Abstract
The widespread presence of plasmid-mediated quinolone resistance determinants, particularly qnr genes, has become a current issue. By protecting DNA-gyrase from quinolones, Qnr proteins confer a low level quinolone resistance that is not sufficient to explain their emergence. Since Qnr proteins were hypothesized to act as DNA-binding protein regulators, qnr genes could have emerged by providing a selective advantage other than antibiotic resistance. We investigated host fitness of Escherichia coli isogenic strains after acquisition of the qnrA3 gene, inserted either alone onto a small plasmid (pBR322), or harbored on a large conjugative native plasmid, pHe96(qnrA3) found in a clinical isolate. The isogenic strains were derived from the susceptible E. coli CFT073, a virulent B2 group strain known to infect bladder and kidneys in a mouse model of pyelonephritis. In vitro experiments included growth analysis by automatic spectrophotometry and flow cytometry, and competitions with CFU enumeration. In vivo experiments included infection with each strain and pairwise competitions in absence of antimicrobial exposure. As controls for our experiments we used mutations known to reduce fitness (rpsL K42N mutation) or to enhance fitness (tetA deletion in pBR322). E. coli CFT073 transformed with pBRAM(PBR322-qnrA3) had significantly higher maximal OD than E. coli CFT073 transformed with pBR322 or pBR322ΔtetA, and in vivo competitions were more often won by the qnrA3 carrying strain (24 victories vs. 9 loss among 42 competitions, p = 0.001). In contrast, when pHe96(qnrA3) was introduced by conjugation in E. coli CFT073, it exerted a fitness cost shown by an impaired growth observed in vitro and in vivo and a majority of lost competitions (33/35, p<0.0001). In conclusion, qnrA3 acquisition enhanced bacterial fitness, which may explain qnr emergence and suggests a regulation role of qnr. However, fitness was reduced when qnrA3 was inserted onto multidrug-resistant plasmids and this can slow down its dissemination without antibiotic exposure.
Introduction
Fluoroquinolones are antibacterial drugs that bind to type II topoisomerases (DNA gyrase and topoisomerase IV) and inhibit DNA re-ligation after enzyme cut [1], [2]. These drugs are very useful, especially for treatment of urinary tract infections due to Enterobacteriaceae [1]. Fluoroquinolone resistance rate has increased much for the last years and is mostly due to their large use [3], [4]. Classical mechanisms of resistance are chromosomal mutations in the genes encoding the quinolone targets or causing enhanced efflux [1], [5]. More recently, plasmid-mediated resistance determinants have been reported encoding for different proteins: the Qnr proteins which belong to the pentapeptide repeat family (PRP) [6], [7], the acetyltransferase AAC(6′)-Ib-cr [8] and the QepA active efflux pump [9]. Enterobacteriaceae with plasmid-mediated quinolone resistance (PMQR) due to qnr genes have been worldwide described with an increase in their prevalence [10]. This rapid widespread is surprising because the acquisition of a qnr gene only confers a low-level resistance to fluoroquinolones [6], [11], [12]. Although this low-level resistance can be clinically relevant and probably contributed to qnr dissemination [13], [14], this does not fully explain the emergence of the qnr genes.
In clinical strains, qnr genes were essentially found on multi-drug resistance (MDR) plasmids, but chromosomal qnr genes have been also described in environmental bacterial species that are the supposed reservoir of these genes [15]–[18]. The native function of Qnr proteins still remains unknown [10]. They bind to type II topoisomerases and thus protect them from quinolone binding and action [7], [19], [20]. Functional and crystallography analyses of PRPs closed to Qnr are in favor of a role of Qnr proteins in topoisomerase regulation [21]–[25].
Most antibiotic resistance mechanisms, particularly fluoroquinolone resistance mutations, are associated with a fitness cost [26]–[29]. However, fitness cost of horizontal transferable resistance genes is often compensated by the regulation of transcription factors encoded by other genes harbored onto the same plasmid [30]. Nonetheless, interplay between resistance and fitness are not always concordant, and bacteria can reverse the cost induced by resistance acquisition. Several mutations could provide both enhanced fitness and increased resistance [29], [31]. Emergence of resistance can be driven by Darwinian selection for improved fitness, and not only by the antibiotic use [26]. We hypothesized that Qnr proteins have an effect on bacterial growth and fitness, which may have contributed to the emergence of qnr genes in commensal bacteria.
The aim of our study was to evaluate the impact of the qnr gene acquisition on bacterial fitness. Therefore we compared the fitness of isogenic strains of Escherichia coli with and without the qnrA3 gene, whether alone onto a small plasmid or carried onto a large conjugative multi-drug-resistant native plasmid. Growth and competitive performances were studied in vitro and in vivo using a mouse model of pyelonephritis.
Results
Description of the isogenic systems expressing qnr or not
Two systems of isogenic strains were derived from E. coli CFT073, a virulent strain belonging to the phylogenetic group B2 and whose genome has been sequenced [32]. This strain was originally used to set the murine model of pyelonephritis used in this study [33]. We selected a streptomycin resistant mutant (SmR) of E. coli CFT073 in order to have a resistance marker for the recipient strain after acquisition of the plasmid pHe96, which is a multidrug resistant plasmid not mediating streptomycin resistance. This mutant was selected using 160 µg/ml streptomycin at a proportion of ca.10−9 and harbored a rpsL K42N mutation which is consistent with its high level of resistance (MIC>512 µg/ml) and the stability of this resistance. Although pHe96 contains an ant3″-I gene known to confer streptomycin resistance [34], this gene is truncated and we confirmed that pHe96 does not confer streptomycin resistance by transferring pHe96 into E. coli J53. The MIC of streptomycin was 4 mg/l for this transconjugant and was stable.
The first isogenic system included five strains: E. coli CFT073, E. coli CFT073 (pBR322) and E. coli CFT073 transformed with three other plasmids derived from pBR322 and described in Figure S1: pBRΔtetA where the tetracycline resistance gene (tetA) was deleted, pBRAM1 where the qnrA3 gene was cloned including the 24-bp DNA motif upstream from qnrA3, and pBRAM2 where qnrA3 was cloned including the 233-bp DNA motif upstream. In both pBRAM1 and pBRAM2, qnrA3 was inserted into pBR322 by inactivating the tetA gene. Minimal inhibitory concentrations (MIC) of quinolones performed on the five strains showed that qnrA3 expressed quinolone resistance equally (Table 1) with an increase of 4-, 8-, 10- and 16-fold for nalidixic acid, ofloxacin, ciprofloxacin and norfloxacin, respectively.
Table 1. Minimal Inhibitory Concentrations (MIC) of quinolones against the strains of the two isogenic systems derived from E. coli CFT073.
| E. coli strains | MICa (µg/ml) | |||||
| NAL | NOR | OFX | CIP | AMK | TOB | |
| E. coli CFT073 | 2 | 0.064 | 0.094 | 0.012 | 1.5 | 0.75 |
| E. coli CFT073(pBR322) | 2 | 0.064 | 0.094 | 0.012 | 1.5 | 0.5 |
| E. coli CFT073(pBRΔtetA) | 2 | 0.064 | 0.094 | 0.012 | 1.5 | 0.5 |
| E. coli CFT073(pBRAM1) | 8 | 1 | 0.75 | 0.125 | 1 | 0.5 |
| E. coli CFT073(pBRAM2) | 8 | 1 | 0.75 | 0.125 | 1.5 | 0.5 |
| E. coli CFT073-SmR | 2 | 0.064 | 0.094 | 0.012 | 1.5 | 0.75 |
| E. coli CFT073-SmR(pHe96) | 6 | 3 | 0.75 | 0.75 | 48 | 32 |
| E. coli CFT073-SmR(pHe96) “R42” | 6 | 3 | 0.75 | 0.75 | 48 | 32 |
Minimal inhibitory concentrations measured by E-test for quinolones and aminoglycosides. NAL, nalidixic acid; NOR, norfloxacin; OFX, ofloxacin; CIP, ciprofloxacin; AMK, amikacin; TOB, tobramycin.
