Abstract
The plant hormone auxin regulates diverse aspects of plant growth and development. We report that in Arabidopsis, auxin response is dependent on a ubiquitin-ligase (E3) complex called SCFTIR1. The complex consists of proteins related to yeast Skp1p and Cdc53p called ASK and AtCUL1, respectively, as well as the F-box protein TIR1. Mutations in either ASK1 or TIR1 result in decreased auxin response. Further, overexpression of TIR1 promotes auxin response suggesting that SCFTIR1 is limiting for the response. These results provide new support for a model in which auxin action depends on the regulated proteolysis of repressor proteins.
Keywords: auxin, TIR1, SCF, ubiquitin-ligase, proteolysis, Arabidopsis
The plant hormone indole-3-acetic acid (IAA or auxin) has a crucial role in diverse aspects of plant growth and development (Davies 1995). At the cellular level, auxin affects cell division (John et al. 1993), cell expansion (Gray et al. 1998), and cell differentiation (Fukada 1996). Despite the importance of auxin in regulating these fundamental processes, little is known about the mechanisms involved in auxin signaling and response.
The isolation of mutations that confer resistance to applied hormone has been a powerful tool for identifying the factors involved in hormone perception and signaling (Hartwell 1980; Rabindran et al. 1987; Li and Chory 1997). In Arabidopsis thaliana, this approach has led to the identification of several genes required for auxin response (Hobbie and Estelle 1994; del Pozo and Estelle 1999). Recessive mutations in the AXR1, AXR4, and TIR1 genes confer diminished auxin response and a variety of corresponding auxin-related growth defects. Genetic evidence suggests that these factors act in the same, or overlapping pathways (Hobbie and Estelle 1995; Ruegger et al. 1998; W.M. Gray and M. Estelle, unpubl.). In addition, mutations in the SAR1 gene were isolated as suppressors of axr1 suggesting that the SAR1 gene product acts in this pathway (Cernac et al. 1997).
Molecular analysis of the TIR1 and AXR1 genes has implicated the ubiquitin pathway in auxin action. Ubiquitin-mediated proteolysis regulates a number of cellular events including cell cycle transitions, metabolic regulation, stress responses, and differentiation. The first step in the ubiquitin conjugation pathway is the formation of a thiolester linkage between an internal cysteine residue of the ubiquitin-activating enzyme (E1) and the carboxyl terminus of ubiquitin. The ubiquitin moiety is then transesterified to a ubiquitin-conjugating enzyme (E2), and with the assistance of a ubiquitin-ligase enzyme (E3), covalently attached to a target protein by an isopeptide linkage between a lysine residue and the ubiquitin carboxyl terminus. Both the E1 and E2 enzymes are encoded by families of related genes. In contrast, E3 enzymes are quite diverse. The precise definition of E3 enzymes is rather ambiguous, but minimally, the E3 facilitates transfer of ubiquitin from the ubiquitin-conjugating enzyme to the substrate protein. In some cases, the E3 enzyme forms a catalytic intermediate with ubiquitin (Scheffner et al. 1995), whereas in others the function of the E3 may simply be to bring the E2 enzyme and the target protein into close proximity.
The TIR1 gene encodes an F-box protein containing 16 degenerate leucine-rich repeats (LRRs) (Ruegger et al. 1998). Work in yeast and mammalian systems indicates that F-box proteins interact with the Skp1 and Cdc53 (cullin) proteins to form ubiquitin ligase complexes called SCFs. (for recent reviews, see Krek 1998; Patton et al. 1998). The F box is a 40-residue domain implicated in binding to Skp1 (Bai et al. 1996). Since several distinct F-box proteins can form SCF complexes with identical Skp1 and cullin subunits, the F-box component has been proposed to provide substrate specificity to the E3 complex (Patton et al. 1998). The sequence of the TIR1 protein and the phenotype of the tir1 mutants suggest that TIR1 may be a component of an SCF complex involved in auxin signaling.
AXR1 encodes a protein related to the amino-terminal half of E1 (Leyser et al. 1993). Recent work has demonstrated that AXR1 interacts with a second protein called ECR1, to activate the ubiquitin-related protein RUB for conjugation (del Pozo et al. 1998). RUB is a member of a conserved family of ubiquitin-related proteins present in fungi, plants, and mammals (NEDD-8) (Kumar et al. 1993; Rao-Naik et al. 1998). Like ubiquitin, RUB is conjugated to target proteins. The Cdc53/Cullin component of SCF complexes is the only known target for RUB/NEDD8 modification in yeast and mammals (Lammer et al. 1998; Osaka et al. 1998). Although the functional significance of this modification is unclear, genetic studies in yeast suggest that RUB modification regulates some aspect of SCFCdc4 function (Lammer et al. 1998). An Arabidopsis Cdc53-related protein called AtCUL1 has been identified, and this protein is also a substrate for RUB conjugation (J.C. del Pozo and M. Estelle, unpubl.). These results suggest a model in which the AXR1–ECR1 E1-like dimer activates RUB for conjugation to AtCUL1. This modification may alter the assembly or function of an SCF complex containing the TIR1 protein.
In this paper, we demonstrate that TIR1 physically interacts with the Arabidopsis Skp1-like proteins ASK1 and ASK2 and the cullin, AtCUL1 to form a complex called SCFTIR1. Plants containing a mutation in the ASK1 gene exhibit reduced auxin response demonstrating the requirement for additional components of the SCF in the auxin response pathway. We also show that overexpression of TIR1 in transgenic plants results in enhanced auxin response including an increase in auxin-dependent gene expression and diverse morphological changes. These findings indicate that the SCF paradigm can be extended to higher plants, and that auxin signaling is mediated by SCFTIR1.
Results
Expression of the TIR1 gene
The AXR1 auxin response gene is expressed primarily in actively dividing and elongating cells (del Pozo et al. 1998; J.C. del Pozo and M. Estelle unpubl.). Because genetic evidence suggests that TIR1 and AXR1 act in the same pathway, we hypothesized that TIR1 would also be expressed in dividing and elongating cells. Northern hybridization analysis detects a single TIR1 transcript in all tissues examined including roots, rosette leaves, stems, and flowers (Fig. 1A). To more precisely determine the pattern of TIR1 expression, we constructed a reporter gene consisting of 4 kb of TIR1 5′- sequence fused to the bacterial β-glucuronidase gene (gus). Several independent transgenic lines were generated and analyzed by histochemical staining for β-glucuronidase activity. Strong TIR1–gus expression was detected in the root apical meristem and the expanding cotyledons and hypocotyls of young seedlings (Fig. 1B). Expression in the cotyledons and developing leaves is reduced in older seedlings, with TIR1–gus expression in the shoot being strongest in the stipules with some expression also seen in the shoot apical meristem (Fig. 1D). TIR1–gus expression remains strong in the root apical meristem (Fig. 1C), and strong expression is also detected in developing lateral root primordia and mature lateral root meristems (Fig. 1E–G). Staining was also evident in vascular elements. In adult plant organs, the TIR1–gus reporter was strongly expressed in the floral stigma, anther filaments, and abscission zones, with some expression also observed throughout the vasculature (Fig. 1H). In situ RNA hybridization experiments verified and extended the results obtained with the TIR1–gus reporter. TIR1 RNA was most abundant in meristematic zones (Figure 1I,K,L,M). Additionally, the in situ hybridization analysis detected abundant expression in developing embryos (Fig. 1O). The sense strand control hybridizations are shown in Figure 1J,N,P.
Figure 1.
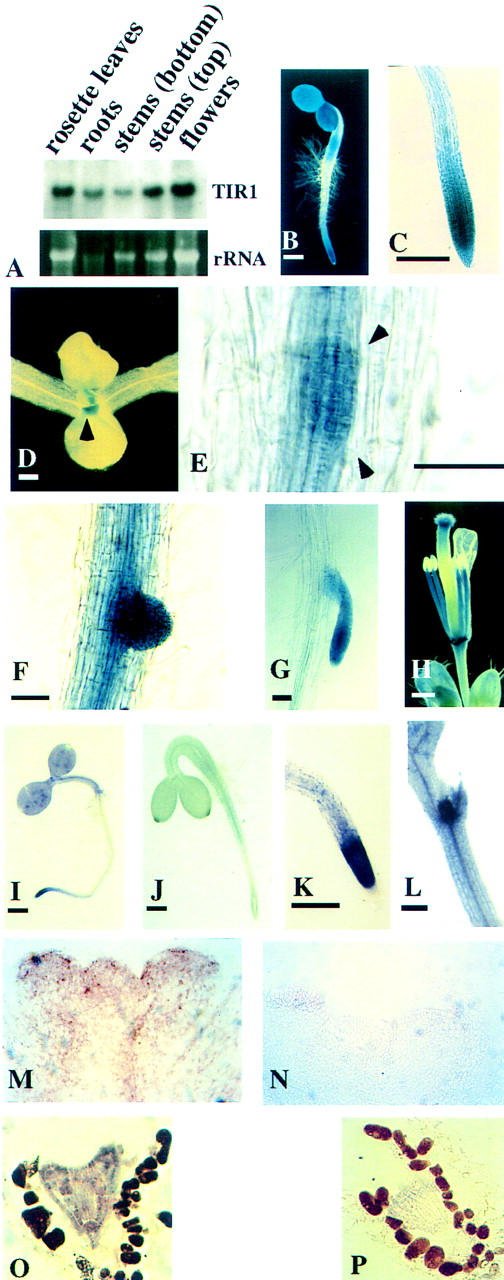
Analysis of TIR1 expression. (A) Northern blot analysis of TIR1 with total RNA isolated from various tissues. TIR1–gus expression in 3-day-old seedling with staining apparent in the root tip, hypocotyl, and cotelydons (B); root tip of 10-day-old seedling (C); shoot of 10-day-old seedling with staining visible in stipules (arrowheads) and the apical meristem (D); an early lateral root primoridium (between arrowheads) (E). Vascular tissues are also stained; (F) an emerging lateral root; (G) emerged lateral root; (H) unfertilized flower. In situ RNA hybridization with TIR1 of 3-day-old seedling (I); 3-day-old seedling (J); root tip of 5-day-old seedling (K); shoot of 5-day-old seedling (L); longitudinal sections (10 μm) through shoot apical meristem (M,N); and heart-stage embryo (O,P). Sense strand control hybridizations are shown in J, N, and P. Size bars, 0.5 mm in B, D, H, I, and J, and 0.1 mm in all other panels.
