Abstract
Sugars have signaling roles in a wide variety of developmental processes in plants. To elucidate the regulatory components that constitute the glucose signaling network governing plant growth and development, we have isolated and characterized two Arabidopsis glucose insensitive mutants, gin5 and gin6, based on a glucose-induced developmental arrest during early seedling morphogenesis. The T-DNA-tagged gin6 mutant abrogates the glucose-induced expression of a putative transcription factor, ABI4, previously shown to be involved in seed-specific abscisic acid (ABA) responses. Thus, ABI4 might be a regulator involved in both glucose- and seed-specific ABA signaling. The characterization of the gin5 mutant, on the other hand, reveals that glucose-specific accumulation of ABA is essential for hexokinase-mediated glucose responses. Consistent with this result, we show that three ABA-deficient mutants (aba1-1, aba2-1, and aba3-2) are also glucose insensitive. Exogenous ABA can restore normal glucose responses in gin5 and aba mutants but not in gin6 plants. Surprisingly, only abi4 and abi5-1 but not other ABA-insensitive signaling mutants (abi1-1, abi2-1, and abi3-1) exhibit glucose insensitivity, indicating the involvement of a distinct ABA signaling pathway in glucose responses. These results provide the first direct evidence to support a novel and central role of ABA in plant glucose responses mediated through glucose regulation of both ABA levels by GIN5 and ABA signaling by GIN6/ABI4.
Keywords: Glucose insensitive, Arabidopsis thaliana, glucose regulation, ABA, abi, hexokinase
Sugars act as regulatory molecules that control many aspects of physiology and development in both prokaryotes and eukaryotes (Grupe et al. 1995; Saier et al. 1995; Koch 1996; Smeekens and Rook 1997; Gancedo 1998; Gibson and Graham 1999; Roitsch 1999; Sheen et al. 1999). In higher plants, sugar signals modulate many important processes such as germination, seedling development, leaf and root morphogenesis, flowering, embryogenesis, wounding and pathogen responses, and senescence, in addition to gene expression (Koch 1996; Moore and Sheen 1999; Sheen et al. 1999; Wobus and Weber 1999).
Although sucrose is the major carbon form translocated in plants and is likely to mediate sucrose-specific signaling pathways (Chiou and Bush 1998; Rook et al. 1998), hexoses seem to play a major role in plant sugar regulation (Sturm and Tang 1999). Different lines of evidence point to the involvement of hexokinase (HXK) as a critical component in one of the sugar sensing and signaling pathways in plants (Jang and Sheen 1997; Smeekens and Rook 1997; Pego et al. 1999; Sheen et al. 1999). Analysis of transgenic Arabidopsis plants with either increased or decreased HXK levels show a hyper- or hyposensitivity to high glucose concentrations during germination and seedling development (Jang et al. 1997). Overexpression of AtHXK1 in transgenic tomato also alters photosynthesis, growth, and senescence (Dai et al. 1999). Recent studies with an AtHXK1 null mutation indicate its predominant role in many glucose responses and the uncoupling of glucose signaling from glucose metabolism (Moore and Sheen 1999). This HXK-dependent sugar signaling pathway has been shown to be responsible for the regulation of genes including several photosynthetic genes, nitrate reductase, and others (Jang and Sheen 1997; Sheen et al. 1999). In addition, through HXK-independent pathways, glucose also positively regulates the expression of another set of genes (Koch 1996; Smeekens and Rook 1997; Roitsch 1999; Sheen et al. 1999). The evidence thus far supports the existence of multiple glucose sensing and signal transduction pathways in higher plants (Smeekens 1998; Roitsch 1999; Sheen et al. 1999).
With the use of biochemical and molecular approaches, putative components of various sugar signal transduction pathways have been identified. There is evidence for the participation of protein phosphatases, calcium-dependent protein kinases, mitogen-activated protein kinases, and SNF1-related protein kinases (SnRK) (for review, see Smeekens and Rook 1997; Sheen 1999). However, the precise roles of most of these molecules in the sugar signal transduction pathways remain to be elucidated.
To dissect the multiple sugar signaling pathways, two general strategies have been employed to isolate sugar response mutants in Arabidopsis. In one, genetic screenings based on sugar-regulated gene repression or activation have yielded sucrose-uncoupled (sun), reduced sucrose responses (rsr), and low and high β-amylase (lba, hba) mutants (Dijkwel et al. 1997; Martin et al. 1997; Mita et al. 1997a,b). The alternative strategy takes advantage of the striking developmental arrest imposed by high concentrations of hexoses or sucrose on early seedling development. Sugar response mutants selected by this method include glucose insensitive (gin), glucose oversensitive (glo), carbohydrate insensitive (cai), sucrose insensitive (sis), and mannose-insensitive germination (mig) (Smeekens and Rook 1997; Zhou et al. 1998; Sheen et al. 1999). The characterization of these mutants has started to provide important information about novel cross-talk between sugar and other signaling pathways mediated by phytohormones, nitrogen, and light signals (Sheen 1999). For example, an Arabidopsis pleiotropic mutant, prl1, displays enhanced sensitivity to sucrose and affects the responses to several plant hormones (Németh et al. 1998). The PRL1 gene encodes a WD-protein that potentially interacts with multiple signaling components, including the Arabidopsis SNF1 homologs (Bhalerao et al. 1999). The analysis of gin1 (Zhou et al. 1998), on the other hand, revealed an interesting link between the HXK-dependent glucose and the ethylene signal transduction pathways.
To further reveal the complex signaling network that connects glucose and other signaling pathways in plants, we are characterizing new gin mutants in Arabidopsis thaliana. We report here the physiological, genetic, and molecular analyses of two recessive glucose-insensitive mutants, gin5 and gin6. Both mutants demonstrate an essential role for ABA in the glucose signaling pathway controlling vegetative development. We have cloned the T-DNA-tagged gin6. Interestingly, this gene corresponds to ABI4, a putative transcription factor important for ABA signaling in seeds, carrying an APETALA2 (AP2) domain (Finkelstein et al. 1998). The analysis of this new abi4 allele showed that ABI4 is required in the glucose-mediated developmental arrest during vegetative morphogenesis. ABI4 is positively regulated by glucose at the transcriptional level. The participation of ABI genes in the glucose signal transduction pathway is specific for ABI4 and ABI5 but not for other ABA signaling genes, ABI1, ABI2, and ABI3. The characterization of the gin5 mutant, on the other hand, has revealed that a glucose-specific up-regulation of ABA levels is a prerequisite of glucose signaling during seedling development. Consistent with this result, we show that the ABA biosynthesis-deficient mutants aba1-1, aba2-1, and aba3-2, are also glucose insensitive. In addition, the altered glucose response of the gin5 mutant can be reverted by the addition of exogenous ABA. Thus, GIN5 is important for the regulation of ABA levels in response to glucose signals.
Although the participation of ABA during seed development, germination, and stress responses is well established, our data provide the first direct evidence to support a novel and central role of ABA in the plant glucose signal transduction pathway governing postembryonic development. Apparently, glucose regulates ABA levels through GIN5 and involves ABA signaling through GIN6/ABI4. Both events are responsible for HXK-mediated responses and constitute part of the glucose signaling network in plants.
Results
Isolation and genetic analyses of the gin5 and gin6 mutants
A genetic screen based on developmental arrest in the presence of high glucose levels has resulted in the isolation of glucose insensitive (gin) mutants in A. thaliana (Zhou et al. 1998). With the aim to obtain additional components of the glucose sensing and signaling pathways, an extensive screening for gin mutants was performed using a T-DNA-mutagenized A. thaliana collection provided by DuPont Co. A total of 204,400 seeds from 7300 initial independent transformants of this T-DNA collection were screened. Several new gin mutants were selected from this screen. Two of them, referred to as gin5 and gin6, that displayed robust glucose insensitivity were chosen for further characterization.