The second isogenic system included three strains: E. coli CFT073-SmR, E. coli CFT073-SmR(pHe96), and a variant of this transconjugant named E. coli CFT073-SmR(pHe96) “R42”, obtained after one passage in the mouse and which showed improved growth in vitro and in vivo and higher plasmid stability (see below). The evolved variant “R42” had the same phenotype for antibiotic resistance than the original strain CFT073-SmR(pHe96) including for streptomycin resistance. The acquisition of pHe96 conferred, as described previously [16], a 62- and 50-fold increase in the MIC of ciprofloxacin and norfloxacin (Table 1), respectively, because this plasmid harbored the aac6′-Ib-cr gene in addition to qnrA3 [16]. In contrast, ofloxacin and nalidixic acid MICs were the same for the transconjugants CFT073-SmR(pHe96) and for E. coli CFT073(pBRAM1) and E. coli CFT073(pBRAM2) confirming that aac6′-Ib-cr has no effect on these quinolones, and showing that the expression of qnrA3 was similar whether it was harbored on the small plasmid derived from pBR322 or on the large clinical plasmid from which qnrA3 originated [11], [16]. The “R42” variant showed similar sensitivity to quinolones as it parental strain.
Clinical E. coli isolates carrying a conjugative multidrug resistant plasmid harboring a qnr gene were also tested in the in vitro experiments along with the strain E. coli J53, a K-12 derivative used as a recipient strain for conjugation [35], [36]. Description of the strains, their qnr allele and the quinolone resistance conferred (increase in quinolone MICs observed for the transconjugants) was done previously [11], [13], [35], [37]. E. coli J53 transconjugants harboring qnr-positive plasmids were studied in similar in vitro experiments as were the two isogenic systems based on E. coli CFT073 (Table S1).
E. coli CFT073-SmR was used as a negative control for host fitness since it has been shown that streptomycin resistance, and especially the rpsL mutation K42N, had a fitness cost [26], [38], [39]. E. coli CFT073(pBRΔtetA) was used as a positive control with regard to E. coli CFT073(pBR322) since the tetA gene has been shown to have a cost for growth when carried by pBR322 and constitutively expressed [40], [41].
In vitro growth capacity of strains harboring qnrA3
In vitro growth capacity was measured by automated spectrophotometry for the calculation of the maximal growth rate, the doubling time and the maximal optical density, and by flow cytometry for the size of bacterial cells.
Growth parameters of E. coli CFT073 and its four isogenic strains harboring the plasmid pBR322 or one of its derivates (pBRΔtetA, pBRAM1 and pBRAM2) are shown in Table 2. Maximal OD was significantly higher for all strains harboring a qnrA3 carrying plasmid. This gain was not due to the inactivation of tetA, since the increase in maximal OD was significantly higher for E. coli CFT073 transconjugants harboring pBRAM1 or pBRAM2 (acquisition of qnrA3 at the place of tetA) than for the transconjugant harboring pBRΔtetA. No significant difference was seen in the doubling time between strains where qnrA3 was present (E. coli CFT073 transformed with pBRAM1 or pBRAM2) and those where qnrA3 was absent (E. coli CFT073 transformed with pBRΔtetA or pBR322).
Table 2. In vitro growth parameters for the strains of the first isogenic system based on E. coli CFT073 carrying pBR322 derivatives harboring qnrA3 or not.
| E. coli strains | Maximal growth rate in log(OD)/h. | Doubling time in min | Maximal OD | |||
| mean | p a | mean | p a | mean | p a | |
| CFT073(pBRΔtetA) | 0.96 (+/−0.02) | - | 18.9 (+/−0.3) | - | 1.07 (+/−0.02) | - |
| CFT073(pBR322) | 0.94 (+/−0.02) | 0.21 | 19.2(+/−0.3) | 0.21 | 1.05 (+/−0.06) | 0.6 |
| CFT073(pBRAM1) | 0.94 (+/−0.01) | 0.17 | 19.2 (+/−0.3) | 0.17 | 1.11 (+/−0.02) | 0.005 |
| CFT073(pBRAM2) | 0.94 (+/−0.02) | 0.13 | 19.2 (+/−0.5) | 0.13 | 1.10 (+/−0.02) | 0.01 |
Data obtained from automatic OD measurements (n = 15) in Trypticase Soy Broth and expressed as mean and confidence interval 95%.
The comparison was performed with a Wilcoxon test between each strains and the corresponding control qnr-negative strain CFT073(pBRΔtetA). p<0.05 was considered significant.
Acquisition of the plasmid pHe96 by E. coli CFT073-SmR was responsible for a significant lengthening of doubling time and decrease in maximal OD (Table 3), at the opposite of what was shown above for pBRAM plasmids. Similar results were obtained in brain heart infusion and minimal media (data not shown). The “R42” variant of E. coli CFT073-SmR(pHe96) showed significantly better growth parameters than the original transconjugant strain but stayed below the control strain E. coli CFT073-SmR. Comparing results between E. coli CFT073-SmR with its parental strain E. coli CFT073 confirmed that the streptomycin resistance due to the rpsL K42N mutation [38] was associated with a decrease in maximal growth rate (0.95 logOD/h+/−0.02 for E. coli CT073 vs. 0.7 logOD/h+/−0.01 for E. coli CFT073-SmR) and increase in the doubling time (18.9 h+/−0.74 for E. coli CFT073 vs. 25.9 h+/−0.5 for E. coli CFT073-SmR). Overall, the decrease in growth capacity conferred by the acquisition of pHe96 seemed to reduce fitness less than the rpsL mutation did.
Table 3. In vitro growth parameters for the strains of the second isogenic system based on acquisition of the multidrug resistant qnrA3-positive plasmid pHe96.
| E. coli strains | Maximal growth rate in log(OD)/h. | Doubling time in min | Maximal OD | |||
| mean | p a | mean | mean | p a | mean | |
| CFT073-SmR | 0.70 (+/−0.01) | - | 25.9 (+/−0.5) | - | 1.08 (+/−0.01) | - |
| CFT073-SmR(pHe96) | 0.67 (+/−0.02) | 0.02 | 27.1 (+/−0.9) | 0.02 | 0.94 (+/−0.03) | <0.0001 |
| CFT073-SmR(pHe96) R42 | 0.70 (+/−0.01) | 0.8 | 26.1 (+/−0.8) | 0.8b | 0.97 (+/−0.02) | <0.0001 |
The comparison was performed with a Wilcoxon test between each strains and the corresponding control qnr-negative strain CFT073-SmR. p<0.05 was considered significant.
The difference between the “R42” variant and E. coli CFT073-SmR (pHe96) was significant (p = 0.04).
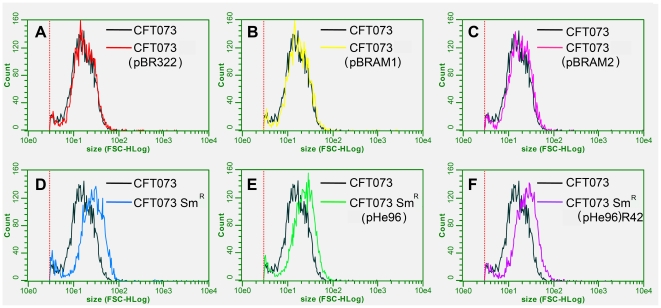
Results (arbitrary unit with 95% CI) of flow cytometry are presented in the Figure 1. Sizes measured by flow cytometry were 20.6 [18.1–23.1], 19.1 [18–20.2] 19.9 [19–20.8], 20.6 [20–21.2] for the E. coli CFT073 wild type strain and its transformants with pBR322 (tet+, qnr−), with pBRAM1 (tet−, qnr+), and with pBRAM2 (tet−, qnr+), respectively. For E. coli CFT073-SmR, its transconjugant with pHe96 and the variant isolate R42 of this transconjugant, sizes were 32.8 [30.8–34.8], 28.9 [26.8–31], and 30.1 [27.9–32.3], respectively, which were all 34.5% higher than those of E. coli CFT073. This showed that the acquisition of qnrA3 onto pBRAM1 or pBRAM2 or onto pHe96 did not change significantly the size of bacteria, contrarily to the mutation of the recipient strain E. coli CFT073-SmR, which was responsible for the increase in bacterial cell size.
Figure 1. Cell size measured by flow cytometry for the strains of the two isogenic systems.
Each graphic compares size measurement for bacterial cell populations consisting of the reference susceptible strain E. coli CFT073 (in black) and of the tested strain : E. coli CFT073(pBR322) (qnr-negative) in red (part A), E. coli CFT073(pBRAM1) (qnrA3-positive) in yellow (part B), E. coli CFT073(pBRAM2) (qnrA3-positive) in pink (part C), E. coli CFT073-SmR (qnr-negative) in blue (part D), E. coli CFT073-SmR(pHe96) (qnrA3-positive) in green (part E) and its variant R42 in purple (part F). Measurements were made separately, not in a competitive assay.
