Abstract
The Iroquois complex (Iro-C) homeodomain proteins allow cells at the proximal part of the Drosophila imaginal wing disc to form mesothoracic body wall (notum). Cells lacking these proteins form wing hinge structures instead (tegula and axillary sclerites). Moreover, the mutant cells impose on neighboring wild-type cells more distal developmental fates, like lateral notum or wing hinge. These findings support a tergal phylogenetic origin for the most proximal part of the wing and provide evidence for a novel pattern organizing center at the border between the apposed notum (Iro-C-expressing) and hinge (Iro-C-nonexpressing) cells. This border is not a cell lineage restriction boundary.
Keywords: Iroquois complex, imaginal wing disc, thorax development, Drosophila
Much is known of the genes and genetic interactions that govern appendage formation in insects and vertebrates (Blair 1995; Brook et al. 1996; Irvine and Vogt 1997; Shubin et al. 1997; Carroll 1998; Schwabe et al. 1998). In Drosophila, selector homeotic genes define large territories (compartments) of the imaginal wing disc, the epithelium that gives rise to the mesothoracic body wall and wing. Thus, engrailed (en) subdivides the disc into anterior (A) and posterior (P) compartments, and apterous (ap) into dorsal (D) and ventral (V) ones, by being expressed, respectively, in the P and D compartments. Compartments are delimited by cell-lineage restriction boundaries (García-Bellido et al. 1976). Interactions between apposed cells of different compartments, mediated by the signaling molecules Hedgehog (A/P boundary), and Serrate and Delta (D/V boundary) give rise, respectively, to gradients of BMP/Decapentaplegic (Dpp) and Wnt/Wingless (Wg), which appear to organize growth and patterning of the wing (Lecuit et al. 1996; Nellen et al. 1996; Zecca et al. 1996; Neumann and Cohen 1997). This needs the participation of vestigial (vg), a gene that specifies wing fate (Kim et al. 1996). Although compartments have not been found in vertebrates, homologous molecules and similar genetic hierarchies control vertebrate limb development (Blair 1995; Brook et al. 1996; Irvine and Vogt 1997; Shubin et al. 1997; Carroll 1998; Schwabe et al. 1998).
In contrast, much less is known of the genes and signals that specify Drosophila body wall development. In the case of the mesothoracic body wall, ap, which is expressed in most of the notum anlage, is largely dispensable for notum development (Cohen et al. 1992) and wg seems only necessary for the generation of late pattern elements like some bristles (Phillips and Whittle 1993). Genes such as extradenticle (exd), vein, or pannier are important for notum development (Ramain et al. 1993; González-Crespo and Morata 1995; Simcox et al. 1996), but they do not appear to specify a notum fate. In contrast, the genes of the Iroquois complex (Iro-C) araucan (ara), caupolican (caup), and mirror (mirr), which encode highly related homeodomain proteins that are members of the prepattern that controls proneural and provein genes (Gómez-Skarmeta et al. 1996; McNeill et al. 1997), are candidates to perform a basic function in the formation of the notum. Their earliest expression in the wing disc is restricted to the notum territory, and their absence in clones of cells induces malformations in the notum but only relatively minor defects in the patterning of the wing (Gómez-Skarmeta et al. 1996; Leyns et al. 1996). Here we show that the Iro-C genes are necessary for notum specification, as their absence transforms notum cells into hinge cells. Moreover, similarly to ap and en in the wing (Blair 1995; Brook et al. 1996; Irvine and Vogt 1997; Carroll 1998), the Iro-C genes establish a signaling system that appears to organize development in the notum and the dorsal wing hinge territories. (In this paper we refer to the hinge in a strict sense (Bryant 1978), meaning the part of the wing articulation characterized by the presence of the tegula and sclerites.)
Results and Discussion
At the notum, clones of cells homozygous for the Iro-C deletions Df(3L)iroDFM1 or Df(3L)iroDFM3 and induced during the first and second larval instars were always associated with extensive malformations. The notum cuticle was replaced by a mostly naked, corrugated cuticle with sclerotized structures. In 52 of 116 cases [Df(3L)iroDFM3 clones], these structures were clearly identifiable as components of an ectopic wing hinge, for example, axillary sclerites (Fig. 1A,E) and tegula-like cuticle, with characteristic bristles and sensilla trichoidea and campaniformia (Fig. 1B,E). The multiple wing hairs (mwh) marker indicated that these sensilla arose within homozygous mutant tissue (Fig. 1B). However, the mwh and forked (f) markers, which affect trichomes and/or bristles, were uninformative (Lindsley and Zimm 1992) for the other ectopic structures and for the nonstructured corrugated cuticle (as they lack cuticular processes) and, consequently, the extent of homozygous mutant tissue within the malformations could not be determined. Forty three of the 52 malformations affected the lateral notum. The ectopic axillary sclerites were always arranged in a mirror-image disposition with respect to the extant ones (Fig. 1E). Malformations at more medial regions of the notum, which did not affect the lateral notum, most frequently contained disorganized groups of tegula-like sensila trichoidea and campaniformia (7 of the 52 malformations; data not shown) and, in 2 cases, almost complete ectopic tegulae (Fig. 1C,D). Malformations reaching the central-most regions of the notum caused defects in the fusion of heminota, which were separated by an undefined cuticle (not shown). Clones not associated with malformations appeared in flies irradiated 96–120 hr after egg laying (AEL). They developed normally in the central notum or induced invaginating cuticle vesicles in the lateral regions (not shown). Altogether, these results demonstrate an early requirement of the Iro-C for notum specification, as its absence changes the fate of its cells to wing hinge or impedes their terminal differentiation.
Figure 1.
