Abstract
The small GTPase Ran enzyme regulates critical eukaryotic cellular functions including nuclear transport and mitosis through the creation of a RanGTP gradient around the chromosomes. This concentration gradient is created by the chromatin bound RCC1 (regulator of chromosome condensation) protein which recruits Ran to nucleosomes and activates Ran’s nucleotide exchange activity. While RCC1 has been shown to bind directly with the nucleosome, the molecular details of this interaction were not known. We have determined the crystal structure of the RCC1-nucleosome core particle complex at 2.9 Å resolution, providing the first atomic view of how a chromatin protein interacts with the histone and DNA components of the nucleosome. Our structure also suggests that the Widom 601 DNA positioning sequence present in our nucleosomes forms a 145 bp and not the expected canonical 147 bp nucleosome core particle.
Mitotic spindle formation, the transport of macromolecules between the cytoplasm and the nucleus, and nuclear envelope formation are critical functions of a eukaryotic cell. These seemingly disparate functions are all regulated by a concentration gradient of the small GTPase Ran in its GTP bound state (RanGTP) around the chromosomes1–3. This RanGTP gradient signal is generated when the Ran guanine-exchange factor (RanGEF) also known as RCC1 (regulator of chromosome condensation), binds to the nucleosome repeating unit of chromatin, recruits Ran to chromatin and activates Ran’s nucleotide exchange activity.
The binding of RCC1 to the nucleosome is central to the formation of this spatial signal within the nucleus. Structurally, RCC1 is a β-propeller protein with an N-terminal tail extension4. Previous studies have shown that the RCC1 β-propeller domain binds the histone H2A/H2B dimer component of the histone octamer and that this interaction does not require the N-terminal tail implicated in DNA-binding5. An unusual post-translational modification, N-terminal α-methylation, of human RCC1 appears to further regulate RCC1’s association with chromatin6,7. Despite these important findings, we currently lack a structural description of how RCC1 interacts with the nucleosome.
The inadequate understanding of how RCC1 binds the nucleosome reflects a more fundamental deficit of structural information for how chromatin enzymes and factors recognize the nucleosome. The nucleosome core particle is an assembly of 145–147 bp of DNA wrapped around an octamer of histone proteins (2 copies each of the 4 core histone proteins H2A, H2B, H3 and H4). The crystal structure of the nucleosome core particle was determined at 2.8 Å thirteen years ago8, and since then structures of nucleosome core particles containing histone proteins from different species and variant DNA sequences of the original human α-satellite sequence have provided important structural insight9–17. However, with the exceptions of a viral peptide and artificial ligands bound to the nucleosome18,19, little structural information is available for the molecular recognition of the nucleosome despite the central role of such recognition in chromatin biology and gene regulation.
We have grown crystals of Drosophila RCC1 (Bj1) bound to recombinant nucleosome core particles and used these crystals to determine the structure of the 300 kDa RCC1/nucleosome core particle complex at 2.9 Å resolution. We find that loops within the RCC1 β-propeller domain interact with the histone component and unexpectedly, with the DNA component of the nucleosome core particle. This first atomic view of a chromatin protein/nucleosome complex also provides the first structure of a nucleosome core particle containing the Widom 601 nucleosome DNA positioning sequence selected for high affinity histone octamer association20 and commonly used in chromatin biology studies in vitro. Our results indicate that 145 bp of the Widom 601 DNA wraps around the histone octamer to form the nucleosome core particle instead of 147 bp of DNA in the canonical human α-satellite nucleosome core particle.
Overview of the complex
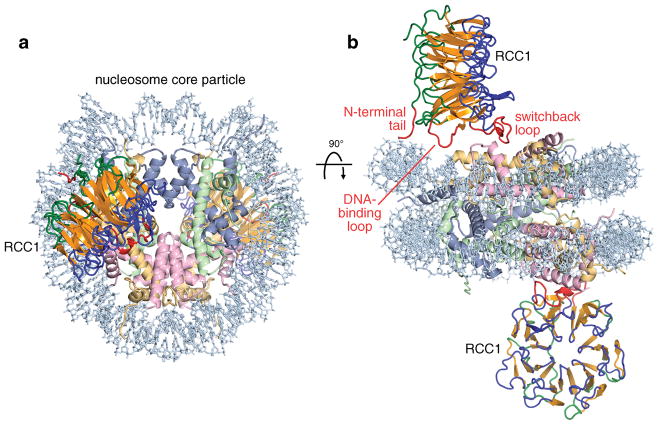
The RCC1/nucleosome core particle crystal structure shows pseudo-twofold symmetry: one RCC1 molecule interacts with each of the two histone faces of the nucleosome (Fig. 1). The RCC1 β-propeller domains extend from each side of the disk-shaped nucleosome core particle, increasing the dimension along the helical axis of the nucleosome from 60 Å to 160 Å. The complex can be likened to the front wheel of a tricycle, with the nucleosome core particle forming the wheel and each RCC1 molecule forming a pedal on the side of the wheel. Each RCC1 molecule is positioned with its β-propeller wheel perpendicular to the nucleosome histone face, and oriented approximately 90° with respect to each other. The two RCC1 molecules make essentially identical interactions with the nucleosome. RCC1 employs loops in the fourth blade of its β-propeller to interact with histones and nucleosomal DNA, while the N-terminal tail is positioned for further interactions with nucleosomal DNA. The RCC1-histone contacts are made with the H2A/H2B histone dimer surface of the nucleosome core particle; no interactions with histones H3 or H4 occur. The binding of each RCC1 molecule to the nucleosome excludes about 910 Å2 of solvent accessible area on the nucleosome surface, with interactions with histone and DNA contributing 77% and 23% of solvent excluded surface area respectively.
