Abstract
In the Rhodobacter (Rba.) species of photosynthetic purple bacteria, a single transmembrane α-helix, PufX, is found within the core complex, an essential photosynthetic macromolecular assembly that performs the absorption and the initial processing of light energy. Despite its structural simplicity, many unresolved questions surround PufX, the most important of which is its location within the photosynthetic core complex. One proposed placement of PufX is at the center of a core complex dimer, where two PufX helices associate in the membrane and form a homodimer. Inability for PufX of certain Rba. species to form a homodimer is thought to lead to monomeric core complexes. In the present study, we employ a combination of computational and experimental techniques to test the hypothesized homodimerization of PufX. We carry out a systematic investigation to measure the dimerization affinity of PufX from four Rba. species, Rba. blasticus, Rba. capsulatus, Rba. sphaeroides, and Rba. veldkampii, using a molecular dynamics-based free-energy method, as well as experimental TOXCAT assays. We found that the four PufX helices have substantially different dimerization affinities. Both computational and experimental techniques demonstrate that species with dimeric core complexes have PufX that can potentially form a homodimer, whereas the one species with monomeric core complexes has a PufX with little to no dimerization propensity. Our analysis of the helix-helix interface revealed a number of positions that may be important for PufX dimerization and the formation of a hydrogen bond network between these GxxxG-containing helices. Our results suggest that the different oligomerization states of core complexes in various Rba. species can be attributed, among other factors, to the different propensity of its PufX helix to homodimerize.
Introduction
Compared to algae and plants, bacterial photosynthesis, while similar in its chemical principles of energy conversion, is a lot simpler in the structure and organization of the associated protein-pigment assemblies. Nonetheless, there are many unknown features regarding the macromolecular arrangement of some of the most critical photosynthetic complexes, one example being the core complex of purple photosynthetic bacteria. The photosynthetic core complex is a combination of two major transmembrane (TM) protein-pigment complexes that carry out the initial steps of the photosynthetic process: light-harvesting complex 1 (LH1) and the reaction center (RC). In some species of purple bacteria, most notably the Rhodobacter (Rba.) genus, the core complex contains an additional TM protein that is largely α-helical, and is named PufX for Rhodobacters. Some Rhodobacter core complexes can form dimers,1,2 resulting in a large assembly with a dimension of approximately 20 nm×10 nm in the membrane plane (Figure 1).1,3-10
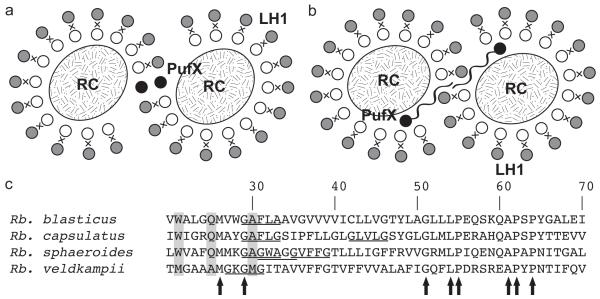
Figure 1.
Proposed models for the protein organization of a dimeric photosynthetic core complex. (a) Model based on atomic force microscopy imaging studies of Rba. sphaeroides and Rba. blasticus photosynthetic membrane,4,6 PufX is placed at the dimerization interface in the center of the core complex, and is itself also thought to be dimerized. (b) Model based on the highest resolution structural data to date of the dimeric Rba. sphaeroides core complex,7 with PufX situated near the gap of the open LH1 ring, and association of PufX is facilitated through a long loop at the N-terminal region.7,11,12 In (a) and (b), PufX helices are represented by black circles, while LH1 helices are shown as gray circles (outer helices, known as LH1β) and white circles (inner helices, known as LH1α), with the embedded pigments between the outer and inner helices denoted by “X”. RC is shown as an oval. (c) Aligned sequences of the central region of PufX from four Rhodobacter species investigated in the present study. Conserved amino acids are indicated by arrows, and amino acids conserved in Rba. blasticus, Rba. capsulatus, and Rba. sphaeroides, but not in Rba. veldkampii, are shaded in gray.13 GxxxG and GxxxA motifs are underlined.
The TM protein PufX is known to be crucial in the formation of dimeric photosynthetic core complexes in Rba. sphaeroides,14 and deletion of this protein leads to monomeric core complexes.2,9,15 Yet, as the location of PufX is still being debated, the molecular mechanism with which PufX determines the oligomerization state of the core complex is still an active topic of discussion.15 Two models have been proposed for the placement and organization of PufX, each model involving a different mechanism for the PufX-assisted dimerization of the core complex (Figure 1). Figure 1a depicts a central placement of PufX, and the dimerization of the TM region of PufX “fuse” the two core complex monomers together.4,6,16,17 In contrast, Figure 1b shows a placement of PufX near the gap of the two open LH1 rings,7 and in this scheme PufX is thought to induce core complex dimerization via interaction of its long N-terminal region in the cytoplasmic space.7,11,12 A crystallographic structure of a dimeric core complex is not yet available to determine unambiguously the validity of either model, although it has also been speculated that different species of Rba. bacteria might have different protein organizations in the core complex.15,18
Interestingly, the oligomerization states of different Rba. core complexes are not the same. Through atomic force microscopy (AFM) imaging of the Rba. blasticus photosynthetic membrane, dimeric core complexes have been identified, though monomeric core complexes were also observed at an approximately 3:1 dimer to monomer ratio.6 The Rba. sphaeroides core complex has also been shown to form dimers,2,4,7,9 with monomeric core complexes present as well at a 1:1 dimer to monomer ratio.18 Unlike Rba. blasticus and Rba. sphaeroides, the Rba. veldkampii core complex was observed to be monomeric in a structural and functional analysis,19 and microscopy studies also reported no sighting of dimeric core complex in the Rba. veldkampii photosynthetic membrane,13,17,20 suggesting that Rba. veldkampii core complex is unable to dimerize. While there is no structural information available for the Rba. capsulatus core complex, its PufX can replace that of Rba. sphaeroides, and the resulting Rba. sphaeroides is still photosynthetically viable,15 prompting the idea that Rba. sphaeroides and Rba. capsulatus core complexes are likely very similar, and that the core complex of Rba. capsulatus is also capable of dimerizing.
Examining the sequences of PufX in four Rba. bacteria, it can be noted that some sequence similarities exist (Figure 1c).13,15 In fact, it has been suggested that the GxxxG motif found in Rba. sphaeroides PufX between amino acids 31 and 35 (the N-terminal Met = 0 convention is adopted here) might serve as the dimerization region,17,21 similar to that in glycophorin A (GpA).22 Computational investigations have subsequently shown that a Rba. sphaeroides PufX dimer appears to be stable in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) membrane.23 However, the GxxxG motif between locations 31 and 35 is only present in Rba. sphaeroides, not in Rba. blasticus or Rba. capsulatus, which also have a dimeric core complex. In addition, mutation of the glycines in this motif does not appear to abolish the ability for Rba. sphaeroides core complex to dimerize, as shown both computationally23 and experimentally.18 Furthermore, as shown in Figure 1c, GxxxG motifs are also present in Rba. capsulatus and Rba. veldkampii, although not between positions 31 and 35. A purely sequence-based argument for the dimerization affinity of PufX and the variability in core complex oligomerization state, thus, seem to be still inconclusive and require further investigation.
In an effort to provide new insight into the potential dimerization of the PufX TM region, a prerequisite for the validity of the core complex organization shown in Figure 1a, and to relate dimerization of PufX segments to the core complex oligomerization state, we employed both computational and experimental methods to measure the dimerization affinity of four species of PufX: Rba. blasticus, Rba. capsulatus, Rba. sphaeroides, and Rba. veldkampii. We first constructed monomeric and dimeric PufX models for Rba. blasticus, Rba. capsulatus, and Rba. veldkampii, and then probed the stability of these structures in a membrane environment using all-atom molecular dynamics (MD), similar to the strategy previously followed for Rba. sphaeroides PufX.23 Subsequently, TOXCAT24 was performed on the four PufX TM segments to quantitatively measure the strength of helix-helix association. To complement the experiment, we also computed the apparent dimerization free energy for the four PufX helices using an MD-based free-energy protocol. Our data reveal a compelling trend on the strength of PufX dimerization: species capable of forming a dimeric core complex have PufX helices that show higher propensity to self-associate. Conversely, Rba. veldkampii, which is observed with only monomeric core complexes, has a PufX that exhibits very little propensity towards homodimerization. These results strongly indicate that differences in PufX dimerization affinity is an important factor for the variability of oligomerization states in Rba. photosynthetic core complexes.