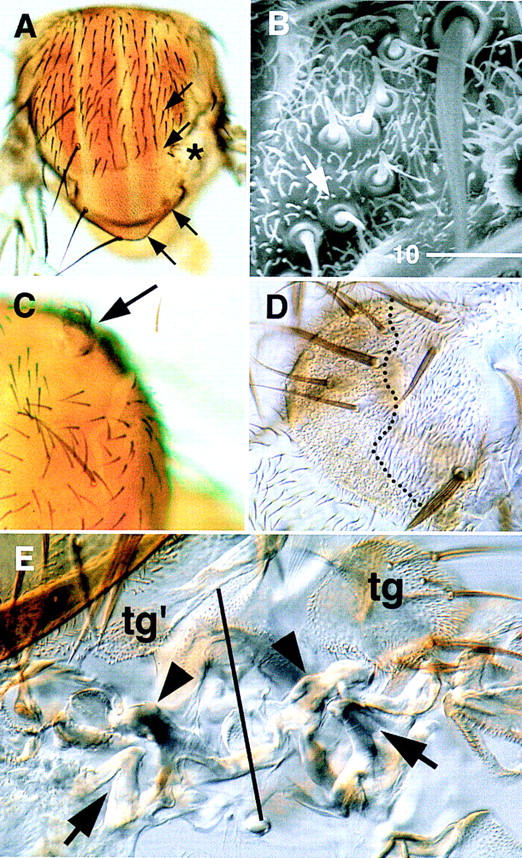
Phenotypes associated with Df(3L)iroDFM3 cell clones. (A) Notum with a malformation caused by mwh Df(3L)iroDFM3 cells (*). In the Df(3L)iroDFM3/+ territory, the dorsocentral and scutellar macrochaetae (arrows) are absent, which suggests an interference of the mutant clone with the patterning of the wild-type tissue. This nonautonomous effect was observed in 72 of 84 clones examined. (B) Ectopic group of sensilla trichoidea (one of them arrowed), similar to those of the tegula (tg in E), within mwh Df(3L)iroDFM3 territory. (C) Ectopic tegula structure (arrowed) arising on the anterior notum. (D) Same tegula structure shown after mounting the cuticle and at higher magnification. Only the tissue to the left of the dotted line is mwh Df(3L)iroDFM3. (E) Cuticle of a malformation similar to that in A. The solid line runs along the approximate axis of symmetry that separates wild type (right) from duplicated, ectopic hinge structures (left). (tg, tg′) Tegula and tegula-like cuticle with ectopic sensilla trichoidea. (Arrowheads) Wild-type and ectopic first axillary sclerites; (arrows) extant and ectopic second axillary sclerites.
In the wild-type third instar wing disc, the notum and the wing hinge territories are separated by a fold in the epithelium (Figs. 2A and 3A,B; Bryant 1978). Remarkably, Iro-C− clones within the notum territory induced a similar fold around themselves (Fig. 2A). Moreover, appropriately positioned clones rerouted the notum–hinge fold, and this became continuous with the clone-induced fold (Fig. 2D–F). In contrast, the border of clones contacting hinge cells did not form a fold and had normal, wiggly outlines (Fig. 2E).
Figure 2.
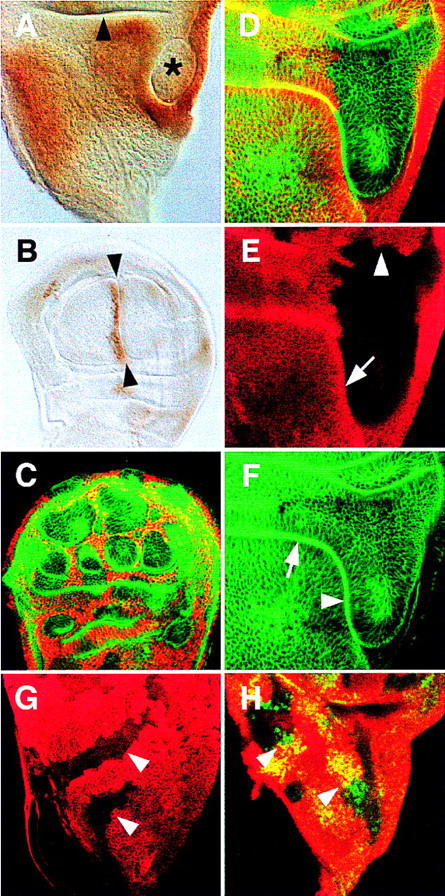
Cell clones lacking [Df(3L)iroDFM3; A, D–F] and overexpressing (UAS–caup; C) Iro-C genes in late third instar wing discs. All discs are oriented anterior to the left and ventral to the top. (A) Mutant clone (*) delimited by an ectopic fold. The disc is stained with anti-Caup antibody. Apparent staining within the clone is due to flattening of the disc upon mounting. The arrowhead points at the notum/hinge fold. (B) Disc overexpressing UAS–caup driven by dpp–GAL4. An ectopic fold (arrowheads) appears on the side where cells with high levels of Caup abut cells without the protein, i.e., the A/P compartment border. (C) Confocal section of a disc with clones of cells overexpressing UAS–caup (anti-Caup antibody, red). Clones contact each other, their cells are arranged in filaments separating large, roundish islands of nonexpressing cells, and create ectopic folds (phalloidin staining, green) that surround the nonexpressing cells. Folds were visible with light transmission optics (not shown). (D–F) Confocal section of a clone in the notum–hinge region that reroutes the normal fold between these regions. The absence of anti-Myc antibody staining (E) marks the homozygous Df(3L)iroDFM3 cells. The clone border is smooth along the fold (arrow) that separates it from notum cells and wiggly in the wing hinge region (arrowhead). Phalloidin staining (F) shows the smooth continuity between the extant (arrow) and ectopic (arrowhead) folds. (G) Clones (arrowed, absence of anti-Myc antibody staining, red) of homozygous Df(3L)iroDFM3 cells in the notum region of a UAS–ara/ap–GAL4 disc. The absence of a fold surrounding these clones was evidenced by phalloidin (not shown) and Myc stainings (cf. with E) and transmission optics. (H) Similar clones stained with anti-Sc antibody (green and yellow). The presence of a clone did not disturb the extant Sc proneural clusters (arrowheads). It should be pointed out that the ubiquitous UAS–ara expression slightly modified the wild-type pattern of sc proneural clusters in the notum (Campuzano and Modolell 1992), but the clones did not modify it further. The UAS–ara expression did not prevent notum formation and bristle development, as observed in pharate individuals (data not shown).