TIR1 functions early in lateral root formation
Lateral root meristems develop from G2-arrested cells in the pericycle layer of Arabidopsis roots (Blakely and Evans 1979). Previous genetic analysis has determined that lateral root development is a process involving at least two steps: (1) the initiation of pericycle cell division resulting in lateral meristem formation, and (2) continued cell division leading to lateral root growth (Celenza et al. 1995). Both of these steps appear to require auxin. Consistent with a role in auxin response, tir1 mutants have a reduced number of lateral roots (Ruegger et al. 1998), and expression analysis revealed that TIR1 is expressed early in lateral root development (Fig. 1E–G).
To determine whether TIR1 functions early in meristem formation, cyc1At expression was examined in tir1-1 mutant plants carrying a cyc1At–gus reporter. The cyc1At gene encodes a mitotic cyclin that is expressed in pericycle cells in the zone of lateral root initiation prior to the occurrence of any morphological changes or cell divisions (Hemerly et al. 1992; Ferreira et al. 1994). Similarly, in a transgenic line carrying the cyc1At–gus reporter, GUS staining is observed before the earliest division to form the lateral root primordium. If TIR1 is required after cyc1At is expressed in primordia, we expected a similar number of β-glucuronidase staining primordia in both wild-type and tir1-1 mutant roots. In contrast, if TIR1 functions prior to cyc1At expression, tir1-1 mutants should display a reduced number of primordia expressing the cyc1At–gus reporter. tir1-1 mutant seedlings grown on unsupplemented nutrient medium displayed a reduction in cyc1At–gus expression compared to wild-type that corresponded to the reduction in the number of lateral roots formed in tir1-1 seedlings (Fig. 2A). When seedlings were grown on medium containing auxin to promote lateral root formation, cyc1At–gus expression was restricted to the few lateral roots that develop in tir1-1 seedlings (Fig. 2A). In wild-type roots, GUS staining of single cells was observed 4 hr after auxin treament, whereas in tir1-1 far fewer cells were stained (Fig. 2B). These results indicate that TIR1 is required prior to the expression of cyc1At in lateral root development, and suggest that TIR1 is necessary for pericycle cells to overcome G2 arrest.
Figure 2.
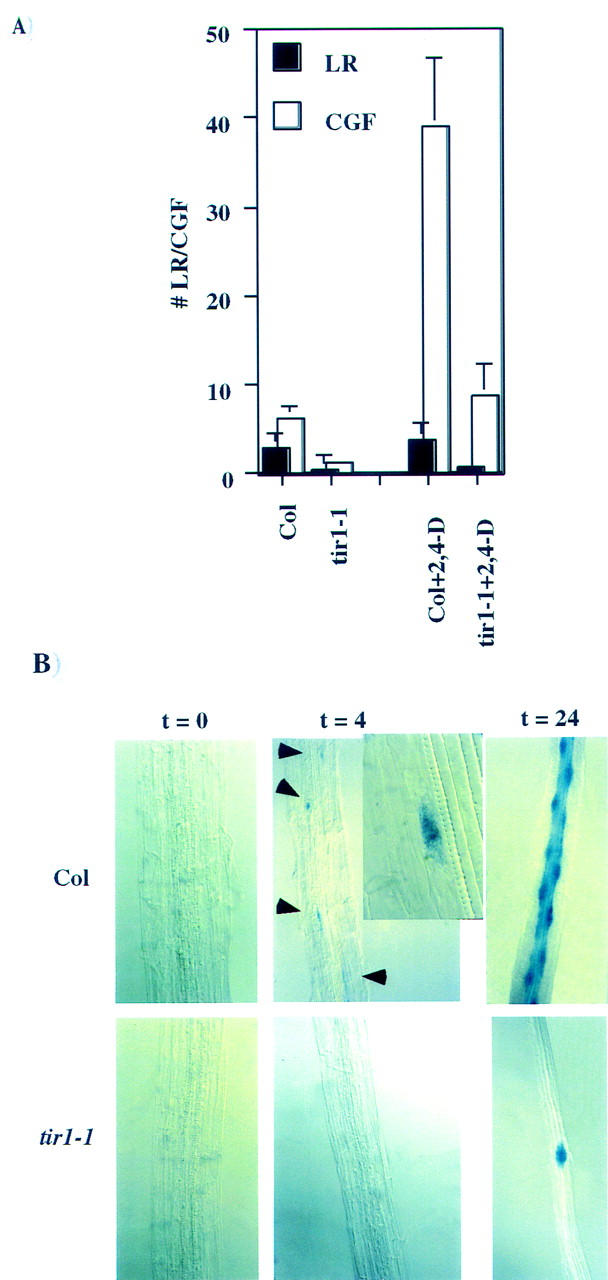
TIR1 functions prior to the expression of cyc1At in lateral root development. (A) Number of emerged lateral roots (LR, solid bars) and the number of cyc1At–gus foci (CGF, open bars) in wild-type and tir1-1 seedlings in the presence and absence of exogenous auxin. For this study CGF are defined as the number of foci of cyc1At–gus staining observed along the root excluding the primary root meristem. This ranges from single cells expressing the cyc1At–gus reporter to mature lateral root meristems. Presumably these foci are all lateral root meristems at various stages of development. (B) cyc1At–gus expression in the roots of 10 day-old wild-type and tir1-1 seedlings. Nine-day-old seedlings were treated with 0.25 μm 2,4-D for various times and stained for gus activity. Root segments shown are located ∼5 mm from the root tip. At this location in wild-type roots, auxin treatment induces a high percentage of pericycle cells to differentiate into lateral root primordia. (Inset) High magnification of a single pericycle cell expressing the cyc1At–gus reporter prior to the first cell division of lateral root development.
Identification of TIR1-interacting proteins
The presence of the F-box motif in the TIR1 protein suggested that TIR1 may function as a component of an SCF ubiquitin–ligase complex. Therefore, TIR1 may interact with other SCF components as well as the SCF substrate. To identify TIR1 interacting proteins, we constructed a Gal4 DNA-binding domain–TIR1 fusion protein (DBD–TIR1) to use as bait in a yeast two-hybrid screen. This construct was introduced into the yeast strain YPB2 and used to screen an expression library in which the Gal4 activation domain (AD) was fused to cDNAs prepared from A. thaliana plants at various stages of development. Approximately 7.5 × 106 library transformants were screened for enhanced growth on medium lacking histidine and supplemented with 15 mm 3AT. Sixty-one candidates were identified, and 33 of these were found to also activate the GAL43 × 17-mer–lacZ reporter gene. DpnII restriction analysis of the PCR-amplified cDNA inserts from these clones revealed two distinct classes of clones, with one class represented by 26 clones and the other by 7 clones.
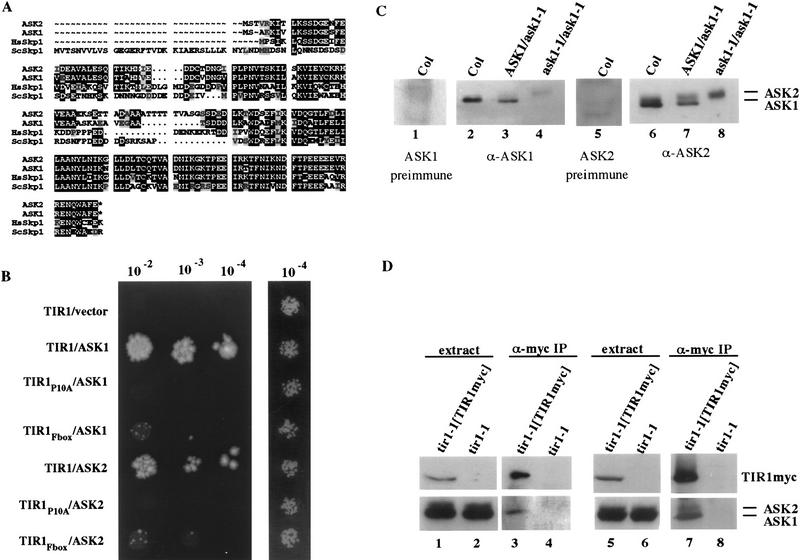
Sequence analysis of representative members of the two classes of DBD–TIR1 interacting clones revealed that the first is identical to the ATskp1 gene (Porat et al. 1998), whereas the second is a highly related gene that we have designated ASK2 (Arabidopsis SKP1-like) (Fig. 3A). ATskp1 was initially identified by its sequence similarity to Skp1 (Porat et al. 1998). ATskp1 and ASK2 are members of a multigenic family of Arabidopsis SKP1-related genes. At least eight additional members of this family have been identified as Arabidopsis ESTs or isolated in separate two-hybrid-screens (E. Risseeuw and W.L. Crosby unpubl.). Because Arabidopsis contains a multigenic family of SKP1-like genes, we have renamed ATskp1 as ASK1 and designated the remaining family members ASK3 through ASK10. The predicted proteins encoded by these genes are all very highly related to one another, with pairwise comparisons between any two members displaying 31% to 77% amino acid identity. ASK1 and ASK2 are slightly more similar to each other than either is to the other members of this family (data not shown).
Figure 3.
TIR1 interacts with the Skp1-like proteins ASK1 and ASK2. (A) Sequence alignment of the A. thaliana ASK1 and ASK2 (GenBank accession no. AF059295) proteins as well as human and S. cerevisiae Skp1p with PileUp (Genetics Computer Group, Inc., Madison, WI). Identical amino acids are boxed in black. Similar amino acids are boxed in gray. (B) Serial dilutions of yeast strain YPB2 carrying the indicated two-hybrid plasmids. Cells were plated onto 3-AT selection medium (left) or control (right) medium and incubated at room temperature. The 3-AT plate was photographed after 10 days and the control plate after 3 days. (C) Western blot analysis of seedling extracts with polyclonal antisera raised against ASK1 (lanes 2–4) or ASK2 (lanes 6–8). (D) Western blot analysis of crude seedling extracts (lanes 1,2,5,6) or α-myc immunoprecipitates (lanes 3,4,7,8). (Bottom lanes 1–4) was probed with α-ASK1 antisera. (Bottom, lanes 5–8) Probed with α-ASK2 antisera.