Figure 1.
Crystal structure of RCC1-nucleosome core particle complex. (a) View of complex looking down the DNA superhelix axis. RCC1 is shown in yellow-orange (central strand region), blue and green (loop regions) and red (N-terminal tail, DNA-binding loop and switchback loop), while the histone H3, H4, H2A, H2B and DNA components of the nucleosome core particle are shown in cornflower blue, light green, wheat, pink and light blue respectively. (b) View of the complex perpendicular to that in (a). The two RCC1 molecules make equivalent interactions on either side of the nucleosome core particle.
RCC1-histone interactions
β-propeller proteins often use the loops on either face of the β-propeller toroid to bind to interacting proteins21–23. Since RCC1 employs loops on one such face to bind to Ran24, it was expected that RCC1 might use loops on the opposite face to bind to the nucleosome4,6,7. Based on biochemical and modeling studies, we have recently proposed instead that the RCC1 β-propeller uses what we have termed the switchback loop (between strands 4C and 4D, Suppl. Fig. 1) to bind to the nucleosome25. This switchback loop is located not on the face opposite to Ran, but instead borders the RCC1 β-propeller on the same face that interacts with Ran4,24.
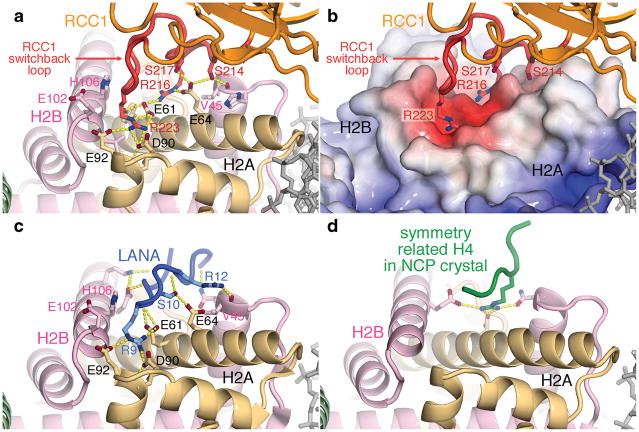
In our crystal structure, we find that RCC1 does, in fact, utilize its switchback loop to bind to an acidic patch on the histone dimer (Fig. 1, 2a. electron density for this region provided in Suppl. Fig. 2a). The direct interaction of RCC1 with the histone dimer was previously observed by Nemergut and colleagues5, and we had further predicted an interaction with the acidic patch of the histone dimer based on our finding that the Kaposi’s sarcoma-associated herpesvirus LANA (latency-associated nuclear antigen) peptide that binds to this acidic patch competes with RCC1 for binding to the nucleosome in vitro18,25. The histone H2A/H2B dimer acidic patch on the nucleosome can be thought of as two depressions separated by a shallow ridge in between (Fig. 2b). RCC1 uses arginines from its switchback loop (Arg216 and Arg223) to bind to each depression formed in part by the triad of acidic histone H2A residues Glu61, Asp90 and Glu92. An extensive network of hydrogen bonds alternating between RCC1 and histone side chains as well as van der Waals contacts mediate this interaction (Fig. 2a and detailed in Supplementary Information). These structural results are supported by biochemical findings. In particular, the critical role of RCC1 Arg223 was predicted in our study which identified the corresponding human RCC1 residue, Arg217, as necessary for the RCC1 β-propeller to interact with nucleosomes25.
Figure 2.