Enhanced fitness after acquisition of qnrA3 onto pBR322 derived plasmids
In vitro competitive assays were run six times by cultivating E. coli CFT073(pBRAM1) (qnrA3+) or E. coli CFT073(pBRAM2) (qnrA3+) with E. coli CFT073(pBR322) (tetA+) or E. coli CFT073 in a 1∶1 ratio. Population increase was measured by CFU counting. Relative Fitness was calculated as the ratio of the increase of each population (see material and methods for details) [42]–[45]. Comparing with E. coli CFT073(pBR322), estimated Relative Fitness (RF) was close to 1 (1.03+/−0.19), meaning that none of the two competing strains had a selective advantage upon the other in vitro. Comparing with E. coli CFT073, RF was estimated at 1.12+/−0.2, showing that the entire plasmid had no significant biological cost. The mean plasmid loss was less than 3% after 30 generations for all the strains harboring pBR322-derivatives.
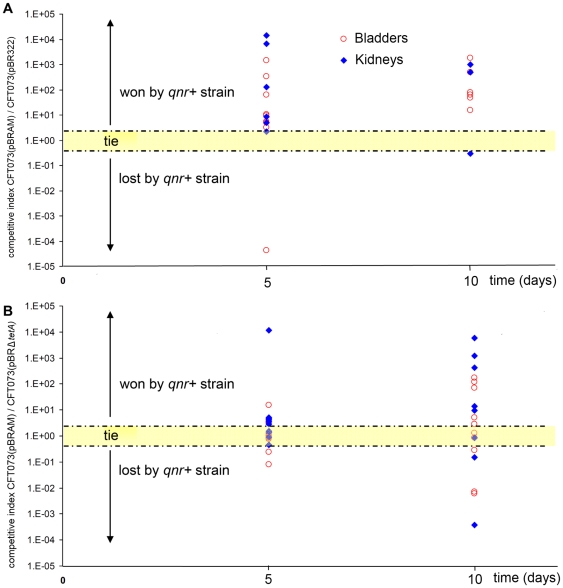
We performed in vivo experiments with the strain E. coli CFT073(pBRAM2) rather than with E. coli CFT073(pBRAM1) in order to be closer to clinical conditions (233-bp vs. 24-bp upstream sequence). In competitive infections (Figure 2A), E. coli CFT073(pBRAM2) (qnrA3+) had a significant advantage upon E. coli CFT073(pBR322) (tetA+) with 22 organs/25 where it had takeover versus only 2/25 where it had lost competition (p<0.0001). This selective advantage was confirmed when competitions were run against E. coli CFT073(pBRΔtetA) (qnr−) (Figure 2B). Indeed among 37 competitions, 24 were won by the qnrA3-positive strain, 4 were judged as tie because the ratio of the two populations was 1 (+/−0.2) and 9 were lost (p = 0.01). At day 10, a median competitive index was calculated for qnr-positive strains as described previously [29]. It was 1.27+/−0.16 in bladders and 13.9+/−7.2 in kidneys.
Figure 2. Enhanced fitness observed in competitive infections for E. coli CFT073 after qnrA3 acquisition onto pBR322.
Each symbol represents the bacterial ratio (number of CFU for the qnr-positive strain/number of CFU for the qnr-negative isogenic strain) measured in organs (blue diamond = kidneys, red circle = bladder) collected five and ten days after inoculation of a 1∶1 mix of the two strains. When the ratio was equal to 1+/−0.2, it was considered as tie. Part A: competitions experiments opposing E. coli CFT073(pBR322) (qnr−, tetA+) and E. coli CFT073(pBRAM2) (qnrA3+, tetA−). Fifteen mice were inoculated, 15 bladders and 10 pairs of kidneys were efficiently infected. Competition was won 22 times by E. coli CFT073(pBRAM2) (qnrA3+, tetA−), was lost 2 times, and one was tie (p<0.0001). Part B: competitions opposing E. coli CFT073(pBRΔtetA) (qnr−, tet−) and E. coli CFT073(pBRAM2) (qnrA3+, tet−). Twenty-three mice were inoculated, 20 bladders and 17 pairs of kidneys were efficiently infected. Competition was won 24 times by E. coli CFT073(pBRAM2) (qnrA3+, tetA−), was lost 9 times, and 6 was tie (p<0.0001).
Stability of pBR322-derived plasmids in in vitro and in vivo experiments
In vitro, no plasmid loss was observed after daily culture of E. coli CFT073(pBR322), E. coli CFT073(pBRAM2) and E. coli CFT073(pBRΔtetA). In vivo, the mean loss of the plasmids pBR322 and pBRAM2 in single strain infections (data not shown) was 47% and 67%, respectively, which was not statistically different. In the competitive experiments, the global plasmid loss was higher when we used E. coli CFT073(pBR322) as a control (37% at day 5 and 80% at day 10) than when we used E. coli CFT073(pBRΔtetA) (3% at day 5 and 11% at day 10). This suggests that the high plasmid loss observed in the competition with tetA-bearing cells corresponded mainly to the loss of pBR322.
In the competitive assays where E. coli CFT073(pBRΔtetA) was used for control, among 45 organs studied, 4 (9%) organs showed only ampicillin-susceptible colonies corresponding to a plasmid loss of 100%. When E. coli CFT073(pBR322) was used as control, it was 10/35 (29%) organs showing only ampicillin-susceptible colonies. Although we excluded these organs in the final counting, it showed that pBR322-derived plasmids were more stable in vivo when they carried a qnrA3 gene that when they did not (p = 0.038), even considering the biological cost of tetA.
Fitness cost due to acquisition of conjugative multidrug resistance plasmids harboring qnr
Reduced growth capacity of E. coli after pHe96 acquisition suggested a fitness cost. This was confirmed by the results of the in vitro competitive assays (eight repeated experiments growing together E. coli CFT073-SmR and E. coli CFT073-SmR(pHe96)) where relative fitness index (RF) of E. coli CFT073-SmR(pHe96) was significantly less than 1 (0.67+/−0.17). The relative fitness of the “R42” variant was similar with an index of 0.68+/−0.14. Plasmid loss measured for E. coli CFT073-SmR(pHe96) was 65% after 30 generations. In contrast, plasmid loss was only 3% for its variant isolate R42 suggesting a compensatory mutation in the chromosome or the plasmid. To investigate whether the reduced growth observed with pHe96 exists for other qnr genes and other qnr-positive MDR plasmids, we studied the in vitro growth capacity of E. coli J53 and of its transconjugants carrying five different qnr-positive and MDR plasmids previously described [13], [35], [37]: pHm13 (qnrA1), pHm477 (qnrA1), pHe96 (qnrA3), pPS105 (qnrS1), and pU1696 (qnrB4). No difference was seen between growth parameters of the strain E. coli J53 and those of the five qnr-positive transconjugants (Table S1).
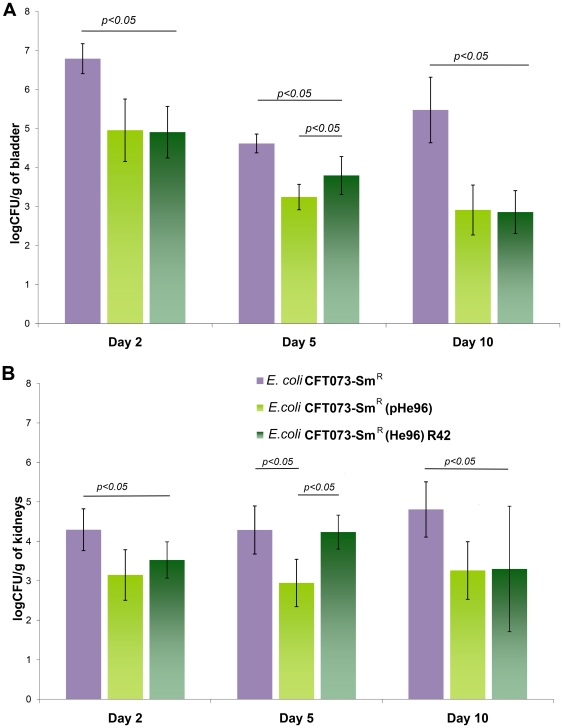
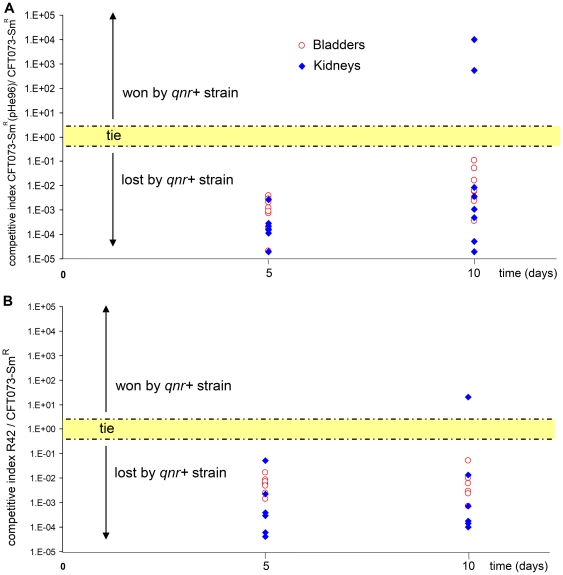
When mice were inoculated by the strain E. coli CFT073-SmR (pHe96), the bacterial density in bladders and kidneys was significantly lower than with the control strain, either at day 2, day 5 or day 10 (Figure 3). In addition, the number of mice where the strain failed to induce a persistent UTI was higher with the transconjugant than with the parental strain: 3 versus 0 for bladders and 7 versus 2 for kidneys (p = 0.003). The results were similar for the “R42” variant carrying the plasmid pHe96 but less than with the original transconjugant. This suggests that a compensatory mutation might have occurred in this variant when it was cultivated into the mouse for the first passage. In competitive experiments, E. coli CFT073-SmR(pHe96) lost the competition in 33 out of 35 organs (p<0.05) with regard to the control strain E. coli CFT073-SmR confirming the fitness cost of pHe96 acquisition (Figure 4A). A similar rate of lost experiments was found for the “R42” variant (Figure 4B). The mean loss of pHe96 was less than 20% for the strains E. coli CFT073-SmR(pHe96) and its variant “R42”, which was expected since these plasmids were coming from clinical isolates. The median value of the competitive index for the transconjugants carrying pHe96 was 0.0012+/−0.001. No competition has been run opposing the two pHe96 transconjugants because it was not possible to distinguish them.