Figure 3.
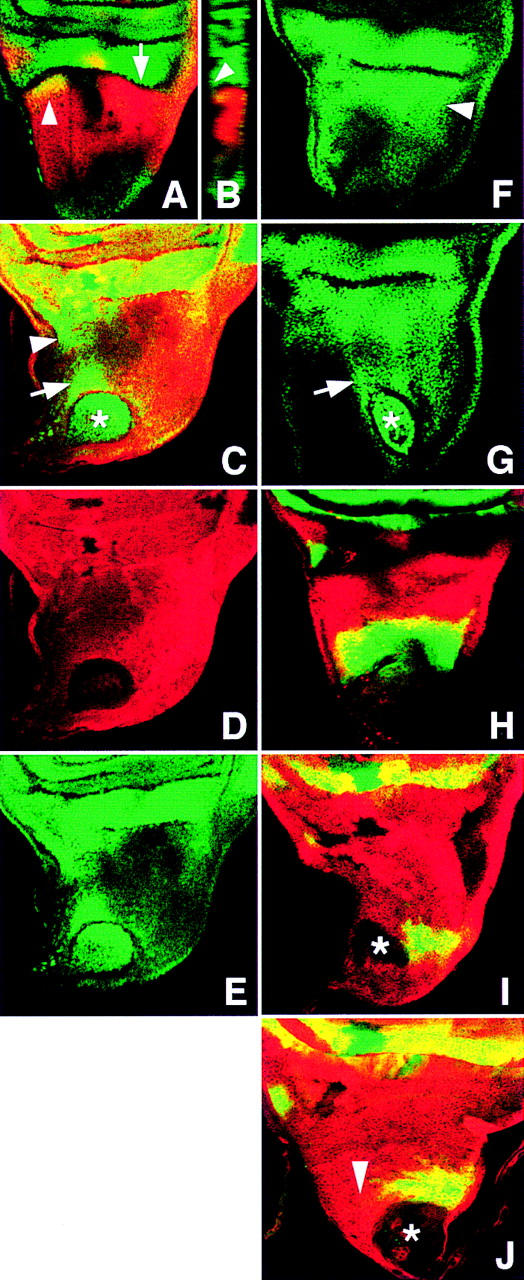
Cell identity changes associated with Iro-C− clones in the presumptive notum. Clones are revealed by the absence of Myc expression (red, C,D,I,J). (A,B) l(2)09261/CyO disc showing caup (red) and lacZ (green) expression. Within the presumptive notum, these expressions overlap only at the NP region (arrowhead). The arrow in A points to the notum/wing hinge fold and at the plane of the z-axis confocal section shown in B. In late third instar discs, cells expressing Caup can form up to approximately half of the fold (B). The arrowhead marks the bottom of the fold. (C–E) l(2)09261/+; Df(3L)iroDFM1 clone (*) with ectopic expression of lacZ (green channel, cf. with A). lacZ is also expressed in cells adjacent to the clone (arrow) and closest to the NP region (arrowhead). (F,G) Notum region of a wild-type disc and a disc bearing a Df(3L)iroDFM3 clone (*) stained with anti-Tsh antibody. Note the strong Tsh accumulation in the lateral notum (arrowhead) and the derepression of tsh in wild-type cells adjacent to the clone (arrow). (H) Notum region of a wg–lacZ/CyO disc stained with anti β-galactosidase (green) and anti-Caup (red) antibodies. (I,J) wg–lacZ/+;Df(3L)iroDFM1 clones (*) in discs stained with anti-β-galactosidase (green) and anti-Myc (red) antibodies. wg–lacZ is not expressed within the clones (I) and in part of the surrounding cells (J, arrowhead, cf. with H).
To ascertain that the phenotypes associated with the Iro-C deficiencies were caused by the absence of the homeodomain Iro proteins, and not by the removal of other transcription units included in the deficiencies (see Materials and Methods), Df(3L)iroDFM3 clones were induced in a background overexpressing the Ara protein [UAS–ara transgene (Gómez-Skarmeta et al. 1996) driven in the dorsal compartment of the disc by ap–GAL4 (Calleja et al. 1996)], and they were examined for a rescue of their phenotype. Figure 2G shows that within the notum territory these UAS–ara-expressing, Iro-C− clones no longer induced a fold around themselves and had normal, wiggly outlines consistent with their maintaining a notum identity (16 of 16 large clones examined). This was the case, as a characteristic marker of notum specification such as the highly resolved pattern of expression of the gene scute (sc) in proneural clusters (Campuzano and Modolell 1992) was not affected within the clones (Fig. 2H). Moreover, clones of mirre48, a small deletion (1 kb) of the mirr promoter that greatly reduces transcription of this gene (McNeill et al. 1997), in the lateral notum also induced malformations that contained ectopic hinge structures (axillary sclerites and tegula-like sensilla; data not shown). Taken together, these results indicate that a reduction in the levels of Iro homeoproteins, which may replace one another functionally (Gómez-Skarmeta et al. 1996), is responsible for the transformation from notum to hinge fate observed in Iro-C− cells.
