by Clustal analysis using DNASTAR software (Lasergene). Identical residues are boxed. Both sequences show substantial sequence identity in the first 450 amino acids, corresponding to the conserved motor domain. Regions of sequence identity also extend into the carboxy-terminal domains. Overall, PAV-KLP shows 34.3% identity with MKLP-1. (B) A phylogenetic tree illustrating the sequence homology between mitotic KLPs. PAV–KLP is related more closely to MKLP-1 than any other KLP shown. Other than these two KLPs, the remaining members of this superfamily that are shown are Drosophila NOD; Drosophila KHC (kinesin heavy chain); S. cerevisiae KIP1; Drosophila KLP61F; Xenopus XKLP1, and Drosophila NCD (Yang et al. 1988; McDonald et al. 1990; Zhang et al. 1990; Hoyt et al. 1992; Heck et al. 1993; Vernos et al. 1995).
PAV–KLP associates with the spindle poles early in mitosis and with the spindle interzone from late anaphase
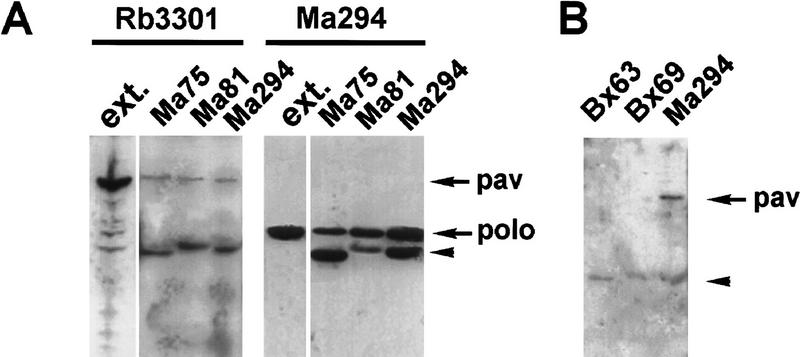
We wished to study the subcellular distribution of the PAV–KLP protein throughout the cell cycle and study its potential association with other molecules. To this end, we raised an antibody against a 450-amino-acid peptide from the central region of the protein (residues 208–686) expressed in Escherichia coli. The antibody recognizes not only the bacterially expressed protein but also one predominant band at ∼100 kD in Western blots of Drosophila embryo extracts (Fig. 6A). It immunostains wild type and pavB200+/ and embryos at cycle 16 to give distinct punctate nuclear staining in interphase cells in the ventral epidermis. In mitotic cells, bright bands of staining are seen equidistant from separating telophase nuclei, presumably at the site of the spindle interzone. Bright spots are also seen that appear to correspond to midbodies persisting from earlier divisions (Fig. 6B and see below). In contrast, pavB200 homozygous mutant embryos lack all nuclear and central spindle staining at cycle 16, except for some punctate spots likely to correspond to maternally contributed protein in remnants of putative midbodies from earlier cycles (Fig. 6C). Thus, the immunostaining is specific for PAV–KLP.
Figure 6.
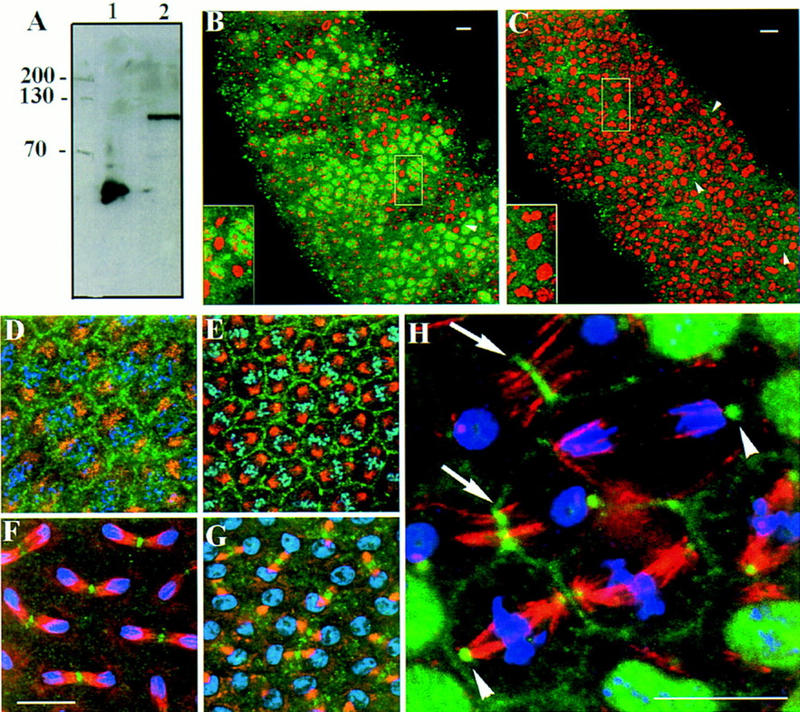
Subcellular localization of PAV–KLP at division. (A) Western blot analysis of E. coli cells induced to express the construct PpavEC2, and 0- to 4-hr Drosophila embryo extracts probed with the rabbit antiserum Rb3301. The antiserum recognizes the 52-kD polypeptide against which it was raised (lane 1) and a single protein of ∼100 kD in the embryo extracts (lane 2). We were unable to carry out Western analysis on homozygous mutant embryos, as it proved difficult to distinguish these from their heterozygous siblings on the basis of the phenotype revealed by DNA staining alone. We could distinguish homozygotes from heterozygotes carrying a balancer chromosome expressing lacZ, but neither immuno- nor histochemical staining methods to detect β-galactosidase preserved proteins sufficiently well for Western analysis. (B) Homozygotes and heterozygotes can, however, be distinguished by immunostaining with the anti-pav antibody, even though this antibody is not sensitive enough to detect levels of protein from single embryos on Western blots. TM6B/TM6B or pavB200/TM6B embryos show strong nuclear staining in segmented bands of interphase cells using the antibody Rb3301 (green; DNA staining is in red). Bright regions of staining are also seen equidistant from separating anaphase cells (arrowhead and inset). Smaller punctate staining is also visible scattered throughout the embryo. (C) pavB200 homozygotes show neither nuclear staining at interphase nor equatorial staining at anaphase using the Rb3301 antibody (arrowheads and inset). However, there are some spots of staining distributed throughout the embryo, likely to correspond to the staining of residual maternally contributed protein. (D–G) Syncytial wild-type type embryos were stained to reveal PAV–KLP with Rb3301 (green), tubulin with monoclonal antibody YL1/2 (red), and DNA with TOTO-3 (blue). At prophase (D), PAV–KLP is diffusely distributed but is beginning to locate to the furrows separating the mitotic nuclei. PAV–KLP localization to the furrows persists through metaphase (E). Upon initiation of anaphase (F), PAV–KLP relocates to the spindle interzone region. At late telophase (G) PAV–KLP staining is concentrated at the midbody region of the spindle, and diffuse staining is reappearing in the cytoplasm. (H) A cellularized embryo at cycle 14 showing cells at various stages of mitosis. PAV–KLP is visible at the centrosomes in metaphase and anaphase cells (arrowheads) but does not appear at the spindle midzone until telophase, where it clearly colocalizes with microtubules (arrows). Interphase cells show punctate nuclear staining.