Figure 3. Single strain urinary tract infections with the isogenic system of E. coli CFT073-SmR harboring or not the multidrug resistance plasmid pHe96 (qnrA3).
Part A: Bacterial density (log10CFU/g of tissue) in bladders collected two, five and ten days after inoculation by E. coli CFT073-SmR (qnr−, purple plot), E. coli CFT073-SmR(pHe96) (qnrA3+, light green plot) and E. coli CFT073-SmR(pHe96) R42 variant (qnrA3+, dark green plot). At day 2, bacterial density was 6.79+/−0.35, 4.95+/−0.8 (p<0.0001), and 4.9+/−0.65 (p<0.0001), respectively; at day 5, it was 4.61+/−0.25, 3.24+/−0.3 (p<0.0001) and 3.79+/−0.5 (p = 0.03), respectively; and at day 10, 5.47+/−0.8, 2.91+/−0.6 (p = 0.0004) and 2.85+/−0.5 (p = 0.001), respectively. At least 10 mice were studied per group. Part B : Bacterial density (log10CFU/g of tissue) in kidneys collected two, five and ten days after inoculation by E. coli CFT073-SmR (qnr−, purple plot), E. coli CFT073-SmR(pHe96) (qnrA3+, light green plot) and E. coli CFT073-SmR(pHe96) R42 variant (qnrA3+, dark green plot). Results were respectively 4.29+/−0.53, 3.14+/−0.64 (p = 0.005) and 3.53+/−0.46 (p = 0.053) at day 2; 4.29+/−0.61, 2.94+/−0.6 (p = 0.02), and 4.23+/−0.43 (p = 0.06) at day 5; and 4.81+/−0.6, 3.26+/0.63 (p = 0.012 and 3.3+/−1.59 (p = 0.04) at day 10. At least 10 mice were studied per group.
Figure 4. Reduced fitness observed after pHe96 acquisition in competitive infections in absence of antimicrobial exposure.
Each symbol represents the ratio (number of CFU for the qnr-positive strain/number of CFU for the qnr-negative isogenic strain) in organs (blue diamond = kidneys, red circle = bladder), collected five and ten days after inoculation of a 1∶1 mix of the two strains. Part A: competition experiments opposing E. coli CFT073-SmR (qnr−) and E. coli CFT073-SmR(pHe96) (qnrA3+). Twenty mice were inoculated, 19 bladders and 16 pairs of kidneys were efficiently infected. Competition was lost 33 times by E. coli CFT073-SmR(pHe96) (qnrA3+), and won only 2 times (p<0.0001). Part B: competition experiments opposing E. coli CFT073-SmR (qnr−) and E. coli CFT073-SmR(pHe96) variant “R42” (qnrA3+). The R42 variant was selected from kidneys that were infected by E. coli CFT073-SmR(pHe96). Twenty mice were inoculated, 18 bladders and 16 pairs of kidneys were efficiently infected. Competition was lost 33 times by the qnrA3-positive strain with only one won (p<0.0001).
We also tested, in competitive experiments against the E. coli CFT073 strain and using the same UTI mouse model, two qnr-positive uropathogenic clinical isolates: E. coli Hm13 (qnrA1) and E. coli PS105 (qnrS1) (Figure S2). In these non-isogenic competitions, the susceptible strain E. coli CFT073 won 53 of 57 competitions (kidneys and bladders) upon E. coli Hm13 (qnrA1) and 40 of 65 competitions upon E. coli PS105 (qnrS1) (p<0.001). This confirmed that the fitness cost attributed to the acquisition of pHe96 was not specific to this plasmid but was similarly observed with other qnr-positive MDR plasmids.
Discussion
The interplay between resistance and fitness is a challenging issue when increase of antibiotic resistance is observed worldwide [26]. Selection pressure due to the increase in antibiotic prescription explains the increase in resistance in most of the settings [3] and quinolone resistance was so far associated to a fitness cost when it was due to mutations either in the topoisomerase genes or in the efflux operons [27]–[29], [45]–[49]. The emergence of plasmid-mediated resistance determinants questioned about links between acquisition of resistance and selective advantage. Although plasmid acquisition brings a fitness cost, the stability of native plasmids varied according to the host and the presence of drug addiction systems may compensate this plasmid cost [50], [51]. For some antibiotic resistance determinants described recently, such as CTX-M beta-lactamases [52], [53], we know that they have been transferred from environmental bacteria harboring the resistance genes as chromosomal-borne, to human commensal bacteria, mainly E. coli, through mobile elements such as plasmids [54]. For quinolone resistance, such transfer may have occurred since similar qnr genes are chromosome-borne in environmental bacteria [18]. However, the persistence of qnr genes on plasmids is not fully explained by the quinolone selective pressure since there are numerous other effective mechanisms to obtain quinolone resistance such as stepwise chromosomal mutations in the target genes [55] and in the several efflux systems present in E. coli [56]. We hypothesized that qnr genes confer a selective advantage outside the quinolone exposure.
To study the impact on fitness of qnr acquisition, we compared in vitro growth curves, in vitro pairwise competition, and in vivo single culture and pairwise competition, which are usual methods found in literature [26], [28], [29], [42]–[45], [57]. Pairwise competitions are usually more sensitive than single cultures to reveal a fitness change, and in vivo assays are more relevant than in vitro assays since the complex growth environment is closer to reality [26]. This justifies the conclusions we drew on qnr impact on fitness, which were based on the results of in vivo competitions assays. We chose the mouse model of pyelonephritis for in vivo experiments because it was well validated and recently used for studying the interplay in fluoroquinolone resistance mutations and bacterial fitness [29]. Marcusson and colleagues showed that although the first quinolone resistance mutations (mostly gyrA) had fitness cost, latest compensatory parC mutations could provide increase of both resistance and fitness, suggesting that a higher level of resistance could be selected in absence of antimicrobial exposure [29].
Our findings, obtained using an isogenic system in E. coli where the qnr gene was cloned onto a simple replicative plasmid such as pBR322, showed that qnr acquisition enhanced the bacterial fitness in vitro and in vivo. Indeed when the only difference between two E. coli strains was the presence of qnr with its flanking region, the strain that acquired the qnr gene took over the susceptible strain in absence of quinolone exposure. Several studies [13], [14] suggested that the low level quinolone resistance is relevant when the host is exposed to quinolone in clinical situations. This could give a partial explanation for widespread but not for emergence. Several environment bacterial species, such as Shewanella algae or Vibrio splendidus, have been shown to harbor chromosomal qnr-like genes and are supposed to be the reservoir of qnrA3 and qnrS1, respectively [15]–[18]. The enhancement of fitness we observed after the acquisition by E. coli CFT073 of qnrA3 without antimicrobial exposure could explain how qnr had first emerged. This could have been the following scenario: the transfer from the chromosome of S. algae to a mobile element replicating in E. coli, or another member of the Enterobacteriaceae family, due to qnr giving a selective advantage. qnrD is the last qnr gene that was discovered in 2009 and it is carried onto a small non conjugative plasmid [58]. Although we did not test the fitness conferred by this small replicative qnr-positive plasmid, similar to pBR322, we hypothesized that the first step of the qnr emergence could have been through mobilization onto small plasmids, which may have resulted in an enhanced fitness as we showed in our in vivo experiments. In addition, there might be low concentrations of quinolones in the environment at some places which could have contributed to the resilience of these bacteria harboring qnr genes [59].