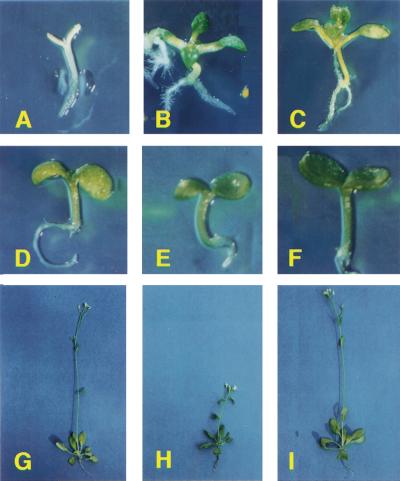
In contrast to wild-type seedlings (Fig. 1A), the gin5 and gin6 mutants develop green and expanded cotyledons when grown in the presence of 7% glucose in the medium (Fig. 1B,C, respectively). This developmental difference between wild-type plants (Fig. 1D) was not observed when both mutants, gin5 (Fig. 1E) and gin6 (Fig. 1F), were grown in the presence of 7% mannitol. In contrast to glucose, mannitol is not efficiently metabolized by plants, causing a constant osmotic stress that might affect plant growth. However, the effect of mannitol is similar in wild-type and gin mutants suggesting that the developmental arrest is glucose-specific and not linked to osmotic stress as reported (Zhou et al. 1998). We have also observed that gin5 plants exhibit a significantly smaller size and darker green leaves (Fig. 1H) compared with wild-type plants (Fig. 1G). This phenotype cosegregated with gin5 and was maintained throughout the life cycle of the plant. This characteristic is not observed in the gin6 mutant (Fig. 1I), whose size and leaf color do not show any appreciable differences compared with wild-type plants.
Figure 1.
Phenotype of gin5 and gin6 mutants. Seedlings of WS wild-type (A,D), gin5 (B,E), and gin6 (C,F) grown for 10 days under 16:8 hr photoperiod in GM medium supplemented with 7% glucose (A–C) or 7% mannitol (D–E). Phenotype of 4-week-old WS wild-type (G), gin5 (H), and gin6 (I) plants grown in Metro Mix 200 in 15:9 photoperiod.
Genetic analyses were conducted to define the Mendelian character of the gin5 and gin6 mutations. A back-cross with the WS wild-type plants showed that the mutant phenotypes were absent in the F1 plants and present in a 1:3 ratio in the F2 families. These results indicate that gin5 and gin6 behave as single recessive mutants (Table 1). Another glucose-insensitive mutant (gin1-1), that exhibits similar phenotypes has been reported previously (Zhou et al. 1998). Allelism tests between these three gin mutants show that they define three different complementation groups (Table 1).
Table 1.
Genetic segregation and complementation tests
| Cross
|
Family
|
Total
|
Observed number
|
χ2 (3:1)
|
|
|---|---|---|---|---|---|
| mutant
|
wt
|
||||
| GIN5/GIN5 × gin5/gin5 | F1 | 87 | — | 87 | |
| GIN5/GIN5 × gin5/gin5 | F2 | 2925 | 773 | 2150 | 3.25a |
| GIN6/GIN6 × gin6/gin6 | F1 | 11 | — | 11 | |
| GIN6/GIN6 × gin6/gin6 | F2 | 600 | 159 | 441 | 0.72a |
| gin5/gin5 × gin1-1/gin1-1 | F1 | 57 | — | 57 | |
| gin6/gin6 × gin1-1/gin1-1 | F1 | 43 | — | 43 | |
| gin5/gin5 × gin6/gin6 | F1 | 16 | — | 16 | |
Mutant phenotypes were scored from seedlings grown in GM plates containing 7% glucose.
Not significant at P = 0.05.
In the case of the gin6 mutant, the segregation analyses of F3 families from 33 independent self-pollinated F2 glucose-insensitive plants showed that the kanamycin-resistant phenotype was always linked to glucose insensitivity. This suggests that the mutant phenotype is likely due to a T-DNA insertion. Southern blot analysis confirmed the presence of one T-DNA insertion in the gin6 mutant genome (data not shown). In contrast, the tagging status of the gin5 mutant was uncertain, and thus genetic mapping was carried out to determine its chromosomal location. A mapping population was generated by crossing gin5 homozygous mutant plants with the WT Ler ecotype. The chromosomal location of gin5 was determined through the analysis of the mutant F2 DNA, using simple sequence length polymorphism (SSLP) markers (Bell and Ecker 1994). The GIN5 gene is located on chromosome 1. The results from the analysis of 280 chromosomes indicated that the gin5 mutation mapped 20 cm centromeric from the SSLP marker nga59.
Molecular cloning of the GIN6 gene
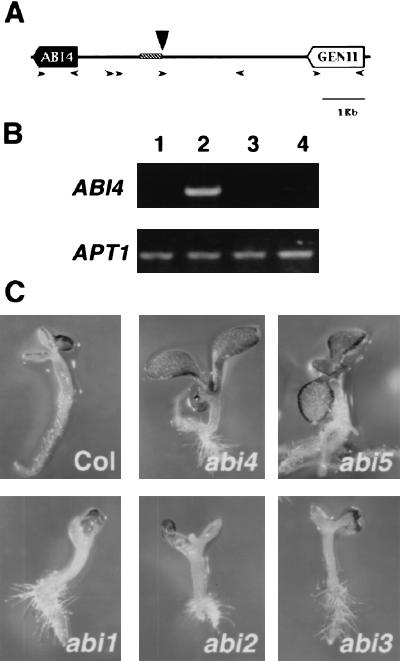
To isolate sequences flanking the T-DNA insertion site in the gin6 mutant plant, thermal asymmetric interlace (TAIL)–PCR was performed (Liu et al. 1995). A single PCR fragment of ∼450 bp was isolated after four enrichment cycles as described in Materials and Methods. The PCR fragment was cloned and sequenced revealing the presence of 48 bp that perfectly matched the left border of the T-DNA and 373 bp of novel sequence. Genomic DNA blot analysis of gin6 and wild-type plants confirm that the isolated PCR fragment was indeed interrupted in the gin6 mutant plants by the T-DNA (data not shown). BLAST database searches revealed that this fragment shares 100% identity with a sequence from a cluster of ABA-regulated genes contained in the BACT07M07 (Wang et al. 1999). As shown in Figure 2A, the insertion site was located at position 44568 of the BAC sequence, in a postulated intragenic region 2 kb upstream of a previously reported gene, ABI4, and 3 kb downstream of a putative gene called gene 11. The sequence around the insertion site did not reveal any open reading frames that could be affected by this insertion. To establish the integrity of the sequences flanking the T-DNA insertion site and to exclude any possible rearrangements that might be the cause of the mutant phenotype, multiple primers were designed to perform PCR and sequencing analysis of a 4-kb region surrounding the T-DNA insertion site in mutant and wild-type plants (Fig. 2A). Based on the sizes of the PCR fragments amplified within this region, no appreciable differences were observed between gin6 and WS wild-type plants (data not shown). This demonstrates that no major rearrangements occur in the gin6 mutant. To verify whether the T-DNA insertion affects the expression of the flanking genes, RT-PCR was carried out using specific primers for the coding region of ABI4 and gene 11. As shown in Figure 2B, an increase was observed in the expression level of the ABI4 gene transcript in wild-type plants when grown in the presence of 7% glucose. In contrast, the APT1 gene that codes for the adenine phosphoribosyltransferase (Moffatt et al. 1994) used as a control is not regulated by glucose. More importantly, this glucose induction was strongly diminished in gin6 plants. No expression of gene 11 was detected either in wild-type or mutant plants in any of the conditions tested so far. In conclusion, we have demonstrated that the ABI4 gene responds positively to glucose and that in the gin6 mutant the T-DNA insertion affects the expression levels of the ABI4 gene in response to glucose.
Figure 2.
