Abstract
FOXO genes are involved in many aspects of development and vascular homeostasis by regulating cell apoptosis, proliferation, and the control of oxidative stress. In addition, FOXO genes have been showed to inhibit Wnt/β-catenin signaling by competing with T cell factor to bind to β-catenin. However, how important of this inhibition in vivo, particularly in embryogenesis is still unknown. To demonstrate the roles of FOXO genes in embryogenesis will help us to further understand their relevant physiological functions. Zebrafish foxo3b gene, an orthologue of mammalian FOXO3, was expressed maternally and distributed ubiquitously during early embryogenesis and later restricted to brain. After morpholino-mediated knockdown of foxo3b, the zebrafish embryos exhibited defects in axis and neuroectoderm formation, suggesting its critical role in early embryogenesis. The embryo-developmental marker gene staining at different stages, phenotype analysis and rescue assays revealed that foxo3b acted its role through negatively regulating both maternal and zygotic Wnt/β-catenin signaling. Moreover, we found that foxo3b could interact with zebrafish β-catenin1 and β-catenin2 to suppress their transactivation in vitro and in vivo, further confirming its role relevant to the inhibition of Wnt/β-catenin signaling. Taken together, we revealed that foxo3b played a very important role in embryogenesis and negatively regulated maternal and zygotic Wnt/β-catenin signaling by directly interacting with both β-catenin1 and β-catenin2. Our studies provide an in vivo model for illustrating function of FOXO transcription factors in embryogenesis.
Introduction
Forkhead box O (FOXO) transcription factors, homologues of DAF-16 (the Caenorhabditis elegans ortholog), including FOXO1, FOXO3a, FOXO4 and FOXO6 in mammalian, are important effectors in insulin/PI3K/Akt signaling pathway. Akt (also known as protein kinase B) can phosphorylate FOXO proteins at three conserved residues upon growth factors stimulation, and result in nuclear exclusion of FOXO proteins, thereby inhibiting FOXO-dependent transcription [1]. On the contrary, increased cellular oxidative stress localizes FOXO proteins to the nucleus, where FOXO proteins bind as monomers to their cognate DNA targeting sequences.
FOXO proteins function as master signaling integrators and participate in a series of dynamic gene expression programs upon various environmental stimuli. FOXO genes are reported to be involved in the regulation of apoptosis, cell cycle progression, and control of oxidative stress, DNA damage repair and cellular differentiation [2]. Most of these environmental stimuli result in post-translational modifications of FOXO proteins, which include but are not limited to phosphorylation, ubiquitylation and acetylation, and affect nuclear/cytoplasmic trafficking of FOXO proteins [1].
Knockout of FOXO1 in mice causes embryonic lethality due to vascular defects, while knockout of FOXO3a only renders the FOXO3a-/- female mice to have an age-dependent reduced fertility and knockout of FOXO4 does not have an obvious phenotype [3]. These observations suggest that FOXO genes have quite diverse physiological roles during vertebrate embryogenesis. However, the underling mechanisms are still not well defined.
Wnt/β-catenin signaling has been revealed participating in the formation of the vertebrate embryonic axes and neuroectoderm during embryogenesis [4]. The role of Wnt/β-catenin signaling during embryogenesis has been well characterized by zebrafish model. In zebrafish, maternally Wnt/β-catenin signaling is essential for the formation of organizer (also known as “shield”). But zygotic Wnt/β-catenin signaling is activated by Wnt ligands after MBT (mid-blastula transition) to antagonize the organizer and be involved in anterior-posterior patterning of the neural axis. Zebrafish embryos homozygous for mutation in the wnt8 locus show significant expansion of the shield and almost absent expression of ventro-lateral mesoderm markers [5], similar to the phenotype observed in morpholino-mediated wnt8 knockdown morphants. These data demonstrate that wnt8 functions to promote the ventro-lateral fate and antagonize the organizer. In addition, the repression of organizer by wnt8 is mediated by Wnt direct target genes, including ved, vent and vox [6], [7]. During the establishment of rostral-caudal compartments of the vertebrate neural tube, exaggerated Wnt signaling leads to loss of rostral neural domains [7]. Over-expression of wnt8 in zebrafish leads to changes in expression of its target genes and results in anterior defects [8], while wnt8 mutants and wnt8 morphants display expanded forebrain marker expression. Wnt inhibitors, such as cerberus, frzb1 and dickkopf1, all function as head inducers [9], [10], [11]. Moreover, as a DNA binding factor in β-catenin transcriptional complex, tcf3 is essential for forebrain formation by repressing the caudal genes induced by Wnt ligands in zebrafish [12], [13]. Its dominant negative form, dnTCF, lacking the β-catenin binding domain and acting exclusively as a repressor, can efficiently promote anterior nervous system [12], and induce the late ectopic expression of dorsal-specific genes in marginal region [14]. In addition, its N-terminal β-catenin–binding domain (tcfBD) is its another dominant negative construct, which inhibits Wnt/β-catenin signaling by depleting the functional β-catenin protein pool. In embryos with ectopic expression of tcfBD, most embryos display ectopic dorsal-specific genes expression and low ventral marker gene eve1 (even-skipped 1) expression [14].
Interestingly, it has been reported that β-catenin can directly bind to FOXO and enhance FOXO transcription activity [15]. On the contrary, FOXO competes with TCF for interaction with β-catenin, thereby inhibiting TCF transcriptional activity [16]. These facts raise the possibility that FOXO might affect vertebrate embryogenesis through inhibiting Wnt/β-catenin signaling.
While investigating the function of zebrafish eaf1/2 during embryogenesis, we identified foxo3b [initially named as zFKHR/foxO5 [17], now named as foxo3b in ZFIN] as one of strongly suppressed genes [18]. Foxo3b is an orthologue of mammalian FOXO3. To enrich our understanding about the function of vertebrate FOXO genes during embryogenesis, we are interested in figuring out the role of foxo3b during early embryogenesis by taking advantage of zebrafish model.
Materials and Methods
Maintenance of Fish Stocks and Embryo Collection
Breeding wild-type zebrafish (Danio rerio) (AB) were maintained and embryos raised under standard library conditions [19]. Embryos were collected and staged as described [20].
Cloning of Zebrafish Foxo3b
Zebrafish foxo3b gene (GenBank accession numbers NM_131085) was amplified using the primers 5′-CACGCTCTAGAATGGCAGAGACAACCCT-3′ and 5′-ATATGGATCCTCAGCCTGGCACCCAACT-3′. Total RNA was isolated from zebrafish embryos using TRIzol reagent (Invitrogen), and cDNA was synthesized by using the RevertAid™ first strand cDNA synthesis Kit (Fermentas). The complete coding sequence was PCR amplified and subcloned into the pCGN-HAM vector (provided by William Tansey), and then sequence verified.
Whole Mount In-situ Hybridization
The probes for identifying zebrafish foxo3a, foxo3b, six3b, opl, cdx4, pax6 and foxi1 were amplified from cDNA pools using the appropriate sets of primers (Table S1). The probes for ved, vox, vent, and flh were generous gifts from Dr Y. Sun (Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan). The probes for sqt, wnt8a, tbx5 and bmp2b were kindly provided by Dr Z. Yin (Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan). Dr. T. Whitefield (Medical Research Council Centre for Developmental and Biomedical Genetics, Sheffield, UK) generously provided the probe for nkx5.1. The probe for gata2 was kindly provided by Dr Tingxi Liu (Institute of Health Sciences, Shanghai). The probes for gsc and bmp4 were described previously [18]. The procedure for whole-mount in situ hybridization was performed as described previously [21].
Morpholino and mRNA Injection and Rescue Experiments
The morpholino antisense oligonucleotides (MOs) were obtained from Gene Tools: foxo3b -ATG-MO (5′-TGGCTCCAGGfGTTGTCTCTGCCATC-3′); foxo3b-SP-MO (5′- TGGAGATGCACTGCGCTTACCTTCC-3′); STD (standard)-MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′); β-catenin1-MO (5′- CTGGGTAGCCATGATTTTCTCACAG -3′); β-catenin2-MO (5′- CCTTTAGCCTGAGCGACTTCCAAAC -3′). They were resuspended and injected as described previously [18].
A fragment of foxo3b containing 5′-UTR and N-terminus was cloned into pEGFP-N1 (Clontech) to generate wild-type foxo3b to validate the efficiency of foxo3b-MO. The primers were: 5′-ACATCTCGAGCACTGCCTATCTAACTTCGACC-3′ and 5′-ACATGGATCCCCATGCATTCCTCCTTGAAGAT-3′. The GFP-tagged mutated foxo3b was generated by PCR using a forward primer with 6 mismatched nucleotides: 5′-ACATCTCGAGATGGGTGAATCTACTCTGGAGCCACTGT-3′ (mismatched nucleotides are underlined). To further validate the specificity of foxo3b-MO and avoid quenching effect of foxo3b mRNA, we used a primer to introduce 5 mismatched nucleotides in the zebrafish foxo3b mRNA without changing the amino acid sequence, the primer was 5′-CACGAAGCTTATGGCTGAAACTACATTGGAGCCACTG-3′ (mismatched nucleotides are underlined). DnTCF construct was kindly provided by Dr Y. Sun (Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan). DnTCF is a dominant negative form of TCF3, deleting the first 47 amino acids of β-catenin-binding domain. Capped mRNAs were synthesized using the AmptiCap SP6 High Yield message maker kit (Epicenter Biotechnologies). The synthetic mRNAs were diluted into different concentrations, and co-injected with morpholino to determine the optimal concentration that could rescue the defects of the morpholino-injected embryos effectively. capped mRNAs were injected into one-cell stage embryos at 1–2 ng. All of the microinjection was performed using a Harvard Apparatus PLI-100.
Semi-quantitative RT-PCR
Total RNA was isolated from 30 whole embryos using TRIzol reagent (Invitrogen) at different developmental stages. Oligo-dT-primed cDNA was synthesized by using RevertAid™ first strand cDNA synthesis Kit (Fermentas) and random primers were used to reverse transcribe 2 µg RNA. Separate reactions were set up with primer pairs for foxo3b and 18s RNA in the presence of SYBR green. All amplifications were performed using a two step temperature profile with annealing and extension at 60°C. Each sample was run in triplicate. Differences were calculated according to the ΔΔCt relative quantitation method (Applied biosystems) using the 18s RNA as calibrator. The primers for the zebrafish foxo3b were 5′-CCAAGCACCTCTACATCTC-3′ and 5′-CTGTGAGAGACCAGCGAAT-3′. The primers for zebrafish 18s were 5′-GAGAAACGGCTACCACATCC-3′ and 5′-CACCAGACTTGCCCTCCAA-3′.
Validation of Splice Morpholino
Total RNA was isolated from 50 whole embryos using TRIzol reagent (Invitrogen) at bud stage. Oligo-dT-primed cDNA was synthesized by using RevertAid™ first strand cDNA synthesis Kit (Fermentas) and random primers were used to reverse transcribe 2 µg RNA. PCR was performed with the following primer sets: Foxo3b-p1-F, GTGAGTTACTGCTGGTGATGC; Foxo3b-p2-R, TCTTCAAGGAGGAATGCATG; β-actin-F, GATGATGAAATTGCCGCACTG; β-actin-R, ACCAACCATGACACCCTGATGT. β-actin was used as an internal control.