To investigate if their impact on fitness could also explain the widespread of qnr genes, we measured the fitness after acquisition of qnrA3 when included in a multidrug resistant plasmid as those usually observed in clinical qnr-positive isolates [10], [11], [36], [60]. When the gene was acquired with the whole multi drug resistant plasmid pHe96, the host fitness was lessened. This could be explained by the presence of another fitness impairing gene on the plasmid. However, this fitness cost was not specific of pHe96 since competitive experiments between E. coli CFT073 and two other E. coli clinical strains with qnr containing MDR plasmids showed the same fitness cost. Our experiments with an evolved clone of the E. coli CFT073(pHe96) transconjugant suggest that the fitness cost can be reduced by the host. Further explorations are needed to investigate the mechanism of this compensation. The difference in the impact on fitness when qnr is included into a large plasmid or into a small non conjugative plasmid could also be a consequence of a different level of qnr expression. However, MICs of quinolones were the same in the two plasmids.
Whatsoever, this would mean that fitness gain could contribute to the emergence of qnr-bearing plasmids among the bacterial communities living in the environment (soil, water) and their transfer to other bacteria such as those of the human commensal flora [54]. For the widespread observed in the following years, the fluoroquinolone resistance conferred and co selection with other resistance determinant (particularly expanded-spectrum-beta-lactamases, ESBL) would be the predominant selection factor [61], [62].
Native functions of Qnr proteins are still unknown. They bind type II topoisomerases even without quinolone binding to their targets [19], [20]. Although known Qnr proteins usually do not interfere with E. coli gyrase activity at the concentration they prevent fluoroquinolone inhibition [10], [19], [21], Qnr-like proteins inhibit catalytic activity of topoisomerases from their natural host [21], [63]. Crystallographic data on the structure of Qnr-like PRPs such as MfpA [22], EfsQnr [23] and AhQnr [24], revealed a right handed β helix structure that mimics a double-strain helix DNA. This could permit interaction with other DNA-linked proteins with a possible regulating function of the activity of these proteins. Mimicking DNA structure could have consequences on cell cycle and replication. Thus, this could explain the impact on fitness.
Limitations
Plasmid instability was significant for some of our plasmid constructions, even the native plasmid pHe96. However our analysis took into account this plasmid loss. Overall, artificial plasmids constructed from pBR322 were less stable than plasmids naturally isolated from clinical strains.
Materials and Methods
Ethics Statement
Animal experiments were performed in accordance with prevailing regulations regarding the care and use of laboratory animals by the European Commission. The experimental protocol was approved by the Departmental Direction of Veterinary Services in Paris, France (agreement N° A 75-18-05, 2005). The procedures are conformed to the Amsterdam protocol on animal protection and welfare and Directive 86/609/EEC on the protection of animals used for experimental and other scientific purposes.
All manipulation of animals were be carried out by qualified personnel. All protocols will be done in accordance with the local institutional review board (Comité d'Ethique en expérimentation animale, Ile de France, Paris Comité 3).
Bacterial strains and plasmids
E. coli CFT073 strain is a clinical isolate susceptible to all antibiotics [32], which was used previously to set the murine model of pyelonephritis [13], [33]. The plasmid pBR322 vector (4,361 bp) carries a bla gene encoding ampicillin resistance and a tetA gene encoding tetracycline resistance [64], [65]. The plasmid pHe96 was previously isolated in a Klebsiella pneumoniae cultivated from feces in a patient with cancer. The strain was described before [16] and the plasmid was sequenced for the 10 kb DNA fragment flanking qnrA3 (GenBank accession number EU495237). qnrA3 is a gene homologous to the chromosome-borne qnr allele found in S. algae strains [18] and was only described as plasmid-borne in the plasmid pHe96 [16].
E. coli J53 strain is a recipient strain widely used for conjugation with plasmid carrying qnr alleles [6], [36]. E. coli J53 transconjugants were obtained after conjugation with the following clinical strains: E. coli pHm477 (qnrA1) [11], K. pneumoniae pHe96 (qnrA3) [16], E. coli pPS105 (qnrS1) [13], E. coli pHm13 (qnr A1) [13] and E. coli pU1696 (qnrB4) [37]. These two latter strains, which were uropathogenic clinical isolates harboring multi drug resistance plasmids containing qnrA1 and qnrS1, respectively, were used for competitive experiments against susceptible E. coli CFT073.
qnrA3 cloning into pBR322
Cloning of the qnrA3 gene and its flanking region was done including either the 24 nucleotides (M24) upstream from qnrA3, resulting in the plasmid pBRAM1, or the 233 nucleotides (M233) upstream, resulting in the plasmid pBRAM2, and 29 nucleotides downstream for both. These upstream regions M24 and M233 are known to be conserved in qnrA containing plasmids [16] and are supposed to contain the promoter region [36], [66]. Amplification was performed using a sense primer containing an EcoRV restriction site (underlined) (5′-ATACTTCCGATATCCCCCTCCCTGATT-3′ for the fragment included in pBRAM1 and 5′-CTGACTGATATCCCCAAATCCAACACT-3′ for the fragment included in pBRAM2) and a SalI restriction site (underlined) in the reverse primer (GCAAGCGTCGACTCAAGTGATATTTGC for both DNA amplified fragments). Plasmid DNA of K. pneumoniae He96 was used as the template. PCR was performed in a 25 µl mixture including 10 µl forward and reverse primers, 5 µl DNA template and one Illustra™ pure Taq™ Ready-To-Go™ PCR bead (GE Healthcare, Buckinghamshire, UK). PCR was run using the following program : 94°C for 4 min; 94°C for 1 min; 55°C for 1 min; 72°C for 1 min for 30 cycles; and 72°C for 10 min. The amplified fragment was purified using Qiaquick® PCR purification kit (Qiagen®), digested with EcoRV and SalI (Invitrogen™), and then ligated into the tetracycline resistance tetA gene downstream of the promoter of pBR322 (Invitrogen™) previously digested with EcoRV and SalI. The ligation product was electroporated into competent E. coli CFT073. Transformants for E. coli CFT073 with pBRAM1 or pBRAM2 were selected on ampicillin (100 µg/ml)-containing Mueller-Hinton (MH) plates and replicated on tetracycline (20 µg/ml)-containing plates to identify inserts.
The control strains were E. coli CFT073(pBR322), where the plasmid pBR322 was electroporated into competent E. coli CFT073, and E. coli CFT073(pBRΔtetA) where the tetA gene in the vector pBR322 was inactivated as follows: pBR322 was digested with EcoRV and NruI, ligated without any insert, and the product was electroporated in competent E. coli CFT073. Susceptibility to tetracycline was tested to check tetA inactivation. Figure S1 summarizes in a scheme the methods used to obtain the pBR322 derivatives.
pHe96 containing strains and controls
A streptomycin-resistant mutant E. coli CFT073-SmR strain was selected in vitro from E. coli CFT073 by plating 109 bacteria onto MH agar containing streptomycin concentration from 10 to 160 µg/ml. The transconjugant E. coli CFT073-SmR(pHe96) was obtained after conjugation between E. coli CFT073-SmR and K. pneumoniae He96 [16] after 40 min of mating into MH broth as previously described [13]. After incubation, transconjugants were selected by plating the conjugation mixture on MH agar supplemented with ampicillin (100 µg/ml) plus streptomycin (100 µg/ml). PCR experiments (RAPD and PCR qnrA) were performed as already described to confirm the success of the conjugation [11], [67].
E. coli CFT073-SmR(pHe96) “R42” is a clone of E. coli CFT073-SmR(pHe96), which has been selected from kidneys of an infected mouse. This clone was studied because it caused an infection with a bacterial load in organs higher than with the other clones of E. coli CFT073-SmR(pHe96).