Methods
Molecular Dynamics
Construction of monomeric and dimeric PufX
As there is currently no structural data available for PufX from Rba. blasticus, Rba. capsulatus, and Rba. veldkampii, the monomeric PufX models in simulations blasticus-Monomer-POPE, capsulatus-Monomer-POPE, and veldkampii-Monomer-POPE (Table 1) were constructed based on that of Rba. sphaeroides PufX, for which two solution structures have been reported11,21 and were used in previous modeling studies.23,25-29 All PufX monomers were modeled with an integral TM helix with the same length as that of Rba. sphaeroides PufX.21 Since we are only interested in the TM interaction of PufX helices, the N- and C-terminal residues that are thought to form loops11,21 were not included. These monomeric PufX structures were then placed in a POPE membrane patch, with addition of neutralizing Na+ and Cl− ions at a total ionic strength of 300 mM, as shown in Figure 2a-c. For comparison, the Rba. sphaeroides PufX monomer, constructed previously,23 is shown in Figure 2d.
Table 1.
List of molecular dynamics simulations performed in the present study.
| Simulation Name | Type | Num. Atoms | Time (ns) |
|---|---|---|---|
| blasticus-Monomer-POPE | EQ | 32,812 | 15 |
| capsulatus-Monomer-POPE | EQ | 34,004 | 15 |
| veldkampii-Monomer-POPE | EQ | 32,904 | 15 |
|
| |||
| blasticus-Dimer-POPE | EQ | 31,801 | 50 |
| capsulatus-Dimer-POPE | EQ | 30,949 | 50 |
| veldkampii-Dimer-POPE | EQ | 32,336 | 100 |
|
| |||
| blasticus-Dimer-DODE-1 | EQ | 20,486 | 10 |
| capsulatus-Dimer-DODE-1 | EQ | 20,487 | 20 |
| veldkampii-Dimer-DODE-1 | EQ | 20,557 | 10 |
|
| |||
| blasticus-Dimer-DODE-ABF-1 | ABF | 20,486 | 220 |
| capsulatus-Dimer-DODE-ABF-1 | ABF | 20,487 | 285 |
| veldkampii-Dimer-DODE-ABF-1 | ABF | 20,557 | 185 |
|
| |||
| blasticus-Dimer-DODE-2 | EQ | 20,496 | 20 |
| capsulatus-Dimer-DODE-2 | EQ | 20,559 | 20 |
| veldkampii-Dimer-DODE-2 | EQ | 20,503 | 20 |
|
| |||
| blasticus-Dimer-DODE-ABF-2 | ABF | 20,496 | 55 |
| capsulatus-Dimer-DODE-ABF-2 | ABF | 20,559 | 105 |
| veldkampii-Dimer-DODE-ABF-2 | ABF | 20,503 | 70 |
|
| |||
| sphaeroides-Dimer-DODE-ABF | ABF | 20,584 | 170 |
Total simulation time: 1.435 μs
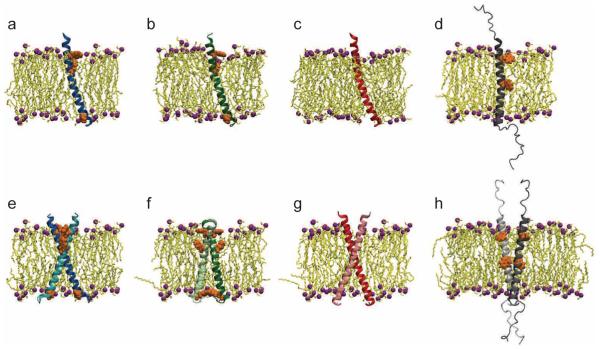
Figure 2.
Simulated molecular systems with POPE lipid bilayers. Protein-membrane systems with monomeric PufX for (a) Rba. blasticus, (b) Rba. capsulatus, and (c) Rba. veldkampii. (d) Monomeric Rba. sphaeroides system is also shown for comparison; the simulation was performed previously.23 PufX helix is shown in blue ((a) Rba. blasticus), green ((b) Rba. capsulatus), red ((c) Rba. veldkampii), and gray ((d) Rba. sphaeroides), lipid is shown in yellow with purple spheres representing the headgroups; polararomatic residues tyrosine and tryptophan of PufX are shown in orange. For clarity, water and ion molecules included in all simulations are not shown. (e-g) Protein-membrane systems with modeled homodimeric PufX for (e) Rba. blasticus, (f) Rba. capsulatus, and (g) Rba. veldkampii. (h) Dimeric Rba. sphaeroides system is also shown for comparison; the simulation was performed previously.23 PufX helices in this and subsequent figures are shown with N-termini pointing upward.
Equilibrium MD simulations were carried out for the three PufX monomer systems for 15 ns each. The final conformations of the PufX helices resulting from these monomer simulations were used to construct the corresponding PufX dimer models. Each PufX helix was replicated, and the two copies of PufX were placed facing each other by mapping them onto the GpA dimer structure,22 as previously done for Rba. sphaeroides PufX.23 Since Rba. blasticus, Rba. capsulatus, and Rba. veldkampii do not have a GxxxG motif at position 31-35 (which Rba. sphaeroides possesses), amino acids 29-33 of Rba. blasticus, Rba. capsulatus, and Rba. veldkampii PufX were mapped onto the GxxxG portion of GpA. Choice of position 29-33 is based on the observation that Rba. blasticus, Rba. capsulatus, and Rba. sphaeroides all have a GxxxA or GxxxG motif at this segment, while Rba. veldkampii does not. In fact, Gly29 is conserved in all four species as shown in Figure 1c.
All PufX dimers were also placed in a POPE membrane patch, and similarly neutralized with additional ions at a total ionic strength of 300 mM, as shown in Figure 2e-g. The Rba. sphaeroides PufX dimer system23 is shown in Figure 2h for comparison. An equilibrium MD simulation was performed for each of the resulting PufX dimer systems, designated as blasticus-Dimer-POPE, capsulatus-Dimer-POPE, and veldkampii-Dimer-POPE in Table 1, for at least 50 ns.
Equilibrium molecular dynamics
All simulations were performed using the MD package NAMD30 with the CHARMM27 force field,31,32 including CMAP corrections.33 Water molecules were described with the TIP3P model.34 Long-range electrostatic forces were evaluated by means of the particle-mesh Ewald (PME) summation approach with a grid spacing of < 1 Å. An integration time step of 2 fs was used in the framework of the Verlet r-RESPA algorithm.35 Bonded terms and short-range, non-bonded terms were evaluated every time step, and long-range electrostatics every other time step. Constant temperature (T = 310 K) was maintained using Langevin dynamics,36 with a damping coefficient of 1.0 ps−1. A constant pressure of 1 atm was enforced using the Langevin piston algorithm37 with a decay period of 200 fs and time constant of 50 fs.