Plasmid Construction
The vectors including pCGN-HAM (provided by William Tansey), pCMV-Flag2C (Stratagene), pEGFP-N1, pM-RFP and pM (Clontech) were used for cloning. Full-length cDNAs of zebrafish foxo3b, β-catenin1 and β-catenin2 (provided by Eric Weinberg) were subcloned into pCGN-HAM, pCMV-Flag2C, pEGFP-N1, pM-RFP or pM vectors to generate HA-foxo3b, HA-β-catenin1, HA-β-catenin2, Flag-β-catenin1, Flag-β-catenin2, pM-β-catenin1, pM-β-catenin2, GFP-β-catenin1, GFP-β-catenin2, RFP-foxo3b. All constructs were verified by sequencing.
Luciferase Reporter Assays
Human embryonic kidney 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (HyClone). Cells were seeded for 24h before transfection in 24-well plates, and were transfected with the mixture of plasmids (200 to 400 ng) by Lipofectamine 2000. pTK-Renilla luciferase reporter (10 ng) was used as an internal control. Similarly, the embryos were injected with the combined plasmids (250 pg per embryo) for luciferase assays. pFR-luc vector was purchased from Stratagene. The luciferase activity was determined at 24–30 hours post transfection or 11 hours after injection using the Dual-luciferase Reporter Assay System (Promega). The relative light units were measured using a luminometer (Sirius, Zylux Corporation, Oak Ridge, TN). Data were normalized by Renilla luciferase enzyme activity. Data are reported as mean ± SEM of three independent experiments performed in triplicate. The statistical analysis (paired t-test) was performed using GraphPad Prism 5.
Fluorescence Microscopy
Human HeLa cells were transfected with different combinations of zebrafish RFP- foxo3b, GFP-β-catenin1, GFP-β-catenin2 or empty RFP vectors. 24–30 hours after transfection, cells were directly observed under a Nikon T-2000 Eclipse inverted fluorescent microscope (Nikon Instruments, Melville, NY).
Immunoprecipitation and Western Blot
Human embryonic kidney 293T cells and mouse L cells (with constitutively expressed wnt3a) were cultured in Dulbecco's modified Eagle's medium (DMEM). For immunoprecipitation assays, 293T cells were transfected with different combinations of HA-foxo3b, Flag-β-catenin1, Flag-β-catenin2 or empty vectors (2–10 µg). 7 hours after transfection, the condition medium of mouse L cells was collected and added appropriately to 293T cells. After 24 hours, the 293T cells were washed with ice-cold PBS buffer and then lysed in RIPA (radioimmune precipitation) buffer containing 50 mM Tris, pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 1 mM EDTA, pH 8.0, 150 mM NaCl, 1 mM NaF, 1 mM PMSF (phenylmethylsulphonyl fluoride), 1 mM Na 3VO4 (sodium orthovanadate) and 1∶100 dilution of protease inhibitor cocktail (Sigma). After incubation on ice for 1 h, lysates were centrifuged for 15 min at 10,000 g at 4°C, and supernatant was incubated with monoclonal anti-HA agarose conjugate beads (sigma) for 6 h or over-night at 4°C. The immunoprecipitates were washed 3 times with RIPA buffer. Immunoprecipitates (IP) and whole cell lysate (WCL) were boiled with 1x SDS sample buffer, separated on SDS-PAGE and transferred to PVDF membrane (Millipore). Western blot analysis was performed as described previously using the indicated antibodies [22].
Results
Expression of Foxo3b during Zebrafish Embryogenesis
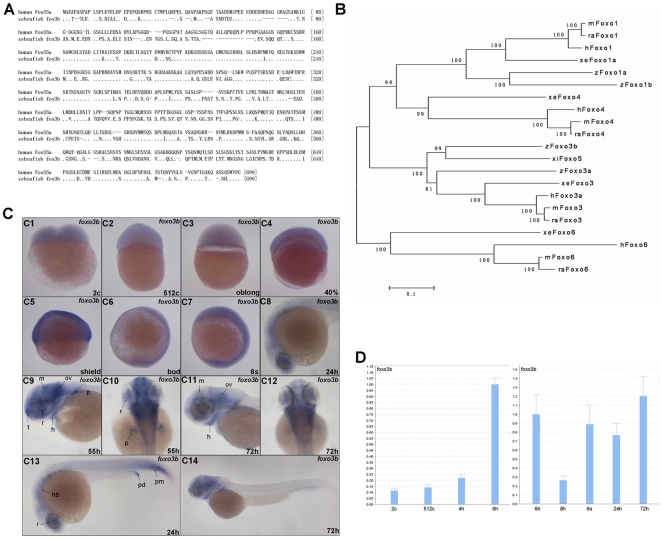
There are two orthologues of mammalian FOXO3 gene in zebrafish, foxo3a and foxo3b. Zebrafish foxo3b was initially identified and named as zFKHR/foxo5 [17]. Zebrafish foxo3b protein shared 55% identity to that of human FOXO3a (Fig. 1A). Neighbor joining phylogenetic analysis of vertebrate FOXO protein sequences showed that zebrafish foxo3b was part of a highly conserved branch including human, mouse, rat, Xenopus, and Xiphophorus FOXO3 gene. Zebrafish foxo3b and Xiphophorus foxo5 branched off somewhat earlier than zebrafish foxo3a (Fig. 1B).
Figure 1. Sequence comparison of zebrafish foxo3b with other FOXOs and developmental expressing patterns of zebrafish foxo3b.
(A) Sequence alignment of zebrafish foxo3b and human FOXO3a protein. (B) Neighbor-Joining Analysis of vertebrate FOXO protein sequences. Phylogenetic analysis was conducted using MEGA version 5 (Tamura, Peterson, Stecher, Nei, and Kumar 2011). hFoxo3a (Homo sapiens, Accession number NM_001455), hFoxo6 (XM_002342102), hFoxo4 (NM_005938), hFoxo1 (NM_002015), mFoxo3 (Mus musculus, NM_019740), mFoxo6 (NM_194060), mFoxo4 (NM_018789), mFoxo1 (NM_019739), raFoxo3 (Rattus norvegicus, NM_001106395), raFoxo6 (XM_001057233), raFoxo4 (NM_001106943), raFoxo1 (NM_001191846), zFoxo3b (Danio rerio, NM_131085), zFoxo3a (NM_001009988), zFoxo1a (NM_001077257), zFoxo1b (NM_001082857), xeFoxo3 (Xenopus laevis, NM_001092949), xeFoxo6 (NM_001159282), xeFoxo4 (FJ811896), xeFoxo1a (NM_001092948), xifoxo5 (Xiphophorus maculates, AY040320). NJ bootstrap values were shown on the branches. (C) The expression pattern of zebrafish foxo3b during embryogenesis. (C1–C3) The expression of foxo3b was detected at 2-cell stage embryos, and became weaker at oblong stage. (C4, C5) Foxo3b was ubiquitously expressed at 40% epiboly stage, a stronger expression was observed at shield stage. (C6, C7) Foxo3b became weaker at bud stage; at 6-somite stage, its expression level almost recovered to that of shield stage embryos. (C8, C13) Foxo3b was observed in the developing eye, hindbrain and posterior mesoderm by 24 hpf. (C9, C10) By 55 hpf, foxo3b expression was confined to the anterior central nervous system (CNS), with weak expression in the heart. (C11, C12, C14) By 72 hpf, foxo3b expression became weaker, but continued in the CNS and heart. C1–C4, lateral view; C5, lateral view with dorsal to the right; C6, C7, lateral view with anterior on top; C8, C9, C11, C13, C14, lateral views with anterior to the left; C10, C12, dorsal views with anterior on top; r, retina; p, pectoral fin bud; hb, hindbrain; pm, posterior mesoderm; pd, pronephric duct; h, heart; t, telencephalon; ov, otic vesicle; m, mesencephalon; c, cell; s, somite; h, hours post-fertilization (hpf). (D) Relative RNA expression levels as determined by semi-quantitative RT-PCR. For each stage, oligo dT-primed cDNA was used as template for three separate PCR amplifications using primers for foxo3b and 18s (internal control), foxo3b reached a high expression level at shield stage. For quantitative purpose, mRNA expression levels were normalized to 6 hpf (1.00).
Firstly, we employed in situ hybridization to check foxo3b expression pattern during embryogenesis and found that the foxo3b transcripts could be detected at 2-cell stage (Fig. 1C1). Its expression sustained until oblong stage, then a little bit decreased (Fig. 1C3). At 75% epiboly, another dramatic reduction of foxo3b expression was observed (data not show). Until 6-somites stage, the foxo3b transcripts were distributed ubiquitously among the whole embryo (Fig. 1C7). By 24 hpf, the expression of foxo3b was observed in brain (mainly in retina and hindbrain) and posterior mesoderm (Fig. 1C8 and C13). By 55 hpf, the strongest signals were predominantly detected in the central nervous system, including brain, retina, otic vesicle, and floor plate (Fig. 1C9 and C10), and became weaker by 72 hpf (Fig. 1C11, C12 and C14). Moreover, we employed semi-quantitative RT-PCR method to further determine relative levels of foxo3b transcripts at several key stages during embryogenesis. The results indicated that foxo3b transcripts could be detected at 2-cell stage (2c), but the expression level was relative low (column 1 from left to right in Fig. 1D). Foxo3b expression reached a higher level at 6 hpf, and dropped dramatically at 8 hpf. These results were consistent with that revealed by in situ hybridization. The detection of pre-MBT foxo3b expression implied that foxo3b might have important role in early embryogenesis.