Growth experiments and antibiotic susceptibility testing
Growth curves of each strain in TS broth were drawn using automatic optical density measurements obtained by reader infinite® M200 (Tecan). After an overnight culture in TS broth at 37°C, bacterial suspensions were diluted in order to reach an Optical Density of 0.5, and then diluted 100 times. Microplates were inoculated with the obtained suspensions. Each combination of strain and medium was run in triplicate. The plates were incubated at 37°C for 24 hours and subject to shaking during 60 s every two minutes. Optical density (OD) at 600 nm was measured every 5 minutes during this 24-hours period. The log10 of each value of OD was calculated. To estimate the doubling time, a slope was calculated between every two consecutive values. After discarding artifacts, a mean of the six highest slopes was calculated, corresponding to the maximal growth rate. Doubling time was calculated as log10 2 divided by the maximal growth rate [57], [68]. Maximal OD increase was also calculated as the mean of the ten highest values of OD reached during stationary phase. This has been performed five times for each strain.
The Minimal Inhibitory Concentrations (MICs) of nalidixic acid, norfloxacin, ofloxacin and ciprofloxacin were determined for each constructed strain and control strain by Etest®, according to the instructions of the manufacturer. MICs of amikacin and tobramycin were also measured in order to evaluate the expression of aac(6′)-Ib-cr. Etest® were performed on MH agar plates and the plates were incubated 18 h at 37°C.
Plasmid stability
Since all the plasmids tested carried an ampicillin resistant determinant, plasmid stability of the transconjugants E. coli CFT073-SmR(pHe96), E. coli CFT073(pBRAM1), E. coli CFT073(pBRAM2), E. coli CFT073(pBR322) and E. coli CFT073(pBRΔtetA) was measured in vitro by daily subculture during ten days in antibiotic-free Trypticase Soy (TS) broth with plating onto TS agar containing 100 µg/ml ampicillin and antibiotic free TS agar plates. In vivo, at the end of each experiment, organs were also spread onto ampicillin-containing and antibiotic-free TS agar. Plasmid loss was calculated for each organ as the ratio ampicillin resistant CFU/total number of CFU.
Flow cytometry
In order to check that OD variations were not caused by a change of cell size, all the studied strains were also studied in flow cytometry as already described [69]. Flow cytometric measurements were performed on a Guava EasyCyte Plus flow cytometer (Millipore, St Quentin en Yvelines, France) equipped with a 488 nm laser. Overnight cultures were diluted to a cell density of ca. 105 CFU/ml. Cells were collected on the forward scatter with logarithmic amplifiers for 5,000 events. The experiment was replicated four times for each strain.
In vitro competitions experiments
Pairwise competition experiments were performed to evaluate the in vitro relative fitness of each qnr-positive strain compared to the qnr-negative control strain. We also performed competition with the wild type strain E.coli CFT073. Each strain of its isogenic system was incubated separately for 18 h at 37°C in TS broth. The cell densities of the suspensions were estimated by flow cytometry [70]. An aliquot containing 108 bacteria was taken from each suspension and mixed two by two in a ratio of 1∶1 to inoculate 10 ml of TS broth and then incubated for 24 h at 37°C for a competitive growth. The initial and the final densities of each competitive strain were estimated from CFU data by diluting and plating population samples onto TS agar with and without antibiotic. Tetracycline was used to distinguish E. coli CFT073(pBR322) from E. coli CFT073(pBRAM1) and E. coli CFT073(pBRAM2) in the first isogenic system Ampicillin was used to distinguish E. coli CFT073 from E. coli CFT073(pBRAM1) and E. coli CFT073(pBRAM2). In the second isogenic system, ampicillin was used to distinguish E. coli CFT073-SmR(pHe96) from E. coli CFT073-SmR. With these densities, the Malthusian parameter of each competitor could be calculated. The Relative Fitness (RF) was calculated as the ratio of the two Malthusian parameters [42]–[45] (RF = (logS1d1−logS1d0)/log (S2d1−logS2d0), where S1d1 and S1d0 are the CFU densities of the qnr+ strain respectively at the end and at the beginning of the competition experiment, and S2d1 and S2d0 their equivalent values for the control strain. A RF score >1 meant that the qnr+ strain had a selective advantage upon the control strain whereas a score <1 represented a fitness cost for qnr acquisition.
Mouse model of urinary tract infection
The ascending unobstructed mouse model of UTI was used as previously described [13], [33]. Animal experiments were performed in accordance with prevailing regulations regarding the care and use of laboratory animals by the European Commission. The experimental protocol was approved by the Departmental Direction of Veterinary Services in Paris, France. Eight-week-old immunocompetent female CBA mice (weight, 20 to 23 g) were used. Inocula of different strains were obtained by overnight incubation in BHI broth, washing of the cells by centrifugation at 15,000× g for 15 min, and resuspension in TS broth to a final inoculum of 1010 CFU/ml. Pyelonephritis was induced during general anaesthesia (with an intraperitoneal administration of Ketamin 150 mg/kg and Xylazin 0.5 mg/kg) by injecting 50 µl (i.e., 5×108 CFU) into the bladder through a urethral catheter.
In vivo growth experiments: single-strain infections
In order to evaluate their potentials to perform persistent UTI and to reach a high bacterial density in bladder and kidneys, mice were inoculated separately by each of the following strains: E. coli CFT073-SmR, E. coli CFT073-SmR(pHe96) and E. coli CFT073-SmR(pHe96) “R42”. No antimicrobial was administrated. Mice were sacrificed at two, five and ten days after inoculation with at least 8 mice per group, and bladders and kidneys were aseptically taken out, weighed and homogenized in 1 ml of saline solution. Bacterial densities were estimated from CFU data by diluting and plating organs samples onto TS agar and were expressed as the number of CFU/g of tissue. The growth rate of each strain was estimated by the bacterial densities in bladder and kidneys and could be compared to the control strain. Plasmid loss on days 5 and 10 was assessed for strains harboring plasmid pHe96 by comparing the number of CFU growing onto MH plates containing or not ampicillin (100 µg/ml).
In vivo fitness measurements: competitive infections
The in vivo selective value of the qnr-positive strain compared to the control qnr-negative strain was determined by competitive experiments between the two isogenic strains using the same mouse model of UTI. Inocula were prepared the same way that for single-strain infection experiments, except that before mixing the two suspensions, bacterial density was measured by flow cytometry. Proportions of each suspension were adjusted so that inoculum contained the two strains in a 1∶1 ratio and that cell density was at least 1010 bacteria/ml. Inoculum was spread on antibiotic containing plate to count the initial ratio. At least 20 mice were inoculated for each competitive experiment, and were sacrificed at five or ten days after inoculation. After removing and homogenizing bladders and kidneys, they were spread onto TS agar plates without antibiotics and containing the appropriate antibiotic to select one of the two competitive strains among the population. Tetracycline (20 µg/ml) was used to distinguish between strains harboring pBR322 or pBRAM2, and ciprofloxacin (0.05 µg/ml) to distinguish strains harboring pBRAM2 or pBRΔtetA. To count the number of pBRΔtetA-bearing cells, we subtracted the number of cells growing onto ciprofloxacin-containing plates from the number of cells growing onto ampicillin-containing plates. Ampicillin (100 µg/ml) was used to distinguish E. coli CFT073-SmR from its transconjugants, E. coli CFT073-SmR(pHe96) and E. coli CFT073-SmR(pHe96) R42. For competitions between strains carrying pBR322 or its derivatives, bladders and kidneys were also spread onto ampicillin-containing plates, in order to evaluate plasmid loss. A competitive index was calculated for each organ as the ratio of the number of qnr-positive cells/number of qnr-negative cells, corrected by the initial ratio. Organs where no CFU grew were excluded.
Statistical analysis
Results were expressed as mean and 95% confidence interval for continuous variables. Continuous variables were compared by nonparametric (Wilcoxon) tests. Discontinuous variables were compared by Fisher's exact test. A p value less than 0.05 was considered significant. Analysis was performed using the online free software Biostatgv (http://www.u707.jussieu.fr/biostatgv).
Supporting Information
Scheme of qnrA3 cloning from its native plasmid pHe96 into pBR322 and resulting qnr -positive (pBRAM1 and pBRAM2) and control plasmids (pBRΔtetA). M24 and M233 are the fragments of 24- and 233-bp upstream from qnrA3 and described in pHe96 [16]. Primers (see text for sequences) Fw2, Fw1 and R contain the EcoRV and SalI restrictions sites and were used to amplify the qnrA3 DNA fragments.