A closer examination of the localization of PAV–KLP revealed that in wild-type syncytial embryos, the protein is diffuse during interphase (Fig. 6D), but during prophase and prometaphase it becomes localized in the actin-containing furrows that separate nuclei and maintain nuclear spacing (Fig. 6E). During anaphase, the central, interzonal region of the mitotic spindle is stained (Fig. 6F). Anaphases showing staining at both sites are very rarely seen, even though the slight asynchrony between dividing nuclei in the blastoderm embryo allows all stages of anaphase to be observed in a single embryo. At telophase, staining intensifies at the spindle interzone, and as nuclei enter the next interphase the staining becomes diffuse once again (Fig. 6G).
In cellularized cycle 14 embryos (Fig. 6H) staining is nuclear during interphase. It is then possible to detect centrosomal staining in cells at metaphase and anaphase, and at late anaphase the protein begins to concentrate at the central region of the spindle. As telophase progresses, staining remains associated with the spindle during its constriction by the contractile apparatus.
As the septin homolog Peanut has been proposed to connect the central spindle with the contractile ring, we examined its distribution with respect to that of PAV–KLP. At late anaphase, Peanut becomes concentrated at the spindle interzone, and colocalizes with PAV–KLP (data not shown). At telophase, we found that PAV–KLP remains associated with microtubules at the midbody, whereas peanut staining extends further and is concentrated at either side of the midbody closer to the plasma membrane. Thus, PAV–KLP and Peanut do not fully colocalize at this stage. However, they do colocalize in the previously identified small punctate regions of staining. As Peanut is known to be expelled from the cell at the end of mitosis (Neufeld and Rubin 1994), the presence of PAV–KLP in the same location supports the notion that these spots are remains of midbodies from previous divisions.
The localization of polo protein kinase is disrupted in pav embryos
It has been reported previously that MKLP1 can be detected in an immunoprecipitate of murine plk (Lee et al. 1995). In light of our results this association of the two proteins would be consistent with a role for plks in cytokinesis in metazoans, as has been shown in fission yeast (Ohkura et al. 1995). We therefore carried out immunoprecipitation experiments on extracts of Drosophila embryos using antibodies against Polo protein kinase. Western blot analysis indicates that PAV–KLP is present in the Polo immunoprecipitates obtained with either of three different anti-polo monoclonal antibodies but not with antibodies that recognize β-tubulin or the centrosomal component CP190 (Fig. 7). We therefore undertook immunostaining experiments to determine the extent to which these proteins colocalize throughout the mitotic cycle. Using the mAb 294 anti-Polo antibody we were able to observe that in cells of wild-type embryos, Polo protein kinase localizes at centrosomes early in mitosis and, simultaneously, some staining is detected associated with the metaphase chromosomes (Fig. 8b). The immunostaining of the centrosomes by the anti-Polo antibody is progressively lost during anaphase–telophase concurrently with an increase in the staining of the midbody. By the end of telophase Polo protein seems to be associated only with the midbody. There is extensive colocalization of PAV–KLP to the centrosome and subsequently to the midbody during mitosis (Fig. 8a,b,c). In pav mutant embryos, on the other hand, no centrosome staining is observed either with the anti-PAV–KLP or the anti-Polo antibodies (Fig. 8d,e,f). The midbody staining is also dramatically changed in the mutant embryos, being completely lost with the anti-PAV–KLP antibody and dramatically reduced with the anti-Polo antibody. Thus it would appear that the localization of Polo protein kinase at the spindle poles and in the central spindle requires the function of PAV–KLP.