Interactions of RCC1, LANA peptide and H4 peptide with the nucleosome histone dimer acidic patch. (a) RCC1-histone interactions in ribbon representation. RCC1 employs side chains in its switchback loop to bind to the histone dimer. Hydrogen bonds are depicted as yellow dotted lines. (b) RCC1 recognizes acidic histone H2A/H2B dimer surface. APBS49-calculated electrostatics (−3 to +20 kT) were mapped onto histone surfaces using same view as in (a). (c) LANA-histone interactions in LANA-nucleosome crystal structure (PDB id 1ZLA). Key side chains of the LANA peptide (blue) are shown. (d) Histone H4-histone crystal contacts in nucleosome core particle (NCP) crystal structure (PDB id 1KX5).
The interaction of Drosophila Arg223 with the H2A acidic triad is very similar, though not identical, to the interactions of the LANA peptide Arg9 with the same triad of H2A residues18 (Fig. 2a and 2c). Both RCC1 and LANA peptide interact extensively with the histone dimer acidic patch, providing a straightforward structural explanation for the observed competitive binding of RCC1 and LANA peptide to the nucleosome. Both RCC1 and LANA employ two arginine side chains to interact with histone dimer residues, both make van der Waals contacts with H2B Glu102, Leu103, His106 and Val45, and both contain serine side chains that interact with H2A Glu64. However, whereas side chain to side chain hydrogen bonds pervade the polar interactions between RCC1 and the histone dimer, both side chain to side chain and side chain to main chain hydrogen bonds characterize the LANA-histone dimer interface. It should be noted that the histone dimer acidic patch recognized by RCC1 is part of the same acidic patch that histone H4 tail binds to via crystal packing interactions in the nucleosome core particle crystals8,12 (Fig. 2d). The interaction between the H4 tail and the histone dimer acidic patch has been proposed to play an important role in higher order chromatin structure formation26,27. It is a striking coincidence that the available atomic level descriptions of interactions with the histone octamer surface of the nucleosome core particle all involve the same acidic patch.
RCC1-nucleosome DNA interactions
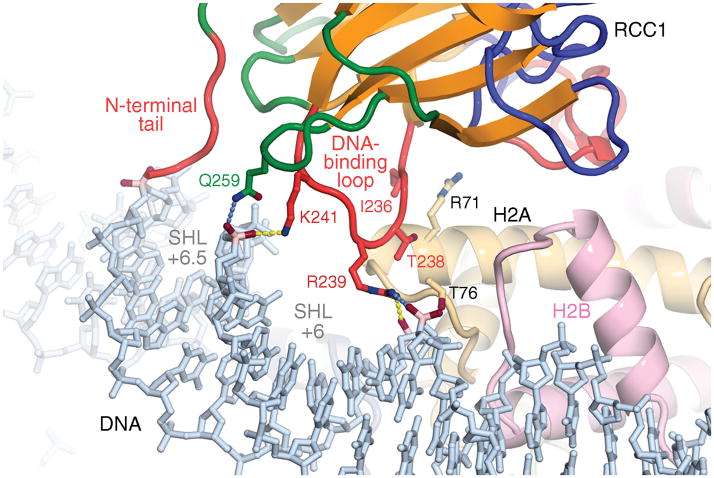
The RCC1-nucleosome complex crystal structure shows that the RCC1 loop between strands 4D and 5A (the “DNA-binding loop”, Suppl. Fig. 1) binds nucleosomal DNA via phosphate interactions (Fig. 3). This interaction had not been anticipated in previous studies. In comparison to the extended hydrogen bonding network between RCC1 and the histone dimer acidic patch, the interactions between RCC1 and nucleosomal DNA are relatively modest: RCC1 Lys241 and Arg239 bridge across the major groove near superhelical location (SHL) 6, about 1.5 turns from the end of the nucleosome core particle DNA, to hydrogen bond with phosphate groups. In addition, the NH1 atom of RCC1 Arg239 interacts with the DNA phosphate group either through charged interactions or possibly through a hydrogen bond (Arg239 NH1 is 3.5 Å from the Guanine131 O2P atom). Besides these polar interactions by RCC1’s DNA binding loop to nucleosomal DNA, Gln259 in an adjacent loop is in position to hydrogen bond to the same phosphate contacted by Lys241 (Gln259 NE2 atom is 3.4 Å from Guanine13 O1P atom). These contacts with the DNA phosphate backbone are consistent with RCC1’s role as a non-DNA-sequence specific chromatin factor. For example, Koerber et al have shown that the yeast RCC1 ortholog, Srm1/Prp20, binds across the genome to most nucleosomes without sequence specificity28. Complementing the RCC1-DNA interactions are a limited set of van der Waals interactions between residues in the RCC1 DNA-binding loop and histone H2A (Fig. 3 and detailed in Supplementary Information).
Figure 3.