Molecular characterization of the GIN6 gene and phenotypic analysis of abi mutants. (A) The GIN6 locus. A region (shaded box) of the BAC T07M07 shows a 100% sequence identity with the genomic flanking sequence of the T-DNA insertion site (vertical arrow) of the gin6 mutant. The horizontal arrows indicate the different primers used to determine the integrity of the sequences flanking the insertion site. The ABI4 gene begins at position 42,623 and gene 11 ends at position 48,323. (B) Glucose induction of the ABI4 transcript is diminished in gin6. Ethidium bromide-stained RT-PCR product (974 bp) of ABI4 from WS wild-type grown on 2% (lane 1) or 7% glucose (lane 2) and from the gin6 mutant grown on 2% (lane 3) or 7% glucose (lane 4). The RT-PCR product of the APT1 gene (478 bp) was used as a control. (C) Glucose insensitivity is detected in abi4-1 and abi5-1 but not inabi1-1, abi2-1, or abi3-1. Seedlings were grown on GM medium containing 7% (abi5) or 7.5% (others) glucose for one week under 16:8 hr light:dark photoperiod.
Glucose signaling is mediated by specific ABI genes
Previous studies report that the ABI4 gene might be a putative transcription factor that participates in a seed-specific ABA signaling network but does not seem to contribute to vegetative ABA responses, despite its expression in shoots and roots (Finkelstein 1994; Finkelstein et al. 1998). Our results indicate that ABI4 participates in the glucose signaling pathway during vegetative growth. Hence, it was of interest to determine the effect of the original abi4 mutation on glucose responsiveness. This original abi4 allele consists of a frameshift mutation that causes an early translation termination (Finkelstein et al. 1998). Thus Col-0 wild-type and abi4 mutant plants were germinated in the presence of 7.5% glucose. As shown in Figure 2C, abi4 plants were able to continue their development in spite of the presence of high glucose concentrations and they behaved like a glucose-insensitive mutant. We conclude that the ABI4 gene, aside from its role in seed development and germination, participates in the glucose signal transduction pathway and that the gin6 mutant represents a new allele of this gene, affected in its glucose-mediated regulation. In addition to ABI4, mutations in other genes (ABI1, ABI2, ABI3, and ABI5) involved in the ABA signal transduction pathway have been isolated (Koornneef et al. 1984; Finkelstein 1994; Nambara et al. 1995). The participation of these four genes in glucose signaling was also investigated based on each mutant's ability to develop in the presence of high glucose concentrations. Surprisingly, only abi5-1 (Fig. 2C) was glucose insensitive; abi1-1, abi2-1, and abi3-1 displayed a glucose-sensitive phenotype similar to their wild-type counterparts. ABI5 has been recently cloned and demonstrated to encode a basic leucine zipper transcription factor that regulates the expression of several genes (Finkelstein and Lynch 2000). It appears that the ABA-mediated glucose signaling pathway belongs to a new branch in which only the ABI4 and ABI5 genes participate as signaling molecules.
gin5 is affected in ABA-mediated responses
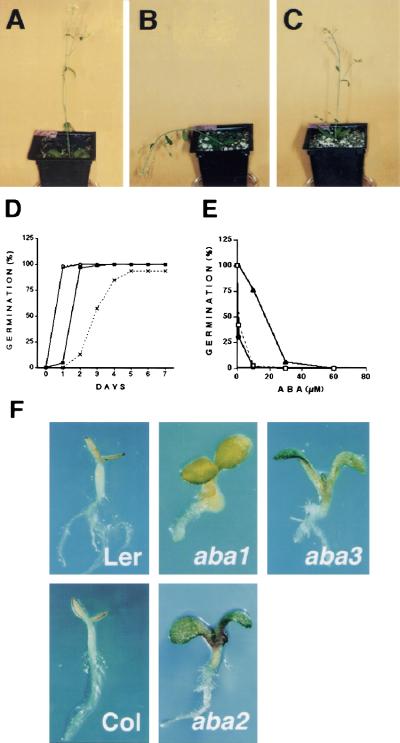
The most striking characteristic of the gin5 mutant in comparison to wild-type plants (Fig. 3A) is its enhanced drought sensitivity (Fig. 3B). Several mutants affected either in the biosynthesis or in the sensitivity to ABA have a wilty phenotype (MacRobbie 1991). This wilty phenotype was never detected in the gin6 mutant, but it was linked to the glucose insensitivity in gin5 plants after several generations, suggesting that the same locus is responsible for both traits. Interestingly, as shown in Figure 3C, the addition of 3μm of ABA to gin5 plants prevents the wilty phenotype observed under mild water stress conditions (Fig. 3B).
Figure 3.
ABA-mediated responses in gin5 and gin6 mutants. gin5 and wild-type plants were grown under well-watered conditions for 3 weeks and then subjected to drought stress by withholding water for 5 days. The plants were then sprayed with 10 ml of either water (A,B) or 3μm of ABA water solution (C) on days 3 and 5 of the stress treatment. (D) Dormancy analysis. Germination of seeds from WS wild-type (broken line) and the gin5 mutant (solid line) were examined without cold treatment (crosses; ▪) or with a 24-hr cold treatment (○, ▴). Germination was scored when the radicle tip had fully emerged from the seed coat. These data represent the average of three independent experiments evaluating 80 seeds. The standard error was smaller than the symbol size. (E) ABA dose response in germination. Wild-type (broken line), gin5 (solid line, circles) and gin6 (solid line, triangles) seeds were analyzed. The data correspond to the average of two different experiments each containing around 50 seeds per data point. The number of germinated seeds was expressed as the percentage of fully germinated seeds of the total seeds plated. (F) Phenotype of aba mutants in the presence of high glucose. Ler and Col wild-type seedlings, aba1-1, aba3-2, and aba2-1 were grown in the presence of 7% glucose on the GM medium for 7 days. The two wild-type ecotypes were included as control for the corresponding aba mutants.
It has been established that ABA plays an important role in the promotion of seed dormancy (Koornneef and Karssen 1994). As shown in Figure 3D, gin5 seeds were less dormant than wild-type. The gin5 seeds had 90% seed germination two days after imbibition without cold treatment. In contrast, only 12% germination was observed in wild-type seeds. Exposure to cold treatment prior to germination resulted in 100% germination in both the gin5 mutant and wild-type plants because the treatment eliminates the endogenous ABA levels and breaks dormancy. In contrast, the gin6 mutant did not show alterations in seed dormancy (data not shown).
We also examined the sensitivity of seeds to ABA during germination in both mutants. Although gin5 seeds display a sensitivity to the ABA inhibition of germination similar to that of wild-type seeds (Fig. 3E), gin6 mutant seeds can germinate in the presence of high ABA concentrations. These results suggest that gin5 plants are able to sense and respond to the exogenous application of ABA, whereas gin6 behaves as an ABA-insensitive mutant.
gin5 is insensitive to glucose regulation of ABA levels
Contrary to the gin6 mutant, the drought sensitivity and decreased seed dormancy in the gin5 mutant indicate that this plant may contain low endogenous ABA levels. To test this hypothesis, the endogenous levels of ABA were measured in 15-day-old seedlings of gin5, gin6, and wild-type grown in different conditions. As shown in Table 2, gin5 mutant plants contain lower ABA levels (7 ng/gram fresh weight) than both gin6 (19 ng/gram fresh weight) and wild-type (14 ng/gram fresh weight) seedlings when grown on GM medium. Interestingly, a dramatic increase (seven- to ninefold) in the ABA levels was observed in gin6 and wild-type plants but not in gin5 plants when grown with 7% glucose in the medium (10 ng/gram fresh weight). Thus, gin6 mutant and wild-type plants respond to the presence of 7% glucose by increasing their endogenous ABA level and the gin5 mutant does not seem to be able to promote such a response. This glucose induction of ABA levels is independent of the plant morphological status as both gin6 mutant and wild-type plants respond similarly in spite of the fact that the wild-type plants are arrested at an early developmental stage. The ABA levels were also similarly enhanced by glucose at an early developmental stage (Table 2) in wild-type (threefold) and gin6 mutant seedlings (eightfold) but not in gin5 mutant plants.