Free-energy calculations
To assess computationally the dimerization affinity of the PufX helices, adaptive biasing force (ABF) calculations38-40 were performed to determine free-energy as a function of helix-helix distance.40,41 Prior to conducting ABF simulations, the PufX TM segments were equilibrated in a dodecane patch in a solvent environment neutralized with ions at 300 mM ionic strength. Use of dodecane as a lipid mimetic is dictated by the slow relaxation times of natural lipid molecules compared to affordable MD timescales.23,40,42 The TM segments of PufX were blocked at the N– and C–termini by Ac– and –NHMe groups, respectively. Two sets of PufX dimer-dodecane systems were constructed (blasticus-Dimer-DODE-1, capsulatus-Dimer-DODE-1, veldkampii-Dimer-DODE-1, blasticus-Dimer-DODE-2, capsulatus-Dimer-DODE-2 and veldkampii-Dimer-DODE-2 in Table 1), using slightly different TM segments to test if inclusion of different residues would alter significantly the results of free-energy calculations. Each protein-dodecane system was subject to equilibrium MD for at least 10 ns. An example setup of the dodecan-PufX system is shown in Figure S1 in Supporting Information.
ABF calculations were carried out subsequently in the framework of NAMD30 for the six dodecane systems (blasticus-Dimer-DODE-ABF-1, capsulatus-Dimer-DODE-ABF-1, veldkampii-Dimer-DODE-ABF-1, blasticus-Dimer-DODE-ABF-2, capsulatus-Dimer-DODE-ABF-2 and veldkampii-Dimer-DODE-ABF-2 in Table 1). The TM portion of a modeled Rba. sphaeroides PufX dimer was previously equilibrated in a dodecane patch,23 and an ABF calculation was also performed for Rba. sphaeroides PufX, designated as sphaeroides-Dimer-DODE-ABF in Table 1. For each ABF simulation, the model reaction coordinate, ξ, is defined as the distance separating the center of mass of the two helices, in the interval 4.5 ≤ ξ ≤ 27 Å. A small ξ indicates that the PufX helices are associated, with a large ξ indicating their separation. In the course of an ABF simulation, average forces applied on the PufX helices in an unconstrained MD simulation are projected onto ξ, and a “biasing force” is calculated and applied to the helices to overcome local energy barriers.38-40 The free-energy profile along ξ is then obtained by integrating the average force, with a standard error estimated according to Rodriguez-Gomez et al.43
TOXCAT
Vectors and constructs
The TOXCAT vector, pccKAN, and positive controls containing the TM domain of wild type GpA (pccGpA-WT) and the G83I disruptive mutant (pccGpA-G83I) have been described previously.24 DNA coding for the TM domains of the PufX proteins (Table 2), flanked by 5′NheI and 3′BamHI restriction sequence, were purchased as synthetic genes (IDT). The sequences were ligated in-frame to NheI and BamHI sites of the pccKAN vector.
Table 2.
Sequences of the TM regions of ToxR’(TM)MBP constructs. Boldface residues represent the TOXCAT construct anking regions including those containing the restriction site codons used for sub-cloning into the TOXCAT construct.
| Species | Amino Acid Sequence |
|---|---|
| Rba. blasticus | NRASQMVWGAFLAAVGVVVVICLLVGTGIL |
| Rba. capsulatus | NRASQMAYGAFLGSIPFLLGLGLVLGSGIL |
| Rba. sphaeroides | NRASQMMKGAGWAGGVFFGTKKKIGFFGIL |
| Rba. veldkampii | NRASAMGKGMGITAVVFFGTVFFVVALGIL |
Expression of ToxR’(TM)MBP constructs
Plasmids encoding ToxR’(TM)MBP chimerae were transformed into Escherichia coli MM39 cells (provided by D. M. Engelman) and plated onto Luria Bertani (LB) plates (with 100 μg/mL ampicillin and 25 μg/mL streptomycin); colonies were inoculated into LB medium (with 100 μg/mL ampicillin) and stored as glycerol stocks at −80°C. LB cultures (with 100 μg/mL ampicillin) were inoculated from frozen glycerol stocks and grown overnight (approximately 18 hrs). 3 mL LB cultures (with 100 μg/mL ampicillin) were inoculated using 50 μL overnight cultures and grown to A420 1.0, and 1 mL of cells were harvested by centrifugation and resuspended in 0.5 mL lysis buffer (25 mM Tris-HCl, 2 mM EDTA, pH 8.0). Cells were then lysed by probe sonication. The lysate was clarified by centrifugation at 17,000×g, and the supernatant was stored on ice until the spectrophotometric assay was performed.
Spectrophotometric CAT assay
The colorimetric assay used to detect chloramphenicol acetyltransferase activity in cell lysates was described previously.44,45 Absorbance was measured using a PerkinElmer Lambda 25 UV/Vis spectrophotometer. 40 μL of lysate was mixed with 1 mL of reaction buffer (0.1 mM acetyl-coA, 0.4 mg/mL 5,5′-dithiobis-(2-nitrobenzoic acid), 0.1 M Tris-HCl, pH 7.8) and absorbance at 412 nm was measured for a period of 2 min with intervals of 3 sec to establish a basal rate of acetyl-coA hydrolysis in the absence of substrate. At 2 min, 40 μL of 2.5 mM chloramphenicol was added with mixing and the absorbance was measured at 412 nm for another 2 min in 3 sec intervals. CAT activity was calculated as the slope of 412 nm absorbance, after subtracting the basal rate prior to substrate addition. Lysates were assayed in triplicate and the reported data are the result of three separate experiments.
Maltose complementation assays
To confirm correct membrane insertion, E. coli MM39 cells expressing the ToxR’(TM)MBP constructs were grown overnight in LB (with 100 μg/mL ampicillin) and then streaked onto M9 minimal media plates containing 0.4% maltose as the only carbon source and incubated for 3 days at 37°C.
Western blot analysis
TOXCAT protein expression levels were verified by western blot analysis. Cell lysate were mixed with 2× SDS-PAGE sample buffer, heated to 70°C for 10 min, run on pre-cast 12% polyacrylamide gels (Invitrogen), transferred onto an Immobilon-P polyvinylidene fluoride membrane (Millipore) and detected with rabbit anti-MPB primary antibodies (New England Biolabs) and anti-rabbit horseradish peroxidase conjugate secondary antibodies (Millipore).
Results and Discussion
Below our computational and experimental results are discussed. First, we report the stability of different species of PufX helices in monomeric and homodimeric conformations as probed by equilibrium MD simulations. Then, we present the dimerization affinity of PufX helices as measured using the TOXCAT assay. Finally, complementing TOXCAT experiments with atomic resolution and quantitative assessment, we report MD-based free-energy calculations conducted to estimate the apparent dimerization free energy, ΔGapp, of PufX helices.
Equilibrium Molecular Dynamics
PufX monomers
All three PufX monomers were seen to be structurally stable during their respective equilibrium MD simulations. Similar to Rba. sphaeroides, the PufX helices of Rba. blasticus, Rba. capsulatus, and Rba. veldkampii were seen to tilt with respect to the normal of the lipid bilayer during simulations (see Movies S1a, S1b, and S1c in Supporting Information). Examining the sequence content of PufX from the different species, it was observed that Rba. veldkampii is the only case without either a tyrosine or a tryptophan residue. Tyrosine and tryptophan residues are known to reside preferentially at the lipid-water interface,46 and might contribute to the anchoring of a TM helix to the lipid headgroups.47-49 As can be seen in Figure 2a-d, for Rba. blasticus, Rba. capsulatus, and Rba. sphaeroides PufX that contain tyrosine and tryptophan, most of these residues appear near the membrane-solvent interface.
The helical structures of all PufX models persisted throughout the simulations, as shown in Figure 3a. Structural stability of each PufX monomer is consistent with the two-stage model of membrane-protein folding, which postulates that TM helices act as independent stable domains and are pre-formed prior to their association into large protein complexes50,51
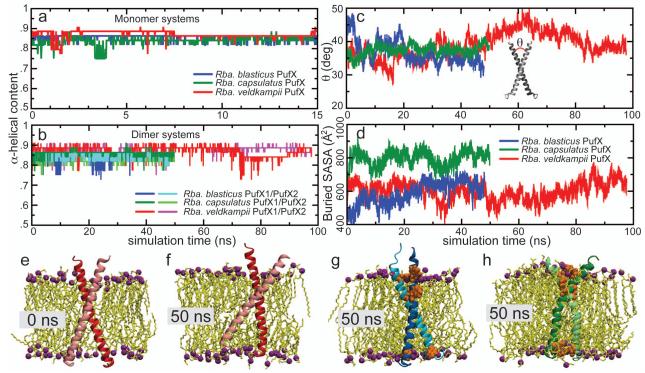
Figure 3.