To demonstrate that the F-box domain (amino acids 9–48) of TIR1 mediates interaction with ASK1 and ASK2, the highly conserved proline at amino acid 10 was changed to an alanine residue. This mutation (TIR1P10A) completely abolished the ability of TIR1 to interact with the Skp1-related proteins in a two-hybrid assay (Fig. 3B). Furthermore, the TIR1P10A derivative does not complement the auxin resistance phenotype of tir1-1 mutants, indicating that the F-box domain is essential for TIR1 function in planta (data not shown). The TIR1 F-box domain alone [DBD–TIR1(1–105)] also interacts with ASK1 and ASK2, albeit at a much reduced level compared with DBD–TIR1, demonstrating that the F-box domain is both necessary and sufficient to mediate interactions with ASK1 and ASK2 in the yeast two-hybrid system (Fig. 3B).
TIR1 forms SCF-like complexes in planta
The ASK1 and ASK2 proteins were expressed in Escherichia coli, purified as 6xHis-fusion proteins, and used to raise polyclonal antibodies. Western blot analysis of Arabidopsis extracts with the α-ASK1 antisera detected one predominant band at ∼25 kD (Fig. 3C, lanes 1–4). This band corresponds to the ASK1 protein because it is not recognized by the preimmune sera and is absent from extracts prepared from ask1-1 mutant seedlings. The ask1-1 mutation consists of a Ds transposon insertion within the ASK1 gene (M. Yang and H. Ma, unpubl.). The α-ASK2 antisera also detected the ASK1 protein as well as a protein that migrated slightly slower than ASK1 (Fig. 3C, lanes 5–8). This larger band is presumably the ASK2 protein. However, we cannot definitively rule out the possibility that it corresponds to one of the other Skp1-like proteins found in Arabidopsis.
The TIR1–ASK1 and TIR1–ASK2 interactions were examined in planta by co-immunoprecipitation. Crude extracts were prepared from 7-day-old seedlings expressing a c-myc epitope-tagged version of TIR1 and from control seedlings that did not express TIR1–myc. The TIR1–myc protein was immunoprecipitated with monoclonal c-myc antibody, and the resulting precipitates were immunoblotted and probed with the α-ASK1 or α-ASK2 antisera. The ASK1 protein and the distinct protein recognized by the α-ASK2 antisera were both present in the α-myc immunoprecipitates of extracts that contained the TIR1–myc fusion protein but were absent from immunoprecipitates of control extracts lacking TIR1–myc (Fig. 3D).
In yeast and mammals, F-box proteins and Skp1p interact with a member of the Cdc53p/cullin family of proteins to form SCF complexes (Patton et al. 1998). We identified an Arabidopsis ORF in the GenBank database (locus 2281115 on AC002330) encoding a cullin-like protein that we have designated AtCUL1. This protein has 25% and 38% amino acid identity to the Saccharomyces cerevisiae Cdc53 and human Cul-4A proteins, respectively (Fig. 4A). Antiserum raised against AtCUL1 detected two predominant bands on immunoblots of Arabidopsis seedling extracts (Fig. 4B, lanes 1–4). These proteins migrated at ∼84 kD, near the predicted molecular mass of AtCUL1 (86 kD), and were not recognized by the preimmune serum. The presence of two immunoreactive bands suggests that AtCUL1 may be present in multiple isoforms in seedlings. Alternatively, the α-AtCUL1 antisera may cross-react with a protein related to AtCUL1. Multigenic families of cullins are found in many organisms, and highly related sequences are found in the Arabidopsis EST database.
Figure 4.
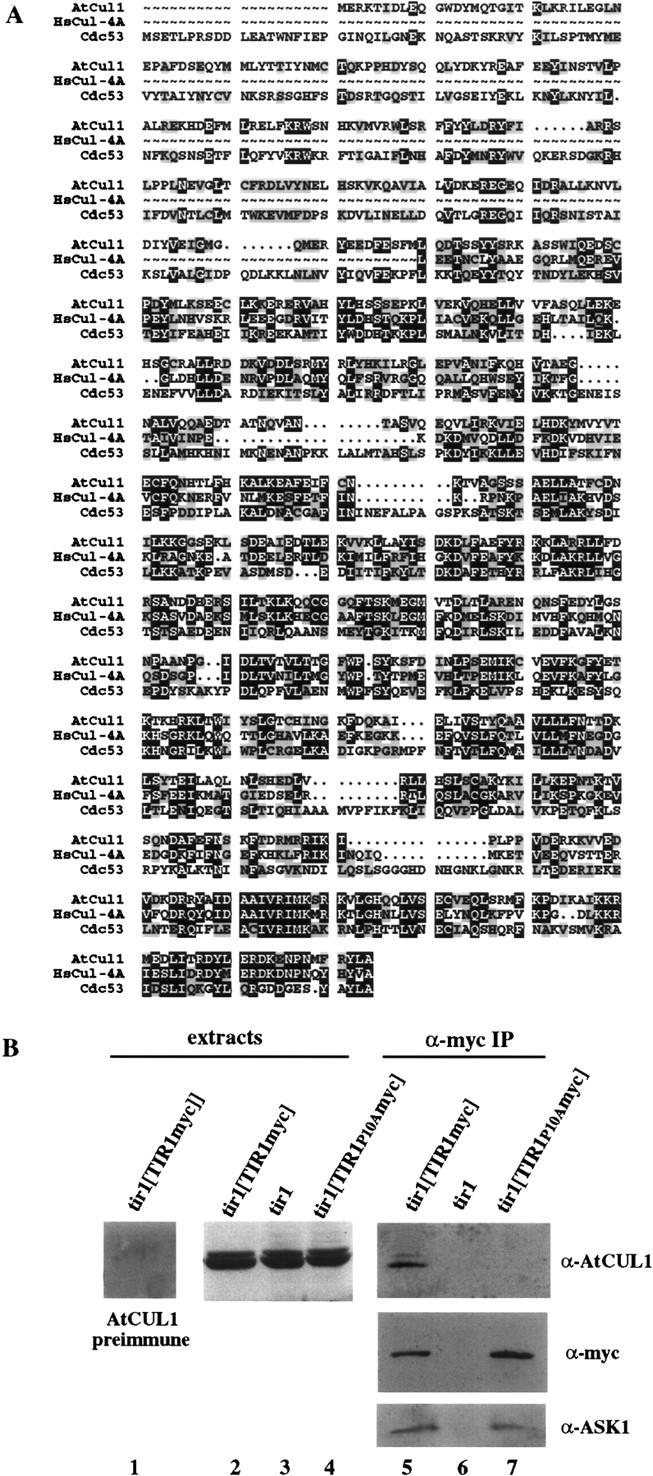
The A. thaliana cullin, AtCUL1, interacts with TIR1 in planta. (A) Sequence alignment of AtCUL1 with the human and S. cerevisiae cullins, Cul4A and Cdc53p, with PileUp (Genetics Computer Group, Inc., Madison,WI). Identical amino acids are boxed in black. Similar amino acids are boxed in gray. (B) Western blot analysis of crude seedling extracts (lanes 1–4) or α-myc immunoprecipitates (lanes 6–7).
To determine whether AtCUL1 associates with TIR1 in planta, α-myc immunoprecipitates from TIR1–myc seedlings were immunoblotted and probed with the affinity purified α-AtCUL1 antiserum. Both of the α-AtCUL1 reactive bands observed in crude extracts were also coimmunoprecipitated with the TIR1–myc protein (Fig. 4B, lanes 5–6). The interaction between TIR1 and AtCUL1 appeared to be dependent on the TIR1 F-box as AtCUL1 was absent from immunoprecipitates from seedlings expressing the myc-tagged TIR1P10A protein. Surprisingly, the ASK1 and ASK2 proteins still coimmunoprecipitated with the TIR1P10A protein even though the F-box mutation prevented these interactions in the yeast two-hybrid system (Fig. 4B, lane 7).
ASK1 is required for normal auxin response
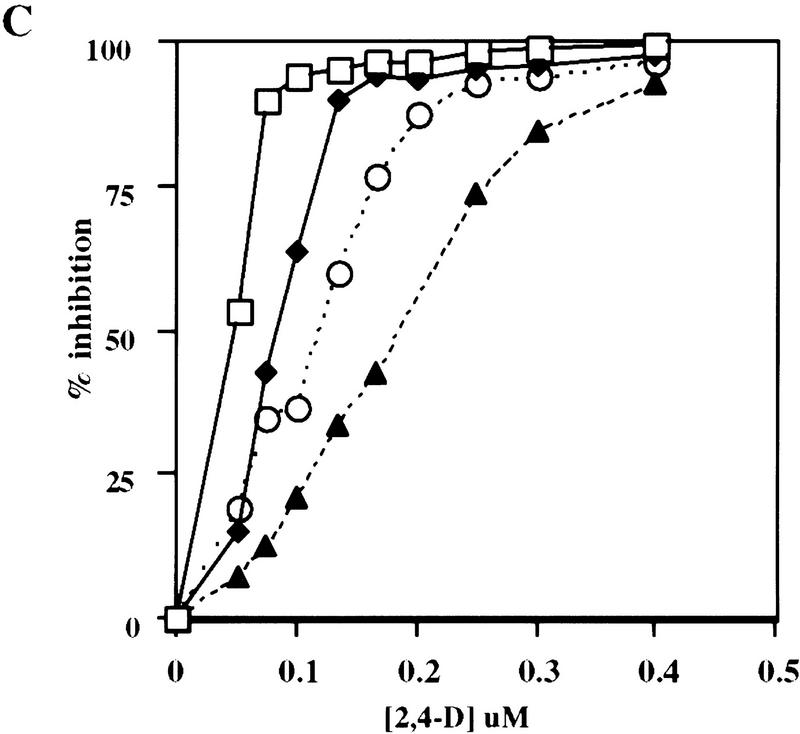
A mutant allele of ASK1 has been isolated recently by Ma and colleagues (M. Yang and H. Ma, unpubl.). This mutant, designated ask1-1, exhibits male sterility as a result of chromosome nondisjunction during meiosis I of microsporogenesis. The ask1-1 mutant also displays a reduction in organ size suggesting that cell division and/or cell expansion might be affected by the ask1-1 mutation. Because we found that the TIR1 protein can interact with ASK1 in planta, we investigated whether or not ASK1 functions in auxin response. Four-day-old seedlings obtained from an ASK1/ask1-1 heterozygote were transferred to medium containing the synthetic auxin 2,4-D. Exogenous auxin in the growth medium inhibits root elongation of wild-type seedlings, but this inhibition is reduced in mutants with impaired auxin response. Thirty-three of the 191 ASK1/ask1-1 progeny tested exhibited resistance to the root growth inhibition of exogenous auxin (Fig. 5A). All resistant seedlings also exhibited male sterility (31/31) indicating that the two phenotypes cosegregate. Also consistent with a reduced auxin response, ask1-1/ask1-1 seedlings develop fewer lateral roots than wild-type seedlings (Fig. 5B). A fraction of the ask1-1 mutants did not display auxin resistance, suggesting that this aspect of the phenotype is not completely penetrant. Incomplete penetrance is also seen for the reduced organ size phenotype of the ask1-1 mutant (D. Zhao, M. Yang, and H. Ma, unpubl.).