This transformation was also manifest using wing disc markers. The enhancer trap line l(2)09261, a hinge and wing marker that is expressed only weakly in the prospective notum (Figs. 3A,B and 4C), was strongly derepressed in notum Iro-C− cells (Fig. 3C–E). teashirt (tsh), a gene strongly expressed in the hinge and the lateral-most part of the prospective notum, but weakly expressed in the more central parts of the notum (Fig. 3F), was also similarly derepressed in the centrally located clones (Fig. 3G). In contrast, vg, optomotor blind and nubbin (nub), genes that are expressed primarily in the wing and minimally or not expressed in the hinge (Fig. 4A,B; Williams et al. 1991; Ng et al. 1995; Grimm and Pflugfelder 1996), were not expressed in the notum clones (not shown), consistent with the transformation of Iro-C− cells toward a wing hinge identity.
Figure 4.
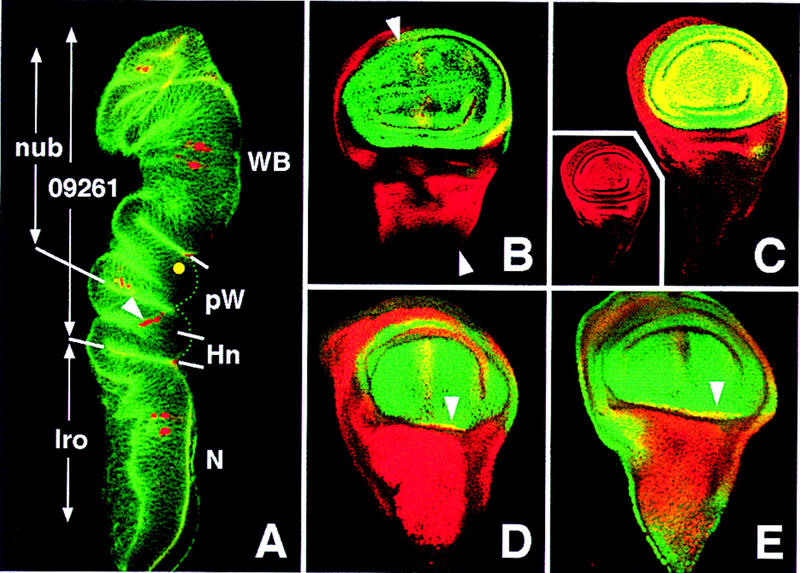
Spatial domains of expression of nub, l(2)09261, and ara–caup in late third instar discs. The main domains of expression of these genes are indicated in A, a transverse confocal section of a neuralized–lacZ (A101) disc stained with phalloidin (green) and anti-β-galactosidase antibody (red), which reveals neural precursors. The approximate plane of the section is shown in B. (Yellow dot) Position of the precursor of the giant sensillum of the radius, as seen in a different section. Abbreviations for presumptive territories: (WB) Wing blade; (pW) proximal wing; (Hn) dorsal hinge; (N) notum. The extent of the dorsal hinge territory is deduced from the fate map of the disc (Bryant 1978), the border of Iro expression, and the position of the precursors of the proximal sensilla of the dorsal radius (arrowhead). Note that the notum/hinge border is defined, most probably by Iro expression in younger discs; it may not correspond strictly to its late third instar expression. (B,C) Wild-type discs stained for Nub (green) and Iro or l(2)09261, respectively (red). (Inset in C) Red channel [l(2)09261 domain]. nub domain comprises the wing pouch and extends into part of the proximal wing, as deduced from a z-axis section of these discs, whereas l(2)09261 domain is more extensive, includes the whole proximal wing and the dorsal hinge, reaching up to the notum (Fig. 3B). These domains are indicated in A. (D,E) UAS–ara/ap–GAL4 discs stained for Iro (red) and Nub or l(2)09261, respectively (red). nub and l(2)09261 expressions become coextensive at the distal limit of Ara ectopic expression (arrowheads), suggesting the absence of specification of the dorsal hinge territory.
As the above results indicated that the Iro-C is necessary for notum specification, we examined whether its early ectopic expression imposed a notum fate on non-notum cells. UAS–ara, driven in the dorsal compartment by ap–GAL4 removed the dorsal hinge territory, as defined by the expression of l(2)09261 and the nonexpression of nub (Fig. 4C,D). The resulting pharate individuals lacked all dorsal hinge elements (axillary sclerites and tegula), had strongly reduced and distorted wings, but ectopic notum structures were not discerned (not shown). Similar adult phenotypes were observed with UAS–caup or UAS–mirr transgenes (Gómez-Skarmeta et al. 1996; McNeill et al. 1997), and phenotypes consistent with these were also found by using drivers dppdisk1–GAL4 (Staehling-Hampton et al. 1994) or Gal4 line C-765 (Gómez-Skarmeta et al. 1996). Simultaneous expression of UAS–mirr and either UAS–ara or UAS–caup did not modify the results. Moreover, imaginal disc cells strongly overexpressing UAS–caup at the wing pouch still expressed vg or nub (not shown). These results indicate that the Iro proteins cannot impose a notum fate on every wing disc cell, although, if present, they prevent the normal development of the wing hinge.
