Abstract
DDC2 is a novel component of the DNA integrity checkpoint pathway, which is required for proper checkpoint response to DNA damage and to incomplete DNA replication. Moreover, Ddc2 overproduction causes sensitivity to DNA-damaging agents and checkpoint defects. Ddc2 physically interacts with Mec1 and undergoes Mec1-dependent phosphorylation both in vitro and in vivo. The phosphorylation of Ddc2 takes place in late S phase and in G2 phase during an unperturbed cell cycle and is further increased in response to DNA damage. Because Ddc2 phosphorylation does not require any other known tested checkpoint factors but Mec1, the Ddc2–Mec1 complex might respond to the presence of some DNA structures independently of the other known checkpoint proteins. Our findings suggest that Ddc2 may be the functional homolog of Schizosaccharomyces pombe Rad26, strengthening the hypothesis that the mechanisms leading to checkpoint activation are conserved throughout evolution.
Keywords: checkpoints, Ddc2, DNA damage, Mec1, S. cerevisiae
Eukaryotic cells ensure genetic integrity after DNA damage or inhibition of DNA replication through a complex network of surveillance mechanisms known as checkpoints, which provide the cells with the capacity to survive genotoxic insults (Weinert and Hartwell 1988). These protective mechanisms are signal-transduction pathways specialized in detecting abnormal DNA structures. Their activation leads to delay of cell cycle progression, preventing replication or segregation of damaged DNA molecules. Checkpoint pathways are conserved from yeast to human cells, and failure to respond properly to DNA damage allows the cells to replicate and segregate damaged DNA molecules, resulting in increased mutagenesis and genetic instability, which may lead to cancer in multicellular organisms (for review, see Hartwell and Kastan 1994). Studies in different organisms, including the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, have allowed the partial dissection of the checkpoint pathways. In S. cerevisiae, the activation of the checkpoint-mediated response to DNA damage leads to a delay of the G1–S transition (Siede et al. 1993), slows down progression through S phase (Paulovich and Hartwell 1995), and delays nuclear division (Weinert and Hartwell 1988; Weinert et al. 1994), when DNA is damaged in G1, during DNA synthesis, or in G2, respectively. This response requires the proteins encoded by the RAD9 gene and by the genes of the RAD24 epistasis group, including RAD17, RAD24, MEC3, and DDC1 (Weinert et al. 1994; Longhese et al. 1996; Longhese et al. 1997; Paulovich et al. 1997a; de la Torre-Ruiz et al. 1998). This subfamily of checkpoint proteins is thought to act at an early step of the pathway by recognizing changes in DNA structure and initiating the signal-transduction cascade (for review, see Longhese et al. 1998; Weinert 1998; Lowndes and Murguia 2000). The finding that Ddc1, Rad17, and Mec3 interact physically with each other provides the evidence that these putative sensor proteins, which were inferred from genetic studies to operate in the same pathway, do indeed interact biochemically (Paciotti et al. 1998; Kondo et al. 1999). Whereas Rad24 has been shown to have homology with and to interact with subunits of replication factor C (RFC) (Griffiths et al. 1995; Lydall and Weinert 1997; Green et al. 1999), both Rad17 and Ddc1 have been reported to be structurally related to PCNA (proliferating cell nuclear antigen) (Thelen et al. 1999). It has been suggested that the Rad24–RFC complex might have a DNA structure-specific activity allowing the loading of PCNA-like checkpoint proteins on particular DNA structures (Thelen et al. 1999; Caspari et al. 2000).
Central to the checkpoint-mediated responses to DNA damage and to incomplete DNA replication are highly conserved phosphatidilinositol-related protein kinases (PIKs), including Mec1 in S. cerevisiae, the gene product of rad3+ in S. pombe, and human ATM (for review, see Carr 1997). Since Mec1 is required for Rad9 and Ddc1 phosphorylation, it has been proposed that Mec1 might participate with Ddc1, Rad9, and, possibly, with Rad17, Rad24, and Mec3 at an early step of the DNA damage recognition process (Emili 1998; Paciotti et al. 1998; Sun et al. 1998; Vialard et al. 1998). Moreover, the finding that cell cycle progression in the presence of irreparable DNA damage is controlled by Mec1, but does not require Rad9 and the Rad24 group of proteins, suggests that Mec1 plays a primary role in the S-phase damage-sensing pathway (Neecke et al. 1999). Altogether these data suggest that Mec1 acts in the DNA structure recognition step, and its activity may be modulated by the association with regulatory subunits (for review, see Longhese et al. 1998; Weinert 1998). Recent support for this hypothesis has come from studies of the fission yeast Mec1 homolog Rad3. The Rad3 protein was found to be associated with Rad26, whose DNA damage-induced phosphorylation depends on Rad3, but not on the other known checkpoint proteins, suggesting that the Rad3–Rad26 complex may function at early steps in the DNA damage recognition process (Edwards et al. 1999).