Figure 5.
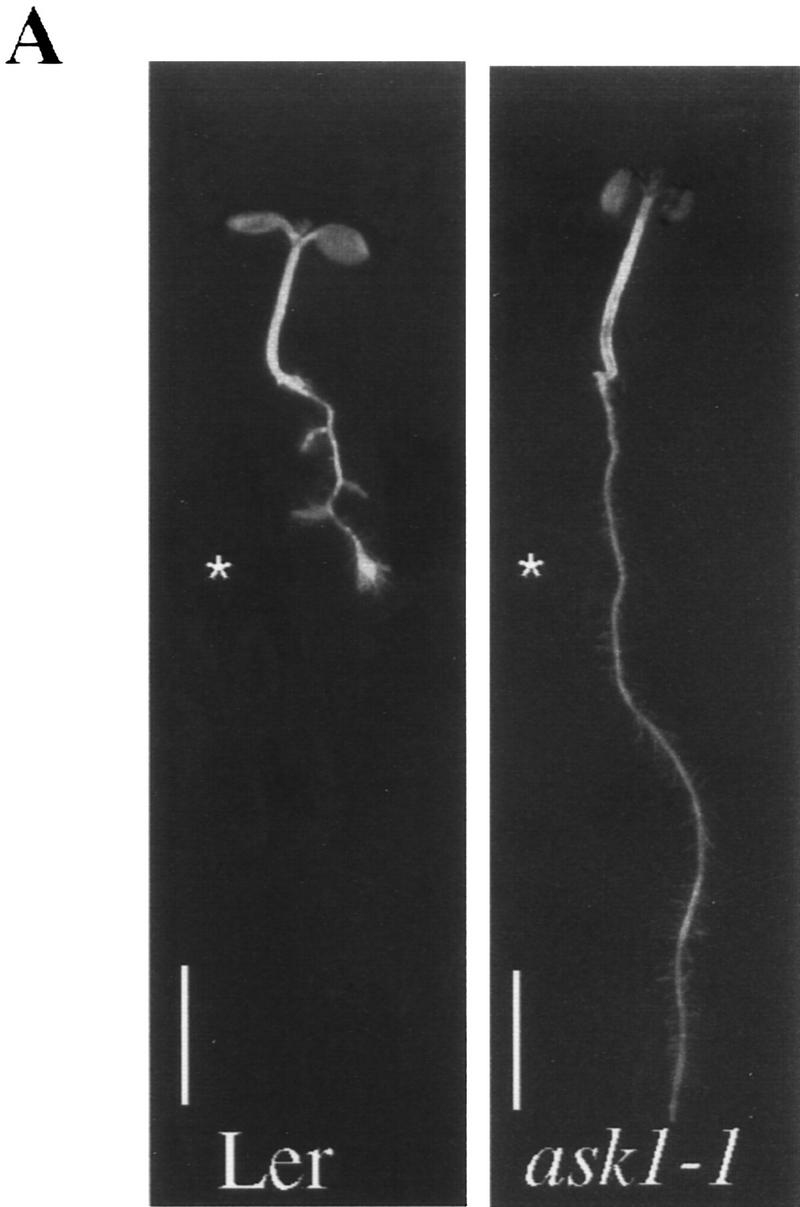
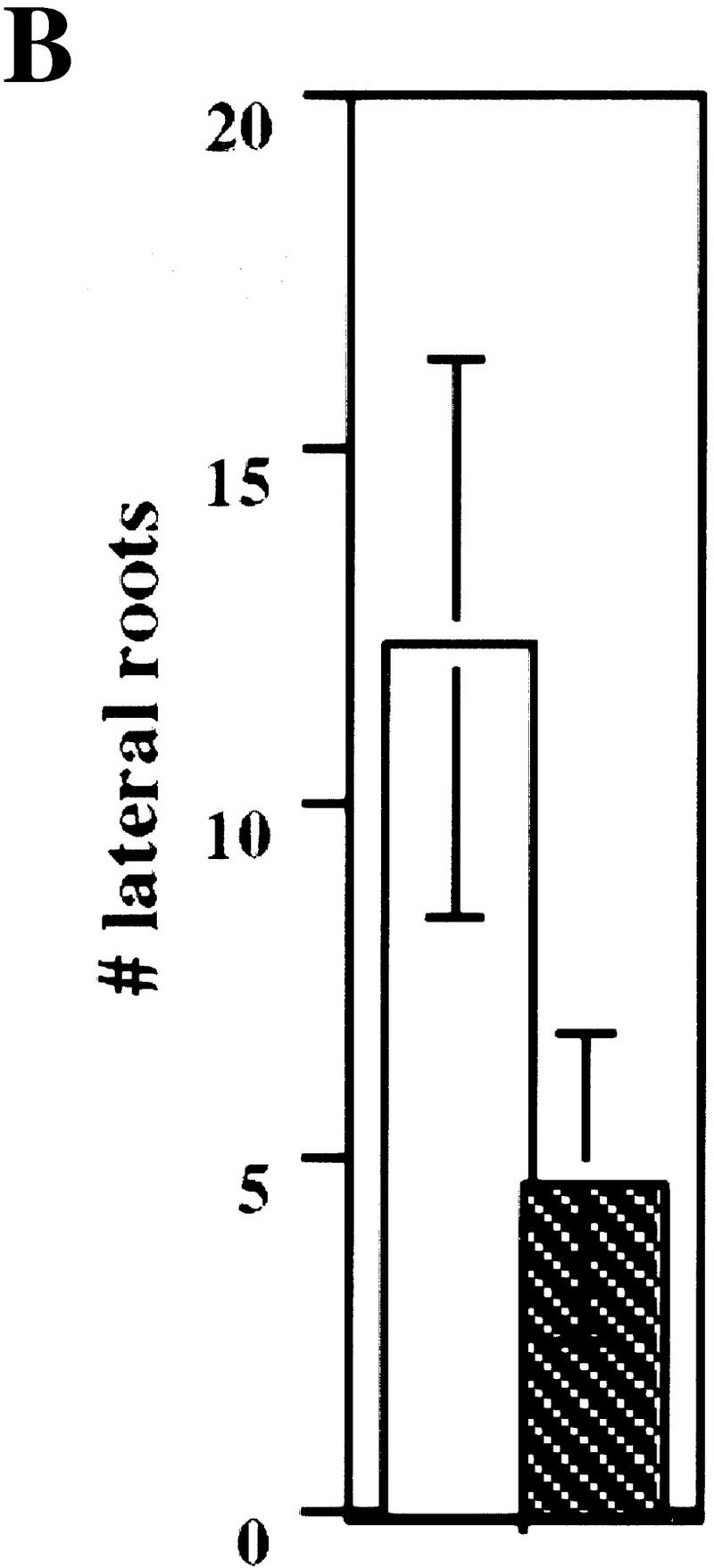
The ASK1 gene is required for normal auxin response. (A) ask1-1 and wild-type Ler seedling were grown on nutrient medium for 4 days and then transferred to medium supplemented with 0.085 μm 2,4-D and grown an additional 5 days. (Asterisk) Position of the root tips at the time of transfer. Size bars, 5 mm. (B) Mean number of lateral roots formed by wild-type and ask1-1 seedlings grown on unsupplemented medium for 11 days. (Open bar) ASKI; (hatched bar) ask1. Bars, standard error. (C) Dose response curve for wild-type, tir1-1, ask1-1, and tir1-1 ask1-1 mutants. Inhibition of root growth is expressed relative to growth on unsupplemented medium. Because of the incomplete penetrance of the ask1-1 mutation, only auxin resistant seedlings are included in this data set. Each value represents the mean of 10 seedlings. (□) Ler; (♦) tir1-1; (○) ask1-1; (▴) tir1-1, ask-1. For each data point, s.e≤10%.
The auxin resistance phenotype of the ask1-1 mutant is consistent with the hypothesis that the ASK1 gene product acts with TIR1 in the auxin response pathway. The resistant ask1-1 seedlings display a slightly more severe auxin resistance phenotype than tir1 mutants. Analysis of tir1-1 ask1-1 plants revealed that double-mutant seedlings exhibited a more severe auxin response defect than either of the single mutants (Fig. 5C).