Iro-C− cells affected the surrounding wild-type tissue. Thus, mutant cells that differentiated as ectopic tegula recruited wild-type cells to form part of this ectopic structure (Fig. 1D). This evidenced a change of fate of the wild-type cells from notum to tegula. Nonautonomous effects were also observed in the imaginal disc. Thus, the Iro-C− clones in the notum territory induced neighboring wild-type cells to express strongly the l(2)09261 marker (Fig. 3C,E). Interestingly, the expressing cells were located nearest to the notopleural (NP) region; consequently, their spatial disposition with respect to the clone was a mirror-image correlate of the NP cells expressing the marker with respect to the notum/hinge fold (Fig. 3C). This nonautonomous effect was weaker in more posterior clones and was not observed in the postnotum territory (not shown). A nonautonomous derepression of tsh also occurred in the wild-type cells nearest the lateral notum (Fig. 3G). Another nonautonomous effect concerned the expression of wg, whose product accumulates in an A/P band that runs near the dorsocentral region of the prospective notum (Baker 1988; Phillips and Whittle 1993) but not in the hinge (except for a small domain at the prospective tegula, Fig. 3H). The Iro-C− cells in the notum, which did not express wg autonomously (Fig. 3I) (in accordance with their transformation toward hinge cells), caused their wild-type neighbors closest to the NP region also not to accumulate Wg protein (Fig. 3J). The nonautonomous repression of wg and activation of l(2)09261 and tsh are consistent with the transformation of these cells toward NP or other lateral notum cells.
Interactions between the Iro-C− and Iro-C+ cells were also revealed by the ectopic fold that surrounded the notum Iro-C− clones (Fig. 2A), as this fold was formed by mutant and wild-type cells (not shown). Moreover, similar interactions evidently occurred at borders where cells with ectopic high levels of Iro proteins confronted cells without or with minimal levels of them, as ectopic folds also formed at these borders (Fig. 2B,C). Therefore, the fold separating the prospective notum and wing hinge (Figs. 2A and 3A,B) is probably induced in early third instar discs by the juxtaposition of Iro-C expressing and nonexpressing cells (Fig. 5B). Other folds or grooves may be similarly induced, as mirr is expressed in the Drosophila embryo, at the dorsal folds, and at the anterior border of each segment (McNeill et al. 1997). Interestingly, the Xiro1 and Xiro2 genes, Xenopus homologs of ara and caup (Gómez-Skarmeta et al. 1998), are expressed at rhombomeres 1, 3, and 5 and may be involved in generating their borders (J.L. Gómez-Skarmeta, unpubl.).
Figure 5.
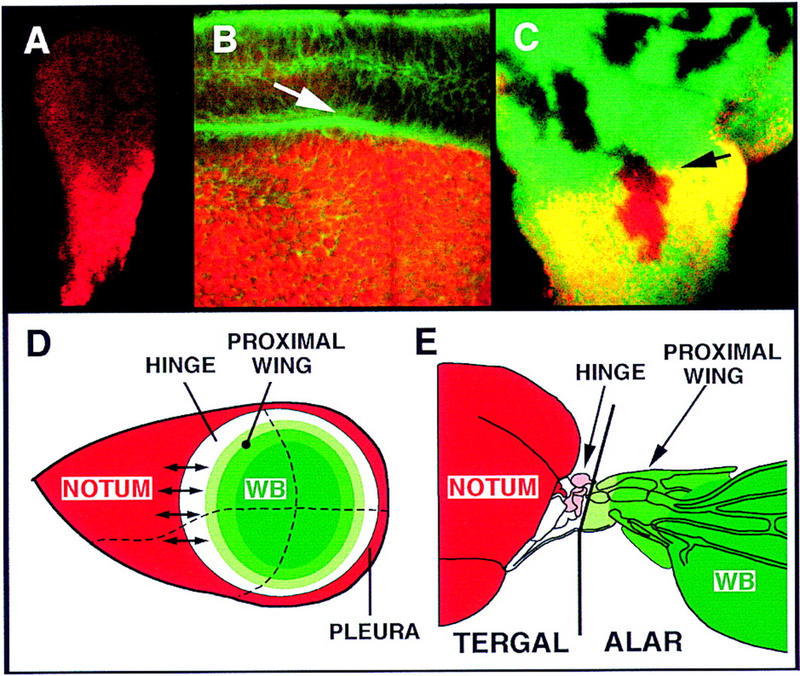
(A) Second instar disc. Caup (red) is present in the whole proximal region, the presumptive notum. During early third instar, expression extends into the presumptive pleura (not shown). (B) High magnification of notum/hinge area of mid-third instar disc showing the close spatial association between the Caup accumulating cells (red) and the developing notum/hinge fold (green, arrowed, phalloidin staining). (C) M+ cell lineage clones (absence of β-galactosidase marker, green) induced 72–96 hr AEL. A clone comprises cells at both sides of the Iro-C expression border (arrowhead). (D) Model of an idealized wing disc showing different concentric domains revealed by the expression patterns like the vg quadrant enhancer (wing blade, dark green), nub (wing blade and part of proximal wing, dark and medium green), l(2)09261 (wing hinge, white, plus wing, green), and Iro-C (notum and pleura, red) (see text). Small arrows indicate tissue-organizing properties of the border of Iro-C-expressing and nonexpressing cells, as suggested by the nonautonomous effects of the Iro-C− clones. Dashed lines represent anterior/posterior and dorsal/ventral compartment borders. (E) Scheme of the notum, wing hinge, and wing blade of an adult fly showing, as suggested by palaeontological (Kukalova-Peck 1978) and our data, extents of the tergal territory, which comprises the notum, hinge, and mesopleura (not shown), and the alar territory. Tegula and first and second sclerites are highlighted in pink.