Foxo3b is Required for the Formation of Axis and Brain
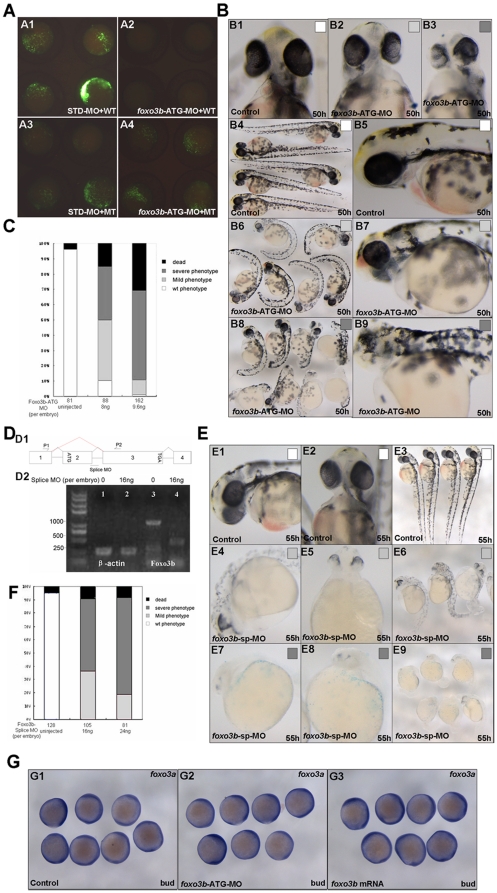
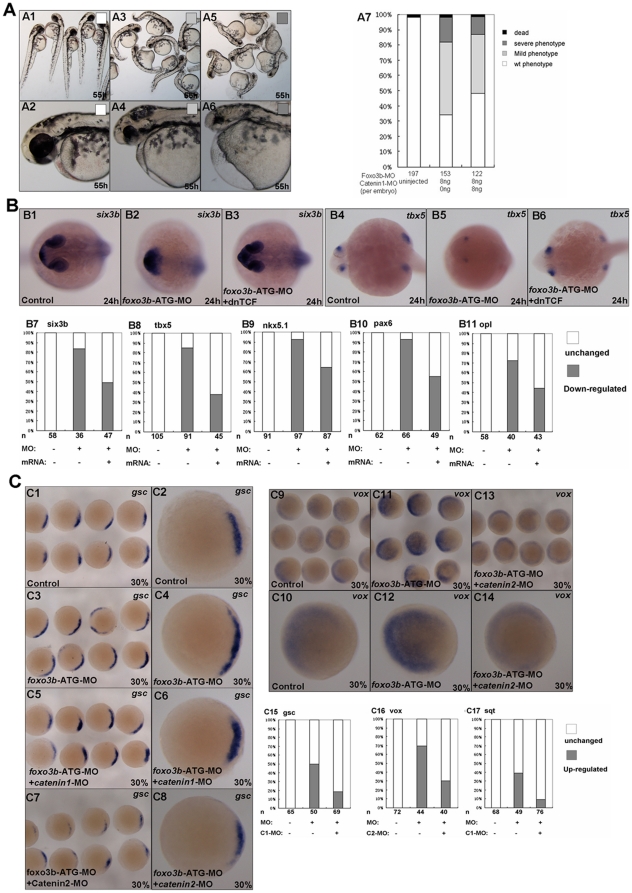
To determine the roles of foxo3b during zebrafish embryogenesis, we knocked down its expression using a morpholino targeting the translation initiation region of zebrafish foxo3b (foxo3b-ATG-MO). Firstly, we evaluated the efficiency of foxo3b-ATG-MO by injecting 1-cell stage zebrafish embryos with either STD-MO (standard morpholino) or foxo3b-ATG-MO, together with a vector expressing the truncated target protein tagged with GFP at the carboxyl terminus (WT) (Fig. 2A1 and A2). We also injected the indicated morpholinos with a vector expressing a mutated form of GFP-tagged truncated foxo3b (MT) (Fig. 2A3 and A4), which had 6 mismatched nucleotides in the foxo3b-ATG-MO targeted sequence. The results showed that foxo3b morpholino (8 ng per embryo) successfully blocked expression of foxo3b-GFP (WT) (Fig. 2A2), but not the expression of the mutated foxo3b-GFP (MT) (Fig. 2A4).
Figure 2. Knockdown of foxo3b results in defects in body axis and brain.
(A) Validation of foxo3b ATG-blocking morpholino (foxo3b-ATG-MO). A1, embryos were injected with STD-MO (8 ng per embryo, control) and a wild-type foxo3b-GFP fusion protein expression vector (WT) and then examined by fluorescence microscopy; A2, embryos were injected with foxo3b-ATG-MO (8 ng per embryo) and a wild-type foxo3b-GFP fusion protein expression vector (WT); A3, embryos were injected with STD-MO and a mutated foxo3b-GFP fusion protein expression vector (MT); A4, embryos were injected with foxo3b-ATG-MO and a mutated foxo3b-GFP fusion protein expression vector (MT). A1-A4, bud stage. (B, C) Morphology of representative morphants in foxo3b-ATG-MO injected embryos. The morphants had shorter body length, abnormal brain and heart at 50 hpf. Black box, dead embryos at 24 hpf; B3, B8, B9, dark gray box, embryos with defects at 50 hpf characterized by severe phenotype: no blood circulation, severely reduced body length and thinner brain; B2, B6, B7, light gray box, embryos with mild phenotype; B1, B4, B5, white box, un-injected wild-type embryos. B1-B3, front views; B4, B6, B8, lateral views; B5, B7, B9, lateral views with anterior to the left. (D) Validation of foxo3b splice-blocking morpholino (foxo3b-SP-MO). D1, Foxo3b exon/intron structure. Foxo3b-SP-MO can alter splicing of foxo3b mRNA, which results in the production of an aberrantly spliced message (as showed by red line). D2, The injection of foxo3b-SP-MO results in the production of a truncated mRNA (440 bp). The embryos were collected at bud stage, and β-actin was used as an internal control. (E, F) Morphology of splice-MO injected embryos. By 55 hpf, the morphants showed defects similar to that of foxo3b-ATG-MO injected embryos. (G) The expression level of foxo3a was not altered in foxo3b-knockdown or foxo3b over-expressed embryos. Embryos were injected with 8 ng foxo3b-ATG-MO (G2) or 1 ng foxo3b mRNA (G3) at 1-cell stage. Wild-type embryos were used as control. G1-G3, bud stage, lateral views.
The embryos injected with foxo3b-ATG-MO showed the phenotypes characterized with shortened body axis and thinner brain by 50 hpf (hours post fertilization) (Fig. 2B). The morphants could be classified into two classes: class I (moderate defect) showing shortened body axis, smaller eyes and thinner head (Fig. 2B2, B6 and B7); class II (serious defect) showing serious abnormality of brain and eyes, some even without forebrain and eyes (Fig. 2B3, B8 and B9). To determine if the changes in the morphology occurred in a dose-dependent manner, we injected 1-cell stage embryos with varying concentrations of foxo3b-ATG-MO (8 ng and 9.6 ng per embryo respectively). We then evaluated the embryos for viability by 24 hpf and for defects by 50 hpf. The results showed that foxo3b-ATG-MO had a dose-dependent effect (Fig. 2C).
To further confirm the specificity of phenotypes exhibited in foxo3b-ATG-MO morphants, we targeted the foxo3b gene with splice-blocking MO (foxo3b-SP-MO), and tested whether it could cause phenotypes similar to that exhibited in foxo3b-ATG-MO morphants. Foxo3b exon/intron structure was showed in Figure 2D1. Foxo3b-SP-MO was designed based on 25 base pair sequence complementary to the linkage site of exon 2 and exon 3 in foxo3b. Firstly, we tested whether the splice-blocking MO could indeed alter splicing of foxo3b mRNA as expected. By RT-PCR, we found that injection of foxo3b-SP-MO into 1-cell stage embryos resulted in production of an aberrantly spliced message with the size matched to the prediction (440 bp) (Fig. 2D1 & D2). The phenotypes exhibited in foxo3b-SP-MO morphants could also be classified into two classes as that of the foxo3b-ATG-MO morphants (Fig. 2E4-E9). In addition, foxo3b-SP-MO had a dose-dependent effect and produced more severe defects with higher concentration (16 ng and 24 ng per embryo respectively) (Fig. 2F). Thus, the phenotypes exhibited in foxo3b-SP-MO morphants phenocopied to that of foxo3b-ATG-MO morphants. Those observations suggested that both foxo3b-ATG-MO and foxo3b-SP-MO could specifically knockdown endogenous foxo3b expression efficiently.
In addition, foxo3b-ATG-MO morphants and foxo3b-SP-MO morphants displayed similar phenotypes implied that both maternal and zygotic foxo3b functioned importantly during embryogenesis. However, two orthologues of mammalian FOXO3, foxo3a and foxo3b, existed in zebrafish genome, we wonder whether the phenotype of foxo3b morphants resulted from the expression change of foxo3a in embryos, so we detected the expression of foxo3a after either foxo3b knockdown or over-expression. As shown in Figure 2G, the expression level of foxo3a was not altered in either foxo3b-knockdown or over-expressed embryos. These results further suggested that the phenotype of foxo3b morphants was specifically caused by foxo3b knockdown, which was further refined by the following rescue experiments.
Foxo3b is Required for Neuroectoderm Formation and Neural Tube Patterning
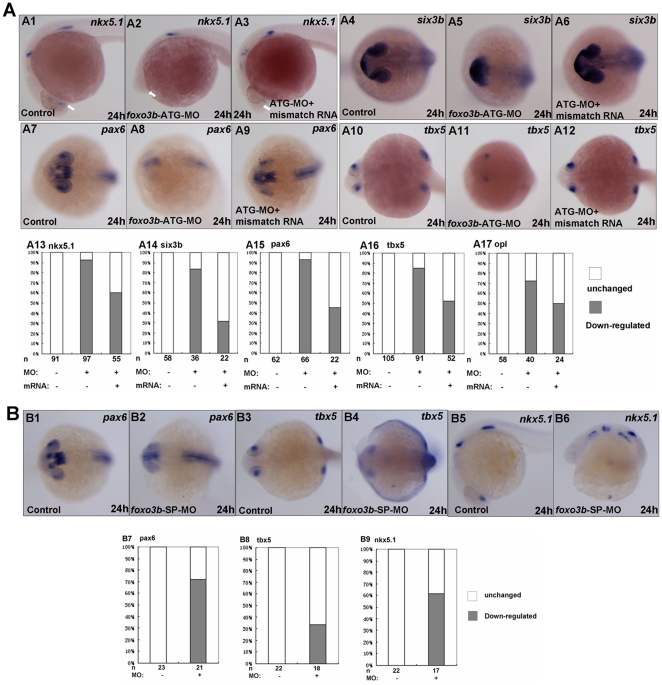
In foxo3b morphants, the embryos displayed abnormality in head formation, such as smaller forebrain and eyes. This fact prompted us to examine whether foxo3b knockdown would cause defects in neural tube formation. We examined several markers to test whether foxo3b had an essential role in neural induction. Nkx5.1 (also known as homeobox 3 gene), is expressed in the central nervous system. A remarkable loss of nkx5.1 expression at the telencephalon was observed in foxo3b morphants (indicated by white arrows, Fig. 3A2). Another forebrain neural keel marker six3b (sine oculis homeobox homolog 3b), specifically expressed at the telencephalon and eyes, exhibited an abnormal expression pattern in foxo3b morphants (Fig. 3A5). Moreover, neuroectoderm marker pax6 (paired box gene 6a), expressed dominantly in forebrain and eyes, was reduced dramatically in foxo3b knockdown embryos (Fig. 3A8). Tbx5 (T box gene 5) is expressed in the eyes, heart and pectoral fins during embryogenesis [23]. As revealed by tbx5 staining, some foxo3b morphants failed to develop one or two eyes, some developed smaller eyes in foxo3b morphants (Fig. 3A11). In addition, the foxo3b morphants seemed to have much thinner head (Fig. 3A).
Figure 3. Loss of foxo3b function results in anterior defects.