TIR1 overexpression promotes auxin response
The effect of TIR1 overexpression on plant growth and development was examined by generating transgenic plants containing a glucocorticoid-inducible TIR1 expression construct. Transgenic lines expressing the TIR1P10A derivative containing the F-box mutation were also obtained. Northern analysis revealed that treatment with the synthetic glucocorticoid dexamethazone resulted in a large increase in TIR1 expression (Fig. 6A). When TIR1 expression was induced in light-grown seedlings, growth of the primary root was inhibited, root tips became agravitropic, and lateral root development was promoted (Fig. 6B). Closer examination of these seedlings revealed increased proliferation of root hairs at the primary root tip. In contrast, dexamethazone treatment had no obvious effects on untransformed control seedlings. Overexpression of the TIR1P10A mutant derivative also did not confer the phenotypes seen in the TIR1-overexpressing plants. Examination of several additional transgenic lines for both overexpression constructs yielded similar results. TIR1 overexpression in dark-grown seedlings severely inhibited hypocotyl elongation and promoted deetiolation (Fig. 6C). The hypocotyls of TIR1-overexpressing seedlings were also moderately agravitropic (data not shown). As observed with light-grown seedlings, overexpression of the TIR1P10A mutant protein had little if any effect on skotomorphogenesis.
Figure 6.
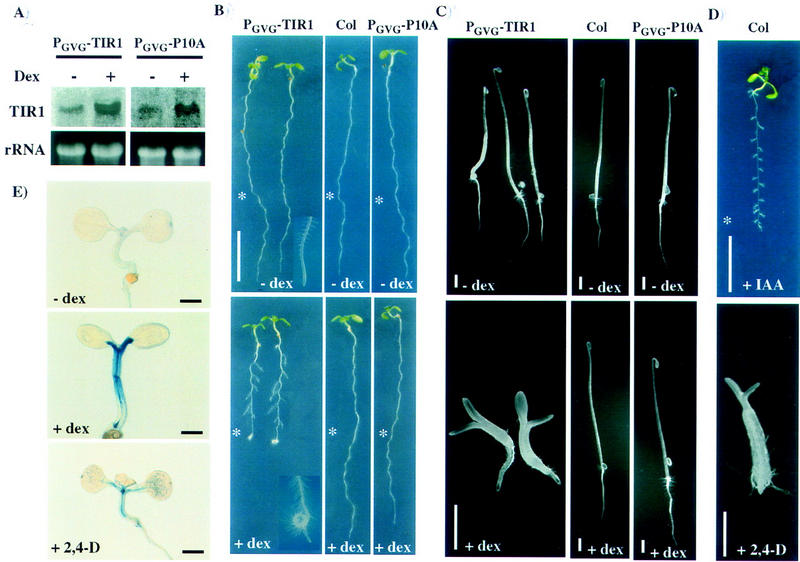
TIR1 overexpression analysis. (A) Northern analysis of T2 seedlings transformed with the glucocorticoid inducible TIR1 or TIR1P10A expression constructs. TIR1 expression was induced by treating with dexamethazone for 24 hr. (B) TIR1 expression was induced in 7-day-old transgenic PGVG–TIR1 and PGVG–TIR1P10A seedlings by transferring seedlings to nutrient medium supplemented with dexamethazone and grown an additional 2 days. Untransformed Columbia seedlings are shown as a control. (Asterisk) Position of the root tips at the time of transfer. Root tips shown in insets are not from the same seedling pictured. Size bars, 10 mm. (C) Transgenic and control seedlings were germinated and grown for 4 days in the dark on nutrient medium containing dexamethazone. Size bars, 1 mm. (D) Wild-type seedlings grown as described in B and C with auxin in the growth medium instead of dexamethazone. The light-grown seedling (top) was transferred to medium containing 1 μm IAA. Size bar, 10 mm. The dark-grown seedling (bottom) was grown on medium supplemented with 1 μm 2,4-D. Size bar, 1 mm. (E) pIAA4–gus expression in PGVG–TIR1 transgenic seedlings in the absence (top) and presence (center) of dexamethazone. Wild-type pIAA4–gus seedlings treated with 0.2 μm 2,4-D are shown at bottom. Seedlings were stained for β-glucuronidase activity 48 hr after induction with dexamethazone or 2,4-D.
The effects of TIR1 overexpression are very similar to the effects of exogenous auxin on wild-type seedlings. In the light, auxin treatment inhibits primary root elongation and promotes lateral root development, whereas in the dark, exogenous auxin inhibits hypocotyl elongation and promotes deetiolation (Fig. 6D). To more directly examine whether TIR1 overexpression promotes auxin signaling, expression of the auxin-inducible pIAA4–gus and SAUR–AC1–gus reporters were examined in lines overexpressing TIR1. A considerable increase in pIAA4–gus expression was observed after a 48-hr induction of TIR1 expression (Fig. 6E). TIR1 overexpression also promoted some ectopic pIAA4–gus expression (Fig. 6E; cf. middle and bottom panels). Similar results were obtained with the SAUR–AC1–gus reporter (data not shown). In contrast, no change in pIAA4–gus staining was observed when control seedlings were treated with dexamethazone (data not shown). This result indicates that the physiological effects of TIR1 overexpression are due to an increase in auxin signaling.
Discussion
Previous studies have suggested that the ubiquitin protein conjugation pathway may regulate auxin response. Ruegger et al. (1998) identified the F-box protein TIR1 as a factor required for normal auxin response in Arabidopsis. These authors suggested that TIR1 may be a component of an SCF ubiquitin–ligase complex that targets one or more regulators of auxin signaling for ubiquitination. In this study, we identify Arabidopsis Skp1p- and Cdc53p-related proteins that interact with TIR1 to form an SCF complex called SCFTIR1. The phenotype of plants deficient in one of the Skp1-related proteins, called ASK1, as well as the effects of overexpression of TIR1 in transgenic plants, confirm that SCFTIR1 function is central to auxin response.
Identification of SCFTIR1
The ASK1 and ASK2 genes were isolated in an extensive two-hybrid screen with TIR1 and encode two highly related Skp1-like proteins. Both of these genes have also been identified in two-hybrid screens with the Arabidopsis UFO and COI1 F-box proteins as bait (W.L. Crosby, unpubl., J. Turner pers. comm.). The in planta significance of the TIR1–ASK1 and TIR1–ASK2 two-hybrid interactions was established by demonstrating that the ASK1 and ASK2 proteins coimmunoprecipitate with epitope-tagged TIR1. At present, 10 ASK genes have been identified in the Arabidopsis genome, suggesting some specificity may exist in the interactions between F-box and ASK proteins. Consistent with this idea, we have found that TIR1 does not interact with ASK3 in a yeast two-hybrid assay (data not shown). In addition, the LRF2 protein, which exhibits 60% identity with TIR1 (Ruegger et al. 1998), does not interact with ASK1 or ASK2 in a two-hybrid test, whereas another family member, LRF1, interacts very weakly with ASK1 and ASK2 (W.M. Gray and M. Estelle, unpubl.). The possibility that LRF1 and LRF2 interact with other members of the ASK family to form SCF complexes that function in auxin signaling is being investigated. The presence of a family of related SCF complexes exhibiting functional redundancy could explain why mutations in TIR1 and ASK1 lead to relatively mild defects in auxin response when compared with mutations in AXR1.
Members of the cullin family of proteins comprise the third subunit of an SCF complex. We identified an Arabidopsis cullin member, designated AtCUL1, that coimmunoprecipitates from plant extracts along with TIR1. AtCUL1 is most closely related to several partial ESTs in the Arabidopsis database and the human cullins, Cul-4A and Cul-4B. Like the yeast Cdc53p and human Cul-4A cullins, AtCUL1 is probably a substrate for modification by RUB. AtCUL1 can be modified by RUB in vitro, and preliminary results suggest that it is also modified in vivo (J.C. del Pozo and M. Estelle, unpubl.). Although the function of this modification is unclear, it would seem to play an important role in auxin signaling because mutations in the RUB-activating enzyme AXR1 dramatically impair the auxin response pathway. Hochstrasser (1998) suggested that Rub1 modification of the yeast Cdc53p protein may regulate the partitioning of Cdc53p among different SCF complexes or affect the activity or substrate specificity of an SCF. An alternative possibility is that RUB modification controls the subcellular localization of an SCF complex in a manner similar to the regulation of protein localization by the SUMO family of ubiquitin-related proteins (Johnson and Hochstrasser 1997). However, neither of the two predominant bands detected by the α-AtCUL1 antisera that coimmunoprecipitate with TIR1 appear to contain the RUB modification as both are present in axr1-12 extracts (data not shown). This result suggests that RUB modification of AtCUL1 is not a prerequisite for SCFTIR1 formation.
The F-box domain is essential for TIR1 function in auxin response. The TIR1P10A derivative, which contains a mutation in the F-box, failed to interact with ASK1 and ASK2 in a yeast two-hybrid assay, suggesting that TIR1P10A may not function in planta due to its inability to bind to ASK1 and ASK2. The F-box two-hybrid construct (DBD–TIR11–105) interacts weakly with the ASK proteins, suggesting that TIR1 sequences outside the F-box are also important for ASK binding. Studies with other F box proteins have also demonstrated that sequences outside of the F box are important for association with Skp1 (Li and Johnston 1997). Surprisingly, when expressed in plants, the TIR1P10A mutant protein still coimmunoprecipitated with the ASK proteins. This may be because the TIR1–ASK interaction is more stable in plants, perhaps because of the interaction with additional plant proteins. Alternatively, TIR1 may function as a dimer in the SCF complex. Kominami et al. (1998) recently demonstrated that the Schizosaccharomyces pombe F-box proteins Pop1 and Pop2 form homo- and heterodimers. If TIR1P10A dimerizes with a second F-box protein or with the endogenous tir1-1 mutant protein (the tir1-1 mutation is in the LRR domain), the ASK proteins may be present in TIR1P10A–myc immunoprecipitates via their association with the dimer partner. Our observation that the TIR1P10A mutation prevents association with AtCUL1 in planta suggests that either F-box sequences are directly involved in mediating this interaction, or that the TIR1P10A mutation sufficiently alters the stability or conformation of the TIR1–ASK complex such that AtCUL1 can no longer bind efficiently.