Table 2.
Endogenous ABA levels in wild-type and gin5 and gin6 plants
| Growth condition
|
WS
|
gin5
|
gin6
|
|---|---|---|---|
| 15-day-old | |||
| GM | 14 ± 1 | 7.2 ± 2 | 19 ± 2 |
| GM 7% glucose | 91 ± 10 | 10 ± 2 | 167 ± 10 |
| Young seedlings | |||
| GM | 10 ± 3a | 11 ± 3a | 12 ± 4a |
| GM 7% glucose | 30 ± 1b | 12 ± 4c | 92 ± 3c |
The ABA concentration is expressed in ng ABA/g of fresh weight. The data were obtained from 15-day-old plants in the indicated media. For comparing young seedlings, plants were harvested at a similar developmental stage. Note that wild-type plants were developmentally arrested when grown on high glucose media. The time for each sample collection is as follows: (a) 48 hr, (b) 7 days, (c) 5 days.
An increase in ABA is required for sugar regulation in plants
To further support the importance of increased ABA levels in glucose responses, we performed the glucose-dependent developmental arrest assay with well-characterized ABA-deficient mutants, such as aba1-1, aba2-1, and aba3-2 (Koornneef et al. 1982; Schwartz et al. 1997). The development of these aba mutants in the presence of high glucose was followed. In contrast to their wild-type counterparts, all of the aba mutants were able to grow in the presence of high glucose (Fig. 3C). This result strongly suggests that the reduced ABA levels present in these mutants decrease their glucose sensitivity.
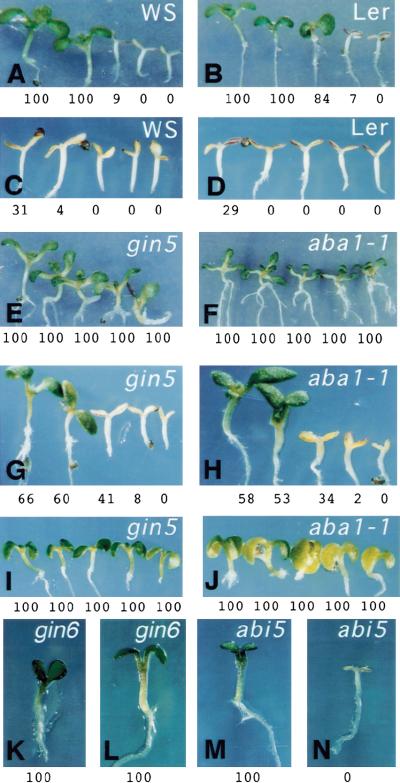
To define whether an increase in ABA is sufficient to promote a normal sugar response in the aba and gin5 mutants, their development in the presence of high glucose and exogenously applied ABA was followed. The ABA concentration used for these experiments was in the nanomolar range (100 nm). We first confirmed that this concentration by itself did not affect plant development in either wild-type or mutant plants (data not shown). As shown in Figure 4 both WS (Fig. 4A) and Ler (Fig. 4B) wild-type plants underwent developmental arrest in the presence of 6% and 6.5% glucose, respectively. When 100 nm ABA was exogenously applied to the medium both WS and Ler wild-type plants became hypersensitive to glucose. In this case ∼70% of these plants became arrested at concentrations as low as 4% glucose (Fig. 4C,D). Using a similar set of conditions we explored the behavior of the gin5 plant. In the presence of 100 nm ABA, 60% of this glucose-insensitive mutant seedlings became sensitive to 6% glucose (Fig. 4G). The glucose-insensitive phenotype of the aba1-1 mutant (Fig. 4F) was also suppressed by the addition of ABA to the medium (Fig. 4H). Similar responses were observed for aba2-1 and aba3-2 mutants (data not shown). To exclude the possibility that this response was caused by an osmotic effect, the phenotype of gin5 and aba1 plants in the presence of 100 nm ABA and mannitol was compared. As shown in Figure 4, I and J, no differences in the development of these plants were detected in the presence of ABA and mannitol.
Figure 4.
The effect of ABA and high glucose on the phenotype of gin5 and aba1-1 mutants. WS (A,C), gin5 (E,G,I), Ler (B,D), and aba1-1 (F,H,J) plants were grown on 4, 5, 6, 6.5, and 7% glucose (A–H) or 4, 5, 6, 6.5 and 7% mannitol (I,J), in the absence (A,B,E,F) or the presence of 100 nm ABA (C,D,G–J) for 10 days. gin6 and abi5-1 mutants were grown in the presence of 7% glucose without (K,M) or with (L,N) a 100 nm ABA. Representative plants are shown in each condition. The number below each plant indicates the percentage of plants displaying a green phenotype for each of the conditions used.
The effect of 100 nm ABA on the mutant phenotypes of gin6, abi4, and abi5-1 was also determined. Based on the endogenous ABA levels of the gin6 mutant and the nature of the ABI4 gene, we would not expect that its mutant phenotype would be reverted by the exogenous application of ABA. Indeed, 100% of the gin6 plants (Fig. 4K) and abi4 (data not shown) maintained the glucose-insensitive phenotype in the presence of 100 nm ABA and 7% glucose (Fig. 4L). Surprisingly, the addition of ABA reverted the glucose-insensitive phenotype of the abi5-1 mutant, in spite of having been isolated as an ABA-insensitive mutant (Fig. 4M,N). These results suggest that the GIN6/ABI4 gene participates downstream of the induction of ABA in the glucose signaling pathway, whereas ABI5 might have a role upstream of this pathway.
Transcription of glucose-regulated genes is altered in gin5 and gin6 mutants
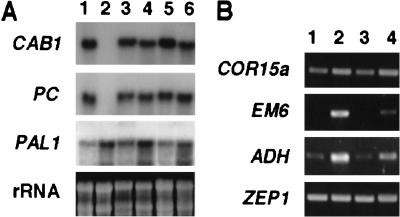
It has been shown that both glucose and ABA regulate the transcription of a variety of genes (Koch 1996; Weatherwax et al. 1996; Shinozaki and Yamaguchi-Shinozaki 1997; Sheen 1998; Sheen et al. 1999). This study has shown that ABA participates in the glucose signaling pathway controlling vegetative development. Thus, it was important to analyze the expression pattern of genes previously reported to be regulated either by glucose or ABA in gin5 and gin6 mutants. For this purpose total RNA from wild-type WS, gin5, and gin6 seedlings, grown in the presence of 2% and 7% glucose, was extracted and analyzed by RNA blot hybridization and RT-PCR. As shown in Figure 5A, lanes 1 and 2, transcript levels of two photosynthetic genes regulated by the HXK-dependent pathway, chlorophyll a/b binding protein (CAB1) and plastocyanin (PC) (Sheen et al. 1999), were strongly repressed in the presence of 7% glucose in wild-type WS plants. This glucose repression pattern was not observed in gin5 or gin6 mutants grown under the same conditions (Fig. 5A, lanes 3–6), indicating that glucose regulation of these photosynthetic genes is altered in both mutants. It has been shown that phenylalanine ammonia-lyase (PAL) transcript is induced by glucose, probably by an HXK-independent pathway (Ehness et al. 1997). We followed the expression pattern of the PAL1 gene in wild-type (Fig. 5A, lanes 1 and 2) and mutant plants (Fig. 5A, lanes 3–6). In contrast to the two photosynthetic genes, the glucose inducibility of PAL1 is observed in the gin5 and gin6 mutants.
Figure 5.