Once DNA perturbations are sensed, checkpoint signals are propagated through the protein kinase Rad53, which becomes phosphorylated and activated in response to genotoxic agents and whose phosphorylation depends on the above-listed checkpoint proteins (Sanchez et al. 1996; Sun et al. 1996). After DNA damage, Rad53 binds specifically to hyperphosphorylated Rad9, raising the possibility that Rad53 kinase activity might be influenced by association with other checkpoint proteins (Emili 1998; Sun et al. 1998; Vialard et al. 1998).
In addition to their involvement in the checkpoint responses, Rad53 and Mec1 are essential for cell viability. Their essential function can be bypassed by increasing expression of genes encoding ribonucleotide reductase (Desany et al. 1998) or by deleting the SML1 gene (Zhao et al. 1998), which negatively affects dNTP pools, possibly through post-translational regulation of ribonucleotide reductase activity.
Although many factors involved in the DNA damage checkpoint pathways have been identified, our knowledge of the molecular details of these mechanisms is still limited. Here, we describe a novel DNA integrity checkpoint protein, which we named Ddc2 (DNA damage checkpoint). We show that Ddc2 is required for all known DNA damage checkpoints and for preventing spindle elongation when DNA synthesis is inhibited. The Ddc2 protein physically interacts with Mec1 and is phosphorylated both during an unperturbed cell cycle and in response to DNA damage. In addition, Mec1 is absolutely required both in vitro and in vivo for Ddc2 phosphorylation. Moreover, Ddc2 is required for phosphorylation of the Ddc1, Pds1, and Rad53 checkpoint proteins. Conversely, Ddc2 phosphorylation does not require any of the other known DNA damage checkpoint proteins, suggesting that the Ddc2–Mec1 complex may respond to DNA insults independently of the other checkpoint factors.
Results
Disruption of DDC2 causes cell lethality, which is suppressed by deletion of SML1
We identified the ddc2-1 allele while screening for mutations causing synthetic lethality when combined with the pri1-2 cold-sensitive allele, which alters the catalytic subunit of DNA primase (Longhese et al. 1996, 1997). Cloning of DDC2 was achieved by screening a yeast genomic DNA library for complementation of the pri1-2 ddc2-1 synthetic lethal phenotype. The minimal complementing DNA fragment identified by this analysis contained only one complete, previously uncharacterized open reading frame (ORF), YDR499W, located on chromosome IV between positions 1447833 and 1450076. The identity between YDR499W and the gene identified by the ddc2-1 mutation was confirmed by complementation and allelism tests. The DDC2 ORF encodes a protein of 747 amino acid residues, with a predicted molecular mass of 86 kD.