Stability of PufX monomeric and homodimeric helices during equilibrium MD simulations. a) α-helical content of the modeled PufX monomer in a full POPE membrane. For each of the three species tested (Rba. blasticus, Rba. capsulatus, and Rba. veldkampii), PufX retains its high α-helical content. Similarly, modeled PufX helices in dimeric conformation also remain largely α-helical, as shown in b). c) Crossingangle between the dimerized helices. d) Buried solvent accessible surface area (SASA) as a measure for helix-helix interaction. e) and f) show the movement of the Rba. veldkampii PufX helices. At 50 ns, one of the Rba. veldkampii helices can be seen to submerge nearly fully into the membrane on the C-terminus. For comparison, g) and h) show the Rba. blasticus and Rba. capsulatus PufX helices also at 50 ns; in these cases the tyrosine and tryptophan residues aided in anchoring the helices in the membrane, and these residues remained at the membrane-solvent interface throughout the simulation.
PufX dimers
Each PufX dimer model was constructed using the final conformation from the equilibrium simulations of monomeric PufX, as described in Methods. All three dimer systems were observed to be structurally robust with consistent α-helical content throughout the simulation (Figure 3b; see Movies S2a, S2b, and S2c in Supporting Information). The dimerized PufX helices also maintained a consistent crossing-angle (Figure 3c), and remained in contact during the simulation as indicated by the measurement of the buried solvent accessible surface area (SASA) (Figure 3d). For all three species, buried SASA of the PufX dimers remained near or above 600 Å2, comparable to that of GpA.52
The most notable motion was seen in the case of Rba. veldkampii PufX, which lacks both tyrosine and tryptophan, and transformed from an originally upright orientation (Figure 3e) to one tilted relative to the membrane at 50 ns (Figure 3f), and with one of the helices submerged in the lipid phase on the C-terminus. This tilted and partially membrane-buried conformation of Rba. veldkampii PufX dimer persisted when the simulation was extended to 100 ns. In comparison, the tyrosine and tryptophan residues in Rba. blasticus and Rba. capsulatus PufX dimers (Figure 3g and h, respectively) remained at the membrane-solvent interface, preventing strong fluctuations in their helix-membrane orientations. Quantitative measurement of helix tilting with respect to the membrane normal during the simulations blasticus-Dimer-POPE, capsulatus-Dimer-POPE, and veldkampii-Dimer-POPE is shown in Figure S2 in Supporting Information. The instability of Rba. veldkampii PufX helices due to the lack of anchorage might be significant in the propensity of the helices to homodimerize. Additionally, it can be seen that the proline residue at position 36 in Rba. capsulatus PufX induces a moderate kink in the helix (10-30 degrees) that persisted throughout the simulations for both the mononeric and dimeric conformations (Figure S3); this residue does not face the dimerization interface in the modeled Rba. capsulatus PufX dimer.
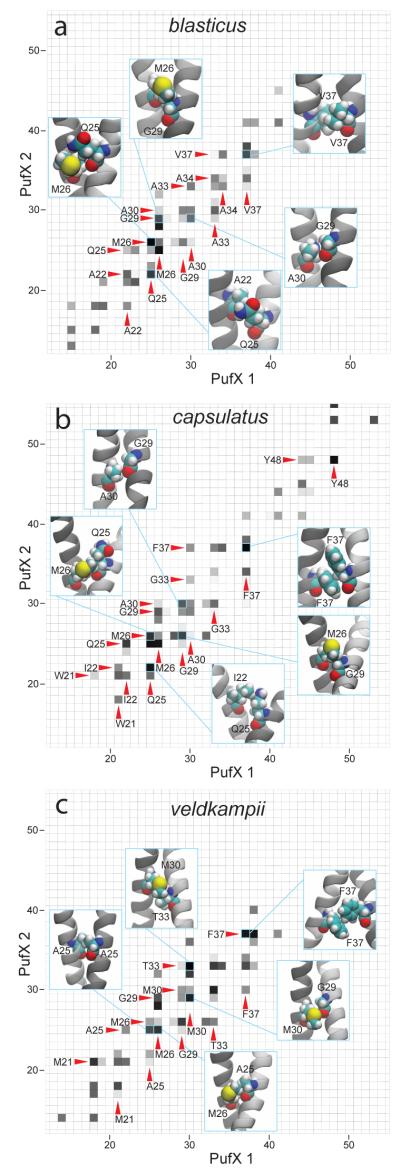
Inter-helical interactions contributing to the stability of PufX dimer models are shown in Figure 4a-c for Rba. blasticus, Rba. capsulatus, and Rba. veldkampii, respectively. For Rba. blasticus and Rba. capsulatus, significant molecular interactions are contributed by residues Gln25 and Met26, which interact with each other, and also with other small amino acids such as glycine and alanine. For example, Gln25 was seen to interact with Ala22, and Met26 interacted with Gly29 in Rba. blasticus PufX dimer (Figure 4a). The Met26-Gly29 interaction was also observed for Rba. capsulatus, and its Gln25 was observed to interact with Ile22 (Figure 4b). Also, helix packing is achieved through close contact between small residues Gly29 and Ala30 for both Rba. blasticus and Rba. capsulatus (Figure 4a and b). Notably, while Gly29 is conserved for all four species investigated in the present study, Rba. veldkampii is the only species that does not contain Gln25 and Ala30 (Figure 1c), two residues that contribute significantly to inter-helical interactions for Rba. blasticus and Rba. capsulatus. For Rba. veldkampii PufX dimer, the helices are held together by a slightly different set of molecular interactions, although Met26, which is conserved in all four PufX sequences (Figure 1c), plays also an important role (Figure 4c). Further away from the dimerization core, bulkier amino acids such as Val37 for Rba. blasticus, and Phe37 for Rba. capsulatus and Rba. veldkampii, provide additional inter-helix contact.
Figure 4.
Interhelical interactions observed during the equilibrium molecular dynamics simulations (a) blasticus-Dimer-POPE, (b) capsulatus-Dimer-POPE, and (c) veldkampii-Dimer-POPE. Highly interacting amino acid pairs are highlighted with darker grids in the interaction map, and five of such pairs are shown in the insets as examples.
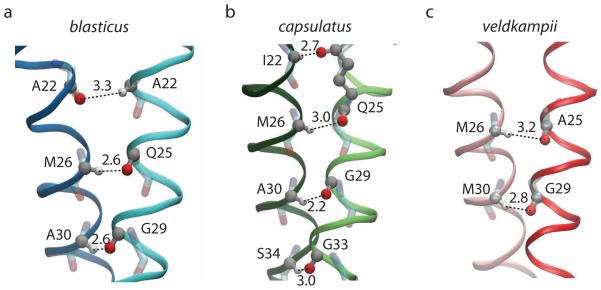
Inter-helical hydrogen bonds are known to be an important factor in mediating helix-helix association in the membrane.53 The close packing of the modeled PufX dimers permitted the formation of several inter-helical hydrogen bonds. In particular, the side chain amide group of Gln25 forms an inter-helical hydrogen bond with the carbonyl of Ala22 in the Rba. blasticus dimer (Figure 5). The same side chain accepts a Cα–H⋯O hydrogen bond from Ile22 in the Rba. capsulatus dimer. As mentioned above, Rba. veldkampii is the only species that does not contain Gln25. Several backbone-to-backbone Cα–H⋯O hydrogen bonds were also observed (illustrated in Figure 5 and Table 3), which are a hallmark of GxxxG mediated transmembrane interactions.53-58 The Gln25-Met26 and Gly29-Ala30 pairs were seen to be sites for the potential formation of Cα–H⋯O hydrogen bonds for the cases of Rba. blasticus and Rba. capsulatus PufX dimers, and locations 33 and 34, which contain small amino acids such as alanine, serine, and glycine, provide additional hydrogen bonding (Figure 5 and Table 3). For the case of Rba. veldkampii PufX dimer, only two Cα–H⋯O hydrogen bonds were observed, and were also formed between amino acid pairs 25-26 (Ala25-Met26), and 29-30 (Gly29-Met30).