Expression of glucose- and ABA-modulated genes in gin5 and gin6 mutants. (A) RNA blot analysis. Total RNA (10 μg) of WS wild-type (lanes 1,2), gin5 (lanes 3,4), and gin6 (lanes 5,6) from 19-day-old seedlings grown in the presence of 2% (lanes 1,3,5) or 7% (lanes 2,4,6) glucose was used for RNA blots. Glucose repression of CAB1 and PC and activation of PAL were examined. Ethidium bromide-stained ribosomal RNA was used as a loading control. (B) RT–PCR analysis. Total RNA (5 μg) from WS wild-type (lanes 1,2) or gin6 (lanes 3,4) seedlings grown in 2% (lanes 1,3) or 7% (lanes 2,4) glucose was used to synthesize cDNA. A portion (1/80) of the synthesized cDNA was used to amplify and quantitate COR15a (accession no. U01377), EM6 (accession no. Z11923 S61761), ADH (accession no. M12196), and ZEP1 (accession no. T45502) gene transcripts. The sizes of PCR products are 210 bp, 302 bp, 398 bp, and 261 bp, respectively.
To further elucidate the function of ABI4 in glucose and ABA signaling during seedling development, we analyzed the expression profile of ABA- and stress-inducible genes in the gin6 mutant by RT-PCR analysis. As shown in Figure 5B, the COR15a gene transcript, which encodes a chloroplast-targeted protein and is induced by ABA, cold, and drought (Wilhelm and Thomashow 1993), is present at similar levels in gin6 and wild-type plants grown in 2% or 7% glucose (Fig. 4B, lanes 1 and 2). This suggests that the regulation of this gene is independent of ABI4 function. In contrast, the expression of AtEm6, an ABA-inducible gene encoding a late embryogenesis-abundant protein (Gaubier et al. 1993), is strongly induced by 7% glucose in wild-type plants (Fig. 4B, lane 2). The glucose induction of AtEm6 in the gin6 mutant is much reduced, suggesting that its expression is ABI4 dependent. This result is consistent with the altered expression of this gene in the original abi4 mutant (Finkelstein 1994), which further supports our finding that gin6 is a new allele of ABI4. We also looked at the expression of the ABA- and stress-inducible alcohol dehydrogenase gene (ADH) (de Bruxelles et al. 1996). We observed that wild-type plants grown in 7% glucose have increased ADH transcript levels, whereas there is only a minor increase in gin6 plants (Fig. 4B). Thus in the gin6 mutant the induction of the ADH gene is diminished but not abolished. These results suggest that a subset of ABA-inducible genes are also regulated by glucose in an ABI4-dependent fashion during vegetative growth. Finally, because we have observed an increase in the endogenous levels of ABA in response to high glucose, we were interested in analyzing the transcript levels of genes involved in the synthesis of this hormone under high glucose conditions. The expression of the ZEP1 gene (zeaxanthin epoxidase) (Cutler and Krochko 1999) showed no difference in the transcript levels in the wild-type and gin6 plants in a high glucose condition (Fig. 4B).
gin5 affects the HKX-mediated glucose signaling pathway
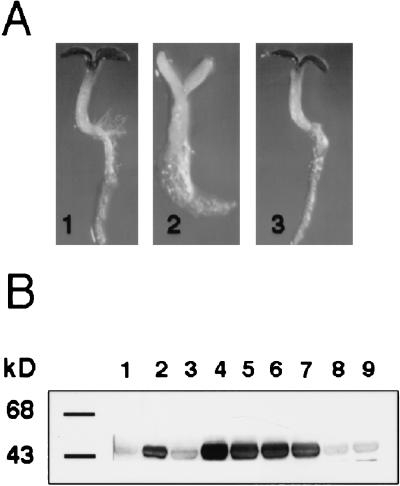
The altered regulation of CAB1 and PC genes in the gin5 and gin6 mutants suggested that these mutations affect the HKX-mediated signal transduction pathway but do not affect genes regulated by the HXK-independent pathway, such as PAL1. To further substantiate this conclusion, a genetic approach was used to generate a double mutant carrying the gin5 mutation for glucose insensitivity and a dominant AtHXK1 gene (Jang et al. 1997) conferring glucose hypersensitivity (Fig. 6A). Plants from the F2 generation were grown on high glucose medium, and it was observed that 24% of these plants (363 out of 1500 seedlings) were insensitive to 7% glucose (Fig. 6A, panel 3). To determine the levels of HXK protein in those glucose-resistant plants, total proteins from six individual plants were extracted and subjected to Western blot analysis. As shown in Figure 6B, four out of six glucose insensitive plants contained higher HXK protein levels (Fig. 6, lanes 4–7) as compared to either wild-type (Fig. 6B, lane 1) or gin5 (Fig. 6B, lane 3) plants. The HXK protein levels of these plants are comparable to the 35S-AtHXK1-overexpressing line (Fig. 6B, lane 2). In contrast, the other two plants from this analysis display comparable HXK levels to wild-type plants (Fig. 6B, lanes 8 and 9). This result demonstrates that plants carrying the 35S-AtHXK1 and the gin5 mutation display a glucose-insensitive phenotype like the gin5 parental plant, suggesting that gin5 participates in the HXK-mediated pathway.
Figure 6.
gin5 acts downstream of the AtHXK gene in the glucose signaling pathway. (A) Analysis of the glucose response. Seedlings of gin5 (panel 1), 35S-AtHXK (panel 2), and 35S-AtHXK gin5 (panel 3) were grown in the presence of 7% glucose for 8 days. (B) Immunoblot analysis. The HXK protein levels of the wild-type (lane 1), 35S-AtHXK transgenic plant (lane 2), the gin5 mutant (lane 3) and independent F2 individuals displaying glucose insensitivity from the 35S-AtHKX gin5 cross (lanes 4–9) are shown. Protein was extracted from 15-day-old seedlings grown on GM medium with 7% glucose.
Discussion
ABA plays a central role in the glucose signaling network controlling postembryonic development
Unlike most animals that follow a predetermined developmental program for growth, postembryonic growth and development in plants displays a remarkable plasticity, which resides in the plant's capacity to integrate and respond to developmental, metabolic, and environmental signals. The most striking example is the regulation by light of photomorphogenesis. Because plant survival relies on light-driven photosynthesis, the dual function of light as a developmental switch and an energy-source makes evolutionary sense. It has been suggested that plant hormones such as auxin, brassinosteroids, cytokinin, gibberellins (GA), ABA, and ethylene play important roles in promoting or antagonizing photomorphogenesis (Fankhauser and Chory 1997; Deng and Quail 1999). Interestingly, light signals regulating plant morphogenesis can be overridden by metabolic signals such as the availability of glucose released from photosynthetic end products or carbon storage sources. For example, in Arabidopsis the availability of abundant glucose can be sensed during germination and can exert a profound influence, resulting in seedling developmental arrest (Jang and Sheen 1997). Although the underlying mechanisms of this glucose-inducible developmental arrest are mostly unknown, a previous analysis of the gin1 mutant has revealed an antagonistic role of the volatile hormone ethylene (Zhou et al. 1998). Thus, glucose responses in multicellular plants are much more complex than in unicellular bacteria and yeast (Saier et al. 1995; Johnston 1999).