Upon confrontation with Iro-C− cells, Iro-C+ cells located near the central region of the notum acquire properties typical of cells located near the hinge border (Fig. 3). This suggests that during development of the wild-type disc, interactions between the Iro-C-expressing and nonexpressing cells at each side of this border (already present in second instar discs; Fig. 5A) establish a signaling system that organizes pattern in the notum and, possibly, the hinge (Fig. 5D). Accordingly, the suppression of notum macrochaetae in wild-type territory induced by the Iro-C− clones (Fig. 1A) may be caused by ectopic signals that emanate from the clone borders and interfere with the extant signals. It should be stressed that in the developing eye, juxtaposition of dorsal Iro-C-expressing and ventral nonexpressing cells also sets up a signaling system, mediated by Delta and Serrate and the receptor Notch (N), that organizes growth and patterning (chirality of the ommatidia) of the whole eye (McNeill et al. 1997; Domínguez and de Celis 1998; Papayannopoulos et al. 1998). Although during notum development this signaling pathway is most important to discriminate between the epidermal and neural fates (Artavanis-Tsakonas et al. 1995), no data are available concerning its possible role in the notum versus hinge decision. The Egf receptor signaling pathway may participate in it, as removal of its ligand Vein impedes notum development (Simcox et al. 1996).
The well-known organizing borders of the wing disc coincide with the A/P and D/V cell-lineage restriction boundaries (Blair 1995; Brook et al. 1996). A similar boundary, located between the notum and the wing hinge, has been inferred after examining lineage clones in the adult cuticle (García-Bellido et al. 1976). However, at the disc level, we have not found evidence for the existence of this cell-lineage restriction. If present, it certainly does not coincide with the border of Iro-C-expressing cells, as cell-lineage clones generated by the Minute technique (Morata and Ripoll 1975) and induced as late as 72–96 hr AEL straddled this border (Fig. 5C). We conclude that upon proliferation, cells that cross the border of Iro-C expression most likely change their fate from notum to proximal hinge or vice versa.
The patterns of expression of Iro and other genes important for development of the wing disc suggest that this disc is organized, in the proximal/distal axis, in concentric domains (Fig. 5D). Iro expression (Fig. 5A; Gómez-Skarmeta et al. 1996) coincides with the outermost domain, which gives rise to the notum and mesothoracic pleura, whereas the vg quadrant enhancer-mediated expression coincides with the innermost one (Kim et al. 1996), essentially the wing blade territory. In between, the expressions of nub (Ng et al. 1995) and l(2)09261 coincide with increasingly larger domains (Fig. 4), and the rings of late expression of wg (Phillips and Whittle 1993) mark concentric subdomains of the proximal wing. The finding that Iro-C promotes the development of notum as opposed to dorsal hinge structures suggests a specific genetic address for the hinge domain, for which we do not know of a corresponding marker. Palaeontological data on the structure of primitive fossil pterygote hinges suggest that the hinge structures (tergal pteralia) originated from the tergal lobes (lateral projections of the tergum) (Kukalova-Peck 1978); that is, phylogenetically, they are probably body wall structures (Fig. 5E). Accordingly, the Iro-C homeodomain proteins help establish a subdivision between two tergal territories and could do so by antagonizing genes specifying hinge fate.
Materials and methods
Fly stocks
Df(3L)iroDFM1 and Df(3L)iroDFM3 are two deficiencies null for ara and caup (Gómez-Skarmeta et al. 1996). The proximal breakpoint of Df(3L)iroDFM3 is 100 bp 5′ of the end of the longest mirr cDNA characterized (R. Diez del Corral and J.L. Gómez-Skarmeta, unpubl.). Homozygous Df(3L)iroDFM3 clones in the notum lack mirr expression, as determined by in situ hybridization. Although the proximal breakpoint of Df(3L)iroDFM1 has not been characterized, this deficiency, at least in the notum, is also most likely null for these genes, as Df(3L)iroDFM1 and Df(3L)iroDFM3 cell clones in the notum have undistinguishable phenotypes. Only two additional transcription units have been found within the DNA eliminated by Df(3L)iroDFM3. They are located in the DNA (73 kb) that separates caup and mirr (Netter et al. 1998). The longest ORF (170 codons) of their cDNAs, as well as the shorter ORFs, seem to be noncoding, as judged by codon and third base usages (R. Diez del Corral and J.L. Gómez-Skarmeta, unpubl.). Enhancer trap line l(2)09261 was a gift from T. Laverty (Howard Hughes Medical Institute, University of California, Berkeley, CA). UAS–caup transgenic line was prepared using an EcoRI fragment of caup cDNA containing the entire ORF subcloned into pUAST (Brand and Perrimon 1993). mirre48 is described in McNeill et al. (1997).
Genetic mosaics
f36a; mwh Df(3L)iroDFM3 (or Df(3L)iroDFM1 or mirre48)/M(3)i55P[f+] (77a) larvae (Gómez-Skarmeta et al. 1996) were irradiated (X rays, 1000 rads) at 48–72, 72-96, and 96–120 hr AEL. In M(3)i55 heterozygous animals, this corresponds to approximately first, second, and early third larval instar, respectively (Ferrús 1975). Notum malformations with hinge structures were also observed using the FLP/FRT system (Xu and Rubin 1993). In this case, recombination was induced by heat treating (37°C, 1 hr) y hsFLP122; mwh iroDFM3 (or iroDFM1 or mirre48) FRT80B/P[mini-w+; hsNM](67B) FRT80B larvae (24–72 hr AEL). Malformations were not observed in flies raised from irradiated control f36a; mwh /M(3)i55 P[f+] (77) larvae. Flies were mounted either in Gary’s magic mountant (Lawrence et al. 1986) or boiled in KOH and mounted in ethanol/lactic acid (1:1). Selected flies were gold sprayed and observed in a Phillips scanning electron microscope. FLP/FRT-induced Iro-C− clones were also examined in wing imaginal discs by the absence of the N-myc marker. Iro-C− clones in UAS–ara-overexpressing background were induced (37°C, 1 hr) in y hsFLP122/+; UAS–ara/ap–GAL4; mwh iroDFM3 FRT80B/P[mini-w+; hsNM](67B) FRT80B larvae (24–48 hr AEL).