Figure 5.
Networks of interhelical Cα–H⋯O hydrogen bond contacts in the three PufX dimer models identified from simulations (a) blasticus-Dimer-POPE, (b) capsulatus-Dimer-POPE, and (c) veldkampii-Dimer-POPE. For the amino acids involved in formation of Cα–H⋯O contacts, carbon atoms are shown in gray, oxygen atoms in red, hydrogen atoms in white, and other backbone atoms are shown in transparent. All H⋯O distances are shown in Å.
Table 3.
Geometry of potential Cα–H⋯O hydrogen bond contacts. De nitions of distances and angles are given as the following:54,57 dH is the distance between the H and O atoms; d is the distance between the Cα and the O atoms; ζ is the Cα-H-O angle; ξ is the H-O-C angle; θ is the elevation angle between the Cα-H vector and the amide plane. All numbers are average values from the last 10 ns of corresponding simulations ( blasticus-Dimer-POPE, capsulatus-Dimer-POPE, and veldkampii-Dimer-POPE). Distances are given in Å, and angles are given in degrees.
| Ideal Values | ||||||
|---|---|---|---|---|---|---|
| Donor | Acceptor | dH (min. value) | d (min. value) | ζ | ξ | θ |
| – | – | ≤2.7 | ≤ 3.8 | 180 | 120 | 0 |
| Rba. blasticus | ||||||
|
| ||||||
| A22 | A22 | 4.04 (3.03) | 4.75 (3.75) | 127.06 | 103.68 | 28.11 |
| M26 | Q25 | 3.65 (2.54) | 4.42 (3.42) | 131.82 | 85.87 | 29.44 |
| A30 | G29 | 3.06 (2.78) | 3.83 (3.13) | 129.28 | 96.57 | 17.14 |
| A34 | A33 | 3.90 (2.76) | 4.69 (3.66) | 132.64 | 96.42 | 39.62 |
|
| ||||||
| Rba. capsulatus | ||||||
|
| ||||||
| I22 | Q25 | 3.01 (2.20) | 4.03 (3.27) | 158.92 | 110.55 | 54.45 |
| M26 | Q25 | 2.86 (2.24) | 3.80 (3.14) | 146.67 | 120.92 | 61.52 |
| A30 | G29 | 2.97 (2.21) | 3.56 (3.01) | 115.23 | 108.14 | 15.58 |
| S34 | G33 | 4.79 (2.81) | 5.58 (3.80) | 133.70 | 115.98 | 38.84 |
|
| ||||||
| Rba. veldkampii | ||||||
|
| ||||||
| M26 | A25 | 2.61 (2.14) | 3.51 (3.07) | 141.31 | 115.86 | 46.91 |
| M30 | G29 | 2.82 (2.17) | 3.65 (3.11) | 135.74 | 104.27 | 13.96 |
TOXCAT
The equilibrium MD simulations of the three PufX dimers conducted here, as well as the one conducted previously for Rba. sphaeroides,23 showed that PufX dimer models for all four species remain associated. Although it appears that Rba. veldkampii PufX dimer has an unstable protein-membrane interaction due to lack of anchorage, no spontaneous disassociation was observed. It is possible that disassociation of PufX requires longer simulation time than currently available due to the slow relaxation time of a full POPE membrane.
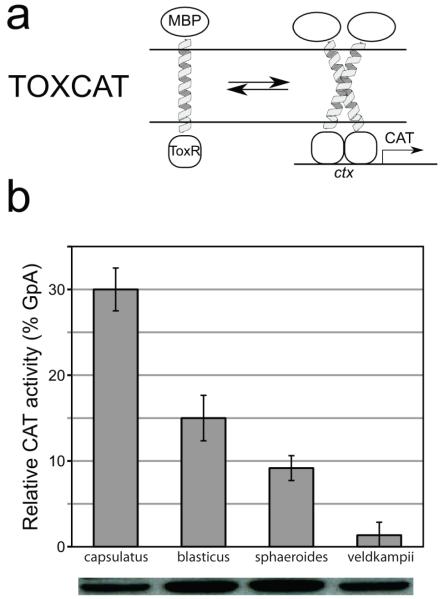
To determine quantitatively the dimerization affinity of the PufX helices, we employed an experimental assay, namely, the TOXCAT method,24 which measures the association of TM helices in a biological membrane. Three TOXCAT measurements were performed on each of the four Rba. species, with the average dimerization affinity for each species shown in Figure 6b as percent of the CAT activity of GpA. Rba. capsulatus is seen to have the highest propensity for homodimerization, with a relative CAT activity at ~ 30%, comparable to a prior measurement reported in Aklujkar and Beatty.59 Rba. blasticus has the second highest CAT activity, albeit only at ~ 15% GpA. Rba. sphaeroides shows even lower propensity to homodimerize, and Rba. veldkampii exhibits no significant CAT activity.
Figure 6.
Quantification of association of TM constructs in E. coli membranes using TOXCAT. a) TOX-CAT24 is an in vivo assay based on a fusion construct consisting of the TM domain under investigation, a maltose binding protein and the ToxR transcriptional activator of V. cholerae. TM association results in the expression of chloramphenicol acetyl transferase (CAT) under the ctx promoter, whose enzymatic activity can be measured. b) TOXCAT data for the PufX TM domains of Rba. capsulatus, Rba. blasticus, Rba. sphaeroides and Rba. veldkampii. The data are reported as percent of the CAT activity of GpA, a strongly dimerizing transmembrane domain.22 The data is the average of three independent replica and the error bars report the standard deviation. Protein expression levels were verified by western blot using anti-MBP antibodies.
Free-Energy Calculations
Concurrent to the experimental measurement of PufX dimerization affinity with TOXCAT, we also employed free-energy calculations to measure the apparent dimerization free energy in silico using the ABF algorithm38-40 for PufX from four Rba. species. The computational treatment is inspired by the atomic resolution of the method, which can reveal great structural details in the dimerization and disassociation pathway.
Two sets of ABF calculations were conducted corresponding to distinct choices of TM residues. In the first set, the same sequences as used in TOXCAT (i.e., those shown in non-boldface in Table 2) were included. Since the TOXCAT experiments contain also flanking residues and is not identical to the setup of the in silico assays, to test if results from ABF are sensitive to the small difference in the sequence of amino acids, a second set of ABF simulations was conducted, using the sequences identified as the TM region from the dimer simulations carried out in the POPE environment (i.e., blasticus-Monomer-POPE, capsulatus-Monomer-POPE, and veldkampii-Monomer-POPE; Table 1). For Rba. sphaeroides PufX, the sequence used in TOXCAT is the same as that identified as the TM region;23 therefore, only one ABF simulation was performed (sphaeroides-Dimer-DODE-ABF in Table 1).