Figure 7.
PAV–KLP coimmunoprecipitates with Polo protein kinase. (A) Polo was immunoprecipitated from Drosophila embryo extracts using three different antibodies (mAb 75, mAb 81, mAb 294), and the immunoprecipitates were analyzed by Western blot as described in Materials and Methods. An aliquot of the embryo extract was loaded in a separate lane as a control (ext.). The Western blot was first probed with mAb Rb3301 to detect PAV–KLP, and after washing, the membrane was reprobed with mAb 294 to detect Polo protein. (B) A comparison of immunoprecipitates obtained using Bx63, a mAb against the centrosomal-associated protein CP190; an anti-β-tubulin mAb Bx69; and the anti-polo mAb Ma294. The Western blot is probed with the anti-PAV-KLP Rb3301. (A,B) The lower band marked by the arrowhead corresponds to the antibodies used for the immunoprecipitations that are detected by the secondary antibody used in the Western blot.
Figure 8.
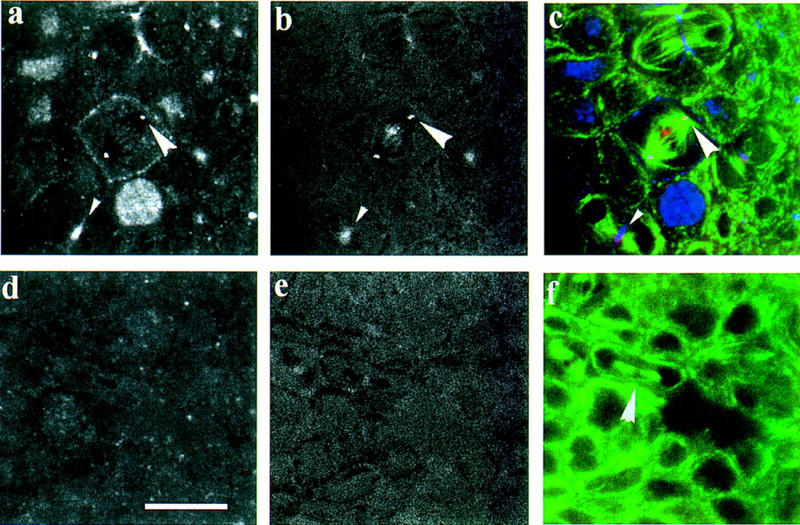
The colocalization of Polo kinase with PAV–KLP at centrosomes and in the central spindle is disrupted in pav mutant embryos. Wild-type (a–c) or pavB200 (d–f) cycle 16 embryos stained for either PAV–KLP (a,d) or Polo (b,e). (e,f) Merged images of PAV–KLP (blue), Polo (red), and tubulin (green). In wild type, Polo colocalizes with PAV–KLP at the centrosome (large arrowhead in a–c) and the midbody (small arrowhead in a–c). In addition, faint Polo staining is apparent at the metaphase plate. However, in pav embryos, neither PAV–KLP (d) nor Polo (e) is present at the centrosomes or midbody of a mutant telophase (arrowhead in f).
Discussion
Pav mutant embryos show cytokinesis defects in virtually all dividing cells at cycle 16. As with other genes required for cell cycle progression in Drosophila, the lethal phase depends on the role and stability of the wild-type protein, the abundance of its maternal provision to the embryo, and the strength of the mutant allele. The majority of cell cycle proteins are supplied maternally, and the rate at which this supply is exhausted depends on the particular protein. The maternal supply of PAV–KLP appears to be depleted early in development, either by degradation or its expulsion from newly divided cells in the remnants of the midbody. Consequently, the lethal phase occurs in embryonic development leading to the previously noted defects in the PNS, embryonic tissues in which cells continue dividing beyond a sixteenth cell cycle. Lethality at this stage is a consequence of the inability of each mutant allele to produce functional gene product, as all alleles of pav show the same lethal phase, whether homozygous or hemizygous against a chromosome deficiency for the region.
As the cell progresses through its division cycle, the distribution of PAV–KLP follows a highly dynamic pattern that is very similar to that of MKLP-1 (Nislow et al. 1990). Both the mammalian and fly proteins show a punctate distribution in the region of the nucleus at interphase. They are present at the centrosomes from metaphase onwards and localize to the spindle midzone at late anaphase and telophase. Many proteins with roles in cytokinesis, including PAV–KLP and Peanut, become concentrated in midbodies, the remnants of which are expelled from the two daughter cells as they separate, and are seen as discrete spots of staining between interphase cells in cellularized embryos.