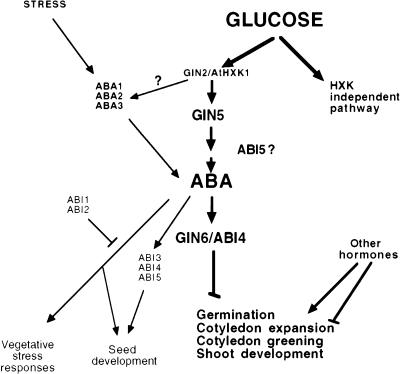
In this work, based on the genetic, physiological, and molecular characterization of two Arabidopsis glucose-insensitive mutants, gin5 and gin6, it was discovered that the plant hormone ABA participates in the HXK-mediated glucose response in plants (Fig. 7). Through the studies of wild-type and gin5 plants, we propose that an increase of ABA levels is involved in the glucose signaling pathway that leads to decrease in gene expression and developmental arrest (Fig. 7). The inability to increase ABA levels results in a glucose-insensitive phenotype manifested in the gin5 mutant. We have substantiated this conclusion further by demonstrating that three ABA biosynthetic-deficient mutants (aba) are also insensitive to high glucose concentrations and that this phenotype can be reverted by the addition of very low levels of exogenous ABA (100 nm). Thus, the intracellular ABA levels seem to determine the glucose sensitivity in all of these mutants. However, it is important to emphasize that an ABA increase is not the only factor mediating the developmental arrest when plants are grown in high glucose conditions. Indeed, during osmotic stress, a substantial increase of ABA levels is detected but seedling development is not arrested. Thus, we hypothesize that at least one other independent component is required in addition to ABA to promote glucose-dependent developmental arrest. Putative candidates are other hormones such as ethylene, which has already been demonstrated to interact with the HXK-mediated glucose signaling pathway (Zhou et al. 1998). It is also possible that the cellular or subcellular distribution of ABA and/or the ABA-responsive cells are different in response to glucose or mannitol signals.
Figure 7.
A model for the role of ABA in the HXK-dependent glucose signaling network.
The increase in ABA levels detected in wild-type plants could be the result of de novo synthesis, release from glucose conjugates, and/or reduced degradation (Cutler and Krochko 1999). Each of these processes is a potential target for the modulation of ABA levels in response to glucose. We have explored the possible glucose regulation on the expression of ZEP1, encoding an enzyme required for ABA biosynthesis (Newman et al. 1994; Cutler and Krochko 1999). This study did not reveal any differential expression of ZEP1 under high glucose conditions. Unfortunately, ZEP1 is currently the only gene identified for the final steps of the ABA biosynthetic pathway in Arabidopsis. This aspect could be better addressed in the future when more ABA biosynthetic and regulatory genes become available. For instance, GIN5 could encode one of the potential molecules involved in the signaling transduction between glucose and ABA. It is likely that the wilty and reduced dormancy phenotypes in gin5 represent pleiotropic effects on other ABA-related processes during seed and vegetative development, in addition to glucose responses. Another gin mutant (gin1), isolated through the use of a similar screening procedure, has been characterized previously (Zhou et al. 1998). Analysis of this mutant revealed an interplay between the ethylene signaling pathway and glucose regulation. Nonallelic gin5 shares very similar traits with gin1 including small size, dark green leaves, fast germination, and wilty phenotype. Both GIN5 and GIN1 have been demonstrated to participate in the HXK-mediated pathway. As HXK has been postulated to initiate the glucose signal (Jang et al. 1997), we propossed that both genes act downstream of this putative sensor. The interaction between gin1 and gin5 and their relationship to the ethylene and ABA signaling pathways will be clarified in the future by analysis of double mutants. In an attempt to integrate all of these data, we propose that the glucose response requires an increase of ABA and a decrease of ethylene. Other plant hormones that interact with ethylene and ABA, such as auxin, cytokinin, and GA, could also be involved in this glucose response (Fig. 7) (Sheen et al. 1999).
ABI4 as a putative transcription factor for glucose signaling
ABA participates in a wide variety of physiological responses that depend on external and internal signals (Shinozaki and Yamaguchi-Shinozaki 1997; Leung and Giraudat 1998). However, the mechanisms by which these signals are discriminated or integrated inside the cell are mostly unknown. The characterization of the gin6 mutant resulted in the identification of ABI4 as a candidate for the transduction of the glucose-specific signal. We found that the ABI4 transcript is induced in wild-type plants in response to high glucose and this induction was diminished in the gin6 mutant. The reduced transcript levels in the gin6 mutant are likely the result of the T-DNA insertion in the promoter region, as no other changes were found in this mutant's ABI4 gene in comparison to the reported amino acid sequence (Finkelstein et al. 1998). We have found that this insertion also affects the function of the ABI4 gene during seed germination, as the gin6 mutant displays an ABA germination-insensitive phenotype. These results suggest that important cis-acting sequences required for the regulation of the ABI4 gene lie at least 2 kb upstream of the start codon. This observation contrasts the data published by Finkelstein et al. (1998), in which the seed-specific phenotype of the abi4 mutant is complemented by a genomic clone containing a promoter region of only 1.3 kb upstream of the start codon but not by a clone containing 1.7 kb upstream. Our results support the idea proposed by Finkelstein et al. (1998) that sequences further upstream of 1.7 kb of the start codon are required for the proper expression of the ABI4 gene. Another possibility is that the T-DNA insertion in the gin6 plant might disrupt the ABI4 gene regulation downstream of the insertion site. Further analysis will be required to clarify these possibilities. We confirmed that the original abi4 allele is also glucose insensitive. The ABI4 gene was identified as a putative transcription factor (Finkelstein et al. 1998), and it shows homology to a family of plant transcription factors with AP2 domains that seem to control a variety of processes (Okamuro et al. 1997). The fact that the glucose-insensitive phenotype is maintained independently of the addition of ABA in gin6 and abi4 mutants suggests that this transcription factor could be acting near the end of the ABA-mediated glucose signaling cascade (Fig. 7). Efforts are currently being directed to find out whether ABI4 can bind directly to CAB1 and PC promoters in high glucose conditions. In addition, the predicted amino acid sequence of ABI4 contains a serine/threonine-rich domain, which is a possible target for protein kinases such as those of the SnRK family. Our gene expression analysis has shown the participation of the ABI4 gene in the expression of both sugar- and stress-responsive genes. The analysis of different alleles or site-directed mutagenesis can be used to further dissect the function of ABI4 in stress and glucose signaling, both in seeds and vegetative tissues.
The glucose responses mediated through ABA involve specific signaling molecules
In addition to ABI4, other ABA signaling components have been characterized. For example, ABI1 and ABI2 belong to the protein phosphatase 2C (PP2C) family and have pleiotropic and overlapping roles affecting ABA signaling in both seeds and vegetative tissues. In contrast, ABI3 and ABI5, as well as ABI4, were initially reported to affect various aspects of seed maturation and seed gene expression (Finkelstein 1994; Bonetta and McCourt 1998). Our results have shown that neither abi1, abi2 , or abi3 mutants are resistant to high glucose. The abi1 and abi2 mutants are dominant gain-of-function alleles with virtually abolished PP2C activity (Leung and Giraudat 1998; Sheen 1998). It is unlikely that these two molecules play any major role in the ABA induction in response to high glucose (Fig. 7). However, our studies demonstrate that in addition to ABI4, ABI5 also has a role in the glucose response. Thus, in spite of the seed-specific roles that have been proposed for ABI4 and ABI5, both molecules have an important role in this newly identified ABA-mediated response in vegetative tissues. In fact, the recent isolation of a salt-tolerant mutant, sañ5 (Quesada et al. 2000), which corresponds to a new abi4 allele, indicates that the participation of ABI4 in vegetative ABA responses might be a more general phenomenon. We have shown that the abi5-1 mutant has a glucose-insensitive phenotype, but surprisingly this phenotype is reverted in the presence of ABA (100 nm). This suggests that this recently cloned transcription factor (Finkelstein and Lynch 2000) might participate in regulating the expression of genes required for transmitting signals from HXK that lead to the ABA increase while GIN6/ABI4 acts downstream of ABA (Fig. 7). Because of the fact that these genes are active in both seed-specific and glucose-mediated signaling pathways, it is possible that each stimulus can trigger multiple signaling pathways that possess complex interconnections. Further analysis will be required to have a better understanding of other convergent points between the glucose and ABA signaling pathways.
In conclusion, we have revealed a fundamental aspect of the glucose signal transduction pathway in plants based on the analysis of two glucose-insensitive mutants. The participation of ABA as part of the glucose signal transduction pathway constitutes an unexpected finding and uncovers the inherent complexity of signaling networks in higher plants.