(A) The foxo3b-ATG-MO injected embryos showed remarkable loss of expression of anterior neural markers by 24 hpf, which could partially be rescued by co-injection of foxo3b mismatch mRNA. (A1–A3, A13) The expression of nkx5.1 at the telencephalon (indicated by white arrows) was dramatically reduced in foxo3b-ATG-MO injected embryos. Co-injection of foxo3b mismatch mRNA could partially restore its expression at the telencephalon. (A4-A6, A14) Six3b was specifically expressed at the telencephalon and eyes. Loss of foxo3b function resulted in abnormal expression pattern of six3b, which was restored by co-injection of foxo3b mismatch mRNA. (A7–A9, A15) Pax6 expression at the forebrain and eyes decreased greatly in foxo3b morphants. Co-injection of foxo3b mismatch mRNA partially restored its expression. (A10–A12, A16) Expression of tbx5 at the retina was dramatically reduced in foxo3b-knockdown embryos. Its expression was rescued by co-injection of foxo3b mismatch mRNA. (A17) Opl expression at the telencephalon was reduced in foxo3b morphants compared to control embryos, which was efficiently rescued by co-injection of foxo3b mismatch mRNA. Embryos were injected with 8 ng foxo3b-ATG-MO or 125 pg foxo3b mismatch mRNA,wild-type embryos were used as control. A1-A3, lateral views with anterior to the left; A4-A12, dorsal views with anterior to the left; A1-A17, 24 hpf. (B) The foxo3b-SP-MO injected embryos exhibited anterior defects similar to that of foxo3b-ATG-MO injected embryos. (B1–B2, B7) The expression of pax6 at the telencephalon and eyes was reduced in foxo3b-SP-MO injected embryos. (B3–B4, B8) Tbx5 expression at the eyes decreased in foxo3b-knockdown embryos. (B5–B6, B9) Loss of zygotic foxo3b function resulted in reduction of nkx5.1 expression at the telencephalon. Embryos were injected with 16 ng STD-MO (control) or 16 ng foxo3b-splice-MO. B1–B4, dorsal views with anterior to the left; B5-B6, lateral views with anterior to the left; B1-B9, 24 hpf.
Meanwhile, we conducted rescue experiments to validate the efficiency and specificity of foxo3b-MO. To avoid quenching, we used foxo3b mismatch mRNA in which the N-terminus of foxo3b mRNA was altered so that it was no longer complementary to the sequence of foxo3b-ATG-MO. As shown in Figure 3A, co-injection of foxo3b mismatch mRNA efficiently restored normal expression of all neuroectoderm markers (nkx5.1, six3b, pax6, tbx5 and opl) at the forebrain and eyes.
As expected, injection of foxo3b-SP-MO resulted in anterior defects similar to that of foxo3b-ATG-MO morphants. Several neuroectoderm markers, tbx5, nkx5.1 and pax6, showed obviously reduction of expression at forebrain and eyes in foxo3b-SP-MO morphants (Fig. 3B). Taken together, these data suggested that foxo3b was required for anterior neuroectoderm formation and neural tube patterning.
However, in the posterior region, particularly in foxo3b splice morphants, the expression of tbx5 increased, the expression of pax6 in posterior neuroectoderm including hindbrain and spinal chord also increased obviously, but the expression of nkx5.1 in posterior neuroectoderm only displayed disorganized, which was not reduced as that in telencephalon (Fig. 3B). As reported, zygotic Wnt8/β-catenin is required for posteriorization of the neuroectoderm and the formation of the posterior mesoderm [24], [25], [26]. These observations indicated that foxo3b might negatively regulate zygotic Wnt8/β-catenin signaling.
Loss of Foxo3b Leads to Defects in Dorsal-ventral Patterning during Early Embryogenesis and Foxo3b Affects Wnt/β-catenin Signaling
The majority of foxo3b morphants displayed a disorganization of the head and shortened body axis, thus, we selected several marker genes involved in DV patterning to further reveal the role of foxo3b in early zebrafish axis formation in this study.
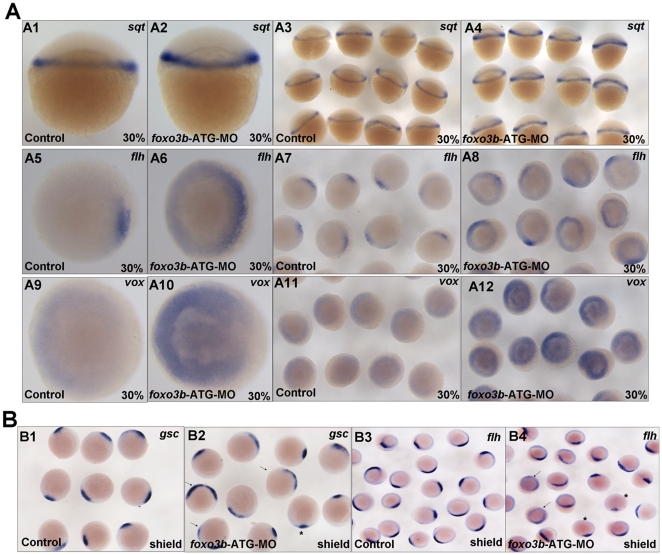
At 30% epiboly, the knockdown of foxo3b caused elevated expression of sqt (squint) (Fig. 4A1-A4), a nodal-related ligand, which is the main direct target of maternal Wnt/β-catenin signaling [27], [28]. Foxo3b morphants also showed expanded expression of organizer marker gene flh (floating head) (Fig. 4A5–A8). These results indicated that knockdown of foxo3b in embryos increased maternal β-catenin signaling activity and promoted organizer formation. In addition, a significant increase of vox expression was also observed in foxo3b morphants at 30% epiboly (Fig. 4A9–A12). As a member of the ventrally expressed homeobox genes, vox gene is a direct target gene of β-catenin [7] and first expressed ubiquitously after the MBT by maternal factors [29]. High level of vox expression is maintained by Wnt8/β-catenin signaling at early gastrulation stage. At later gastrulation, its expression is regulated by BMP signaling. These observations implied that foxo3b knockdown could affect zebrafish dorsal-ventral patterning during early embryogenesis probably through antagonizing Wnt/β-catenin signaling.
Figure 4. Knockdown of foxo3b leads to defects in DV patterning during early embryogenesis.
(A) A1–A4, sqt expression increased in foxo3b-MO injected embryos compared to control embryos. A5–A8, presumptive organizer marker flh expanded at foxo3b-knockdown embryos. A9–A10, foxo3b knockdown caused ectopic vox expression at 30% epiboly. (B) The expression of presumptive organizer marker gsc and flh in foxo3b-MO injected embryos. The arrows identified embryos with expanded expression (B2, 74%, n = 23; B4, 52%,n = 21, respectively) and asterisks identified embryos with decreased expression. Embryos were injected with 8 ng STD-MO (control) or 8 ng foxo3b-ATG-MO. A1–A4, lateral views; A5–A12, B1–B4, animal pole views; A1–A12, 30% epiboly; B1–B4, shield stage.
Wnt/β-catenin signaling is essential for organizer formation and DV patterning [4], [7], [30]. In addition, Wnt/β-catenin signaling can affect discrete domains of gene expression along the anterior-posterior (AP) axis of the neural plate and plays a role in establishing neural tube compartments along the axis [12]. In this study, the fact that the loss of foxo3b could result not only in defects of neuroectoderm formation, neural tube patterning and dorsal-ventral patterning, but also in mis-expression of Wnt target genes, strongly suggested that foxo3b could affect Wnt/β-catenin signaling.
As reported, wnt8 directly regulates the transcription of vent and vox, starting at the blastula/gastrula transition (30/40% epiboly). The maintenance of high levels of vent or vox expression by wnt8 is required for the repression of organizer genes on the ventral side of the embryo [7]. At gastrula stage, β-catenin activity influences in both dorsal and ventro-lateral discrete domains, to mediate Wnt ligand signaling [4]. Thus, the organizer genes expression are affected by both the positive influence of maternal and the negative influence of ventro-lateral zygotic Wnt/β-catenin signaling at 50% epiboly [31]. Consistent with this notion, the foxo3b morphants at shield stage displayed mixed expression of organizer genes flh and gsc (goosecoid): 52% and 74% of morphants showed expanded flh and gsc expression respectively (arrow indicated, Fig. 4B2 and B4), while 10% and 9% of morphants showed reduced flh and gsc expression (asterisk indicated, Fig. 4B2 and B4). However, we could not rule out a possibility that the mixed expression of flh and gsc in shield stage morphants might also resulted from the incomplete penetration of morpholinos or delayed embryogenesis process, which is needed to be further defined.
As showed above, foxo3b was maternally expressed and maternal β-catenin direct target gene sqt displayed enhanced expression. Moreover, other canonical Wnt/β-catenin signaling markers also exhibited mixed expression pattern at shield stage. Taken together, these observations implied that zebrafish foxo3b might serve as a main partner participating in negatively regulating both maternal and zygotic Wnt/β-catenin signaling.
Foxo3b Antagonizes Wnt/β-catenin Signaling during Dorsal-ventral Patterning
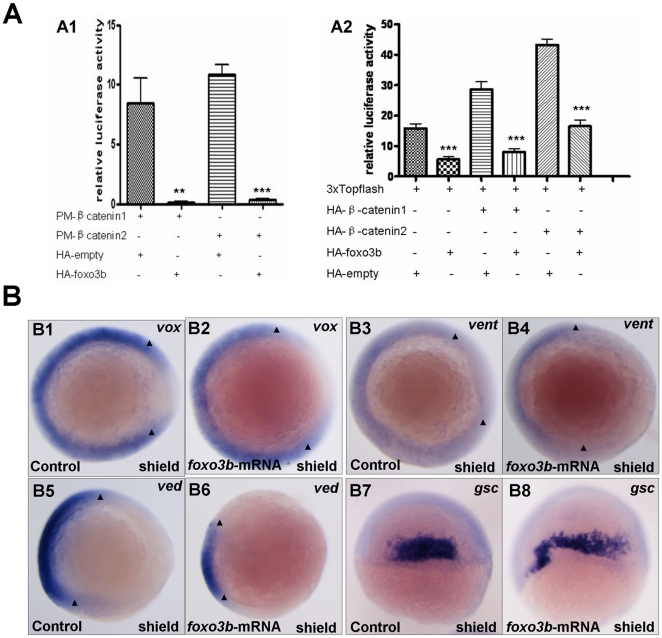
To further verify that foxo3b could inhibit Wnt/β-catenin signaling, we checked whether foxo3b suppressed the transcriptional activity of zebrafish β-catenin. Firstly, we constructed an artificial transcription factor by fusing zebrafish full-length β-catenin1/2 with Gal4 DBD (the corresponding expression plasmids were designated as pM-β-catenin1 and pM-β-catenin2, respectively). Subsequently, we injected 1-cell stage embryos with pM-β-catenin1/2, HA-empty or HA-Foxo3b, together with pFR-luc (a Gal4-dependent promoter linked to the luciferase gene) as a reporter, and pTK-Rellina as an internal control. The luciferase activity was measured 11 hours after injection. The results indicated that foxo3b dramatically inhibited β-catenin transcriptional activity (p = 0.0069 for pM-β-catenin1, p<0.0001 for pM-β-catenin2 in Fig. 5A1).