RCC1-nucleosomal DNA interactions. RCC1’s N-terminal tail approaches the DNA minor groove at superhelical location (SHL) 6.5, while its DNA-binding and adjacent loop binds across the major groove at SHL 6. Hydrogen bonds (<3.2 Å) are shown as yellow dotted lines, while potential hydrogen bonds are shown in blue.
We have tested the prediction that the DNA-binding loop contributes to RCC1-nucleosome binding by mutating basic residues in the human RCC1 DNA-binding loop. We find that the human RCC1 β-propeller domain with Lys232, Arg234 and Arg237 all mutated to Ala is unable to bind to nucleosome core particles (Suppl. Fig. 3). In contrast, negative controls employing RCC1 with triple mutations in other similarly exposed loop regions were unaffected in their nucleosome binding activity. We also prepared RCC1 variants containing individual alanine mutations of Lys232, Arg234 and Arg237 and we find that each of these is able to bind to nucleosomes in the pulldown assay (data not shown). This suggests that multiple interactions within the DNA-binding loop might be necessary to stabilize the RCC1-nucleosome complex.
RCC1’s interactions with nucleosomal DNA is not limited to those made by RCC1 loops. The N-terminal arm of RCC1 has been implicated in DNA-binding and we have recently demonstrated that this arm is involved in nucleosome binding25. Although the Drosophila RCC1 polypeptide present in the crystal includes this N-terminal tail, residues 2–27 are not visible in the electron density map and are presumably disordered in the crystal. Residues 28 and 29 of the N-terminal arm are positioned to enter the DNA minor groove adjacent to the major groove contacted by the RCC1 DNA-binding loop. To address the role of N-terminal arm residues in nucleosome binding, we prepared Drosophila RCC1 N-terminal deletion variants. Drosophila RCC1 lacking residues 2–23 formed stable complexes with nucleosome core particles in size exclusion chromatography experiments, and the resulting RCC1/nucleosome core particle complex produced crystals with similar morphologies to complexes containing full length RCC1 (data not shown). In contrast, Drosophila RCC1 lacking residues 2–29 failed to form a stable complex with nucleosome core particles in parallel experiments. This suggests that at least some of residues 24 to 28 are required for Drosophila RCC1 to bind to the nucleosome core particle. We note that Drosophila RCC1 residues 22–27 (KAKRAR) contain positively charged side chains that could potentially interact with DNA. We further speculate that these N-terminal arm residues bind in the DNA minor groove or with the backbone phosphate in multiple conformations in the crystal, and that the lack of a unique or predominant conformation accounts for the missing electron density despite the apparent importance of this interaction.
601 nucleosome structure
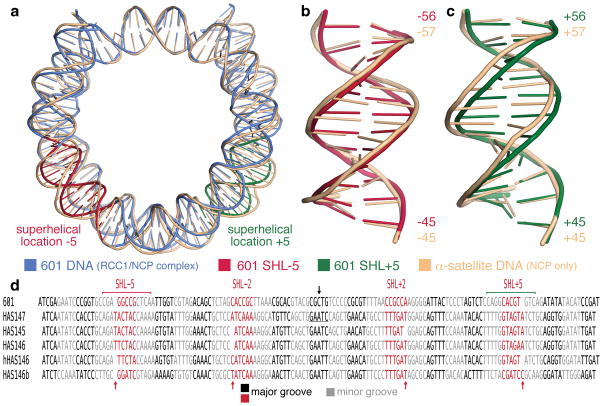
Our structure provides a first view of a 601 sequence nucleosome core particle in contrast to all previous nucleosome crystal structures which contained the human α-satellite DNA sequence or variants thereof. The Widom 601 sequence was selected from a random DNA pool to bind with high affinity to the histone octamer20 and this sequence has become a de facto standard for in vitro nucleosome reconstitutions in chromatin biology research. The Widom 601 nucleosome core particle in our structure adopts a conformation that is overall similar to the 147 bp and other human α-satellite nucleosome core particles (Fig. 4a) despite the different crystal packing from previous nucleosome core particle structures which have all crystallized in the same crystal space group (detailed in Supplementary Information). Most of the direct histone-DNA contacts observed in the 147 bp human α-satellite nucleosome core particle structure are maintained in the 601 nucleosome core particle. The two structures share striking 10 bp periodicities in the tip DNA base parameter and the roll, slide and twist DNA base step parameters throughout the nucleosome (this periodicity is particularly conspicuous when a rolling average is used to plot the parameters) and large alternations of the shift parameter especially within the central 70 bp12 (Suppl. Fig. 4). The DNA of the 601 and the 147 bp human α-satellite nucleosomes share very similar conformations within the central 20 bp around the dyad, consistent with the large number of histone-DNA interactions at superhelical locations (SHL) ±0.5.