Materials and methods
Plant material and growth conditions
Arabidopsis plants used were routinely grown on Metro-Mix 200 (Grace Sierra, Milpitas, CA) soil in controlled growth chambers (24°C, 16:8 hr light:dark photoperiod). Seeds under sterile conditions were surface-sterilized and plated on germination medium (GM) containing 1× Murashige and Skoog basal salt mixture supplemented with B5 vitamins, 0.05% MES, 1% sucrose as carbon source (unless otherwise indicated), and 0.7% of phytoagar. Seeds were incubated at 4°C for 4 days to brake dormancy prior to germination. ABA (Sigma, Inc., St. Louis, MO) was prepared as a 10 mm stock in NaOH and diluted into appropriate concentrations for each experiment.
Glucose-insensitive mutants were isolated in 7% glucose GM medium from T4 pools of 7300 independent T-DNA tagged lines in Wassilewskija (WS) ecotype, kindly donated by DUPONT Co. The different aba, abi mutant and wild-type plants, WS, Landsberg erecta (Ler), and Columbia (Col-0) used in this work were obtained from the Arabidopsis Biological Resource Center (ABRC). The aba1-1, aba3-2, abi1-1, abi2-1, and abi3-1 mutants are in the Ler ecotype, whereas aba2-1 and abi4-1 are in Col-0, and abi5-1 in WS.
Genetic analyses
Backcrosses were performed using mutant plants as the pollen donors and WS-0 wild-type as female plants. F1 plants were allowed to self-pollinate and the glucose-insensitive phenotype was scored in the F1 and F2 population at the seedling stage. Complementation groups between gin1-1, gin5 and gin6 were determined by crossing homozygous plants in reciprocal orientation.
The mapping population was obtained by crossing homozygous gin5 mutant plants to wild-type Ler plants. Homozygous gin5 mutant plants from the F2 progeny were grown on 7% glucose medium and plants displaying green and fully expanded cotyledons were selected. Genomic DNA was prepared from these homozygous gin5 individual plants. The map position was determined by testing the linkage to simple sequence length polymorphism (SSLP) markers (Bell and Ecker 1994).
Phenotypic analyses
The glucose-resistant phenotype was scored by growing plants in GM in the presence of 7% glucose, or 7.5% in the case of Col-0 and Ler because of the higher level of resistance to glucose of these ecotypes. The analysis of seed dormancy was performed using seeds that were never exposed to cold treatment. The wilty phenotype in gin5 mutant plants was observed in 4-week-old plants subjected to mild stress conditions by withholding water for 5 days. To examine drought responsiveness in the presence of ABA, gin5 plants were sprayed with 10 ml of 3μm ABA on days 3 and 5, following the instructions provided by the ABRC for the maintenance of the aba1 mutant. ABA sensitivity tests were done following the protocol reported by Quesada et al. (2000), using agar medium supplemented with ABA (1–60 μm) and scored 5 days after sowing.
Molecular cloning of GIN6
For specific amplification of the gin6 genomic sequence flanking the T-DNA insertion, a TAIL–PCR procedure was followed as described (Liu et al. 1995). The specific primers from the left border and the degenerate primer used are: LB1, 5′-CGATCTGAGGATGAGATGTCATT-3′; LB2, 5′-GACGATATAGAGCAAGATGGAAA-3′; LB3, 5′-CACATCATCTCATTGATGCTTGG-3′; LB4, 5′-AACGTCCGCAATGTGTATTA-3′. We used the reported AD2 degenerate primer (Liu et al. 1995). PCR amplifications were performed using high-quality genomic DNA isolated from the gin6 mutant in a Peltier-Effect Cycling PTC-100 (7.0). Cycle settings used were as recommended (Liu et al. 1995), but with a fourth enrichment super-cycle using the following PCR conditions: (94°C for 30 sec, 54°C for 1.5 min, 72°C for 2.5 min, 94°C for 30 sec, 54°C for 1.5 min, 72°C for 2.5 min, 94°C for 30 sec, 45°C for 1.5 min, 72°C for 2.5 min) for 15 cycles and 72°C for 10 min. DNA fragments were cloned into the pMOS vector and sequenced.
Expression analysis techniques
RNA was isolated using standard protocols (Ausubel et al. 1987) from 19-day-old seedlings grown on plates containing GM supplemented with 2% and 7% glucose. Hybridizations were done at high-stringency conditions with fragments from the CAB1 gene (GenBank accession no. J04098), the PC gene (GenBank accession no. M20937), and the PAL1 gene (GenBank accession no. L33677). cDNA was synthesized using 5 μg of total RNA by oligo(dT)-primed reverse transcription, using Super Script II reverse transcriptase (GIBCO BRL). A fraction (1/80) of the first strand cDNA was used as a template for PCR. Linearity for each amplification was corroborated. The primers used and the corresponding size products and accession numbers are as follows: ABI4 (5′-ATGGACCCTTTAGCTTCCCA-3′, 5′-AAGATGGGATCAATAAAATC-3′; 974 pb; accession no. AF040959); APT1 (5′-TCCCAGAATCGCTAAGATTGCC-3′; 5′-CCTTTCCCTTAAGCTCTG-3′; 478 pb; accession no. Y07681); ADH (5′-GCCAGGAGATCATGTGTTGC-3′; 5′-GCACCAGCGATTCTAGCACC-3′; 398 pb; accession no. M12196); COR15a (5′-GTGACGGATAAAACAAAAGAGG-3′; 5′-GACCCTACTTTGTGGCATCCTT-3′; 210 pb; accession no. U01377); AtEm6 (5′-AAACCATGGCGTCTCAACAAGAG-3′; 5′-GCGGTCGACATCCGTGTGGGGAAGT-3′; 302 pb; accession no. Z11923); ZEP1 (5′-GCGAACACGGAACCTAT-3′; 5′-CTCAAATTATCTGCAAAACCG-3′; 261 pb; accession no. T45502).
Western blot analysis
Crosses between gin5-1 and 35S-AtHXK1 were performed and the F2 generation was obtained. The glucose-resistant phenotype was scored from a total of 1550 plants selected on 7% glucose during 8 days. Total protein extracts were obtained from individual plants. Immunodetection was done using a polyclonal antibody, produced against the HXK following the protocol by Zhou et al. (1998). An anti-rabbit immunoglobulin alkaline phosphatase-conjugate was used as a secondary antibody (Zymed Laboratories, Inc.) and was detected using the BCIP/NBT substrate kit (Zymed Laboratories, Inc.).
ABA determination
To quantify the ABA content of gin5, gin6, and wild-type plants, 15 to 20 mg for the 15-day-old seedlings or 5 to 7 mg for the young seedling stage of fresh tissue was homogenized in 1 ml of ABA-extraction buffer (10 mm HCL, 1% PVPP in methanol). Extraction was done overnight with constant shaking at 4°C. The supernatant was collected and neutralized with 15 μl of 1m NaOH as described (Peña-Cortes et al. 1989). ABA was quantified with a Phytodetek-ABA-kit (AGDIA Inc., IN) using the protocol provided but with the addition of TBS and 0.1% gelatin in all of the samples.