Figure 5. Foxo3b gain-of-function inhibits Wnt/β-catenin signaling in embryos.
(A) Foxo3b inhibited β-catenin/T cell factor activity in embryos. A1, 1-cell stage embryos were injected with a mixture of plasmids as indicated, together with pFR-luc as a reporter gene and pTK-renilla as an internal control; luciferase activity was measured after 11h. Date presented are the average (±SEM) of four independent experiments. A2, 1-cell stage embryos were injected with 3xTOPFlash, and the plasmids as indicated, together with pTK-renilla as an internal control; luciferase activity was measured after 11 h, performed in triplicate. “**” indicates p<0.01; “***” indicates p<0.001. (B) Gain-of-function of foxo3b resulted in suppression of Wnt/β-catenin signaling in embryos. B1-B2, foxo3b over-expressed embryos showed reduced vox expression (arrowheads in B1 and B2) compared to wild-type. B3-B6, the expression of ventral marker vent and ved (domain width indicated by arrowheads) decreased in 70% (n = 20) and 75% (n = 16) of foxo3b-ATG-MO-injected embryos respectively. B7-B10, the expression domain of dorsal marker gsc expanded in most foxo3b over-expressed embryos. Embryos were injected with 2 ng GFP mRNA (control) or 2 ng foxo3b mRNA. B1-B6, animal pole views with dorsal to the right; B7-B8, dorsal views with anterior on top; B1-B8, shield stage.
β-catenin induces transcription of Wnt target genes through binding to lymphoid enhancer factor/T cell factor in the nucleus. To test the inhibition of foxo3b on β-catenin transcriptional activity, we analyzed the effect of foxo3b on TCF-dependent transcription using 3xTOPFlash reporter (a β-catenin-dependent promoter which contains 3 copies of an optimal TCF-binding site) assays. As shown in Figure 5A2, TCF-dependent transcription was activated by endogenous zebrafish β-catenin1/2, and this activation was dramatically suppressed by over-expression of foxo3b (column 1 and 2 from left to right in Fig. 5A2); Over-expression of zebrafish β-catenin1 or β-catenin2 enhanced TCF-dependant transcription, which could also be suppressed by over-expression of foxo3b (column 3-6 from left to right in Fig. 5A2).
To further determine whether foxo3b could indeed antagonize Wnt/β-catenin signaling in vivo, we injected synthetic zebrafish foxo3b mRNA into 1-cell stage embryos, then assayed for Wnt target genes at shield stage. We observed the morphogenesis of embryos with ectopic foxo3b expression firstly, most embryos with ectopic foxo3b expression exhibited expansion of anterior brain, and curved body, partly with cyclopic eye of normal size (Data not shown), which is not totally opposite to the phenotype of foxo3b morphants. FOXO transcription factors are important mediator of the PI3K/Akt pathway and involved in a series of cellular functions [32]. As a transcriptional factor, FOXO can regulate multiple target gene expression, such as dLnR, d4EBP and Bim, which participate in modulating cell apoptosis and cell cycle [33]. In addition, FOXO can bind with other transcription factors, such as C/EBP beta, to affect their function [34]. Therefore, we assumed that ectopic expression of foxo3b might influence multiple signaling pathways in addition to Wnt/β-catenin signaling during early embryogenesis, resulting in complex phenotypes exhibited in foxo3b over-expressed embryos.
Over-expression of foxo3b resulted in reduction of ventral gene expression in most embryos. As shown in Figure 5, in 70% injected embryos (n = 20), vent expression domain was reduced obviously (Fig. 5B3 and B4), and 75% injected embryos (n = 16) displayed reduced ved expression arc (Fig. 5B5 and B6). Similarly, high frequency of foxo3b mRNA injected embryos showed reduced vox expression (Fig. 5B1 and B2). On the contrary, the dorsal marker gene gsc, displayed expanded expression pattern in most foxo3b over-expressed embryos (Fig. 5B7 and B8).
Foxo3b is Required for Posterior Neuroectoderm and Mesoderm
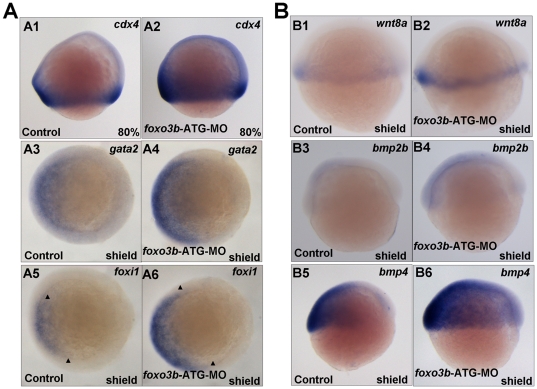
Previous studies revealed that Wnt/β-catenin signaling is required for neural posteriorization by directly regulating cdx4 to promote posterior hox gene expression [35]. As foxo3b morphants displayed increased Wnt/β-catenin signaling, we next examined whether posterior body formation was affected by foxo3b knockdown. Firstly, we checked expression of Wnt target gene cdx4 (caudal type homeobox transcription factor 4/kugelig gene), which was required for posterior body formation. The result showed that almost all foxo3b morphants exhibited expanded expression of cdx4 (Fig. 6A2). In addition, non-neural ectoderm marker gata2 (GATA-binding protein 2a) and pan-ectoderm marker foxi1 (forkhead box I1) expression were up-regulated in foxo3b morphants (Fig. 6A4 and A6), suggesting that foxo3b might also play important roles in regulating cell fate differentiation.
Figure 6. Foxo3b functions in posterior neuroectoderm formation and involves with BMP signaling in DV patterning.
(A) Foxo3b affected posterior neuroectoderm formation and mesoderm induction. A1-A2, at 80% epiboly, cdx4 expression expanded in foxo3b-MO injected embryos compared to control embryos. A3-A4, the expression of non-neural ectodermal marker gata2 was up-regulated in foxo3b-knockdown embryos. A5-A6, ectoderm marker foxi1 expanded in foxo3b morphants. (B) The expression of wnt8 and bmp ligands in foxo3b morphants. B1–B2, wnt8 expression increased in foxo3b morphants at shield stage. B3–B6, the expression of ventral markers bmp2b/bmp4 was up-regulated in foxo3b-knockdown embryos at shield stage. Embryos were injected with 8 ng STD-MO (control) or 8 ng foxo3b-ATG-MO. A1–A2, lateral views with dorsal to the right; A3–A6, animal pole views with dorsal to the right; B1–B6, lateral views with dorsal to the right; A1–A2, 80% epiboly; A3–A6, B1–B6, shield stage.
The above observations have showed that knockdown of foxo3b led to elevated Wnt/β-catenin signaling and resulted in defects in DV (dorsal-ventral) patterning and AP neural tube patterning. DV patterning of vertebrate embryos requires the concerted actions of the BMP and Wnt signaling, and Wnt signals cooperate with Bmp to regulate the formation of the posterior mesoderm [26]. To better understand the role of foxo3b and the underlying mechanism, we checked the expression of wnt8, bmp2b and bmp4 (bone morphogenetic protein 2b and 4) in foxo3b knockdown embryos. Both wnt8 and bmp2b/bmp4 expression were up-regulated in foxo3b morphants (Fig. 6B). In addition, the up-regulation of bmp2b expression in foxo3b morphants could be rescued by co-injection of foxo3b mismatched mRNA (Fig. S1).
In addition, wnt8 was also used as a ventro-lateral and posterior mesoderm marker during zebrafish embryogenesis to monitor the mesoderm induction in embryos [36], [37], [38]. The increased expression of wnt8 in foxo3b morphants was consistent with the observation that cdx4 exhibited expanded expression in posterior position. Wnt8/β-catenin signaling was reported to promote the formation of the ventro-lateral and posterior mesoderm and neuroectoderm [24], [25], so it was possible that enhanced Wnt/β-catenin signaling in foxo3b morphants resulted in increased ventro-lateral mesoderm marker wnt8 expression. Furthermore, the increased wnt8 expression might partially contribute to the anterior defects in foxo3b morphants by activating the canonical Wnt signaling.
Foxo3b Mediated Anterior Neuroectoderm Formation and DV Patterning Through Maternal and Zygotic Wnt/β-catenin Signaling
Based on our above data, we proposed that defects of foxo3b morphants in DV patterning and neuroectoderm formation, might result from up-regulation of Wnt/β-catenin signaling. To verify our hypothesis, we performed rescue experiments by co-injection of β-catenin1-MO, β-catenin2-MO or dnTCF mRNA with foxo3b-ATG-MO. After injection, we scored embryos for expression of the neuroectoderm markers six3b, tbx5, nkx5.1, pax6 and opl using in situ hybridization (Fig. 7B) as well as general morphological characteristics (Fig. 7A). Translation-blocking morpholinos targeting zebrafish β-catenin1 and β-catenin2 are designed as previously described [4]. We found that β-catenin1 morpholino could partially rescue the anterior defects resulting from foxo3b knockdown (Fig. 7A), and β-catenin2 morpholino could also rescue the phenotype of foxo3b morphants (data not show).
Figure 7. Foxo3b inhibits both maternal and zygotic Wnt/β-catenin signaling.
(A) Foxo3b morphants could be rescued by knockdown of β-catenin1. Black box, dead embryos at 24 hpf; A5-A6, dark gray box, embryos with defects at 55 hpf characterized by severe phenotype (as described in Fig. 2B); A3-A4, light gray box, embryos with mild phenotype; A1–A2, white box, un-injected wild-type embryos. A1, A3, A5, lateral views; A2, A4, A6, lateral views with anterior to the left; A1–A7, 55 hpf. (B) Anterior defects caused by loss of foxo3b function could be rescued by co-injection of dnTCF mRNA. (B1–B3, B7) Six3b was expressed abnormally in foxo3b morphants, which could be rescued by co-injection of dnTCF mRNA by 24 hpf. (B4–B6, B8) Tbx5 expression at the eyes was greatly reduced in foxo3b-knockdown embryos. Co-injection of dnTCF mRNA could efficiently restore its expression at the eyes. (B9–B11) Nkx5.1, pax6 and opl expression at the anterior neural plate could also be efficiently rescued by co-injection of dnTCF mRNA. Embryos were injected with 8 ng foxo3b-ATG-MO or 10 pg dnTCF mRNA,wild-type embryos were used as control. B1–B6, dorsal views with anterior to the left; B1–B11, 24 hpf. (C) Foxo3b knockdown resulted in abnormal expression of early Wnt target genes, which could be rescued by co-injection of β-catenin1/2 morpholino. (C1–C8, C15) The morphants showed increased expression of gsc, which could also be rescued by co-injection of β-catenin1 MO. Co-injection of β-catenin2 MO resulted in dramatically reduction of gsc expression (75%, n = 32) compared to control embryos. (C9-C14, C16) The expression level of vox increased dramatically in foxo3b-knockdown embryos, which could be partially rescued by co-injection of β-catenin2 MO. (C17) At 30% epiboly, sqt expression was up-regulated in foxo3b morphants. Co-injection of β-catenin1 morpholino efficiently reduced its expression. Embryos were injected with 8 ng foxo3b-ATG-MO or 8 ng β-catenin1/2 morpholino,wild-type embryos were used as control. C1-C14, dorsal views; C1-C17, 30% epiboly.