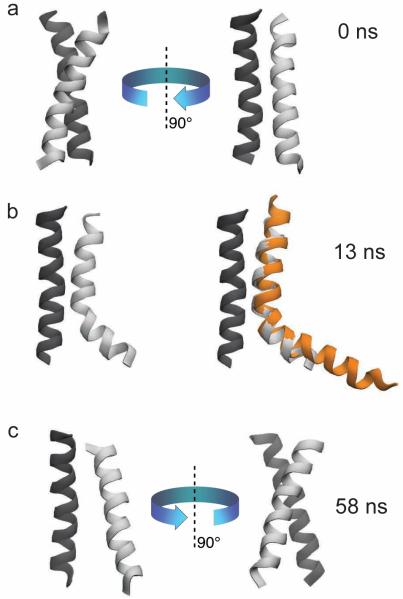
The dimerization pathway of PufX is observed to be more complex than that of GpA,40 with an example shown in Figure 7 for the case of Rba. sphaeroides PufX. At the beginning of the ABF simulation sphaeroides-Dimer-DODE-ABF, the two PufX helices are both straight (Figure 7a). However, spontaneous bending occurred after 13 ns (Figure 7b, left), in agreement with prior in silico observation on the inherent flexibility of the Rba. sphaeroides PufX helix.23 Furthermore, bending of the PufX helix occurs at the same location as that seen for one of the PufX solution structures,11 and, as a result, the bent conformation seen in simulation sphaeroides-Dimer-DODE-ABF is structurally very similar to the solution structure (Figure 7b, right). Helix bending persisted for a few tens of ns, but eventually the helix spontaneously straightened (Figure 7c). Previously, we had estimated that bending of the Rba. sphaeroides PufX only costs a few kcal/mol;23 our present results support such low value. We note that the tendency for Rba. sphaeroides PufX to bend might complicate self-association, but does not completely prohibit it as the helix quickly straightens back and the straight conformation can possibly be stabilized by dimerization.
Figure 7.
Spontaneous bending and straightening of Rba. sphaeroides PufX. a) At the onset of the ABF simulation for Rba. sphaeroides PufX, both helices were straight. b) At 13 ns, one of the helices bent spontaneously. The bending corresponded well to the observed NMR solution structure of PufX (pdb code 2NRG11), and persisted for the next ~40 ns. c) The bent helix was seen to straighten back at 58 ns, suggesting that bending and straightening of the helix occur spontaneously with a low energy barrier, as suggested previously.23
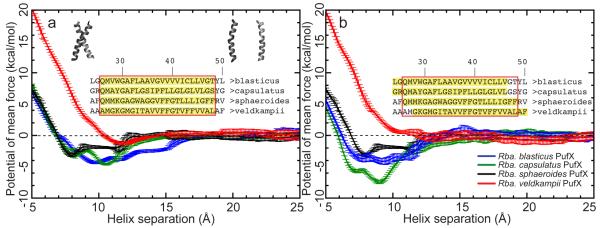
The results from the two sets of ABF calculations are compared in Figure 8 in the form of free-energy profiles as a function of helix-helix distance. For the first set of ABF calculations (Figure 8a), all four species of PufX are seen to have energy minima for an associated, dimerized conformation, albeit with different well depths. Rba. veldkampii has the most shallow free-energy minimum near a helix-helix separation of 12 Å. Rba. sphaeroides has the second-most shallow free-energy minimum. Unlike GpA dimer, which exhibits a well-defined free-energy well,40 the free-energy well of Rba. sphaeroides PufX is seen to span nearly 5 Å, with the minimum occurring near 8 Å. Rba. blasticus and Rba. capsulatus have the deepest free-energy wells and both possess multiple local minima. For Rba. capsulatus, two local free-energy minima are found at helix-helix distances of 8 and 11 Å; for Rba. blasticus, its free-energy well has several less well-defined local minima that stretches up to a helix-helix distance of 15 Å. The much wider free-energy well for Rba. blasticus PufX is possibly due to additional stabilizing inter-helical interactions arising from transient van der Waals contact between the bulkier Leu43, Leu44 and Thr47 residues near the C-terminal end (Figure S4 in Supporting Information). In general, the PufX dimers exhibit more complex free-energy profiles than does the GpA dimer.40 This extra complexity is possibly due to the usage of modeled dimer systems rather than experimentally derived structures. Alternatively, it is also possible that the complex dimerization scheme is intrinsic to PufX due to its difference to GpA.
Figure 8.
Potential of mean force measured in ABF simulations. Highlighted sequences are those included in the ABF simulations; the sequences outlined in the red box are those included in the TOXCAT measurement (Table 2). a) First set of ABF simulations using the same sequence for TM segments of PufX as that in TOXCAT experiments. b) Second set of ABF simulations using the sequence identified as the TM segments in the PufX dimer-POPE membrane simulations.
In the second set of ABF simulations (Figure 8b), Rba. veldkampii appears to have no preference for association. Rba. capsulatus PufX is seen to have a deeper free-energy well than that of Rba. blasticus, and retains the two minima observed in Figure 8a, albeit at closer helix-helix distances (7 and 9 Å). Rba. blasticus PufX again exhibits a wide minimum, with the global free-energy minimum occurring at a helix-helix distance of 9 Å. Although the precise free-energy profiles are different in the two sets of ABF simulations, it is reassuring that distinct features in the Rba. blasticus and Rba. capsulatus free-energy profiles are preserved across the two simulations. Additionally, Rba. veldkampii PufX consistently exhibits the lowest propensity towards dimerization.
From the free-energy profiles in Figure 8, we calculated the apparent disassociation free energy, ΔGapp, for the four species of PufX in two sets of ABF calculations using the expression utilized by Hénin et al.,40 with the results shown in Table 4 and Figure 9. Calculation of ΔGapp allows comparison of PufX dimerization affinity with that of GpA (Figure 9), which has a ΔGapp value of 11.5±0.4 kcal/mol as previously reported using also the ABF method in a dodecane environment.40 It can be seen that the order of PufX dimerization affinity is similar to the experimental results (Figure 6), in the decreasing order of Rba. capsulatus > Rba. blasticus > Rba. sphaeroides > Rba. veldkampii. Differences in experimental and simulation setups might contribute to the consistent overestimate of in silico free energies compared to TOXCAT measurements (Figure 9 and Figure 6). For example, the PufX sequences used in the TOXCAT experiment and the ABF measurements are not exactly identical (Table 2 and Figure 8), and, as shown by comparing Figure 8a and b, small differences in sequence content can lead to varying dimerization affinity. Additionally, while TOXCAT was performed in a biological membrane environment, the ABF measurements were conducted with dodecane. Finally, the reaction coordinate, ξ, chosen in the ABF calculation does not consider the relative orientation of the two helices, including their intrinsic rotation about their longitudinal axis, a degree of freedom important in optimizing helix-helix packing. Considering these factors limiting direct comparison between experiment and simulation results, it is significant that a consensus in the relative strength of dimerization for the four PufX sequences tested here was reached.
Table 4.
Free energy of association. Two ABF calculations were conducted each for Rba. blasticus, Rba. capsulatus, and Rba. veldkampii PufX segments using slightly different amino acid sequences (Figure 8), and one ABF calculation was performed for Rba. sphaeroides.
| Species | ΔGapp from ABF1 (kcal/mol) | ΔGapp from ABF2 (kcal/mol) |
|---|---|---|
| Rba. capsulatus | 6.7 ± 0.3 | 9.4 ± 0.3 |
| Rba. blasticus | 6.8 ± 0.4 | 6.6 ± 0.5 |
| Rba. sphaeroides | 5.2 ± 0.4 | n/a |
| Rba. veldkampii | 3.8 ± 0.3 | 0 |
Figure 9.
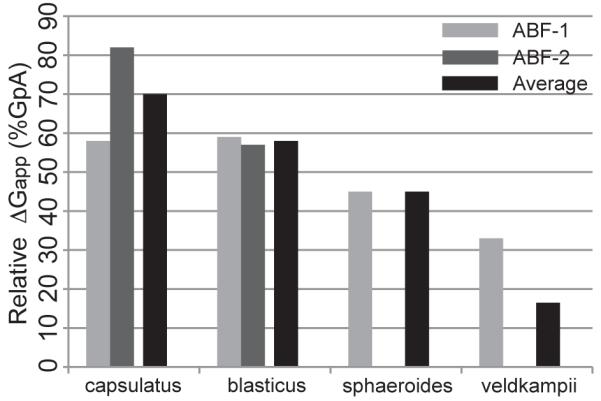
Free energy of association, ΔGapp, for various PufX sequences as a fraction of GpA ΔGapp. GpA ΔGapp was calculated in Hénin et al.40
Concluding Remarks
We have shown through experiments and molecular dynamics simulations that PufX from different Rba. species of purple photosynthetic bacteria exhibit distinct propensities towards homodimerization. This result can explain in part why core complexes have different oligomerization states in different species, i.e., due to the different inherent affinity of the TM regions of PufX to dimerize. In particular, species with PufX shown to be least likely to dimerize, namely, Rba. veldkampii, form only monomeric core complexes.13,17 On the other hand, Rba. blasticus that possesses dimeric core complexes6 is seen to have PufX with a relatively high dimerization affinity.