This study shows that the SCF ubiquitin ligase model can be extended to higher plant systems. The presence of a large family of Skp1-like proteins and multiple cullins in the Arabidopsis EST database suggests that SCFs are widely used by plants. Several uncharacterized F-box proteins have been isolated in two-hybrid screens with members of the ASK protein family (E. Risseeuw and W.L. Crosby, unpubl.). Xie et al. (1998) recently reported that the COI1 gene encodes an F-box protein with leucine-rich repeats that functions in jasmonic acid response. This result suggests that jasmonic acid also controls plant growth via SCF-mediated ubiquitination.
SCFTIR1 is required for auxin response
Like mutations in TIR1, the ask1-1 mutation confers an auxin-resistance phenotype. This finding validates the physiological significance of the interaction between TIR1 and ASK1 by demonstrating that SCF components in addition to TIR1 function in auxin response. tir1-1 ask1-1 double-mutant plants display a more severe auxin resistance phenotype than either tir1-1 or ask1-1 plants. This may be the result of some degree of functional redundancy for TIR1 and ASK1 function in the auxin response pathway. We have shown that TIR1 can interact with the ASK2 protein, suggesting ASK1 and ASK2 display some functional redundancy. Similarly, perhaps ASK1 forms an SCF complex with a protein(s) that exhibits some functional redundancy with TIR1. The leucine-rich repeat f-box proteins LRF1 and LRF2 are highly related to TIR1 (Ruegger et al. 1998), and a weak two-hybrid interaction between LRF1 and ASK1 has been observed supporting this possibility (data not shown).
The TIR1 gene is strongly expressed in regions actively undergoing cell division and elongation. This expression pattern is consistent with TIR1 functioning in the auxin response pathway. Furthermore, this pattern is very similar to that of AXR1 (del Pozo et al. 1998; J.C. del Pozo and M. Estelle, unpubl.) and ASK1 (Porat et al. 1998; E. Risseeuw and W.L. Crosby, unpubl.), consistent with the suggestion that these genes act together in the same pathway.
Overexpression of TIR1 promotes auxin response. Seedlings that overexpress TIR1 have a striking resemblance to auxin-treated plants, and display an increase in auxin-inducible gene expression. Similar effects were observed in plants at later stages in development (W.M. Gray and M. Estelle, unpubl.). These results suggest that TIR1 is a limiting factor in auxin signaling, a conclusion that is supported by genetic studies of the tir1 mutants. Ruegger et al. (1998) showed that heterozygous tir1 plants have an auxin-response defect, indicating response is sensitive to TIR1 levels. Thus, it is possible that auxin response is regulated in part by altering TIR1 levels. In addition to providing a useful tool for studying TIR1 function, the ability to manipulate auxin signaling by regulating TIR1 expression may have important biotechnological applications. Applied auxin is widely used in the agricultural and horticultural fields to control plant growth processes such as root development and fruit ripening. The use of developmental or organ-specific promoters to modulate auxin signaling by increasing TIR1 expression may be a simpler and more precise means of targeting auxin response to specific tissues.
What is the SCFTIR1 substrate?
The putative target protein(s) for SCFTIR1-mediated ubiquitination are unknown. According to the SCF ubiquitin-ligase model (Patton et al. 1998), substrate proteins are recruited to the SCF complex by the F-box protein. In yeast, the stability of several key regulators of the cell division cycle is regulated by SCF-mediated degradation (Patton et al. 1998). Mutations that prevent removal of these regulators result in cell cycle arrest. Auxin is required for plant cell division, and tir1 mutants display defects in some cell division processes. In this study we show that TIR1 is required in root pericycle cells prior to the occurrence of the first cell division during lateral root primordium formation. Thus it is possible that auxin-promoted cell division in the pericycle is achieved by SCFTIR1-facilitated degradation of one or more regulators of the plant cell cycle. However, it is important to note that the tir1 mutants are also defective in cell elongation (Ruegger et al. 1998). Perhaps SCFTIR1 has multiple substrates, some of which integrate auxin signaling with the cell cycle, and others which control distinct auxin responses.
The AUX/IAA genes were identified because their transcription is rapidly induced by auxin (Abel et al. 1995). These genes encode nuclear proteins that have been proposed to function as transcription factors that mediate downstream auxin responses. The expression of the AUX/IAA genes can also be induced by inhibiting protein synthesis with cycloheximide, suggesting that at least this aspect of auxin response is regulated by one or more short-lived repressor proteins (Ballas et al. 1995; Abel and Theologis 1996). Auxin may relieve this repression by promoting the ubiquitin-mediated degradation of these factors via SCFTIR1. Alternatively, certain members of the AUX/IAA protein family themselves may be the targets of SCFTIR1. The available evidence suggests that the AUX/IAA proteins function as both activators and repressors of auxin-regulated genes. At least some AUX/IAA proteins have very short half-lives and have been shown to repress the transcription of certain auxin-inducible genes (Ulmasov et al. 1997). In contrast, gain-of-function mutations in the IAA17/AXR3 and IAA3/SHY2 genes confer a constitutive auxin-response phenotype (Rouse et al. 1998, Tian and Reed 1999). This phenotype could be explained if IAA17/AXR3 and IAA3/SHY2 encode activators of auxin response, and the mutations confer an increased stability on the mutant proteins. Measurement of IAA protein stability in axr1 and tir1 mutant backgrounds is in progress.
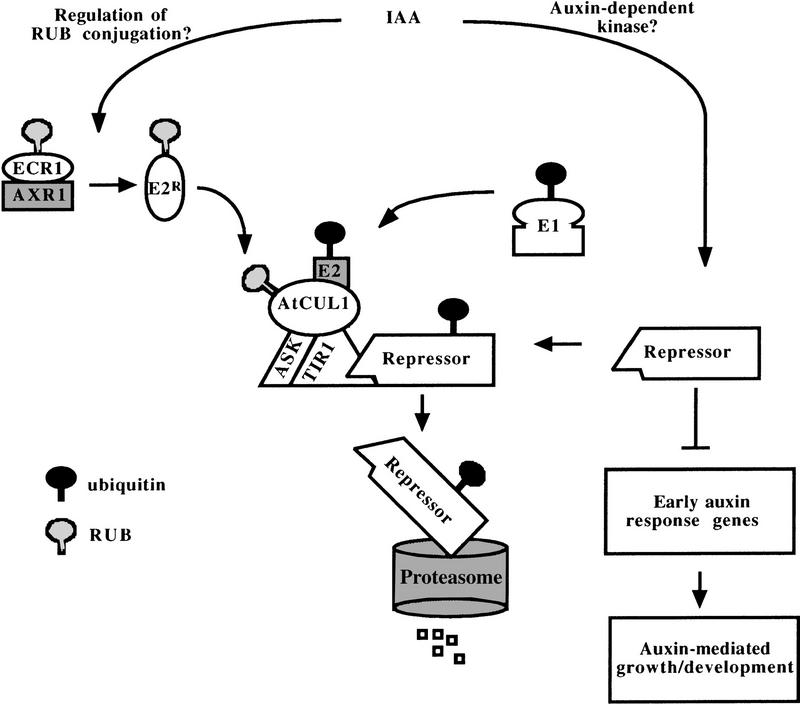
A model for auxin response
A model for SCFTIR1 function in auxin action is outlined in Figure 7. In response to hormone, SCFTIR1 ubiquitinates one or more repressors of the auxin response pathway. This ubiquitination requires the AXR1–ECR1-mediated RUB modification of the AtCUL1 protein. The subsequent ubiquitin-mediated degradation of the SCFTIR1 substrate(s) derepresses the auxin response pathway, including expression of auxin-regulated genes, resulting in changes in plant growth and development. In some respects, this model is similar to the activation of the NF-κB transcription factor in mammalian cells. Cytokine stimulation promotes the ubiquitin-mediated degradation of the NF-κB repressor IκB. This occurs through the activation of the IκB kinase (IKK), which phosphorylates IκB and targets it to an SCF complex containing the β-TrCP/hE3RSIκB F-box protein (Ghosh et al. 1998; Yaron et al. 1998). In the absence of IκB, NF-κB can activate the expression of downstream target genes involved in immune response and other cellular processes.
Figure 7.
Model for auxin response. In this model, TIR1 functions in an SCF ubiquitin ligase that targets one or more negative regulators of the auxin response pathway for ubiquitination and degradation. SCFTIR1 activity requires the RUB modification of AtCUL1 via the AXR1–ECR1 pathway. The ubiquitin–mediated degradation of the repressor(s) derepresses the response pathway resulting in the activation of the early auxin response genes (AUX/IAA genes) that may control downstream events resulting in auxin-regulated growth and development. E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme; E2R, RUB-conjugating enzyme.
A major question yet to be addressed is how auxin regulates the AXR1–SCFTIR1 pathway. Although several proteins capable of binding auxin have been isolated, no convincing candidate auxin receptor has been identified. Equally unclear are the nature and mechanisms involved in signal transmission. MAP kinase cascades, G proteins, phosphatidylinositol, and calcium-based signaling pathways all have been proposed to be involved in auxin signal transduction, however, the question has yet to be resolved. Although the details of auxin perception and signal transmission are unclear, genetic and molecular studies have clearly demonstrated the importance of AXR1 and SCFTIR1 in auxin response. It is possible that the auxin signaling pathway regulates the assembly, localization, or activity of SCFTIR1. The observation that TIR1 is limiting for auxin response is consistent with this model. Effects on SCFTIR1 may occur by regulated RUB modification of AtCUL1 or by an as yet undetermined mechanism (Fig. 7). Alternatively, the regulatory input may be through the substrate protein (Skowyra et al. 1997; Winston et al. 1999). According to the SCF paradigm from yeast and mammalian systems, activation of a protein kinase results in phosphorylation of the SCF substrate protein. The SCF then binds the phosphorylated protein and facilitates ubiquitination and degradation. Thus far, genetic screens for mutants exhibiting reduced auxin response have not uncovered any components of a kinase cascade. Kinases have been identified that are rapidly activated by auxin treatment (Mizoguchi et al. 1994). However, it is unclear whether activation is a direct consequence of auxin treatment or a secondary effect of auxin-mediated growth. Recently, the tobacco MAP kinase kinase kinase (MAPKKK) NPK1 was shown to repress auxin-mediated gene expression when transfected into maize mesophyl protoplasts (Kovtun et al. 1998). This result suggests that NPK1 negatively regulates auxin signaling. Thus, NPK1 or some other factor in the NPK1 pathway could be a potential substrate for SCFTIR1. The cloning and analysis of additional auxin response genes, such as AXR4, and modifiers of AXR1 and TIR1, such as SAR1, should answer many of these questions and further elucidate the long-standing problem of how auxin controls plant growth and development.