Clones strongly overexpressing Caup were obtained by crossing a UAS–caup line with line f36a FLP1.22; abx/Ubx < FRT f+ stop FRT > GAL4–lacZ (de Celis and Bray 1997) and heat-treating larvae (24–48 hr AEL) at 37°C for 15 min. Caup accumulation was severalfold higher than that from the endogenous gene in notum cells (anti-Caup antibody fluorescent labeling).
Cell lineage clones were induced in larvae (24–48, 48–72, and 72–96 hr AEL) of the genotype M(1)Osp, armlacZ FRT18A/sn3FRT18A; hsFLP38, Bc/+ using the FRT/FLP technique (Xu and Rubin 1993). Cell clones were identified by the absence of β-galactosidase accumulation in discs dissected at 120–144 hr AEL.
Histochemistry
N-myc induction and antibody staining of imaginal discs were as in Xu and Rubin (1993). Primary antibodies were rabbit and mouse anti-β-galactosidase (Cappel and Promega), mouse monoclonal anti-Myc (BAbCO), rat polyclonal anti-Caup, rabbit anti-Nub (Cockerill et al. 1993), mouse anti-Tsh (Ng et al. 1996), and rabbit anti-Sc (Skeath and Carroll 1991). Anti-Caup antibody was prepared with a Caup–GST fusion protein (Smith and Johnson 1988) that contained the amino-terminal 324-amino-acid residues of Caup (including the homeodomain). This antiserum cross-reacted with Ara but not with Mirr. Secondary antibodies were anti-mouse, -rabbit, -rat antibodies coupled to fluorescein or lissamine–rhodamine (Jackson). Phalloidin–fluorescein staining to show the apicobasal polarity of the cells in the folds was performed according to Blair (1992). Images were collected in a Zeiss confocal microscope.
Acknowledgments
We thank J. Botas, S. Campuzano, J.F. de Celis, F.J. Díaz-Benjumea, P. Lawrence, A. Martínez-Arias, G. Morata, K. Storey, and colleagues of our laboratory for constructive criticisms on the manuscript, and S. Carroll, S. Cohen, J. Culí, F.J. Díaz-Benjumea, D. Ferres-Marcó, L. García-Alonso, P. Heitzler, T. Laverty, H. McNeill, and The Bloomington Stock Center for suggestions and/or materials. Fellowships from Comunidad Autónoma de Madrid (C.A.M.) to R.D.C. and J.L.G.-S., from Ministerio de Educación y Cultura to F.C., a C.S.I.C. contract to P.A., a University of Cambridge (Department of Zoology) visiting professorship from Fundación Banco Bilbao Vizcaya to J.M., and grants from Dirección General de Investigación Científica y Técnica (D.G.I.C.Y.T.), C.A.M., European Commission (E.C.) and Fundación Ramón Areces are acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jmodol@cbm.uam.es; FAX 34-91-397-4799.
References
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Baker NE. Transcription of the segment-polarity gene wingless in the imaginal discs of Drosophila, and the phenotype of a pupal-lethal wg mutation. Development. 1988;102:489–497. doi: 10.1242/dev.102.3.489. [DOI] [PubMed] [Google Scholar]
- Blair SS. engrailed expression in the anterior lineage compartment of the developing wing blade of Drosophila. Development. 1992;115:21–33. doi: 10.1242/dev.115.1.21. [DOI] [PubMed] [Google Scholar]
- ————— Compartments and appendage development in Drosophila. BioEssays. 1995;17:299–309. doi: 10.1002/bies.950170406. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brook WJ, Díaz-Benjumea FJ, Cohen SM. Organizing spatial pattern in limb development. Annu Rev Cell Dev Biol. 1996;12:161–180. doi: 10.1146/annurev.cellbio.12.1.161. [DOI] [PubMed] [Google Scholar]
- Bryant PJ. Pattern formation in imaginal discs. In: Ashburner M, Wright TRF, editors. The genetics and biology of Drosophila. 2c. London, UK: Academic Press; 1978. pp. 230–335. [Google Scholar]
- Calleja M, Moreno E, Pelaz S, Morata G. Visualization of gene expression in living adult Drosophila. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- Campuzano S, Modolell J. Patterning of the Drosophila nervous system: The achaete-scute gene complex. Trends Genet. 1992;8:202–207. doi: 10.1016/0168-9525(92)90234-u. [DOI] [PubMed] [Google Scholar]
- Carroll SB. From pattern to gene, from gene to pattern. Int J Dev Biol. 1998;42:305–309. [PubMed] [Google Scholar]
- Cockerill KA, Billin AN, Poole SJ. Regulation of expression domains and effects of ectopic expression reveal gap gene-like properties of the linked pdm genes of Drosophila. Mech Dev. 1993;41:139–153. doi: 10.1016/0925-4773(93)90044-x. [DOI] [PubMed] [Google Scholar]
- Cohen B, McGuffin ME, Pfeifle C, Segal D, Cohen SM. apterous, a gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Genes & Dev. 1992;6:715–729. doi: 10.1101/gad.6.5.715. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Bray S. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 1997;124:3241–3251. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- Domínguez M, de Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396:276–278. doi: 10.1038/24402. [DOI] [PubMed] [Google Scholar]
- Ferrús A. Parameters of mitotic recombination in Minute mutants of Drosophila melanogaster. Genetics. 1975;79:589–599. doi: 10.1093/genetics/79.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bellido A, Ripoll P, Morata G. Developmental compartmentalization in the dorsal mesothoracic disc of Drosophila. Dev Biol. 1976;48:132–147. doi: 10.1016/0012-1606(76)90052-x. [DOI] [PubMed] [Google Scholar]