The pattern of distribution of PAV–KLP in syncytial embryos differs in some respects from that seen in individual cells presumably reflecting the absence of cytokinesis. The protein still shows a pronounced association with the central region of the spindle at late telophase but then follows a pattern that suggests association with actin or actin-associated molecules. Although cytokinesis does not take place at this stage of development, the organization of the actin cytoskeleton is important in maintaining the distribution of and spacing between the rapidly increasing numbers of syncytial nuclei. Thus, an actin-containing structure analogous to the contractile ring forms at the position of the former metaphase plate, but in interphase is redistributed to the cortex where it will form a “cap” that sits between the nucleus and plasma membrane. Upon entry into mitosis the cortical actin is reorganized into the “pseudocleavage furrows” that form a network capturing individual metaphase figures. In many respects these are analagous to cleavage furrows except that they are formed earlier in the mitotic cycle. PAV–KLP appears to concentrate in this network but is then liberated to associate with the spindle and move to the central region at late anaphase. Foe et al. (1993) have proposed that actin is continually binding to microtubules at this stage and moving toward their plus ends. One might speculate that PAV–KLP could provide the requisite motile force.
Mutations leading to cytokinesis defects have been described in genes encoding two other KLPs in Drosophila, KLP3A, and KLP38B (Williams et al. 1995; Ohkura et al. 1997). It is difficult to make comparisons between the mutant phenotypes, as in the case of KLP38B, a cytokinesis phenotype is inferred in larval neuroblasts that attain low levels of polyploidy, and from the observation of binucleate follicle cells in the ovary (Ohkura et al. 1997). KLP3A mutants also show a central spindle defect leading to a failure of cytokinesis, but this is seen in male meiosis (Williams et al. 1995). The differing lethal phases of the pav and KLP3A mutants suggest a specialized role for the two proteins for cytokinesis in different cell types, but an overlapping function of PAV–KLP in meiosis cannot be ruled out at the present time. Both PAV-KLP and KLP3A accumulate in the midbody at telophase, although PAV–KLP has the distinctive attribute of being associated with the centrosome.
pav embryos initially show dramatic defects in the morphology of the central spindle at telophase. The simplest model for PAV–KLP function is that it participates in reorganizing the central spindle region after anaphase B has occurred, in a manner that is vital for assembly of the contractile ring. How the central spindle assists formation of the contractile ring is poorly understood. The septin Peanut, itself essential for cytokinesis in Drosophila (Neufeld and Rubin 1994), fails to localize to a ring-like structure in the pav mutants. This may be a direct consequence of the disrupted structure of the telophase spindle, as recent work with Xenopus shows that septins are microtubule-associated proteins, and that the injection of anti-septin antibodies will inhibit contractile ring function (Glotzer and Hyman 1996). The septins may link the central spindle to the cell cortex, and consequently the delocalization of peanut in pav mutants would disrupt the formation of the contractile ring. Anillin and actin also fail to associate with the equatorial region of the cell during anaphase and telophase, and consequently cells fail to divide and become tetraploid.
The role of the spindle poles in cytokinesis is less clear. Do they signal the onset of cytokinesis as suggested by Rappaports experiments (see introductory section) or, rather, direct the positioning of cell cleavage? In Drosophila male meiosis, study of the male sterile mutation asterless reveals that in the absence of astral microtubules cell cleavage occurs to completion but in the wrong place (Bonaccorsi et al. 1996). A normal central spindle is seen in these mutants, indicating that in this system the central spindle may play some role in the recruitment of contractile ring components, whereas astral microtubules dictate the position of the cleavage furrow.
Our finding that pav is required for cytokinesis seems at odds with previous work suggesting that the mammalian homolog might function during either metaphase or anaphase. In vitro assays demonstrated an ability to cross-link and slide apart antiparallel microtubules, leading to the suggestion of a role in spindle pole separation during anaphase B, whereas the injection of anti-MKLP-1 antibodies into mitotic HeLa cells resulted in a metaphase arrest (Nislow et al. 1990, 1992). The in vitro assay may not be sufficiently specific to dissect the exact role of MKLP-1, as the system lacks centrosomes and the ancillary mechanisms necessary to generate a contractile ring, both of which influence microtubule behavior during cytokinesis. Thus, the ability to cross-link and slide along microtubules in vitro may be a property required of the KLP when associated with contractile ring structures. In any event, the pav mutant phenotype shows no indication of a defect in anaphase B, and we find that spindles extend to the same length as in wild-type embryos at late anaphase and telophase. Moreover, the rare binucleate cells that have lost pav function in the previous mitosis can perform anaphase perfectly well. Thus, if PAV–KLP does play a role in anaphase spindle elongation or spindle assembly, it would seem that this role is redundant and can be fulfilled by other KLPs. Although MKLP-1 is the most closely related klp to PAV–KLP, the sequence homology outside the conserved motor domain is not particularly high (20%), so there could be nonidentical functions. The idea that KLPs in the same subfamily can have distinct functions in different organisms is not without precedent. For example, the chromokinesin subfamily of DNA-binding KLPs involved in chromosome congression includes Drosophila KLP3A, which does not appear to bind DNA and is also required for cytokinesis (Williams et al. 1995). Nevertheless, as the size, domain structure, ability to associate with plks, and dynamic localization of pav–KP is so similar to MKLP-1, it would be surprising if it were not a functional homolog.