Materials and methods
Plant material and growth conditions
A. thaliana plants were grown as described by Lincoln et al. (1990). The mutant lines used in this study have been described previously (Ruegger et al. 1998). The FA4 transgenic line containing a translational fusion of the Cyc1At cyclin to β-glucuronidase was generously provided by Drs. A. Colon and P. Doerner (Salk Institute, La Jolla, CA). The BA3 transgenic line containing auxin-responsive promoter elements from the pea IAA4 gene fused to β-glucuronidase has been described previously (Oono et al. 1998). The construction of the TIR1–gus, TIR1–myc, TIR1P10A, and TIR1P10A–myc transgenic lines is described below. All lines used in this study were of the Columbia ecotype with the exception of the ask1-1 mutant which is derived from Landsberg erecta. To construct the tir1-1 ask1-1 double mutant, the tir1-1 mutation was first introduced into the Landsberg erecta background by five successive backcrosses. Pollen from tir1-1 segregants were then used to pollinate emasculated ask1-1 flowers.
In experiments on sterile medium, seeds were sterilized and grown as described by Ruegger et al. (1998). The synthetic auxin 2,4-D and 30 μm dexamethazone were added to autoclaved medium when indicated. To examine sensitivity to auxin, seedlings were germinated and grown for 4 days on nutrient medium. Seedlings were then transferred to 2,4-D supplemented medium, grown an additional 5 days, and root elongation was measured.
Lateral root studies
The cyc1At–gus reporter in wild-type line FA4 was crossed with tir1-1 plants, and F2 seedlings homozygous for both the transgene and tir1-1 mutation identified. tir1-1[cyc1At–gus] and FA4 control seeds were germinated on nutrient medium plates and grown for 9 days at 23°C with a 16 hr light cycle and a light intensity of 45–60 μE/m2/s. Nine-day-old seedlings were transferred to nutrient plates containing 0.25 μm 2,4-D or unsupplemented plates and grown an additional 24 hr. cyc1At–gus expression in lateral roots and developing lateral root primordia was detected by incubating seedlings in an X-Gluc solution for 8 hr at 37°C and destaining with 70% ethanol as described previously (Stomp 1991).
ASK1/ask1-1 progeny seeds were sterilized and plated on nutrient medium. Plates were incubated vertically in a 23°C incubator with a 16 hr light cycle and a light intensity of 45–60 μE/m2/s. After 11 days growth, lateral roots and lateral root primordia were recorded, and the seedlings potted. The ASK1 phenotype was scored when the plants flowered.
Yeast two-hybrid screen
A 2.0-kb SmaI–PvuII fragment containing the TIR1 coding sequence from plasmid pGB14 was cloned into the SmaI site of pBI880 (Kohalmi et al. 1998) to generate the GAL41–147-TIR1 bait plasmid pGB16. This construct was introduced into the yeast strain YPB2 [MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 canR gal4–542 gal80–538 LYS2::GAL1UAS–LEU2TATA–HIS3 URA3::(GAL43 × 17mer)-CYC1TATA–lacZ] and used to screen an Arabidopsis cDNA two-hybrid library. The library was made from poly(A)+ RNA isolated from tissues taken from plants at four different stages of development, ranging from 2-week-old seedlings through plants showing early signs of senescence. The library was directionally cloned as SalI–NotI fragments with the BRL Superscript system and cloned into pBI771 (Kohalmi et al. 1998). The library contained ∼2 × 107 independent clones. The library was transformed into YPB2 by standard methods (Gietz and Schiestl 1995) and transformants plated directly onto synthetic complete nutrient medium lacking leucine and tryptophan (Sherman et al. 1978) and supplemented with 15 mm 3-amino-1′, 2′, 4′ triazole (3-AT). Plates were incubated at room temperature for 7–10 days and 3-AT-resistant transformants were assayed for β-galactosidase activity as described previously (Chevray and Nathans 1992).
DNA was isolated from library transformants that were both 3-AT resistant and expressed the lacZ reporter. Library cDNA inserts were amplified with PCR primers complementary to flanking GAL4 activation domain and ADH1 transcription termination sequences. Amplified inserts were analyzed by DpnII restriction analysis to identify related clones, and representative members were sequenced using the automated Thermo Sequenase Dye Terminator Cycle Sequencing kit (Amersham).
The TIR1P10A two-hybrid plasmid, pGB21, was constructed by introducing a single amino acid change (proline to alanine) in the F-box domain in plasmid pGB16 with the Quickchange site-directed mutagenesis kit (Stratagene) and complementary oligonucleoties with the sequence 5-GCCTTGTCGTTTGCAGAAGAGGTACTAGAGC-3′. The TIR1 ORF of pGB21 was sequenced to confirm that no additional changes occurred during the site-directed mutagenesis procedure.
The TIR1-F-box two-hybrid construct, pGB17, was constructed by cloning a SmaI–PmlI fragment encoding amino acids 1–105 of TIR1 into the SmaI site of pBI880. Junctions were sequenced to confirm the reading frame.
Transgenic lines
The TIR1–gus reporter plasmid, pGB10, was constructed by cloning a 4-kb XhoI fragment containing TIR1 5′ sequences (∼−4000 to −6) from the TIR1 genomic clone, D109ES, into the SalI site of pBI101.2 (Clontech). pGB10 was introduced into Agrobacterium strain GV3101 that was used to inoculate wild-type (Col) plants by vacuum infiltration as described previously (Bechtold et al. 1993). TIR1–gus expression was examined by incubating seedlings/organs in an X-Gluc solution for 2–12 hr at 37°C and destaining with 70% ethanol as described previously (Stomp 1991).
The TIR1 overexpression construct, pGB19, was constructed by cloning the TIR1 cDNA into the XhoI site downstream of the GAL4-binding sites of the plant transformation vector pTA7002. This vector also expresses the glucocorticoid-inducible Gal4–VP16-glucocorticoid receptor (GVG) transgene to drive TIR1 expression (Aoyama and Chua 1997). The PGVG–TIR1P10A expression vector, pGB22, was constructed by mutagenizing the TIR1 F-box as described above and cloning the cDNA into the XhoI–SpeI sites of pTA7002.
TIR1 was epitope tagged by introducing a SmaI restriction site after amino acid 586 of the TIR1 coding sequence in plasmid pGB33 with the Quickchange site-directed mutagenesis kit (Stratagene) and complementary oligonucleotides with the sequence 5′-GACTCAACAATGAGGTTTCCCGGGCAAATCATTACTACTAACG-3′. A DraI–SmaI cassette encoding six copies of the c-myc 9E10 epitope was cloned into this SmaI site. The 5′ TIR1-c-myc epitope junction is in-frame; however, the 3′ junction causes a frameshift that changes the extreme carboxyl terminus of the TIR1 protein (RQIITTNGL* to RANHYY*). A SalI fragment containing the TIR1–myc fusion was cloned into the XhoI site of the glucocorticoid-inducible expression vector pTA7002 (Aoyama and Chua 1997) to form plasmid pGB28. To construct the c-myc tagged TIR1P10A transgene, the c-myc-tagged derivative of plasmid pGB33 described above was mutagenized with the Quickchange site-directed mutagenesis kit (Stratagene) and complementary oligonucleotides with the sequence 5-′GCCTTGTCGTTTGCAGAAGAGGTACTAGAGC -3′. The TIR1P10A mutation was confirmed by sequencing and a SalI–SpeI fragment encompassing the coding region was cloned into the XhoI–SpeI sites of the pTA7002 expression vector. These plasmids were used to transform Columbia and tir1-1 plants as described above.
TIR1 overexpression analysis
The glucocorticoid-inducible TIR1 expression constructs pGB19 and pGB22 were introduced into Columbia and tir1-1 backgrounds by Agrobacterium-mediated transformation as described above. Transgenic seedlings were identified on nutrient medium containing 50 μg/ml hygromycin. Lines that overexpressed TIR1 were identified by Northern blot analysis of T2 seedlings that had been induced with 0.03 mm dexamethazone for 24 hr. Physiological studies were done with T3 seedlings homozygous for the transgenes.
Antibodies
The ASK1 and ASK2 coding sequences were cloned into pBI784 and pBI786 (Kohalmi et al. 1998) and expressed as 6xHis fusion proteins in E. coli strain BL(21) pLysS. The recombinant proteins were purified by standard IMAC chromatography over Ni+ affinity resin (Quagen). New Zealand white rabbits were immunized by subcutaneous injection of ∼200 μg of recombinant protein suspended in complete Freund’s adjuvent, followed by three additional injections at 2-week intervals with protein suspended in incomplete Freund’s adjuvant. Titers were assessed at 6 and 8 weeks, and immune serum recovered from animals exhibiting an appropriate titer (detecting 1 ng of antigen on a dot blot using a 1:10,000 dilution of whole serum). Crude α-ASK1 and α-ASK2 antisera were used at 1:5000 dilutions for Western blot analysis of plant extracts and immunoprecipitates.
A full-length predicted ORF for AtCUL1 was PCR amplified from an Arabidopsis two-hybrid library (Kohalmi et al. 1998) and genomic BAC clone (Accession no. AC002330) with Pfu DNA polymerase (Stratagene) by the vendor’s recommended conditions. Amplicons were cloned into pSL1180 (Pharmacia Biotech) and sequenced.