In addition, we tested whether dnTCF could rescue anterior defects in foxo3b morphants. As a dominant negative form of zebrafish TCF3 (also known as headless), dnTCF is an effective suppressor of Wnt/β-catenin signaling. As expected, the reduction of neuroectoderm markers expression at the forebrain and eyes could be rescued by co-injection of dnTCF mRNA by 24 hpf. As shown in Figure 7B, 83.3% of foxo3b morphants displayed abnormal expression of six3b, this proportion dropped down to 49% after co-injection with dnTCF mRNA. In addition, 84.6% of tbx5 expression at the eyes was greatly reduced in foxo3b morphants, after co-injection with dnTCF mRNA, only 37.8% of embryos displayed the reduced expression of tbx5 at the eyes. Co-injection of dnTCF mRNA could also rescue the expression of nkx5.1, pax6 and opl at the anterior neuroectoderm in foxo3b morphants. These results suggested that the defects of anterior neural domains in foxo3b morphants indeed resulted from up-regulation of Wnt/β-catenin signaling. Besides the anterior defects of foxo3b morphants could be rescued by co-injection of β-catenin1-MO or β-catenin2-MO (Fig. 7A), further proving that the role of foxo3b in affecting anterior neuroectoderm formation was mediated by wnt/β-catenin signaling.
At late blastula period, loss of foxo3b function in embryos resulted in up-regulation of several Wnt targets (sqt, gsc, and vox) (Fig. 7C), which might result from the elevation of maternal wnt signaling. Co-injection of β-catenin1 morpholino successfully reduced dorsal markers genes gsc and sqt to normal level in foxo3b morphants at blastula stage. 38% of foxo3b morphants exhibited up-regulation of gsc expression, when co-injected with β-catenin1 morpholino, the rate of gsc up-regulation was reduced to 18.8% (Fig. 7C6 and C15). Co-injection of foxo3b-ATG-MO with β-catenin2 morpholino resulted in obviously reduced expression of gsc at 30% epiboly (Fig. 7C8). 38.8% of foxo3b morphants with up-regulation of sqt was reduced to 9.2% when co-injected with β-catenin1 morpholino (Fig. 7C17). In addition, co-injection of β-catenin2 morpholino successfully restored ventral marker vox expression to normal level at blastula stage. 69.7% of foxo3b morphants (n = 44) showed up-regulation of vox expression, which was dropped down to 30% after co-injection of β-catenin2 morpholino (Fig. 7C14 and C16).
These results that the abnormal expression of marker genes involved in DV patterning or anterior neuroectoderm defects, resulting from foxo3b knockdown, could be successfully rescued by knockdown of β-catenin1 or β-catenin 2 suggested that foxo3b could inhibit both maternal and zygotic Wnt/β-catenin signaling.
In the rescue experiments, we could partially rescue the foxo3b morphants by injecting with dnTCF mRNA, β-catenin1-MO or β-catenin 2-MO, which further indicated that foxo3b could indeed affect embryogenesis by inhibiting both maternal and zygotic Wnt/β-catenin signaling. FOXO transcription factors act as downstream effectors of many important pathways, such as PI3K/Akt pathway, TGF-β pathway [32], [39], they can mediate multiple biological function by regulating different target genes [33], [34], [40]. Our rescue experiments also implied that foxo3b might affect other signaling pathways in addition to Wnt signaling during zebrafish embryogenesis.
Foxo3b Interacts with β-catenin 1 and β-catenin 2
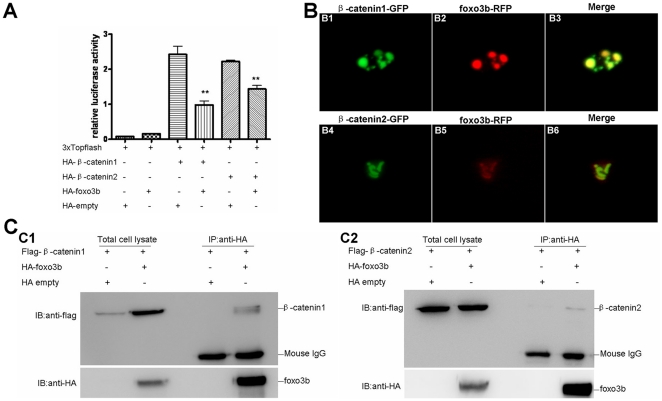
Because foxo3b could inhibit the transactivity of artificial transcription factors, pM-β-catenin1 and pM-β-catenin2, and previous studies had showed that mammalian FOXO competed with TCF to interact with β-catenin [16], these evidences prompted us to check whether foxo3b could interact with β-catenin1 and β-catenin2. Firstly, we analyzed the effect of foxo3b on TCF-dependent transcription activity using 3xTOPFlash reporter in 293T cell. As showed in Figure 8A, over-expression of foxo3b could suppress β-catenin/TCF signaling significantly in 293T cell (p = 0.0049 for β-catenin1, p = 0.0019 for β-catenin2 in Fig. 8A).
Figure 8. Foxo3b interacts with β-catenin1/2 in 293T cell.
(A) Foxo3b inhibited β-catenin/T cell factor activity in 293T cell line. 293T cells were transfected with 3xTOPFlash, and the plasmids as indicated, together with pTK-renilla as an internal control; luciferase activity was measured after 24h. Date presented were the average (±SEM) of three independent experiments, performed in triplicate. “**” indicates p<0.01. (B) Foxo3b co-localized with β-catenin1/2. HeLa cells were transfected with GFP-β-catenin1 (B1-B3) or GFP-β-catenin2 (B4–B6), together with RFP-foxo3b. Transfected cells were then observed using fluorescent microscopy after 24h. (C) Foxo3b interacted with β-catenin1/2. 293T cells were transfected with the indicated expressing plasmids. HA-foxo3b was immunoprecipitated, and binding of Flag-β-catenin1 (C1) or Flag-β-catenin2 (C2) was analyzed by immunoblotting.
Subsequently, we did co-localization assays by co-transfecting RFP-tagged foxo3b together with GFP-tagged β-catenin1 or β-catenin2 into HeLa cells. We observed that foxo3b co-localized with β-catenin1 and β-catenin2 in the nucleus (Fig. 8B). Then, we performed immunoprecipitation (IP) experiments to further confirm the interaction between foxo3b and β-catenin1/2. As showed in Figure 8C, foxo3b could indeed interact with both β-catenin1 and β-catenin2. The interaction between foxo3b and β-catenin1 seemed to be stronger than that between foxo3b and β-catenin2 (line 4 from left to right in Fig. 8C1 and C2). The protein level of β-catenin1 was enhanced after co-transfection with foxo3b (line 2 from left to right in Fig. 8C1), while the protein level of β-catenin2 was not changed after co-transfection with foxo3b (line 2 from left to right in Fig. 8C2). Taken together, these observations suggested that foxo3b interacted with β-catenin1 and β-catenin2 to negatively regulate Wnt/β-catenin signaling. The interaction between foxo3b and β-catenin1 was much stronger than that between foxo3b and β-catenin2 implied that β-catenin1 might act as a major target for foxo3b in regulating Wnt/β-catenin signaling during zebrafish early embryogenesis.
Discussion
Although the function of mammalian FOXO inhibiting β-catenin/T cell factor activity has been revealed through cell-culture system [16], how important of this inhibition in vivo, particularly in embryogenesis is still unclear. In this study, we took advantage of zebrafish model thoroughly exploring the role of foxo3b, an orthologue of mammalian FOXO3, in embryogenesis. We found that zebrafish foxo3b played important roles in axis and neuroectoderm formation. Furthermore, we confirmed that foxo3b could indeed interact with zebrafish β-catenin1 and β-catenin2 to inhibit LEF/TCF-dependent transcription in vitro and in vivo. In summary, we thought that zebrafish foxo3b affected early embryogenesis through negatively regulating maternal and zygotic β-catenin transactivity.
Zebrafish Foxo3b Gene is Expressed Maternally and Required for Early Embryogenesis
The opposite roles of zebrafish maternal and zygotic Wnt/β-catenin signaling during embryogenesis have been recognized [4], [7], [8]. In addition, β-catenin and TCF activate dorsal-specific genes at the blastula stage, but mediate the activity of the ventro-laterally expressed wnt8 to repress the dorsal-specific genes during gastrulation [14]. Herein, the major concern was that whether foxo3b was expressed maternally or at early stage of embryogenesis. In our study, by in situ hybridization, we revealed that high expression of foxo3b could be detected in 2-cell stage embryos, implying that foxo3b was expressed maternally, and distributed ubiquitously as β-catenin1, β-catenin2 and ICAT [4], [41] during the blastula stage (Fig. 1). In addition, semi-quantitative RT-PCR assays also detected foxo3b transcripts as early as 2-cell stage, further confirming that foxo3b was expressed maternally. Thus, zebrafish foxo3b might play important roles during early embryogenesis. Knockdown of foxo3b expression by injections of either foxo3b-ATG-MO or foxo3b-SP-MO, resulted in severe developmental defects including reduced body length, abnormality of brain and eye development, and curved body, which implied that both maternal and zygotic foxo3b function importantly in zebrafish body axis formation.
In zebrafish, Wnt/β-catenin signaling has been reported to affect body axis patterning by opposing effect of maternal and zygotic actions [12]. Exaggerated Wnt signaling after the MBT (mid-blastrula transition) by ectopic expression of wnts, β-catenin [12] or by LiCl treating [10], [42], leads to loss of rostral neural domains, whereas reduced Wnt signaling leads to expansion of rostral neural domains. In this study, the defects of anterior brain were observed in both foxo3b-ATG-MO morphants and splice-MO morphants, suggesting that these morphants harbored elevated Wnt signaling activity after MBT. Interestingly, the phenotypes exhibited in foxo3b morphants were similar to that of the embryos with ectopic expression of wnts, vent, vox and ved, all components of zygotic Wnt/β-catenin signaling [6], [7], [8]. As indicated by cdx4 expansion in foxo3b morphants (Fig. 6), foxo3b knockdown caused high zygotic wnt8/β-catenin signaling to continually suppress the dorsal organizer and promote the posterior neuroectoderm formation. In addition, the exaggerated zygotic wnt8/β-catenin signaling in the ventro-lateral region disturbed the balance of the opposing actions between dorsal organizer genes boz, gsc and ventral genes ved, vent and vox in embryos injected with either foxo3b-ATG-MO or foxo3b-SP-MO, which might cause high rate of anterior neuroectoderm defects (Fig. 2 and Fig. 3). These observations suggested that knockdown of either maternal or zygotic foxo3b in embryos resulted in elevated and constant β-catenin signaling in morphants, resulting in anterior brain defects (Fig. 2, Fig. 3 and Fig. 7).