- Gómez-Skarmeta JL, Diez del Corral R, de la Calle-Mustienes E, Ferrés-Marcó D, Modolell J. araucan and caupolican, two members of the novel Iroquois complex, encode homeoproteins that control proneural and vein forming genes. Cell. 1996;85:95–105. doi: 10.1016/s0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- Gómez-Skarmeta JL, Glavic A, de la Calle-Mustienes E, Modolell J, Mayor R. Xiro, a Xenopus homolog of the Drosophila Iroquois complex genes, controls deve-lopment of the neural plate. EMBO J. 1998;17:181–190. doi: 10.1093/emboj/17.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Crespo S, Morata G. Control of Drosophila adult pattern by extradenticle. Development. 1995;121:2117–2125. doi: 10.1242/dev.121.7.2117. [DOI] [PubMed] [Google Scholar]
- Grimm S, Pflugfelder GO. Control of the gene optomotor-blind in Drosophila wing development by decapentaplegic and wingless. Science. 1996;271:1601–1604. doi: 10.1126/science.271.5255.1601. [DOI] [PubMed] [Google Scholar]
- Irvine KD, Vogt TF. Dorsal-ventral signaling in limb development. Curr Opin Cell Biol. 1997;9:867–876. doi: 10.1016/s0955-0674(97)80090-7. [DOI] [PubMed] [Google Scholar]
- Kim J, Sebrig A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Kukalova-Peck J. Origin and evolution of insect wings and their relation to metamorphosis, as documented by the fossil record. J Morphol. 1978;156:53–126. doi: 10.1002/jmor.1051560104. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P, Morata G. Methods of marking in cells. In: Roberts DB, editor. Drosophila, a practical approach. Oxford, UK: IRL Press; 1986. pp. 229–242. [Google Scholar]
- Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, Cohen SM. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Leyns L, Gómez-Skarmeta JL, Dambly-Chaudière C. iroquois: A prepattern gene that controls the formation of bristles on the thorax of Drosophila. Mech Dev. 1996;59:63–72. doi: 10.1016/0925-4773(96)00577-1. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. San Diego, CA: Academic Press; 1992. [Google Scholar]
- McNeill H, Yang CH, Brodsky M, Ungos J, Simon MA. mirror encodes a novel PBX-class homeoprotein that functions in the definition of the dorso-ventral border of the Drosophila eye. Genes & Dev. 1997;11:1073–1082. doi: 10.1101/gad.11.8.1073. [DOI] [PubMed] [Google Scholar]
- Morata G, Ripoll P. Minutes: Mutants of Drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Netter S, Fauvarque MO, Diez del Corral R, Dura JM, Coen D. white+ transgene insertions presenting a dorsal/ventral pattern define a single cluster of homeo box genes that is silenced by the Polycomb-group proteins in Drosophila melanogaster. Genetics. 1998;149:257–275. doi: 10.1093/genetics/149.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann CJ, Cohen SM. Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development. 1997;124:871–880. doi: 10.1242/dev.124.4.871. [DOI] [PubMed] [Google Scholar]
- Ng M, Díaz-Benjumea FJ, Cohen SM. nubbin encodes a POU-domain protein required for proximal-distal patterning in the Drosophila wing. Development. 1995;121:589–599. doi: 10.1242/dev.121.2.589. [DOI] [PubMed] [Google Scholar]
- Ng M, Díaz-Benjumea FJ, Vincent JP, Wu J, Cohen SM. Specification of the wing by localized expression of the wingless protein. Nature. 1996;381:316–318. doi: 10.1038/381316a0. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V, Tomlinson A, Panin VM, Rauskolb C, Irvine KD. Dorsal-ventral signaling in the Drosophila eye. Science. 1998;281:2031–2034. doi: 10.1126/science.281.5385.2031. [DOI] [PubMed] [Google Scholar]
- Phillips RG, Whittle JRS. wingless expression mediates determination of peripheral nervous system elements in late stages of Drosophila wing disc development. Development. 1993;118:427–438. doi: 10.1242/dev.118.2.427. [DOI] [PubMed] [Google Scholar]
- Ramain P, Heitzler P, Haenlin M, Simpson P. pannier, a negative regulator of achaete and scute in Drosophila, encodes a zinc finger protein with homology to the vertebrate transcription factor GATA-1. Development. 1993;119:1277–1291. doi: 10.1242/dev.119.4.1277. [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Rodríguez-Esteban C, Izpisúa-Belmonte JC. Limbs are moving: Where are they going? Trends Genet. 1998;14:229–235. doi: 10.1016/s0168-9525(98)01477-2. [DOI] [PubMed] [Google Scholar]
- Shubin N, Tabin C, Carroll SB. Fossils, genes and the evolution of animal limbs. Nature. 1997;388:639–648. doi: 10.1038/41710. [DOI] [PubMed] [Google Scholar]
- Simcox AA, Grumbling G, Schnepp B, Bennington-Mathias C, Hersperger E, Shearn A. Molecular, phenotypic, and expression analysis of vein, a gene required for growth of the Drosophila wing disc. Dev Biol. 1996;177:475–489. doi: 10.1006/dbio.1996.0179. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Carroll SB. Regulation of achaete-scute gene expression and sensory organ pattern formation in the Drosophila wing. Genes & Dev. 1991;5:984–995. doi: 10.1101/gad.5.6.984. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K, Jackson PD, Clark MJ, Brand AH, Hoffmann FM. Specificity of bone morphogenetic protein related factors: Cell fate and gene expression changes in Drosophila embryos by decapentaplegic but not 60A. Cell Growth Differ. 1994;5:585–593. [PubMed] [Google Scholar]
- Williams JA, Bell JB, Carroll SB. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes & Dev. 1991;5:2481–2495. doi: 10.1101/gad.5.12b.2481. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. Direct and long range action of a Wingless morphogen gradient. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]