Figure 4.
The Widom 601 sequence forms a 145 bp nucleosome core particle. (a) Superposition of 601 nucleosome core particle DNA (in blue) and human αsatellite nucleosome core particle DNA (peach) in simplified cartoon representation. The major differences between the two DNA structures at SHL +5 and −5 are highlighted in red and green respectively. (b) and (c) Superpositions of the two DNA structures at SHL +5 and −5 respectively. Numbers shown are the base positions from the nucleosome dyad. (d) Alignment of DNA present in nucleosome core particle crystal structures shows 5 bp major and minor grooves blocks. The 5 or 6 bp major groove blocks at SHL ±2 and ±5 accommodates differences in the DNA structures (red arrows). DNA dyad indicated by black arrow. Human α-satellite sequence alignments based on Fig.1 of Ong et al13.
Despite these similarities, the 601 nucleosome structure manifests remarkable differences from the 147 bp human α-satellite nucleosome. In particular, we find that the 601 DNA sequence in our structure unexpectedly forms a 145 bp nucleosome core particle. The disparate DNA lengths within the 601 and α-satellite nucleosome core particles are the consequence of differences localized around SHL ±5 (Fig. 4a, electron density provided in Suppl. Fig. 2b and 2c). At these two locations, the 601 DNA is overwound compared to the α-satellite DNA, resulting in 12 bp of 601 DNA where 13 bp of α-satellite DNA would be (Fig. 4b and 4c). This increase in DNA twist for the 601 DNA around SHL±5 is accompanied by larger DNA slide values compared to the α-satellite DNA nucleosome (Suppl. Fig. 4) and some changes in histone-DNA contacts at this location (Suppl. Fig. 5 and Supplementary Information). Our structure of the 601 nucleosome lends credence to the idea that nucleosomes possess an ability to absorb some variations in DNA lengths at both SHL±2 and SHL±5 (Fig. 4d and Supplementary Information) and this feature may have implications for the mechanism of chromatin remodelling enzymes, including those found to interact with nucleosomal DNA near these locations29–31.
Implications for Ran recruitment
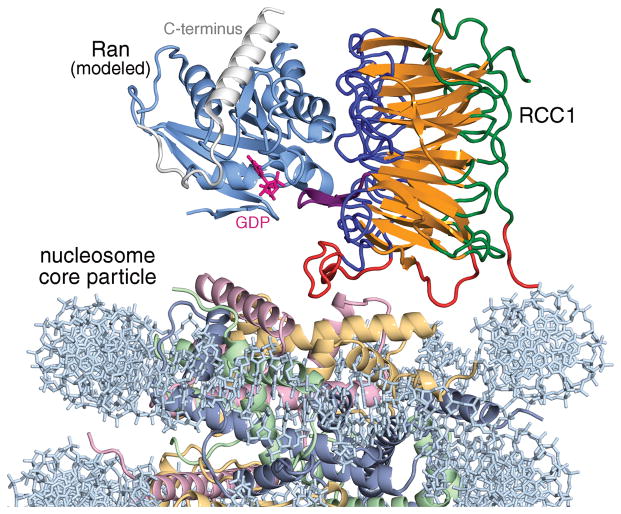
RCC1 recruits and activates the small GTPase Ran to the nucleosome through direct interactions with both Ran and the nucleosome. Our structure of the RCC1/nucleosome core particle complex and the previously determined structure of the RCC1/Ran binary protein complex24 allow us to examine the implications for Ran recruitment to chromatin. Superposition of the two crystal structures via the common RCC1 component shows that Ran would not interact with either histone or DNA components of the nucleosome in this model for the RCC1/Ran/nucleosome triple complex (Fig. 5). Such a model would not explain how binding to nucleosomes increases RCC1’s guanine-exchange activity on Ran5, nor would it explain how Ran is able to interact with the nucleosome both in the presence and absence of RCC132,33.
Figure 5.
Model for Ran-RCC1-nucleosome core particle complex assuming no conformational changes. Ran (light blue) was modeled by aligning the RCC1 component of the Ran-RCC1 complex crystal structure (PDB id 1I2M) with one of the RCC1 subunits in the RCC1-nucleosome core particle structure. The flexible C-terminus of Ran is shown in white. The GDP nucleotide (magenta) was modeled using the nucleotide present in the RanGDP structure (PDB id 3GJ0). If Ran interacts with histones in the Ran-RCC1-nucleosome complex, a conformational change in either RCC1 or Ran presumably must occur.