The AtCUL1 cDNA was cloned in-frame into the 6×His-tagged expression vector pQE31 (Qiagen). 200 ml of XL1-blue cells, carrying the pQE–AtCUL1 plasmid, were grown at 30°C to an OD of ∼1.2 and then were induced with 1.5 mm IPTG for 4 hr. The cells were spun down and resuspended in 10 ml of buffer A (6 m Urea, 100 mm NaPO4, 0.1% Tween 20, adjusted at pH8) and sonicated at 4°C. The lysate was cleared by centrifugation at 10,000 rpm for 20 min at 4°C. The supernatant was incubated with Ni–NTA agarose (Qiagen) for 1 hr at 4°C which was then washed four times with buffer B (6 m urea, 100 mm NaPO4, 0.1% Tween 20, adjusted at pH6). The AtCUL1 protein was eluted from the beads with buffer B + 0.5 m imidazole and checked for purity by SDS-PAGE. The purified protein was dialyzed overnight against 15 liters of 0.5 m urea, 100 mm Tris-HCl (pH7.5), 0.05% Tween 20. A sample of >90% purity was used for serum production, by standard procedures (Cocalico Biologicals Inc., Reamstown, PA). The antisera was immunoaffinity purified against bacterially expressed AtCUL1 bound to immobilon membrane as described (Pringle et al. 1989). Affinity purified α-AtCUL1 was diluted 1:1000 for Western blot analysis.
Monoclonal anti-c-myc 9E10 antibody was purchased from BabCo and used as recommended.
Immunoprecipitations and Western blot analysis
Arabidopsis protein extracts were prepared from 5 to 7-day-old seedlings that were grown under sterile conditions in liquid nutrient medium. Dexamethazone was added 24 hr prior to harvest to induce TIR1–myc expression when indicated. Extracts were prepared by homogenizing seedlings in ice cold buffer C (50 mm Tris.Cl, 150 mm NaCl, 0.5% NP-40, 1 mm PMSF, 5 μg/ml leupeptin, 5 μg/ml pepstatin at pH 7.5). Extracts were cleared by spinning for 15 min in a microcentrifuge.
For immunoprecipitations, 1–2 mg of extract was precleared by incubating with 50 μl of protein A-agarose (Boehringer Mannheim) for 3 hr at 4°C with gentle mixing. A total of 5 μl of anti-c-myc 9E10 antibody was added to the precleared extract and incubated for 1–2 hr at 4°C. Immune complexes were collected by adding 50 μl of protein A-agarose and mixing gently at 4°C for 3 hr followed by brief (∼5 sec) centrifugation. Immune complexes were washed three times for 20 min in 1 ml of Buffer C and resuspended in 50 μl of 2× SDS-PAGE sample buffer. Proteins were resolved on SDS gels containing 10% acrylamide and transferred to nitrocellulose membranes. Proteins were detected by Western analysis with enhanced chemiluminescence as described by the manufacturer (Amersham).
In situ hybridization
Digoxigenin-labeled TIR1 sense and antisense probes (Boehringer Mannheim Corp.) for in situ hybridization were produced from the full-length TIR1 cDNA sequence. Limited alkaline hydrolysis of the probes to 150 bp was performed according to Cox et al. (1984). Whole mount in situ hybridization of 3 to 7-day-old seedlings was performed as described by Ludevid et al. (1992). Inflorescences and siliques were fixed in 4% paraformaldahyde, embedded in paraffin wax, and 10-μm sections were prepared for in situ hybridization according to Jackson (1991).
Acknowledgments
This work was supported by the Department of Energy (DE-FG02-98ER20313 to M.E.), the National Institutes of Health (GM43644 to M.E., GM 18680 to W.M.G.), and the NRC-PBI core program (W.L.C.). The pTA7002 plasmid was kindly provided by Nam-Hai Chua. We thank Jocelyn Turner and Peter Schorr for technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mestelle@bio.indiana.edu; FAX (812) 855-6705.
References
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Ballas N, Wong L-M, Ke M, Theologis A. Two auxin-responsive domains interact positively to induce expression of the early indolacetic acid-inducible gene PS-IAA4/5. Proc Natl Acad Sci. 1995;92:3483–3487. doi: 10.1073/pnas.92.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser III Sci Vie. 1993;316:1194–1199. [Google Scholar]
- Blakely LM, Evans TA. Cell dynamics studies on the pericycle of radish seedling roots. Plant Sci Lett. 1979;14:79–83. [Google Scholar]
- Celenza JL, Jr, Grisafi PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Genes & Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Cernac A, Lincoln C, Lammer D, Estelle M. The SAR1 gene of Arabidopsis acts downstream of the AXR1 gene in auxin response. Development. 1997;124:1583–1591. doi: 10.1242/dev.124.8.1583. [DOI] [PubMed] [Google Scholar]
- Chevray PM, Nathans D. Protein interaction cloning in yeast: Identification of mammalian proteins that interact with the leucine zipper of jun. Proc Natl Acad Sci. 1992;88:9578–9582. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, DeLeon DV, Angerer LM, Angerer RC. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101:485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- Davies PJ. The plant hormones: Their nature, occurrence and functions. In: Davies PJ, editor. Plant hormones: Physiology, biochemistry and molecular biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–12. [Google Scholar]
- del Pozo JC, Estelle M. Function of the ubiquitin-proteosome pathway in auxin response. Trends Plant Sci. 1999;4:107–112. doi: 10.1016/s1360-1385(99)01382-5. [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Timpte C, Tan S, Callis J, Estelle M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- Ferreira PCG, Hemerly AS, de Almeida Engler J, Van Montagu M, Engler G, Inze D. Developmental expression of the Arabidopsis cyclin gene cyc1At. Plant Cell. 1994;6:1763–1774. doi: 10.1105/tpc.6.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada H. Xylogenesis: Initiation, progression, and cell death. Annu Rev Plant Physiol Mol Biol. 1996;47:299–325. doi: 10.1146/annurev.arplant.47.1.299. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. Transforming yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-kB and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Gray W M, Östin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormones. J Cell Biol. 1980;85:811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A, Bergounioux C, Van Montagu M, Inze D, Ferreira P. Genes regulating the plant cell cycle: Isolation of a mitotic-like cyclin from Arabidopsis thaliana. Proc Natl Acad Sci. 1992;89:3295–3299. doi: 10.1073/pnas.89.8.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, Estelle MA. Genetic approaches to auxin action. Plant Cell Environ. 1994;17:525–540. doi: 10.1111/j.1365-3040.1994.tb00147.x. [DOI] [PubMed] [Google Scholar]
- ————— The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 1995;7:211–220. doi: 10.1046/j.1365-313x.1995.7020211.x. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. There’s the Rub: A novel ubiquitin-like modification linked to cell cycle regulation. Genes & Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- Jackson DP. In-situ hybridization in plants. In: Bowles DJ, Gurr SJ, McPherson M, editors. Molecular plant pathology: A practical approach. UK: Oxford University Press; 1991. [Google Scholar]
- John PCL, Zhang E, Dong C, Diederich L, Wrightman F. p34cdc2 related proteins in control of cell cycle progression, the switch between division and differentiation in tissue development, and stimulation of cell division by auxin and cytokinin. Aus J of Plant Physiol. 1993;20:503–526. [Google Scholar]
- Johnson PR, Hochstrasser M. SUMO-1: Ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- Kohalmi SE, Reader LJV, Samach A, Nowak J, Haughn GW, Crosby WL. Identification and characterization of protein interactions using the yeast 2-hybrid system. Plant Mol BiolMan. 1998;M1:1–30. [Google Scholar]
- Kominami KI, Ochotorena I, Toda T. Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF (Skp1-Cullin-1-F-box) ubiquitin ligase. Genes Cells. 1998;3:721–735. doi: 10.1046/j.1365-2443.1998.00225.x. [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu W-L, Zeng W, Sheen J. Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature. 1998;395:716–720. doi: 10.1038/27240. [DOI] [PubMed] [Google Scholar]
- Krek W. Proteolysis and the G1-S transition: The SCF connection. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- Kumar S, Yoshida Y, Noda M. Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem Biophys Res Commun. 1993;195:393–399. doi: 10.1006/bbrc.1993.2056. [DOI] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast CDC53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes & Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Li FN, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: Coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludevid D, Höfte H, Himelblau E, Chrispeels MJ. The expression pattern of the tonoplast intrinsic protein γ-TIP in Arabidopsis thaliana is correlated with cell enlargement. Plant Physiol. 1992;100:1633–1639. doi: 10.1104/pp.100.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Gotoh Y, Eisuke N, Yamaguchi-Shinozaki K, Hayashida N, Iwasaki T, Kamada H, Shinozaki K. Characterization of two cDNAs that encode MAP kinase homologues in Arabidopsis thaliana and analysis of the possible role of auxin in activating such kinase activities in cultured cells. Plant J. 1994;5:111–122. doi: 10.1046/j.1365-313x.1994.5010111.x. [DOI] [PubMed] [Google Scholar]
- Oono Y, Chen QG, Overvoorde PJ, Kohler C, Theologis A. Age mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell. 1998;10:1649–1662. doi: 10.1105/tpc.10.10.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F, Kawasaki H, Aido N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. The new NEDD8-ligating system for cullin-A. Genes & Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: Don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Porat R, Lu P, O’Neill SD. Arabidopsis SKP1, a homologue of a cell cycle regulator gene, is predominantly expressed in meristematic cells. Planta. 1998;204:345–351. doi: 10.1007/s004250050265. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AE, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Rabindran SK, Danielsen M, Stallcup MR. Glucocorticoid-resistant lymphoma cell variants that contain functional glucocorticoid receptors. Mol Cell Biol. 1987;7:4211–4217. doi: 10.1128/mcb.7.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao-Naik C, delaCruz W, Laplaza JM, Tan S, Callis J, Fisher AJ. The Rub family of ubiquitin-like proteins. J Biol Chem. 1998;273:34976–34982. doi: 10.1074/jbc.273.52.34976. [DOI] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser HMO. Changes in auxin response from mutations in an AUX/IAA gene. Science. 1998;279:1371–1373. doi: 10.1126/science.279.5355.1371. [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes & Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thiolester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence CW. Methods in yeast genetics. Cold Sping Harbor, NY: Cold Spring Harbor Laboratory; 1978. [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Stomp A-M. Histochemical localization of β-glucuronidase. In: Gallagher SR, editor. GUS protocols. New York, NY: Academic Press; 1991. pp. 103–113. [Google Scholar]
- Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126:711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes & Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IkappaBalpha- ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]