(TIF)
Reduced fitness observed for clinical isolates of E. coli harboring qnr -positive multi drug resistance plasmids in competitive infections with E. coli CFT073 without antimicrobial exposure. Each symbol represents the ratio (number of CFU for the qnr-positive strain/number of CFU for the qnr-negative isogenic strain) in organs (blue diamond = kidneys, red circle = bladder), collected five to forty days after inoculation of a 1∶1 mix of the two strains. Part A: competition experiments opposing E. coli CFT073 (qnr−) and E. coli Hm13 (qnrA1+). Thirty mice were inoculated, 27 bladders and 30 pairs of kidneys were efficiently infected. Fifty-three competitions were lost by the qnr+ strain and 2 only were won (p<0.0001). Part B: competition experiments opposing E. coli CFT073 (qnr−) and E. coli PS105 (qnrS1+). Thirty-three mice were inoculated, 33 bladders and 30 pairs of kidneys were efficiently infected. Forty-one competitions were lost by the qnr+ strain, and 17 were won (p<0.0001).
(TIF)
In vitro growth parameters of E. coli J53 and its transconjugants with qnr-positive multidrug resistant plasmids.
(DOC)
Acknowledgments
We thank Sandra Palmier, Bruno Harmenil, Sara Dion, Raphaël Lepeule, Géraldine Marcadé, and Jean-Luc Donay for their help in the experiments.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The work was supported with an annual grant from the University Paris Diderot for the team EA3964. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis. 2000;31(Suppl 2):S24–28. doi: 10.1086/314056. [DOI] [PubMed] [Google Scholar]
- 2.Heisig P. Type II topoisomerases–inhibitors, repair mechanisms and mutations. Mutagenesis. 2009;24:465–469. doi: 10.1093/mutage/gep035. [DOI] [PubMed] [Google Scholar]
- 3.Goossens H. Antibiotic consumption and link to resistance. Clin Microbiol Infect. 2009;15(Suppl 3):12–15. doi: 10.1111/j.1469-0691.2009.02725.x. [DOI] [PubMed] [Google Scholar]
- 4.Garau J, Xercavins M, Rodriguez-Carballeira M, Gomez-Vera JR, Coll I, et al. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob Agents Chemother. 1999;43:2736–2741. doi: 10.1128/aac.43.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41(Suppl 2):S120–126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Martinez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 7.Tran JH, Jacoby GA. Mechanism of plasmid-mediated quinolone resistance. Proc Natl Acad Sci U S A. 2002;99:5638–5642. doi: 10.1073/pnas.082092899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med. 2006;12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 9.Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 2007;51:3354–3360. doi: 10.1128/AAC.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009;22:664–689. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cambau E, Lascols C, Sougakoff W, Bebear C, Bonnet R, et al. Occurrence of qnrA-positive clinical isolates in French teaching hospitals during 2002–2005. Clin Microbiol Infect. 2006;12:1013–1020. doi: 10.1111/j.1469-0691.2006.01529.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang M, Sahm DF, Jacoby GA, Zhang Y, Hooper DC. Activities of newer quinolones against Escherichia coli and Klebsiella pneumoniae containing the plasmid-mediated quinolone resistance determinant qnr. Antimicrob Agents Chemother. 2004;48:1400–1401. doi: 10.1128/AAC.48.4.1400-1401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allou N, Cambau E, Massias L, Chau F, Fantin B. Impact of low-level resistance to fluoroquinolones due to qnrA1 and qnrS1 genes or a gyrA mutation on ciprofloxacin bactericidal activity in a murine model of Escherichia coli urinary tract infection. Antimicrob Agents Chemother. 2009;53:4292–4297. doi: 10.1128/AAC.01664-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Martinez JM, Pichardo C, Garcia I, Pachon-Ibanez ME, Docobo-Perez F, et al. Activity of ciprofloxacin and levofloxacin in experimental pneumonia caused by Klebsiella pneumoniae deficient in porins, expressing active efflux and producing QnrA1. Clin Microbiol Infect. 2008;14:691–697. doi: 10.1111/j.1469-0691.2008.02020.x. [DOI] [PubMed] [Google Scholar]
- 15.Cattoir V, Poirel L, Mazel D, Soussy CJ, Nordmann P. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob Agents Chemother. 2007;51:2650–2651. doi: 10.1128/AAC.00070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lascols C, Podglajen I, Verdet C, Gautier V, Gutmann L, et al. A plasmid-borne Shewanella algae Gene, qnrA3, and its possible transfer in vivo between Kluyvera ascorbata and Klebsiella pneumoniae. J Bacteriol. 2008;190:5217–5223. doi: 10.1128/JB.00243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Liard A, Rodriguez-Martinez JM, Nordmann P. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J Antimicrob Chemother. 2005;56:1118–1121. doi: 10.1093/jac/dki371. [DOI] [PubMed] [Google Scholar]
- 18.Poirel L, Rodriguez-Martinez JM, Mammeri H, Liard A, Nordmann P. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob Agents Chemother. 2005;49:3523–3525. doi: 10.1128/AAC.49.8.3523-3525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother. 2005;49:118–125. doi: 10.1128/AAC.49.1.118-125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob Agents Chemother. 2005;49:3050–3052. doi: 10.1128/AAC.49.7.3050-3052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merens A, Matrat S, Aubry A, Lascols C, Jarlier V, et al. The pentapeptide repeat proteins MfpAMt and QnrB4 exhibit opposite effects on DNA gyrase catalytic reactions and on the ternary gyrase-DNA-quinolone complex. J Bacteriol. 2009;191:1587–1594. doi: 10.1128/JB.01205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, et al. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 23.Hegde SS, Vetting MW, Mitchenall LA, Maxwell A, Blanchard JS. Structural and biochemical analysis of the pentapeptide repeat protein EfsQnr, a potent DNA gyrase inhibitor. Antimicrob Agents Chemother. 2011;55:110–117. doi: 10.1128/AAC.01158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong X, Bromley EH, Oelschlaeger P, Woolfson DN, Spencer J. Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: conserved surface loops direct the activity of a Qnr protein from a Gram-negative bacterium. Nucleic Acids Res. 2011;39:3917–3927. doi: 10.1093/nar/gkq1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vetting MW, Hegde SS, Wang M, Jacoby GA, Hooper DC, et al. Structure of QnrB1, a plasmid-mediated fluoroquinolone resistance factor. J Biol Chem. 2011 doi: 10.1074/jbc.M111.226936. 10.1074/jbc.M111.226936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 27.Heisig P, Tschorny R. Characterization of fluoroquinolone-resistant mutants of Escherichia coli selected in vitro. Antimicrob Agents Chemother. 1994;38:1284–1291. doi: 10.1128/aac.38.6.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komp Lindgren P, Marcusson LL, Sandvang D, Frimodt-Moller N, Hughes D. Biological cost of single and multiple norfloxacin resistance mutations in Escherichia coli implicated in urinary tract infections. Antimicrob Agents Chemother. 2005;49:2343–2351. doi: 10.1128/AAC.49.6.2343-2351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcusson LL, Frimodt-Moller N, Hughes D. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 2009;5:e1000541. doi: 10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morosini MI, Ayala JA, Baquero F, Martinez JL, Blazquez J. Biological cost of AmpC production for Salmonella enterica serotype Typhimurium. Antimicrob Agents Chemother. 2000;44:3137–3143. doi: 10.1128/aac.44.11.3137-3143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo N, Pereira S, Sahin O, Lin J, Huang S, et al. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci U S A. 2005;102:541–546. doi: 10.1073/pnas.0408966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch RA, Burland V, Plunkett G, 3rd, Redford P, Roesch P, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labat F, Pradillon O, Garry L, Peuchmaur M, Fantin B, et al. Mutator phenotype confers advantage in Escherichia coli chronic urinary tract infection pathogenesis. FEMS Immunol Med Microbiol. 2005;44:317–321. doi: 10.1016/j.femsim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Shaw KJ, Rather PN, Hare RS, Miller GH. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cesaro A, Bettoni RR, Lascols C, Merens A, Soussy CJ, et al. Low selection of topoisomerase mutants from strains of Escherichia coli harbouring plasmid-borne qnr genes. J Antimicrob Chemother. 2008;61:1007–1015. doi: 10.1093/jac/dkn077. [DOI] [PubMed] [Google Scholar]