We recognize at least two possibilities to resolve these discrepancies. Firstly, a conformational change in Ran might allow Ran to directly interact with the nucleosome. Ran’s C-terminal 20 amino acids adopts different conformations in the 10 or more crystal structures that contain Ran on its own or complexed with partner proteins. This C-terminal region (colored white in Fig. 5) forms a linker and an α-helix which folds back on the globular region of Ran in the GDP-bound state34. However, in the GTP bound state, this C-terminal region can be disordered or the helix can be positioned for interactions with other proteins (as observed in structures of Ran-RanBP1 complexes)35–37. It is possible that Ran employs its C-terminal helix to interact with the nucleosome in the presence of RCC1, perhaps via interactions previously observed for Ran with the histone H3/H4 tetramer32 or perhaps with DNA. A second possibility is that RCC1 modulates its binding to the nucleosome in the presence of Ran. If the RCC1 β-propeller were to pivot about the switchback loop region towards the nucleosome, it could position Ran for direct contact with the nucleosome. Doing so would juxtapose Ran’s guanine nucleotide binding site to the nucleosome, providing a potential mechanistic basis for how RCC1 binding to the nucleosome enhances RCC1’s nucleotide-exchange activity on Ran. In this scenario, the nucleosome contacts made by the RCC1 DNA-binding loop and perhaps the N-terminal arm would need to be broken, but they could be compensated by new Ran-nucleosome interactions. This second model is similar to one that we have proposed for the Ran-RCC1-nucleosome complex25. The two models we describe here differ in whether Ran or RCC1 undergoes conformational change, but the two models are not mutually exclusive. Future biochemical and structural studies should clarify how these two chromatin proteins can interact with the nucleosome in a ternary complex to mediate their critical biological functions.
Methods Summary
The complex of Drosophila RCC1(2–422) and nucleosome core particles containing Xenopus core histones and the 147 bp Widom 601 sequence were purified by size exclusion chromatography and crystallized against 25 mM sodium acetate pH 5.5, 25 mM sodium citrate, 1 mM DTT and 6–7% polyethylene glycol monomethyl ether 2,000 (PEG MME 2,000) at 21°C. Crystals were soaked in 25 mM sodium acetate pH 5.5, 25 mM sodium citrate, 1 mM DTT, 5% ethanol, 10% PEG MME 2,000 containing increasing concentrations of polyethylene glycol 400 (0 to 24% in 2% increments) before flash cooling in liquid nitrogen. Diffraction data were collected using an ADSC Quantum 315 CCD detector at Advanced Photon Source’s NE-CAT beamline 24-ID-E, and the data was processed using the HKL-2000 program suite38. The structure was solved by molecular replacement using Phaser software39 and a search model containing Drosophila RCC1, the histone octamer with tails removed, and the 147 bp human αsatellite DNA, each treated as a rigid body. Crystallographic refinement was carried out using REFMAC540 and PHENIX41 together with manual model building in COOT42. The structure was refined to 2.9 Å resolution with Rwork/Rfree of 17.49/21.55%. All molecular graphics were prepared using PyMOL43.
Methods
Complex preparation and crystallization
Drosophila RCC1(2–422) was expressed in BL21(DE3)pLysS E. coli as an N-terminal hexahistidine fusion at 18°C. The fusion protein was purified by metal affinity chromatography using Talon resin (Clontech), the N-terminal tag removed using tobacco etch virus (TEV) protease, and the cleaved protein further purified by SourceS cation and SourceQ anion exchange chromatography (GE Healthcare). Recombinant Xenopus core histone were expressed, purified, reconstituted with the Widom 601 DNA sequence into nucleosome core particles essentially as described previously44. To generate the RCC1-nucleosome core particle complex, Drosophila RCC1(2–422) protein was mixed with nucleosome core particles in a 2.2:1 molar ratio and purified by Superdex 200 HR size exclusion chromatography (GE Healthcare). Crystals of the complex were grown by mixing 1 μl of complex (~12–18 mg/ml) in 5 mM Tris-Cl pH 7.5, 50 mM NaAc, 1 mM DTT with 1 μl of 25 mM sodium acetate pH 5.5, 25 mM sodium citrate, 1 mM DTT, 6 to 7% PEG2000-MME and then overlaid with 50–75 μl Al’s oil at 21°C in 96 well microtiter plates. Crystals were also grown by hanging drop or sitting drop vapor diffusion method with microseeding.
Post-crystallization soaks
The RCC-nucleosome core particle crystals diffract to about 6.5 Å at room temperature using a laboratory X-ray source when mounted in the mother liquor. Initial cryocooling experiments produced diffraction to about 5.0 Å using a synchrotron X-ray source. To improve the diffraction quality, the crystals were soaked in a base soak solution (25 mM sodium acetate buffer pH 5.5, 25 mM sodium citrate, 1 mM DTT, 5% ethanol, 10% polyethylene glycol monomethyl ether 2,000 [PEG MME 2,000]) at 4°C and then transferred from 0 to 24% PEG 400 in 2% increments with 10–15 minutes between each step. The soaked crystals were then flash frozen in liquid nitrogen for data collection.