Foxo3b Affects Zebrafish DV Patterning Through Negatively Regulating Both Maternal and Zygotic Wnt/β-catenin Signaling
As reported, the specification of vertebrate body axes, such as the dorsal-ventral (DV) and anterior-posterior (AP) axis, is initiated soon after fertilization. The maternal factors are required to locally activate zygotic genes to specify dorsal-ventral polarity [43], [44] and to induce patterning center (such as Nieuwkoop center and Spemann organizer) formation [45]. Maternal Wnt/β-catenin signaling genes play important roles in this process. In this study, we found that foxo3b morphants displayed defects in body axis formation. Further marker gene staining showed that the expression of organizer genes sqt and flh expanded in foxo3b morphants at 30% epiboly (Fig. 3). Sqt is a direct target gene of maternal β-catenin [28]. Thus, these results suggested that foxo3b might affect the specification of zebrafish body axes through influencing maternal Wnt/β-catenin signaling. In addition, the fact that Wnt target genes sqt and gsc increased dramatically in foxo3b morphants at blastula stage also indicated the up-regulation of Wnt/β-catenin signaling (Fig. 4 and Fig. 7). Furthermore, gsc expression could be rescued by co-injection of β-catenin2-MO in foxo3b morphants at 30% epiboly. However, notably, most co-injected embryos displayed dramatically reduced gsc expression (Fig. 7C), which suggested that β-catenin2-MO can successfully suppressed the increased expression of gsc in foxo3b morphants, and β-catenin2 might function downstream of foxo3b in Wnt/β-catenin signaling. Noteworthily, β-catenin2 morphants but not β-catenin1 morphants failed to express organizer genes boz, sqt and gsc, which established the importance of maternal β-catenin2 for organizer formation [4]. Beyond expectation, both β-catenin2-MO and β-catenin1-MO suppressed the increased expression of organizer genes in foxo3b morphants, promoting the possibility that when β-catenin1 protein was reduced in foxo3b morphants, the remained foxo3b protein in morphants could still interact with β-catenin2 efficiently to continuously suppress the β-catenin2 activity effectively, resulting in counteraction of the increased expression of sqt and gsc in foxo3b morphants.
The following two observations might support this point. Firstly, foxo3b interacted with β-catenin1 much more efficiently than with β-catenin2. Thus, if β-catenin1 protein level was the same as that of β-catenin2, the remained foxo3b protein in morphants should be apt to interact with β-catenin1. Secondly, low dosage of foxo3b mismatch mRNA (125 pg per embryo) could rescue the defects of foxo3b morphants efficiently (Fig. 2 and Fig. 3), suggesting that only a little inhibitory effect of foxo3b was required in foxo3b morphants to counteract developmental defects.
Moreover, we found the mixed expression of organizer genes flh and gsc at shield stage in foxo3b morphants (Fig. 4B), which suggested that their expression were affected by both the positive influence of maternal Wnt signaling and the negative influence of ventro-lateral zygotic Wnt/β-catenin signaling. These results strongly supported that zebrafish foxo3b could inhibit both maternal and zygotic Wnt/β-catenin signaling, similar to that observed for Naked1/Naked2 genes [31].
Ventralizing transcriptional repressors in the Vox/Vent family have been proposed to serve as important regulators for DV patterning in the early embryogenesis [46], [47], [48], [49]. In addition, BMP gene family is also shown to be an important type of ventral genes [50], [51], [52]. To date, only maternal runx2 has been identified to induce zygotic vox, vent and ved expression by directly binding to their promoters [29]. As reported, the zygotic inducers of Vox/Vent family are wnt8 at 40-50% epiboly and BMP at 75% epiboly. During the developmental stages, dorsal genes, such as boz and gsc continue to repress vox, vent and ved expression. In this study, we found that vox displayed robust expression at 30% epiboly in foxo3b morphants. At this stage, ventro-lateral wnt8 expression was just initiated, so it was unlikely that zygotic inducer wnt8 up-regulated vox expression. In addition, it was also unlikely that vox expression was up-regulated by the organizer, because the expanded organizer was supposed to suppress expression of Vox/Vent family [7]. Thus, the expansion of vox expression at 30% epiboly in foxo3b morphants might result from directly up-regulation by maternal Wnt/β-catenin signaling. In fact, vox/vent family genes, harboring TCF binding sites in their promoters, have already been shown to be directly regulated by wnt8 in zebrafish embryos [7]. We further validated this point by rescue experiment that β-catenin2-MO could neutralize the increased expression of vox in foxo3b morphants efficiently (Fig. 7).
In foxo3b over-expressed embryos, most embryos displayed expanded gsc expression at shield stage, similar to that of embryos over-expressed with Nkds [31] or Wnt inhibitors [14], [53]. Over-expression of Nkds dramatically reduces the negative influence of zygotic ventro-lateral Wnt/β-catenin signaling on organizer suppression, so foxo3b over-expression might also reflect the influence of foxo3b on zygotic wnt/β-catenin signaling. In this study, apart from dorsal marker genes staining, we also checked the ventro-lateral wnt target genes expression in foxo3b over-expressed embryos. As showed in Figure 5, the results suggested that foxo3b might repress β-catenin activity in embryos to cause the down-regulation of vox/vent gene family, which further resulted in promoting dorsal marker gene expression.
Foxo3b Affects Zebrafish Anterior Neuroectoderm Patterning Through Negatively Regulating Both Maternal and Zygotic Wnt/β-catenin Signaling
Wnt/β-catenin signaling has been identified to define discrete domains of gene expression along the anterior-posterior (AP) axis of the neural plate and then help establish the formation of neural tube compartments along the axis [8], [54], [55]. As reported, TCF3 (headless) is expressed in the anterior neuroectoderm, functioning as a repressor of posterior neural fates in this region [12]. β-catenin signaling activated by wnt8 participates in posteriorizing the neural plate. Loss of TCF3 function leads to the over-activity of the Wnt pathway in the anterior part of neuroectoderm and results in defects in anterior fates [12]. Moreover, bozozok mutant zebrafish shows defects in anterior neural structures, which can be rescued by dn-Xwnt8 [56]. These previous observations strongly support that exaggerated wnt8 activity leads to loss of rostral neural domains. In our study, knockdown of foxo3b resulted in defects in forebrain and eyes. The expression of forebrain markers in foxo3b morphants was relatively reduced (Fig. 3 and Fig. 7). Therefore, in addition to affecting DV patterning, foxo3b could also affect forebrain induction through negatively regulating maternal and zygotic wnt/β-catenin signaling. In fact, the reduced expression of forebrain markers in foxo3b morphants could be rescued by co-injection of dnTCF mRNA (Fig. 7), further verifying that the anterior defects in foxo3b morphants might result from exaggerated Wnt/β-catenin activity and foxo3b was a negative regulator of wnt/β-catenin signaling.
It has been reported that Wnt/β-catenin signaling antagonizes eye specification through activating wnt8b and fz8a [57]. Similar results were obtained by studies of masterblind (axin mutated) and headless zebrafish mutants, which refined the function of wnt/β-catenin signaling in eye formation [12], [58]. In addition, over-expression of wnt8 in zebrafish results in most embryos failing to develop one or both eyes [8]. In foxo3b morphants, eye formation was disrupted to different extent, as indicated by reduction of tbx5, six3b and pax6 expression in retina. These results further highlighted the role of foxo3b in maintenance of anterior neural fates.
Supporting Information
Bmp2b expression was rescued in foxo3b-knockdown embryos co-injected with foxo3b mismatch mRNA. Animal views, shield stage.
(TIF)
The partial primers used for cloning probes.