Acknowledgments
We thank A. Covarrubias, M. Treviño, and S. Reichler for their critical comments on the manuscript; J. Penney for editing the manuscript; A. Cantero and V. Hernández for their expert technical help; S. Smeekens for sharing unpublished information and providing the PC plasmid pPC8.130; and F. Campos for generous supply of AtEm6 primers. Thanks go to the ABRC for providing mutant seeds stocks and the DUPONT Co. for supplying the T-DNA seed pools. A.A. and F.A. were fellows from CONACyT. This research was supported by the CONACyT (31791-N) and DGAPA (IN205697) grants to P.L. and by NSF (IBN-9723610) to J.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL patricia@ibt.unam.mx; FAX 52-(73)-172388 or 139988.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley & Sons; 1987. [Google Scholar]
- Bell CJ, Ecker JR. Assignement of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bakó L, Ökrész L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C. Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinase. Proc Natl Acad Sci. 1999;96:5322–5327. doi: 10.1073/pnas.96.9.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetta D, McCourt P. Genetic analysis of ABA signal transduction pathways. Trends Plant Sci. 1998;3:231–235. [Google Scholar]
- Chiou TJ, Bush DR. Sucrose is a signal molecule in assimilate partitioning. Proc Natl Acad Sci. 1998;95:4784–4788. doi: 10.1073/pnas.95.8.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler AJ, Krochko JE. Formation and brake down of ABA. Trends Plant Sci. 1999;4:472–478. doi: 10.1016/s1360-1385(99)01497-1. [DOI] [PubMed] [Google Scholar]
- Dai N, Schaffer A, Petreikov M, Shahak Y, Giller Y, Ratner K, Levine A, Granot D. Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell. 1999;11:1253–1266. doi: 10.1105/tpc.11.7.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruxelles GL, Peacock WJ, Dennis ES, Dolferus R. Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 1996;111:381–391. doi: 10.1104/pp.111.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Quail PH. Signalling in light-controlled development. Semin Cell Dev Biol. 1999;10:121–129. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- Dijkwel PP, Huijser C, Weisbeek P, Chua N-M, Smeekens SCM. Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell. 1997;9:583–595. doi: 10.1105/tpc.9.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehness R, Ecker M, Godt DE, Roitsch T. Glucose and stress independently regulate source and sink metabolism and defense mechanisms via signal transduction pathways involving protein phosphorylation. Plant Cell. 1997;9:1825–1841. doi: 10.1105/tpc.9.10.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994;5:765–771. [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscicic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubier P, Raynal M, Hull G, Huestis GM, Grellet F, Arenas C, Pages M, Delseny M. Two different Em-like genes are expressed in Arabidopsis thaliana seeds during maturation. Mol Gen Genet. 1993;238:409–418. doi: 10.1007/BF00292000. [DOI] [PubMed] [Google Scholar]
- Gibson SI, Graham IA. Another player joins the complex field of sugar-regulated gene expression in plants. Proc Natl Acad Sci. 1999;96:4746–4748. doi: 10.1073/pnas.96.9.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupe A, Hultgren B, Ryan A, Ma YH, Bauer M, Stewart TA. Transgenic knockouts reveal a critical requirement for pancreatic β cell glucokinase in maintaining glucose homeostasis. Cell. 1995;83:68–78. doi: 10.1016/0092-8674(95)90235-x. [DOI] [PubMed] [Google Scholar]
- Jang JC, Sheen J. Sugar sensing in higher plants. Trends Plant Sci. 1997;115:208–213. [Google Scholar]
- Jang JC, León P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. Feasting, fasting and fermenting glucose sensing in yeast and other cells. Trends Genet. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- Koch KE. Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Karssen CM. Seed dormancy and germination. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor Appl Genet. 1982;61:385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant. 1984;61:377–383. [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- MacRobbie EAC. Effect of ABA on ion transport and stomatal regulation. In: Davies WJ, Jones HG, editors. Abscisic acid, physiology and biochemistry. Oxford, UK: BIOS Scientific Publishers; 1991. pp. 153–158. [Google Scholar]
- Martin T, Hellmann H, Schmidt R, Willmitzer L, Frommer WB. Identification of mutants in metabolically regulated gene expression. Plant J. 1997;11:53–62. doi: 10.1046/j.1365-313x.1997.11010053.x. [DOI] [PubMed] [Google Scholar]
- Mita S, Hirano H, Nakamura K. Negative regulation in the expression of a sugar-inducible gene in Arabidopsis thaliana. A recessive mutation causing enhanced expression of a gene for β-amylase. Plant Physiol. 1997a;114:575–582. doi: 10.1104/pp.114.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, Murano N, Akaike M, Nakamura K. Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for β-amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J. 1997b;11:841–851. doi: 10.1046/j.1365-313x.1997.11040841.x. [DOI] [PubMed] [Google Scholar]
- Moffatt BA, McWhinnie EA, Agarwhal SK, Schaff DA. The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene. 1994;143:211–216. doi: 10.1016/0378-1119(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Moore B, Sheen J. Plant sugar sensing and signaling—a complex reality. Trends Plant Sci. 1999;4:250. doi: 10.1016/s1360-1385(99)01433-8. [DOI] [PubMed] [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S. A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development. 1995;121:629–636. [Google Scholar]
- Németh K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kálmán Z, Stankovic-Stangeland B, Bakó L, Mathur J, Ökrész L, Stabel S, et al. Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes & Dev. 1998;12:3059–3073. doi: 10.1101/gad.12.19.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, et al. Genes galore: A summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Montagu VM, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pego JV, Weisbeek PJ, Smeekens SCM. Mannose inhibits Arabidopsis germination via a hexokinase-mediated step. Plant Physiol. 1999;119:1017–1023. doi: 10.1104/pp.119.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortés H, Sanchez-Serrano J, Martens R, Willmitzer L, Pratt S. Abscisic acid is involved in the wound-induced expression of the proteinase inhibitor II gene in potato and tomato. Proc Natl Acad Sci. 1989;86:9851–9855. doi: 10.1073/pnas.86.24.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL. Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics. 2000;154:421–436. doi: 10.1093/genetics/154.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T. Source-sink regulation by sugar and stress. Curr Opin Plant Biol. 1999;2:198–206. doi: 10.1016/S1369-5266(99)80036-3. [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S. Sucrose-specific signaling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 1998;15:253–263. doi: 10.1046/j.1365-313x.1998.00205.x. [DOI] [PubMed] [Google Scholar]
- Saier MH, Chauvaux S, Deutscher J, Reizer J, Ye JJ. Protein phosphorylation and regulation of carbon metabolism in Gram-negative versus Gram-positive bacteria. Trends Biol Sci. 1995;20:267–271. doi: 10.1016/s0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Léon-Kloosterziel KM, Koorneef M, Zeevaart JAD. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 1997;114:161–166. doi: 10.1104/pp.114.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci. 1998;95:975–980. doi: 10.1073/pnas.95.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— C4 gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:187–217. doi: 10.1146/annurev.arplant.50.1.187. [DOI] [PubMed] [Google Scholar]
- Sheen J, Zhou L, Jang JC. Sugars as signaling molecules. Curr Opin Plant Biol. 1999;2:410–418. doi: 10.1016/s1369-5266(99)00014-x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S. Sugar regulation of gene expression in plants. Curr Opin Plant Biol. 1998;1:230–234. doi: 10.1016/s1369-5266(98)80109-x. [DOI] [PubMed] [Google Scholar]
- Smeekens S, Rook F. Sugar sensing and sugar-mediated signal transduction in plants. Plant Physiol. 1997;115:7–13. doi: 10.1104/pp.115.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Tang GQ. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 1999;4:401–407. doi: 10.1016/s1360-1385(99)01470-3. [DOI] [PubMed] [Google Scholar]
- Wang ML, Belmonte S, Kim U, Dolan M, Morris JW, Goodman HM. A cluster of ABA-regulated genes on Arabidopsis thaliana BAC T07M07. Genome Res. 1999;9:325–333. [PubMed] [Google Scholar]
- Weatherwax SC, Ong MS, Degenhardt J, Bray EA, Tobin EM. The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiol. 1996;111:363–370. doi: 10.1104/pp.111.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm KS, Thomashow MF. Arabidopsis thaliana cor15b, an apparent homolog of cor15a, is strongly responsive to cold and ABA, but not to drought. Plant Mol Biol. 1993;23:1073–1077. doi: 10.1007/BF00021822. [DOI] [PubMed] [Google Scholar]
- Wobus U, Weber H. Sugars as signal molecules in plant seed developmnet. Biol Chem. 1999;380:937–944. doi: 10.1515/BC.1999.116. [DOI] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci. 1998;95:10294–10299. doi: 10.1073/pnas.95.17.10294. [DOI] [PMC free article] [PubMed] [Google Scholar]