Data collection, data processing, model building and refinement
Diffraction data were collected using an ADSC Quantum 315 CCD detector at Advanced Photon Source’s NE-CAT beamline 24-ID-E and the data processed using the HKL-2000 program suite38. One copy of the complex is present in the P21 space group asymmetric unit, corresponding to a Matthews packing density coefficient of 3.34 Å3 Da−1 or 63% solvent content. The structure was solved by molecular replacement using Phaser software39 and a search model containing three rigid bodies: Drosophila RCC1(39–211, 242–415), the histone octamer with histone tails removed, and the 147 bp human α-satellite DNA (PDB id 1KX5) with the DNA bases manually changed to match the Widom 601 sequence (Phaser final log-likelihood gain of 7292). The difference electron density map after one round of restrained refinement of the molecular replacement structure solution showed clear positive electron density for the RCC1 N-terminal region (residues 29–38) and the RCC1 switchback loop region (residues 212–241) omitted in the molecular replacement model. Crystallographic refinement was carried out using REFMAC540 and PHENIX41 together with manual model building in COOT42. The final model contains RCC1 residues 28–418 for chain K, residues 28–421 for chain L; histone H3 residues 37–134 for chain A, residues 40–134 for chain E; histone H4 residues 20–101 for chain B, 17–102 for chain F; histone H2A residues 12–118 for chain C, residues 12–118 for chain G; histone H2B residues 29–121 for chain D, residues 29–121 for chain H; DNA residues −72 to +73 for chain I and residues −72 to +73 for chain J. The histone tails are generally not visible in this structure. The stereochemistry of the protein components was analyzed using the program PROCHECK45. Simulated annealing omit maps were calculated using the CNS software package46.
The complex contains two copies of RCC1 positioned symmetrically about the nucleosome core particle, itself a pseudosymmetric molecule. However, the nucleosome core particle in the complex contains the asymmetric 601 DNA sequence and there does not appear to be sequence-specific contacts between RCC1 and the nucleosome whether within a complex or between complexes in the crystal that would favor one orientation of the DNA. While the electron density for the DNA clearly defines the DNA path around the histone octamer, it is not clear enough here to determine the sequence of DNA at 2.9 Å. Thus we cannot definitively determine the orientation of the DNA, nor can we exclude the possibility that crystal might contain a mixture of nucleosome core particles which differ in the two-fold orientation of the DNA sequence. We have selected one orientation after refining the complex in either orientations because the DNA in this chosen orientation produced a slightly lower real space R-factor (12.33% vs 12.63%) in composite omit maps and because the chosen orientation produced a marginally lower Rfree (21.55% vs 21.79%). It should be noted that the DNA conformation in the two models are very similar, as are the features described for the chosen orientation (RCC1-DNA interactions, stretching of DNA around SHL±5, the base pair and base pair step parameters shown in Suppl. Fig. 4, histone-DNA contacts around SHL±5).
Data analysis and molecular graphics
The CURVES+ software47 was used to analyze DNA parameters. Root mean square differences (rmsd) were calculated using LSQMAN48. All molecular graphics were prepared using PyMOL software43 with electrostatic potentials calculated using APBS49.
Pulldown assays
The pulldown assays in Suppl. Figure 3 used recombinant nucleosome core particles tagged at the N-terminus of histone H2B with the combination Strept-hexahistidine-TEV site affinity tag50. Pulldown assays were performed as described previously25.
Supplementary Material
Acknowledgments
We are grateful to Manoj Saxena for sharing unpublished coordinates of Drosophila RCC1 and to Kristin Wiley, Dylan Schlaich and Michael Porzio for technical assistance. We thank the staff of APS NE-CAT beamline 24-ID-E and Cornell CHESS beamlines A1 and F1 for their assistance during synchrotron data collection and to Neela Yennawar at the Penn Sate Huck Institutes X-ray core facility. We also thank Will Selleck, Melanie Adams, the Tan laboratory and the Penn State Center for Eukaryotic Gene Regulation for discussions, to Todd Stukenberg for advice and encouragement at the initiation of this project, to Jonathan Widom for sending the 601 nucleosome DNA positioning sequence and to the Pew Scholar 20th Reunion Meeting for stimulating this project.