The presence of PAV–KLP and its mammalian counterparts at the centrosomes and the central region of the spindle raises the possibility that it may transport a signaling molecule required for either centrosome function, cytokinesis, or both. The serine–threonine protein kinase encoded by polo is an ideal candidate for such a molecule. polo mutants show abnormal centrosome behavior, including the formation of monopolar spindles, a phenotype that is also seen following the microinjection of anti-plk antibodies into mammalian cells (Sunkel and Glover 1988; Llamazares et al. 1991; Lane and Nigg 1996.) In addition to the formation of monopolar spindles, the additional consequence of disruption of the fission yeast homolog is to prevent cytokinesis (Ohkura et al. 1995). It is not clear whether the polo mutation results in cytokinesis defects in Drosophila as the mitotic cycle is blocked at an earlier stage. Nevertheless, the localization of polo kinase throughout the mitotic cycle would be consistent with a dual role for the enzyme similar to that demonstrated in fission yeast. The association that we have demonstrated between Polo kinase and PAV–KLP leads us to speculate that in addition to a role in assembling the central region of the spindle, PAV–KLP may also be responsible for transporting Polo kinase from one set of centrosome-associated substrates to a second set of substrates in the midzone of the spindle as mitosis progresses.
Materials and methods
Stocks
Stocks were maintained under standard conditions (Roberts 1986). Wild-type embryos used were of the Oregon-R strain. EMS-induced alleles of pav were of genotype yw; +; p[w+]64A pav th cu sr e ca/TM6B–abd lacZ (Salzberg et al. 1994). P-element-induced alleles of pav were of genotype yw; +; p[w+ lacZ]/TM6B abd–lacZ (Deak et al. 1997). The lacZ reporter gene under the control of the abdA promoter allowed homozygous embryos to be identified by an absence of β-galactosidase staining in the posterior half of the embryo.
Analysis of mutants and immunostaining
The pav alleles pavB200, pavM187, pavM547, and pav934/13 are all embryonic lethal and show cytokinesis defects during mitosis 16, either as homozygotes or as hemizygotes. The pavB200 allele showed the greatest penetrance of the phenotype and was used for more detailed analysis. Immunostaining of embryos was performed as described previously (Glover and Gonzalez 1993). Freshly prepared 4% paraformaldehyde in buffer B (45 mm KCl, 15 mm NaCl, 10 mm phosphate buffer at pH 6.8) was found to be the best fixative. The following primary antibodies were used: tubulin (YL/12; 1:20; Harlan SeraLabs); CP190 (Rb188; 1:500; Whitfield et al. 1988); lamin A (T47; 1:20; Frasch et al. 1986); spectrin (anti-chicken spectrin; 1:200; Sigma); Peanut [mAb4C9; 1:10; kind gift of G. Rubin (Neufeld and Rubin 1994)]; anillin [Rb1-271; 1:200; kind gift of C. Field (Field and Alberts 1995)]; actin (C4; 1:200; ICN Biomedicals); Pav (Rb3301; 1:100; this study); Polo (mAb M294; 1:10; Tavares et al. 1996). FITC, Texas Red, and Cy5 secondary antibodies were purchased from Jackson Immunochemicals. DNA was stained using either 1 μg/ml propidium iodide or 5 μg/ml TOTO-3 (Molecular Probes). Specimens were viewed using either an MRC600 or MRC1024 confocal microscope (Biorad). Images were processed using PhotoShop (Adobe Systems) and printed using a Fargo dye-sublimation color printer.
Cloning pav
P elements and flanking genomic sequences were rescued from the strains pav934/13 and pav973/8 following DNA digestion with either EcoRI or BamHI followed by ligation, transformation into E. coli XL1B and selection for ampicillin-resistant colonies. This enabled the isolation of a 3.2-kb EcoRI fragment and a 2.6-kb BamHI fragment flanking both sides of the insertion in pav934/13; and a 10-kb EcoRI fragment of flanking DNA from one side of the insertion in pav973/8. The flanking fragments hybridized in situ to salivary gland chromosome region 64B2-7, the site to which pav has been mapped genetically (Salzberg et al. 1994). The 934/13 EcoRI fragment detected a number of similar cDNAs in 0- to 4-hr and 0- to 8-hr embryonic cDNA libraries (Brown and Kafatos 1988), kindly provided on a gridded filter by J. Hoheisel. The P-element-flanking fragments and the cDNA identified several cosmids when used as hybridization probes to the European Genome Project cosmid library, of which cosmid 113G4 was used as a source of a 10.5-kb SmaI–StuI fragment (termed RC1) for germ-line transformation experiments.