In addition to the ability for dimerization at the TM region, presence of aromatic amino acids with polar groups (tyrosine and tryptophan) appears to aid in stabilizing PufX helices in the membrane. Rba. veldkampii PufX contains no tyrosine or tryptophan, and also shows the lowest tendency for dimerization in its TM region. These two characteristics of Rba. veldkampii PufX, namely, the lack of anchoring residues and a TM region with low likelihood for self-association, make Rba. veldkampii PufX a poor candidate for forming homodimers. Interestingly, a tryptophan residue on the N-terminal end of the TM region is part of the recently identified PufX motif that is missing in Rba. veldkampii, but present in Rba. blasticus, Rba. capsulatus, and Rba. sphaeroides (Figure 1c).13
While Rba. blasticus, Rba. capsulatus, and Rba. sphaeroides PufX show some preference for self-association, their dimerization affinity is significantly lower than that of GpA.52,60 The lower dimerization affinity might be the reason why dimeric and monomeric core complexes are both present in Rba. blasticus and Rba. sphaeroides,4,6,18 as a subset of PufX helices might be in monomeric forms in the photosynthetic membrane, residing in monomeric core complexes. It is also possible that dimerization of PufX requires additional molecular interactions other than those arising from the PufX TM region, or between PufX and the rest of the core complex. For Rba. sphaeroides, there are experimental reports showing that the N-terminal segment of PufX is critical for the formation of dimeric core complexes,12,61 although the molecular role of these residues in PufX-assisted core complex dimerization is unclear. Dimerization of PufX might also be strengthened by interaction between PufX and LH1α helices observed previously for Rba. sphaeroides and Rba. capsulatus,62 or by the binding of the light-absorbing pigment bacteriochlorophyll (shown in Figure 1a and b as crosses) to PufX.59,63,64 Speculation that different Rba. species might have different organizations for their core complexes has also been raised.15,18
While identification of a GxxxG motif in the Rba. sphaeroides PufX sequence at the 31-35 position is intriguing,17,21 the motif by itself does not explain the observed oligomerization states of Rba. core complexes. As alluded to above, Rba. blasticus lacks this particular sequence motif, yet it has been confirmed to contain dimeric core complexes.6 It should be noted, however, that Rba. blasticus, Rba. capsulatus, and Rba. sphaeroides actually all feature a GxxxA/G motif at the 29-33 position that is not present in Rba. veldkampii (Figure 1c), with GxxxA previously suggested as a motif for dimerization of TM helices.55,65-69 The glycine residue at position 29 in PufX is actually conserved across the four Rba. species and an alanine residue is found at position 30 except for Rba. veldkampii PufX (Figure 1c), providing another small amino acid that allows for potential helix-helix interaction. It is conceivable that the combination of presence of protein-membrane anchoring provided by tyrosine or tryptophan, and small amino acids such as glycine and alanine at the helix-helix contact site, is a molecular principle that renders a PufX TM segment more prone to homodimerize.
Supplementary Material
Acknowledgement
We thank C. Neil Hunter and Pu Qian for many helpful discussions, Simon Scheuring for illuminating communications, and Ben Mueller for assistance in performing the TOXCAT assays. This work was supported by Grants NSF MCB-0744057 and NIH P41-RR005969. Computer time was provided by NCSA and TACC via Large Resources Allocation Committee Grant MCA93S028, resources of the Argonne Leadership Computing Facility at Argonne National Laboratory, supported by the Office of Science of the U.S. Department of Energy (Contract DE-AC02-06CH11357), and resources of the Oak Ridge Leadership Computing Facility at Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract DE-AC05-00OR22725.
Footnotes
Supporting Information Available
Complete citation for reference 31, six movies, seven additional figures and one table are available in Supporting Material accompanying this manuscript.
This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- (1).Jungas C, Ranck J, Rigaud J, Joliot P, Verméglio A. EMBO J. 1999;18:534–542. doi: 10.1093/emboj/18.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Siebert CA, Qian P, Fotiadis D, Engel A, Hunter CN, Bullough PA. EMBO J. 2004;23:690–700. doi: 10.1038/sj.emboj.7600092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Loach PA. Proc. Natl. Acad. Sci. USA. 2000;97:5016–5018. doi: 10.1073/pnas.97.10.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Scheuring S, Francia F, Busselez J, Melandris BA, Rigaud J-L, Lévy D. J. Biol. Chem. 2004;279:3620–3626. doi: 10.1074/jbc.M310050200. [DOI] [PubMed] [Google Scholar]
- (5).Scheuring S, Lévy D, Rigaud J-L. Biochim. Biophys. Acta. 2005;1712:109–127. doi: 10.1016/j.bbamem.2005.04.005. [DOI] [PubMed] [Google Scholar]
- (6).Scheuring S, Busselez J, Lévy D. J. Biol. Chem. 2005;280:1426–1431. doi: 10.1074/jbc.M411334200. [DOI] [PubMed] [Google Scholar]
- (7).Qian P, Hunter CN, Bullough PA. J. Mol. Biol. 2005;349:948–960. doi: 10.1016/j.jmb.2005.04.032. [DOI] [PubMed] [Google Scholar]
- (8).Cogdell RJ, Gall A, Köhler J. Quart. Rev. Biophys. 2006;39:227–324. doi: 10.1017/S0033583506004434. [DOI] [PubMed] [Google Scholar]
- (9).Qian P, Bullough PA, Hunter CN. J. Biol. Chem. 2008;283:14002–14011. doi: 10.1074/jbc.M800625200. [DOI] [PubMed] [Google Scholar]
- (10).Scheuring S, Sturgis JN. Photosyn. Res. 2009;102:197–211. doi: 10.1007/s11120-009-9413-7. [DOI] [PubMed] [Google Scholar]
- (11).Tunnicliffe RB, Ratcliffe EC, Hunter CN, Williamson MP. FEBS Lett. 2006;580:6967–6971. doi: 10.1016/j.febslet.2006.11.065. [DOI] [PubMed] [Google Scholar]
- (12).Ratcliffe EC, Tunnicliffe RB, Ng IW, Adams PG, Qian P, Holden-Dye K, Jones MR, Williamson MP, Hunter CN. Biochim. Biophys. Acta. 2011;1807:95–107. doi: 10.1016/j.bbabio.2010.10.003. [DOI] [PubMed] [Google Scholar]
- (13).Liu L-N, Sturgis JN, Scheuring S. J. Struct. Biol. 2010;173:138–145. doi: 10.1016/j.jsb.2010.08.010. [DOI] [PubMed] [Google Scholar]
- (14).Francia F, Wang J, Venturoli G, Melandri BA, Barz WP, Oesterhelt D. Biochemistry. 1999;38:6834–6845. doi: 10.1021/bi982891h. [DOI] [PubMed] [Google Scholar]
- (15).Holden-Dye K, Crouch LI, Jones MR. Biochim. Biophys. Acta. 2008;1777:613–630. doi: 10.1016/j.bbabio.2008.04.015. [DOI] [PubMed] [Google Scholar]
- (16).Scheuring S. Curr. Opin. Cell Biol. 2006;10:387–393. doi: 10.1016/j.cbpa.2006.08.007. [DOI] [PubMed] [Google Scholar]
- (17).Busselez J, Cottevielle M, Cuniasse P, Gubellini F, Boisset N, Lévy D. Structure. 2007;15:1674–1683. doi: 10.1016/j.str.2007.09.026. [DOI] [PubMed] [Google Scholar]