Footnotes
Author Contributions
R.D.M cloned and purified macromolecules, crystallized, collected, processed X-ray data, refined and analyzed the structure; J.R.E. performed pulldown assays and collected X-ray data; H.P.Y. collected and processed X-ray data, S.T. designed the study, cloned and purified macromolecules, crystallized, collected X-ray data, analyzed the results and wrote the paper. All authors commented on the manuscript.
Author Information
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data Bank under accession code 3MVD. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Bibliography
- 1.Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, et al. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- 2.Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- 3.Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renault L, Nassar N, Vetter I, Becker J, Klebe C, et al. The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- 5.Nemergut ME, Mizzen CA, Stukenberg T, Allis CD, Macara IG. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 2001;292:1540–1543. doi: 10.1126/science.292.5521.1540. [DOI] [PubMed] [Google Scholar]
- 6.Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF, et al. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat Cell Biol. 2007;9:596–603. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hao Y, Macara IG. Regulation of chromatin binding by a conformational switch in the tail of the Ran exchange factor RCC1. J Cell Biol. 2008;182:827–836. doi: 10.1083/jcb.200803110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 9.Bao Y, White CL, Luger K. Nucleosome core particles containing a poly(dA.dT) sequence element exhibit a locally distorted DNA structure. J Mol Biol. 2006;361:617–624. doi: 10.1016/j.jmb.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 10.Chakravarthy S, Luger K. The histone variant macro-H2A preferentially forms “hybrid nucleosomes”. J Biol Chem. 2006;281:25522–25531. doi: 10.1074/jbc.M602258200. [DOI] [PubMed] [Google Scholar]
- 11.Clapier CR, Chakravarthy S, Petosa C, Fernandez-Tornero C, Luger K, et al. Structure of the Drosophila nucleosome core particle highlights evolutionary constraints on the H2A-H2B histone dimer. Proteins. 2008;71:1–7. doi: 10.1002/prot.21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 13.Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J Mol Biol. 2007;368:1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 14.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 15.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 16.Tsunaka Y, Kajimura N, Tate S, Morikawa K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005;33:3424–3434. doi: 10.1093/nar/gki663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, et al. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 19.Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, et al. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J Mol Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 20.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 21.Davis TL, Bonacci TM, Sprang SR, Smrcka AV. Structural and molecular characterization of a preferred protein interaction surface on G protein beta gamma subunits. Biochemistry. 2005;44:10593–10604. doi: 10.1021/bi050655i. [DOI] [PubMed] [Google Scholar]
- 22.Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gbetagamma. Science. 2003;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- 23.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 24.Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1) Cell. 2001;105:245–255. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- 25.England JR, Huang J, Jennings MJ, Makde RD, Tan S. RCC1 uses a conformationally diverse loop region to interact with the nucleosome: a model for the RCC1-nucleosome complex. J Mol Biol. 2010;398:518–529. doi: 10.1016/j.jmb.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 27.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 28.Koerber RT, Rhee HS, Jiang C, Pugh BF. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell. 2009;35:889–902. doi: 10.1016/j.molcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edayathumangalam RS, Weyermann P, Dervan PB, Gottesfeld JM, Luger K. Nucleosomes in solution exist as a mixture of twist-defect states. J Mol Biol. 2005;345:103–114. doi: 10.1016/j.jmb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Gangaraju VK, Prasad P, Srour A, Kagalwala MN, Bartholomew B. Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol Cell. 2009;35:58–69. doi: 10.1016/j.molcel.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 32.Bilbao-Cortes D, Hetzer M, Langst G, Becker PB, Mattaj IW. Ran binds to chromatin by two distinct mechanisms. Curr Biol. 2002;12:1151–1156. doi: 10.1016/s0960-9822(02)00927-2. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C, Goldberg MW, Moore WJ, Allen TD, Clarke PR. Concentration of Ran on chromatin induces decondensation, nuclear envelope formation and nuclear pore complex assembly. Eur J Cell Biol. 2002;81:623–633. doi: 10.1078/0171-9335-00288. [DOI] [PubMed] [Google Scholar]
- 34.Partridge JR, Schwartz TU. Crystallographic and biochemical analysis of the Ran-binding zinc finger domain. J Mol Biol. 2009;391:375–389. doi: 10.1016/j.jmb.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran × GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 36.Seewald MJ, Korner C, Wittinghofer A, Vetter IR. RanGAP mediates GTP hydrolysis without an arginine finger. Nature. 2002;415:662–666. doi: 10.1038/415662a. [DOI] [PubMed] [Google Scholar]
- 37.Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- 38.Otwinoski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Delano Warren L. The PyMOL molecular graphics system. Available at http://www.pymol.org.
- 44.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK - a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 46.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 47.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr D Biol Crystallogr. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 49.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan S, Kern RC, Selleck W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr Purif. 2005;40:385–395. doi: 10.1016/j.pep.2004.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.