- 36.Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, et al. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother. 2003;47:2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liassine N, Zulueta-Rodriguez P, Corbel C, Lascols C, Soussy CJ, et al. First detection of plasmid-mediated quinolone resistance in the community setting and in hospitalized patients in Switzerland. J Antimicrob Chemother. 2008;62:1151–1152. doi: 10.1093/jac/dkn295. [DOI] [PubMed] [Google Scholar]
- 38.Schrag SJ, Perrot V. Reducing antibiotic resistance. Nature. 1996;381:120–121. doi: 10.1038/381120b0. [DOI] [PubMed] [Google Scholar]
- 39.Paulander W, Maisnier-Patin S, Andersson DI. The fitness cost of streptomycin resistance depends on rpsL mutation, carbon source and RpoS (sigmaS). Genetics. 2009;183:531SI–532SI. doi: 10.1534/genetics.109.106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SW, Edlin G. Expression of tetracycline resistance in pBR322 derivatives reduces the reproductive fitness of plasmid-containing Escherichia coli. Gene. 1985;39:173–180. doi: 10.1016/0378-1119(85)90311-7. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen TN, Phan QG, Duong LP, Bertrand KP, Lenski RE. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol Biol Evol. 1989;6:213–225. doi: 10.1093/oxfordjournals.molbev.a040545. [DOI] [PubMed] [Google Scholar]
- 42.Balsalobre L, de la Campa AG. Fitness of Streptococcus pneumoniae fluoroquinolone-resistant strains with topoisomerase IV recombinant genes. Antimicrob Agents Chemother. 2008;52:822–830. doi: 10.1128/AAC.00731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hurdle JG, O'Neill AJ, Chopra I. The isoleucyl-tRNA synthetase mutation V588F conferring mupirocin resistance in glycopeptide-intermediate Staphylococcus aureus is not associated with a significant fitness burden. J Antimicrob Chemother. 2004;53:102–104. doi: 10.1093/jac/dkh020. [DOI] [PubMed] [Google Scholar]
- 44.Petersen A, Aarestrup FM, Olsen JE. The in vitro fitness cost of antimicrobial resistance in Escherichia coli varies with the growth conditions. FEMS Microbiol Lett. 2009;299:53–59. doi: 10.1111/j.1574-6968.2009.01734.x. [DOI] [PubMed] [Google Scholar]
- 45.Rozen DE, McGee L, Levin BR, Klugman KP. Fitness costs of fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2007;51:412–416. doi: 10.1128/AAC.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bagel S, Hullen V, Wiedemann B, Heisig P. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob Agents Chemother. 1999;43:868–875. doi: 10.1128/aac.43.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bemer P, Corvec S, Guitton C, Giraudeau C, Le Gargasson G, et al. Biological cost of fluoroquinolone resistance in Escherichia coli implicated in polyclonal infection. Pathol Biol (Paris) 2007;55:288–291. doi: 10.1016/j.patbio.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Johnson CN, Briles DE, Benjamin WH, Jr, Hollingshead SK, Waites KB. Relative fitness of fluoroquinolone-resistant Streptococcus pneumoniae. Emerg Infect Dis. 2005;11:814–820. doi: 10.3201/eid1106.040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kugelberg E, Lofmark S, Wretlind B, Andersson DI. Reduction of the fitness burden of quinolone resistance in Pseudomonas aeruginosa. J Antimicrob Chemother. 2005;55:22–30. doi: 10.1093/jac/dkh505. [DOI] [PubMed] [Google Scholar]
- 50.De Gelder L, Ponciano JM, Joyce P, Top EM. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology. 2007;153:452–463. doi: 10.1099/mic.0.2006/001784-0. [DOI] [PubMed] [Google Scholar]
- 51.Mnif B, Vimont S, Boyd A, Bourit E, Picard B, et al. Molecular characterization of addiction systems of plasmids encoding extended-spectrum beta-lactamases in Escherichia coli. J Antimicrob Chemother. 2010;65:1599–1603. doi: 10.1093/jac/dkq181. [DOI] [PubMed] [Google Scholar]
- 52.Rossolini GM, D'Andrea MM, Mugnaioli C. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin Microbiol Infect. 2008;14(Suppl 1):33–41. doi: 10.1111/j.1469-0691.2007.01867.x. [DOI] [PubMed] [Google Scholar]
- 53.Jacoby GA, Munoz-Price LS. The new beta-lactamases. N Engl J Med. 2005;352:380–391. doi: 10.1056/NEJMra041359. [DOI] [PubMed] [Google Scholar]
- 54.Skippington E, Ragan MA. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol Rev. 2011;35 doi: 10.1111/j.1574-6976.2010.00261.x. 10.1111/j.1574-6976.2010.00261.x. [DOI] [PubMed] [Google Scholar]
- 55.Hooper D. Mechanisms of quinolone resistance. In: Hooper D, Rubinstein E, editors. Quinolone antimicrobial agents. Washington DC: ASM press; 2003. pp. 41–67. [Google Scholar]
- 56.Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 57.Begot CDI, Daudin JD, Labadie JC, Lebert A. Recommendations for calculating growth parameters by optical density measurements. Journal of Microbiological Methods. 1996:225–232. [Google Scholar]
- 58.Cavaco LM, Hasman H, Xia S, Aarestrup FM. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob Agents Chemother. 2009;53:603–608. doi: 10.1128/AAC.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamtam F, Mercier F, Le Bot B, Eurin J, Tuc Dinh Q, et al. Occurrence and fate of antibiotics in the Seine River in various hydrological conditions. Sci Total Environ. 2008;393:84–95. doi: 10.1016/j.scitotenv.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 60.Wang M, Sahm DF, Jacoby GA, Hooper DC. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob Agents Chemother. 2004;48:1295–1299. doi: 10.1128/AAC.48.4.1295-1299.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 62.Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother. 2006;50:2872–2874. doi: 10.1128/AAC.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arsene S, Leclercq R. Role of a qnr-like gene in the intrinsic resistance of Enterococcus faecalis to fluoroquinolones. Antimicrob Agents Chemother. 2007;51:3254–3258. doi: 10.1128/AAC.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, et al. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 65.Watson N. A new revision of the sequence of plasmid pBR322. Gene. 1988;70:399–403. doi: 10.1016/0378-1119(88)90212-0. [DOI] [PubMed] [Google Scholar]
- 66.Mammeri H, Van De Loo M, Poirel L, Martinez-Martinez L, Nordmann P. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob Agents Chemother. 2005;49:71–76. doi: 10.1128/AAC.49.1.71-76.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang G, Whittam TS, Berg CM, Berg DE. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21:5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warringer J, Blomberg A. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast. 2003;20:53–67. doi: 10.1002/yea.931. [DOI] [PubMed] [Google Scholar]
- 69.Chau F, Lefort A, Benadda S, Dubee V, Fantin B. Flow cytometry as a tool to determine the effects of cell wall-active antibiotics on vancomycin-susceptible and resistant Enteroroccus faecalis. Antimicrob Agents Chemother. 2011;55:395–398. doi: 10.1128/AAC.00970-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pinder AC, Purdy PW, Poulter SA, Clark DC. Validation of flow cytometry for rapid enumeration of bacterial concentrations in pure cultures. J Appl Bacteriol. 1990;69:92–100. doi: 10.1111/j.1365-2672.1990.tb02916.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scheme of qnrA3 cloning from its native plasmid pHe96 into pBR322 and resulting qnr -positive (pBRAM1 and pBRAM2) and control plasmids (pBRΔtetA). M24 and M233 are the fragments of 24- and 233-bp upstream from qnrA3 and described in pHe96 [16]. Primers (see text for sequences) Fw2, Fw1 and R contain the EcoRV and SalI restrictions sites and were used to amplify the qnrA3 DNA fragments.
(TIF)
Reduced fitness observed for clinical isolates of E. coli harboring qnr -positive multi drug resistance plasmids in competitive infections with E. coli CFT073 without antimicrobial exposure. Each symbol represents the ratio (number of CFU for the qnr-positive strain/number of CFU for the qnr-negative isogenic strain) in organs (blue diamond = kidneys, red circle = bladder), collected five to forty days after inoculation of a 1∶1 mix of the two strains. Part A: competition experiments opposing E. coli CFT073 (qnr−) and E. coli Hm13 (qnrA1+). Thirty mice were inoculated, 27 bladders and 30 pairs of kidneys were efficiently infected. Fifty-three competitions were lost by the qnr+ strain and 2 only were won (p<0.0001). Part B: competition experiments opposing E. coli CFT073 (qnr−) and E. coli PS105 (qnrS1+). Thirty-three mice were inoculated, 33 bladders and 30 pairs of kidneys were efficiently infected. Forty-one competitions were lost by the qnr+ strain, and 17 were won (p<0.0001).
(TIF)
In vitro growth parameters of E. coli J53 and its transconjugants with qnr-positive multidrug resistant plasmids.
(DOC)