(DOC)
Acknowledgments
We gratefully acknowledge Drs. Randall Moon, Eric Weinberg, Yonghua Sun and Bingyu Mao for the generous gift of reagents. We also thank Moira Hitchens for editing.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: W.X. is supported by “973” grant 2010CB126306, 2007CB815705, NSFC grant 91019008, 31071212, 30971667 and National Transgene Project 2009ZX08010-021B. CAS grant KSCX2-Y-W-N-020. J.L. is supported by innovation project of Chinese Academy of Science KSCX2-EW-Q-12 and NSFC grant 20890113. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 2.Carter ME, Brunet A. FOXO transcription factors. Curr Biol. 2007;17:R113–114. doi: 10.1016/j.cub.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Hosaka T, Biggs WH, 3rd, Tieu D, Boyer AD, Varki NM, et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci U S A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellipanni G, Varga MT, Maegawa S, Imai Y, Kelly C, et al. Essential and opposing roles of zebrafish beta-catenins in the formation of dorsal axial structures and neurectoderm. Development. 2006;133:1299–1309. doi: 10.1242/dev.02295. [DOI] [PubMed] [Google Scholar]
- 5.Lekven AC, Thorpe CJ, Waxman JS, Moon RT. Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev Cell. 2001;1:103–114. doi: 10.1016/s1534-5807(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu T, Yamanaka Y, Nojima H, Yabe T, Hibi M, et al. A novel repressor-type homeobox gene, ved, is involved in dharma/bozozok-mediated dorsal organizer formation in zebrafish. Mechanisms of Development. 2002;118:125–138. doi: 10.1016/s0925-4773(02)00243-5. [DOI] [PubMed] [Google Scholar]
- 7.Ramel MC, Lekven AC. Repression of the vertebrate organizer by Wnt8 is mediated by Vent and Vox. Development. 2004;131:3991–4000. doi: 10.1242/dev.01277. [DOI] [PubMed] [Google Scholar]
- 8.Kelly GM, Greenstein P, Erezyilmaz DF, Moon RT. Zebrafish wnt8 and wnt8b share a common activity but are involved in distinct developmental pathways. Development. 1995;121:1787–1799. doi: 10.1242/dev.121.6.1787. [DOI] [PubMed] [Google Scholar]
- 9.Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis EM. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann's organizer. Nature. 1996;382:595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- 10.Leyns L, Bouwmeester T, Kim SH, Piccolo S, De Robertis EM. Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell. 1997;88:747–756. doi: 10.1016/s0092-8674(00)81921-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, et al. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 12.Kim CH, Oda T, Itoh M, Jiang D, Artinger KB, et al. Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature. 2000;407:913–916. doi: 10.1038/35038097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorsky RI, Itoh M, Moon RT, Chitnis A. Two tcf3 genes cooperate to pattern the zebrafish brain. Development. 2003;130:1937–1947. doi: 10.1242/dev.00402. [DOI] [PubMed] [Google Scholar]
- 14.Pelegri F, Maischein HM. Function of zebrafish beta-catenin and TCF-3 in dorsoventral patterning. Mech Dev. 1998;77:63–74. doi: 10.1016/s0925-4773(98)00132-4. [DOI] [PubMed] [Google Scholar]
- 15.Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, et al. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- 16.Hoogeboom D, Essers MA, Polderman PE, Voets E, Smits LM, et al. Interaction of FOXO with beta-catenin inhibits beta-catenin/T cell factor activity. J Biol Chem. 2008;283:9224–9230. doi: 10.1074/jbc.M706638200. [DOI] [PubMed] [Google Scholar]
- 17.Biggs WH, 3rd, Cavenee WK, Arden KC. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm Genome. 2001;12:416–425. doi: 10.1007/s003350020002. [DOI] [PubMed] [Google Scholar]
- 18.Liu JX, Hu B, Wang Y, Gui JF, Xiao W. Zebrafish eaf1 and eaf2/u19 mediate effective convergence and extension movements through the maintenance of wnt11 and wnt5 expression. J Biol Chem. 2009;284:16679–16692. doi: 10.1074/jbc.M109.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westerfield M, Doerry E, Douglas S. Zebrafish in the Net. Trends Genet. 1999;15:248–249. doi: 10.1016/s0168-9525(99)01741-2. [DOI] [PubMed] [Google Scholar]
- 20.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z, Liu N, Lin S. A zebrafish forebrain-specific zinc finger gene can induce ectopic dlx2 and dlx6 expression. Dev Biol. 2001;231:138–148. doi: 10.1006/dbio.2000.0139. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J, Feng X, Ban B, Liu J, Wang Z, et al. Elongation factor ELL (Eleven-Nineteen Lysine-rich Leukemia) acts as a transcription factor for direct thrombospondin-1 regulation. J Biol Chem. 2009;284:19142–19152. doi: 10.1074/jbc.M109.010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Begemann G, Ingham PW. Developmental regulation of Tbx5 in zebrafish embryogenesis. Mech Dev. 2000;90:299–304. doi: 10.1016/s0925-4773(99)00246-4. [DOI] [PubMed] [Google Scholar]
- 24.Erter CE, Wilm TP, Basler N, Wright CVE, Solnica-Krezel L. Wnt8 is required in lateral mesendodermal precursors for neural posteriorization in vivo. Development. 2001;128:3571–3583. doi: 10.1242/dev.128.18.3571. [DOI] [PubMed] [Google Scholar]
- 25.Szeto DP, Kimelman D. Combinatorial gene regulation by Bmp and Wnt in zebrafish posterior mesoderm formation (vol 131, pg 3751, 2004). Development. 2004;131:4117–4117. doi: 10.1242/dev.01236. [DOI] [PubMed] [Google Scholar]
- 26.Ramel MC, Buckles GR, Baker KD, Lekven AC. WNT8 and BMP2B co-regulate non-axial mesoderm patterning during zebrafish gastrulation. Dev Biol. 2005;287:237–248. doi: 10.1016/j.ydbio.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Kelly C, Chin AJ, Leatherman JL, Kozlowski DJ, Weinberg ES. Maternally controlled (beta)-catenin-mediated signaling is required for organizer formation in the zebrafish. Development. 2000;127:3899–3911. doi: 10.1242/dev.127.18.3899. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu T, Yamanaka Y, Ryu SL, Hashimoto H, Yabe T, et al. Cooperative roles of Bozozok/Dharma and Nodal-related proteins in the formation of the dorsal organizer in zebrafish. Mech Dev. 2000;91:293–303. doi: 10.1016/s0925-4773(99)00319-6. [DOI] [PubMed] [Google Scholar]
- 29.Flores MV, Lam EY, Crosier KE, Crosier PS. Osteogenic transcription factor Runx2 is a maternal determinant of dorsoventral patterning in zebrafish. Nat Cell Biol. 2008;10:346–352. doi: 10.1038/ncb1697. [DOI] [PubMed] [Google Scholar]
- 30.Hoppler S, Brown JD, Moon RT. Expression of a dominant-negative Wnt blocks induction of MyoD in Xenopus embryos. Genes Dev. 1996;10:2805–2817. doi: 10.1101/gad.10.21.2805. [DOI] [PubMed] [Google Scholar]
- 31.Van Raay TJ, Coffey RJ, Solnica-Krezel L. Zebrafish Naked1 and Naked2 antagonize both canonical and non-canonical Wnt signaling. Dev Biol. 2007;309:151–168. doi: 10.1016/j.ydbio.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 33.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–284. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 34.Christian M, Zhang XH, Schneider-Merck T, Unterman TG, Gellersen B, et al. Cyclic AMP-induced forkhead transcription factor, FKHR, cooperates with CCAAT/enhancer-binding protein beta in differentiating human endometrial stromal cells. Journal of Biological Chemistry. 2002;277:20825–20832. doi: 10.1074/jbc.M201018200. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu T, Bae YK, Muraoka O, Hibi M. Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev Biol. 2005;279:125–141. doi: 10.1016/j.ydbio.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Seiliez T, Thisse B, Thisse C. FoxA3 and goosecoid promote anterior neural fate through inhibition of Wnt8a activity before the onset of gastrulation. Developmental Biology. 2006;290:152–163. doi: 10.1016/j.ydbio.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 37.Wilm TP, Solnica-Krezel L. Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development. 2005;132:393–404. doi: 10.1242/dev.01572. [DOI] [PubMed] [Google Scholar]
- 38.Martin BL, Kimelman D. Regulation of canonical Wnt signaling by Brachury is essential for posterior mesoderm formation. Developmental Biology. 2008;319:581–581. doi: 10.1016/j.devcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seoane J, Le HV, Shen LJ, Anderson SA, Massague J. Integration of Smad and Forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 40.Stahl M, Dijkers PF, Kops GJPL, Lens SMA, Coffer PJ, et al. The forkhead transcription factor FoxO regulates transcription of p27(Kip1) and bim in response to IL-2. Journal of Immunology. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 41.Tago K, Nakamura T, Nishita M, Hyodo J, Nagai S, et al. Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev. 2000;14:1741–1749. [PMC free article] [PubMed] [Google Scholar]
- 42.Kim SH, Shin J, Park HC, Yeo SY, Hong SK, et al. Specification of an anterior neuroectoderm patterning by Frizzled8a-mediated Wnt8b signalling during late gastrulation in zebrafish. Development. 2002;129:4443–4455. doi: 10.1242/dev.129.19.4443. [DOI] [PubMed] [Google Scholar]
- 43.Steward R, Govind S. Dorsal-ventral polarity in the Drosophila embryo. Curr Opin Genet Dev. 1993;3:556–561. doi: 10.1016/0959-437x(93)90090-c. [DOI] [PubMed] [Google Scholar]
- 44.Reim G, Brand M. Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development. 2006;133:2757–2770. doi: 10.1242/dev.02391. [DOI] [PubMed] [Google Scholar]
- 45.Moon RT, Kimelman D. From cortical rotation to organizer gene expression: toward a molecular explanation of axis specification in Xenopus. Bioessays. 1998;20:536–545. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 46.Imai Y, Gates MA, Melby AE, Kimelman D, Schier AF, et al. The homeobox genes vox and vent are redundant repressors of dorsal fates in zebrafish. Development. 2001;128:2407–2420. doi: 10.1242/dev.128.12.2407. [DOI] [PubMed] [Google Scholar]
- 47.Gawantka V, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. Antagonizing the Spemann organizer: role of the homeobox gene Xvent-1. EMBO J. 1995;14:6268–6279. doi: 10.1002/j.1460-2075.1995.tb00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onichtchouk D, Gawantka V, Dosch R, Delius H, Hirschfeld K, et al. The Xvent-2 homeobox gene is part of the BMP-4 signalling pathway controlling [correction of controling] dorsoventral patterning of Xenopus mesoderm. Development. 1996;122:3045–3053. doi: 10.1242/dev.122.10.3045. [DOI] [PubMed] [Google Scholar]
- 49.Onichtchouk D, Glinka A, Niehrs C. Requirement for Xvent-1 and Xvent-2 gene function in dorsoventral patterning of Xenopus mesoderm. Development. 1998;125:1447–1456. doi: 10.1242/dev.125.8.1447. [DOI] [PubMed] [Google Scholar]
- 50.Kishimoto Y, Lee KH, Zon L, Hammerschmidt M, Schulte-Merker S. The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning. Development. 1997;124:4457–4466. doi: 10.1242/dev.124.22.4457. [DOI] [PubMed] [Google Scholar]
- 51.Dosch R, Gawantka V, Delius H, Blumenstock C, Niehrs C. Bmp-4 acts as a morphogen in dorsoventral mesoderm patterning in Xenopus. Development. 1997;124:2325–2334. doi: 10.1242/dev.124.12.2325. [DOI] [PubMed] [Google Scholar]
- 52.Hoppler S, Moon RT. BMP-2/-4 and Wnt-8 cooperatively pattern the Xenopus mesoderm. Mech Dev. 1998;71:119–129. doi: 10.1016/s0925-4773(98)00004-5. [DOI] [PubMed] [Google Scholar]
- 53.Kawahara A, Wilm T, Solnica-Krezel L, Dawid IB. Antagonistic role of vega1 and bozozok/dharma homeobox genes in organizer formation. Proc Natl Acad Sci U S A. 2000;97:12121–12126. doi: 10.1073/pnas.97.22.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelly GM, Moon RT. Involvement of wnt1 and pax2 in the formation of the midbrain-hindbrain boundary in the zebrafish gastrula. Dev Genet. 1995;17:129–140. doi: 10.1002/dvg.1020170205. [DOI] [PubMed] [Google Scholar]
- 55.Keynes R, Lumsden A. Segmentation and the origin of regional diversity in the vertebrate central nervous system. Neuron. 1990;4:1–9. doi: 10.1016/0896-6273(90)90438-l. [DOI] [PubMed] [Google Scholar]
- 56.Fekany-Lee K, Gonzalez E, Miller-Bertoglio V, Solnica-Krezel L. The homeobox gene bozozok promotes anterior neuroectoderm formation in zebrafish through negative regulation of BMP2/4 and Wnt pathways. Development. 2000;127:2333–2345. doi: 10.1242/dev.127.11.2333. [DOI] [PubMed] [Google Scholar]
- 57.Cavodeassi F, Carreira-Barbosa F, Young RM, Concha ML, Allende ML, et al. Early stages of zebrafish eye formation require the coordinated activity of Wnt11, Fz5, and the Wnt/beta-catenin pathway. Neuron. 2005;47:43–56. doi: 10.1016/j.neuron.2005.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heisenberg CP, Houart C, Take-Uchi M, Rauch GJ, Young N, et al. A mutation in the Gsk3-binding domain of zebrafish Masterblind/Axin1 leads to a fate transformation of telencephalon and eyes to diencephalon. Genes Dev. 2001;15:1427–1434. doi: 10.1101/gad.194301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bmp2b expression was rescued in foxo3b-knockdown embryos co-injected with foxo3b mismatch mRNA. Animal views, shield stage.
(TIF)
The partial primers used for cloning probes.
(DOC)