Germ-line transformation
NotI linkers (Stratagene) were ligated to both ends of the 10.5-kb SmaI–StuI fragment RC1. Following NotI digestion, RC1 was first subcloned into pKS and then into the NotI site of the pW8 transformation vector pW8 (Klemenz et al. 1987). Transformation was achieved following the injection of this plasmid, pRC1w+, into embryos derived from homozygous w−; p[Δ2-3] flies following standard procedures. A transformed line was established with the insertion on the third chromosome. This was maintained as a stock balanced against TM6B. The transforming P element was mobilized by crossing this stock to one carrying the Δ2-3 P element on chromosome 3 as a source of transposase. Transposition events to the second chromosome in the resulting p[RC1 w+]/p[Δ2-3] Sb ry e males were detected in a subsequent generation by the generation of Sb w+ males in a w− background. Crossing to Tft/CyO females enabled a balanced stock to be selected. Successful rescue was demonstrated by first crossing this p[RC1 w+]/CyO stock to TM3/TM6B to select p[RC1 w+]; TM3 progeny, which were sequentially crossed to pavM547/TM6B and pavB200/TM6B stocks for the rescue of pavM547/pavB200. As a negative control, a second construct, pRC2, was made by digestion of pW8RC1 with Bsu36I followed by recircularization using blunt end ligation to delete a 1.3-kb Bsu36I fragment from within the coding region of the gene. This was transformed into flies using standard procedures to obtain five independent transformant lines. Two lines in which the pW8RC2 construct was carried on the X chromosome were tested for their ability to rescue pav by sequential crosses to pavM547/TM6B and pavB200/TM6B stocks testing for the capabilty to generate w− p[RC2 w+]; pavM547/pavB200 flies. Neither transformant was capable of rescuing pav.
Production of anti-Pav antibodies
An amino-terminally His-tagged protein fragment of PAV–KLP was expressed in E. coli, purified, and used to immunize rabbits. A 1.4-kb BamHI–PstI of pav cDNA coding for amino acids 208–686 was subcloned into pQE31 (Qiagen) to make pEC2. pEC2 was transformed into E. coli M15/REP4 cells. Induced protein was purified on a Ni–agarose column under denaturing conditions according to the manufacturer’s instructions (Qiagen) and then dialyzed in PBS. Protein was injected into rabbits at 1 mg/ml with Freund’s adjuvant. After four boosts, serum was obtained that recognized a single band of 100 kD on Western blots of total Drosophila embryo extracts.
Immunoprecipitations and Western blotting
Polo–kinase immunoprecipitations were carried out using the three previously described monoclonal antibodies mAb 75, mAb 81, and mAb 294 (Llamazares et al. 1991; Tavares et al. 1996) coupled to magnetic beads coated with sheep anti-mouse IgG (DYNAL). Briefly, 0- to 8-hr Drosophila embryos were homogenized in 5 volumes of NEWIP buffer (25 mm Tris at pH 7.5, 1% Triton X-100, 20 mm glycerophosphate, 20 mm NaF, and 5 mm EGTA) supplemented with 1 mm PMSF and 1 μg/ml aprotinin, pepstatin A, and leupeptin, and centrifuged for 10 min at 14000g to remove cellular debris. The supernatant obtained was thoroughly chilled to 4°C to depolymerize microtubules and incubated with the antibodies coupled to Dyna beads for 1.5 hr at 4°C. As a control an aliquot of the extract was incubated with Dyna beads not coupled to anti-polo antibodies. The immunoprecipitates were washed four times with NEWIP buffer and resuspended in SDS sample buffer, and the proteins separated on SDS–10% polyacrylamide gels. The proteins were transferred to Immobilon-P membrane (Millipore), and the Westerns were executed as described previously (Tavares et al. 1996) using the antibody Rb3301 diluted 1/250 to detect PAV–KLP, and the antibody mAb 94 diluted 1/5 to detect Polo.
Acknowledgments
We are indebted to Christine Field and Gerald Rubin for kind donation of antibodies and to J. Hoheisel for provision of a gridded embryonic cDNA library. Yoshihiro Inoue helped with the P-element reversion analysis. We are grateful to Mar Carmena for critical reading of the manuscript. R.A. held a Medical Research Council (MRC) research studentship. Support for the project was provided by grants from the CRC, the MRC, the BBSRC, and the European Union.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL d.m.glover@dundee.ac.uk; FAX 44(0) 1382 344213.