- (18).Crouch LI, Holden-Dye K, Jones MR. Biochim. Biophys. Acta. 2010;1797:1812–1819. doi: 10.1016/j.bbabio.2010.07.007. [DOI] [PubMed] [Google Scholar]
- (19).Gubellini F, Francia F, Busselez J, Venturoli G, Lévy D. Biochemistry. 2006;45:10512–10520. doi: 10.1021/bi0610000. [DOI] [PubMed] [Google Scholar]
- (20).Milhiet P-E, Gubellini F, Berquand A, Dosset P, Rigaud J-L, Grimellec CL, Lévy D. Biophys. J. 2006;91:3268–3275. doi: 10.1529/biophysj.106.087791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wang Z-Y, Suzuki H, Kobayashi M, Nozawa T. Biochemistry. 2007;46:3635–3642. doi: 10.1021/bi0618060. [DOI] [PubMed] [Google Scholar]
- (22).MacKenzie KR, Prestegard JH, Engelman DM. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- (23).Hsin J, Chipot C, Schulten K. J. Am. Chem. Soc. 2009;131:17096–17098. doi: 10.1021/ja905903n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Russ WP, Engelman DM. Proc. Natl. Acad. Sci. USA. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Chandler D, Hsin J, Harrison CB, Gumbart J, Schulten K. Biophys. J. 2008;95:2822–2836. doi: 10.1529/biophysj.108.132852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hsin J, Gumbart J, Trabuco LG, Villa E, Qian P, Hunter CN, Schulten K. Biophys. J. 2009;97:321–329. doi: 10.1016/j.bpj.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Sener MK, Hsin J, Trabuco LG, Villa E, Qian P, Hunter CN, Schulten K. Chem. Phys. 2009;357:188–197. doi: 10.1016/j.chemphys.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Hsin J, Chandler DE, Gumbart J, Harrison CB, Sener M, Strumpfer J, Schulten K. ChemPhysChem. 2010;11:1154–1159. doi: 10.1002/cphc.200900911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Trabuco LG, Schreiner E, Gumbart J, Hsin J, Villa E, Schulten K. J. Struct. Biol. 2011;173:420–427. doi: 10.1016/j.jsb.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).MacKerell AD, Jr., et al. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- (32).Foloppe N, MacKerrell AD., Jr. J. Comp. Chem. 2000;21:86–104. [Google Scholar]
- (33).MacKerell AD, Jr., Feig M, Brooks CL., III J. Comp. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- (34).Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- (35).Tuckerman ME, Berne BJ, Martyna GJ. J. Phys. Chem. B. 1992;97:1990–2001. [Google Scholar]
- (36).Brünger AT, Brooks CL, III, Karplus M. Chem. Phys. Lett. 1984;105:495–498. [Google Scholar]
- (37).Feller SE, Zhang YH, Pastor RW, Brooks BR. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- (38).Darve E, Pohorille A. J. Chem. Phys. 2001;115:9169–9183. [Google Scholar]
- (39).Hénin J, Chipot C. J. Chem. Phys. 2004;121:2904–2914. doi: 10.1063/1.1773132. [DOI] [PubMed] [Google Scholar]
- (40).Hénin J, Pohorille A, Chipot C. J. Am. Chem. Soc. 2005;127:8478–84. doi: 10.1021/ja050581y. [DOI] [PubMed] [Google Scholar]
- (41).Kim H, Hsin J, Liu Y, Selvin PR, Schulten K. Structure. 2010;18:1443–1449. doi: 10.1016/j.str.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Stockner T, Ash WL, MacCallum JL, Tieleman DP. Biophys. J. 2004;87:1650–1656. doi: 10.1529/biophysj.104.045310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Rodriguez-Gomez D, Darve E, Pohorille A. J. Chem. Phys. 2004;120:3563–3578. doi: 10.1063/1.1642607. [DOI] [PubMed] [Google Scholar]
- (44).Shaw WV. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- (45).Sulistijo ES, Jaszewski TM, MacKenzie KM. J. Biol. Chem. 2003;278:51950–51956. doi: 10.1074/jbc.M308429200. [DOI] [PubMed] [Google Scholar]
- (46).Senes A, Chadi DC, Law PB, Walters RFS, Nanda V, DeGrado WF. J. Mol. Biol. 2007;366:436–448. doi: 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]
- (47).Killian JA, von Heijne G. Trends Biochem. Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- (48).Yuen CTK, Davidson AR, Deber CM. Biochemistry. 2000;39:16155–16162. doi: 10.1021/bi0016117. [DOI] [PubMed] [Google Scholar]
- (49).de Planque MRR, Killian JA. Mol. Membr. Biol. 2003;20:271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- (50).Popot J, Engelman D. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- (51).Engelman DM, Chen Y, Chin C-N, Curran AR, Dixon AM, Dupuy AD, Lee AS, Lehnert U, Matthews EE, Reshetnyak YK, Senes A, Popot J-L. FEBS Lett. 2003;555:122–125. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- (52).Fleming KG, Ackerman AL, Engelman DM. J. Mol. Biol. 1997;272:266–275. doi: 10.1006/jmbi.1997.1236. [DOI] [PubMed] [Google Scholar]
- (53).Bowie JU. Curr. Opin. Struct. Biol. 2011;21:42–49. doi: 10.1016/j.sbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Senes A, Ubarretxena-Belandia I, Engelman DM. Proc. Natl. Acad. Sci. USA. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Senes A, Engel DE, DeGrado WF. Curr. Opin. Struct. Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- (56).Arbely E, Arkin IT. J. Am. Chem. Soc. 2004;126:5362–5363. doi: 10.1021/ja049826h. [DOI] [PubMed] [Google Scholar]
- (57).Derewendaf ZS, Lee L, Derewenda U. J. Mol. Biol. 1995;252:248–262. doi: 10.1006/jmbi.1995.0492. [DOI] [PubMed] [Google Scholar]
- (58).Lawrie CM, Sulistijo ES, MacKenzie KR. J. Mol. Biol. 2010;396:924–936. doi: 10.1016/j.jmb.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Aklujkar M, Beatty JT. Photosyn. Res. 2006;88:159–171. doi: 10.1007/s11120-006-9047-y. [DOI] [PubMed] [Google Scholar]
- (60).Duong MT, Jaszewski TM, Fleming KG, MacKenzie KR. J. Mol. Biol. 2007;371:422–434. doi: 10.1016/j.jmb.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Francia F, Wanf J, Zischka H, Venturoli G, Oesterhelt D. Eur. J. Biochem. 2002;269:1877–1885. doi: 10.1046/j.1432-1033.2002.02834.x. [DOI] [PubMed] [Google Scholar]
- (62).Recchia PA, Davis CM, Lilburn TG, Beatty JT, Parkes-Loach PS, Hunter CN, Loach PA. Biochem. 1998;37:11055–11063. doi: 10.1021/bi980657l. [DOI] [PubMed] [Google Scholar]
- (63).Parkes-Loach PS, Law CJ, Recchia PA, Kehoe J, Nehrlich S, Chen J, Loach PA. Biochemistry. 2001;40:5593–5601. doi: 10.1021/bi002580i. [DOI] [PubMed] [Google Scholar]
- (64).Law CJ, Chen J, Parkes-Loach PS, Loach PA. Photosyn. Res. 2003;75:193–210. doi: 10.1023/A:1023982327748. [DOI] [PubMed] [Google Scholar]
- (65).Senes A, Gerstein M, Engelman DM. J. Mol. Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- (66).Schneider D. FEBS Lett. 2004;577:5–8. doi: 10.1016/j.febslet.2004.10.029. [DOI] [PubMed] [Google Scholar]
- (67).Schneider D, Engelman DM. J. Mol. Biol. 2004;343:799–804. doi: 10.1016/j.jmb.2004.08.083. [DOI] [PubMed] [Google Scholar]
- (68).Kairys V, Gilson MK, Luy B. Eur. J. Biochem. 2004;271:2086–2092. doi: 10.1111/j.1432-1033.2004.04107.x. [DOI] [PubMed] [Google Scholar]
- (69).Rath A, Deber CM. Proteins: Struct., Func., Bioinf. 2007;70:786–793. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.