References
- Andreassen PR, Palmer DK, Wener MH, Margolis RL. Telophase disc: A new mammalian mitotic organelle that bisects telophase cells with a possible function in cytokinesis. J Cell Sci. 1991;99:523–534. doi: 10.1242/jcs.99.3.523. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi S, Giansanti MG, Gatti M. Asterless, a Drosophila gene specifically required for the formation of asters during male meiosis. Mol Biol Cell. 1996;7S:574a. [Google Scholar]
- Brown NH, Kafatos FC. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Burton K, Taylor DL. Traction forces of cytokinesis measured with optically modified elastic substrata. Nature. 1997;385:450–454. doi: 10.1038/385450a0. [DOI] [PubMed] [Google Scholar]
- Cao L-G, Wang Y-L. Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol Biol Cell. 1996;7:225–232. doi: 10.1091/mbc.7.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak, P., M.M. Omar, R.D.C. Saunders, M. Pal, O. Komonyi, J. Szidonya, P. Maroy, Y. Zhang, M. Ashburner, P. Benos, C. Savakis, I. Siden-Kiamos, C. Louis, V.N. Bolshakov, F.C. Kafatos, E. Madueno, J. Modolell, and D.M. Glover. 1997. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: Correlation of physical and cytogenetic maps in chromosomal region 86E-87F. Genetics (in press). [DOI] [PMC free article] [PubMed]
- Devore JJ, Conrad GW, Rappaport R. A model for astral stimulation of cytokinesis in animal cells. J Cell Biol. 1989;109:2225–2232. doi: 10.1083/jcb.109.5.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Ainsztein AM, Mackay AM, Goldberg IG, Earnshaw WC. Chromosomal proteins and cytokinesis: Patterns of cleavage furrow formation and inner centromere protein positioning in mitotic heterokaryons and mid-anaphase cells. J Cell Biol. 1997;136:1169–1183. doi: 10.1083/jcb.136.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W, Cooke C. Analysis of the distribution of the INCENPs throughout mitosis reveals the existence of a pathway of structural changes in the chromossomes during metaphase and early events in the cleavage furrow formation. J Cell Sci. 1991;98:443–461. doi: 10.1242/jcs.98.4.443. [DOI] [PubMed] [Google Scholar]
- Field CM, Alberts BA. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, Odell GM, Edgar BA. Mitosis and morphogenesis in the Drosophila embryo: Point and counterpoint. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Frasch M, Glover DM, Saumweber H. Nuclear antigens follow different pathways into daughter nuclei during mitosis in early Drosophila embryos. J Cell Sci. 1986;82:155–172. doi: 10.1242/jcs.82.1.155. [DOI] [PubMed] [Google Scholar]
- Giansanti MG, Bonaccorsi S, Williams B, Williams EV, Santolamazza C, Goldberg ML, Gatti M. Mutations in the profilin-encoding gene chickadee reveal interactions between the actin cytoskeleton and the central spindle in Drosophila male meiosis. Mol Biol Cell. 1996;7S:202a. [Google Scholar]
- Glotzer M, Hyman AA. The biochemistry of Xenopus septins and their role in cytokinesis. Mol Biol Cell. 1996;7S:212a. [Google Scholar]
- Glover DM, Gonzalez C. Techniques for studying mitosis in Drosophila. In: Brookes R, Fantes P, editors. The cell cycle: A practical approach. Oxford, UK: IRL Press; 1993. pp. 163–168. [Google Scholar]
- Gunsalus KC, Bonaccorsi S, Williams E, Verni F, Gatti M, Goldberg ML. Mutations in twinstar, a Drosophila cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Mortimer RK, Culotti J, Culotti M. Genetic control of the cell division cycle in yeast: V. Genetic analysis of the cdc mutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck MMS, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LSB. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess RE, Chang X-J, Edwards GA, Kulkarni S, Aguilers G, Kiehart DP. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell. 1991;65:1177–1189. doi: 10.1016/0092-8674(91)90013-o. [DOI] [PubMed] [Google Scholar]
- Kawamura K. Microdissection studies on the dividing neuroblast of the grasshopper, with special reference to the mechanism of unequal cytokinesis. Exp Cell Res. 1977;106:127–137. doi: 10.1016/0014-4827(77)90249-x. [DOI] [PubMed] [Google Scholar]
- Klemenz R, Weber U, Gehring WJ. The white gene as a marker in a new P-element vector for gene transfer in Drosophila. Nucleic Acids Res. 1987;18:87–93. doi: 10.1093/nar/15.10.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Erikson RL. Plk is a functional homolog of Saccharomyces cerevisiae Cdc5, and elevated Plk activity induces multiple septation structures. Mol Cell Biol. 1997;17:3408–3417. doi: 10.1128/mcb.17.6.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Yuan YO, Kuriyama R, Erikson R. Plk is an M-phase specific protein kinase and interacts with a kinesin-like protein, CHO1/MKLP-1. Mol Cell Biol. 1995;15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, Karess RE, Glover DM, Sunkel CE. polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes & Dev. 1991;5:2153–2165. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, Virgilio CD, Pringle JR. The septins: Roles in cytokinesis and other processes. Curr Biol. 1996;8:106–118. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Lupas A, Vandyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Martineau SM, Andreassen PR, Margolis RL. Delay of HeLa cell cleavage into interphase using dihydrocytochalasin B: Retention of a postmitotic spindle and telophase disc correlates with synchronous cleavage recovery. J Cell Biol. 1995;131:191–205. doi: 10.1083/jcb.131.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar microtubules in PTK cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald HB, Stewart RJ, Goldstein LSB. The kinesin-like ncd protein of Drosophila is a minus end directed microtubule motor. Cell. 1990;63:1159–1165. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- Moore JD, Endow SA. Kinesin proteins: A phylum of motors for microtubule based motility. BioEssays. 1996;18:207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, Rubin GM. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- Nislow C, Sellitto C, Kuriyama R, McIntosh JR. A monoclonal-antibody to a mitotic microtubule-associated protein blocks mitotic progression. J Cell Biol. 1990;111:511–522. doi: 10.1083/jcb.111.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C, L. VA, Kuriyama R, McIntosh JR. A plus end directed motor enzyme that moves antiparallel microtubules in vitro and localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- Ohkura H, Hagan IM, Glover DM. The conserved Schizosaccharomyces pombe kinase, plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes & Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- Ohkura H, Torok T, Tick G, Hoheisel J, Kiss I, Glover DM. Mutation of a gene for a Drosophila kinesin-like protein, Klp38B, leads to failure of cytokinesis. J Cell Sci. 1997;110:945–954. doi: 10.1242/jcs.110.8.945. [DOI] [PubMed] [Google Scholar]
- Rappaport R. Experiments concerning the cleavage stimulus in sand dollar eggs. J Exp Zool. 1961;148:81–89. doi: 10.1002/jez.1401480107. [DOI] [PubMed] [Google Scholar]
- Rieder C, Khodjakov A, Paliulis LV, Fortier TM, Cole RW, Sluder G. Mitosis in vertebrate somatic cells with two spindles: Implications for the metaphase/anaphase transition checkpoint and cleavage. Proc Natl Acad Sci. 1997;94:5107–5112. doi: 10.1073/pnas.94.10.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DB. Drosophila: A practical approach. Oxford, UK: IRL Press; 1986. [Google Scholar]
- Salzberg A, Develyn D, Schulze KL, Lee JK, Strumpf D, Tsai L, Bellen HJ. Mutations affecting the pattern of the PNS in Drosophila reveal novel aspects of neuronal development. Neuron. 1994;13:269–287. doi: 10.1016/0896-6273(94)90346-8. [DOI] [PubMed] [Google Scholar]
- Salzberg, A., S.N. Propkopenko, Y. He, P. Tsai, M. Pal, P. Maroy, D.M. Glover, P. Deak, and H.J. Bellen. 1997. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: Mutations affecting embryonic PNS development. Genetics (in press). [DOI] [PMC free article] [PubMed]
- Schroeder T. The contractile ring. II. Determining its brief existence, volumetric changes and vital role in cleaving Arbacia . J Cell Biol. 1972;53:419–434. doi: 10.1083/jcb.53.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellitto C, Kuriyama R. Distribution of a matrix component of the midbody during the cell cycle in Chinese hamster ovary cells. J Cell Biol. 1988;106:431–439. doi: 10.1083/jcb.106.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel CE, Glover DM. Polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- Swann MM, Mitchison JM. Cleavage of sea urchin eggs in colchicine. J Exp Biol. 1956;30:506–514. [Google Scholar]
- Tavares AAM, Glover DM, Sunkel CE. The conserved mitotic kinase polo is regulated by phosphorylation and has preferred microtubule-associated substrates in Drosophila embryo extracts. EMBO J. 1996;15:4873–4883. [PMC free article] [PubMed] [Google Scholar]
- Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C. Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell. 1995;81:117–127. doi: 10.1016/0092-8674(95)90376-3. [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Wang Y-L. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J Cell Biol. 1996;135:981–989. doi: 10.1083/jcb.135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Borisy GG. On the mechanisms of cytokinesis in animal cells. J Theor Biol. 1983;101:289–316. doi: 10.1016/0022-5193(83)90342-9. [DOI] [PubMed] [Google Scholar]
- Whitfield WGF, Millar SE, Saumweber H, Frasch M, Glover DM. Cloning of a gene encoding an antigen associated with the centrosome in Drosophila. J Cell Sci. 1988;89:467–480. doi: 10.1242/jcs.89.4.467. [DOI] [PubMed] [Google Scholar]
- Williams BC, Riedy MF, Williams EV, Gatti M, Goldberg ML. The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J Cell Biol. 1995;129:709–723. doi: 10.1083/jcb.129.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JT, Saxton WM, Goldstein LSB. Isolation and characterization of the gene encoding the heavy chain of Drosophila kinesin. Proc Natl Acad Sci. 1988;85:1864–1868. doi: 10.1073/pnas.85.6.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Nicklas RB. “Anaphase” and cytokinesis in the absence of chromosomes. Nature. 1996;382:466–468. doi: 10.1038/382466a0. [DOI] [PubMed] [Google Scholar]
- Zhang P, Knowles BA, Goldstein LSB, Hawley RS. A kinesin-like protein required for distributive chromosome segregation in Drosophila. Cell. 1990;62:1053–1062. doi: 10.1016/0092-8674(90)90383-p. [DOI] [PubMed] [Google Scholar]