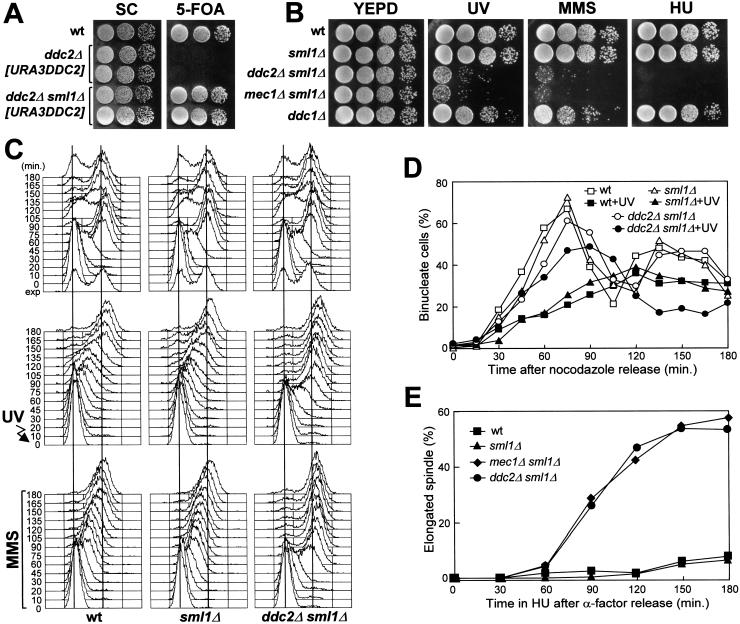
The function of Ddc2 protein is essential for cell viability. In fact, analysis of meiotic tetrads derived from DDC2/ddc2Δ heterozygous strains showed that only two spores were viable in each tetrad, and both carried the wild-type DDC2 allele. Moreover, haploid strains carrying a chromosomal ddc2Δ allele could not lose a URA3 centromeric plasmid carrying the wild-type DDC2 allele (Fig. 1A). The ability of the ddc2Δ strain to lose the DDC2 allele on the plasmid was restored by deletion of the SML1 gene (Fig. 1A). Therefore, similarly to what was observed for the deletion of the MEC1 gene (Zhao et al. 1998), deletion of SML1 suppressed cell lethality caused by the absence of Ddc2. Conversely, deletion of SML1 did not suppress the genotoxic-insult hypersensitivity caused by the ddc2Δ mutation (Fig. 1B). The DNA damage sensitivity of the ddc2Δ sml1Δ strain was indistinguishable from that of a mec1Δ sml1Δ strain, but higher than that caused by deletion of the DNA damage checkpoint gene DDC1 (Fig. 1B).
Figure 1.
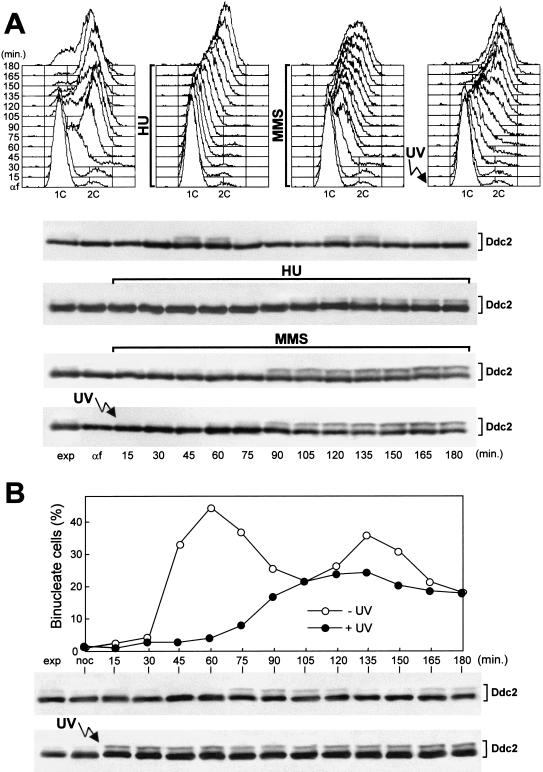
DNA damage hypersensitivity and checkpoint defects of ddc2Δ sml1Δ cells. Strains were as follows: wild type (K699), ddc2Δ [URA3 DDC2] (YLL275), ddc2Δ sml1Δ [URA3 DDC2] (DMP2995/7A), sml1Δ (YLL488) (Longhese et al. 2000), ddc2Δ sml1Δ (DMP2995/1B), mec1Δ sml1Δ (YLL490) (Longhese et al. 2000), and ddc1Δ (YLL244). (A–B) Serial dilution of YEPD-exponentially growing cell cultures were spotted on SC plates with or without 5-FOA (A) or on YEPD plates with or without MMS (0.005%) or HU (5 mm) (B). YEPD plates were made in duplicate, and one of them was UV-irradiated (30 J/m2) (UV). (C) α-Factor-synchronized cells were released from α-factor at time zero in YEPD (top), were UV-irradiated (40 J/m2) prior to the release in YEPD (middle), or were released in YEPD containing 0.02% MMS (bottom). Samples of untreated, UV- and MMS-treated cell cultures were collected at the indicated times after α-factor release and analyzed by FACS. (D) Cell cultures were arrested with nocodazole and were UV-irradiated (50 J/m2). Cell cycle progression was monitored at the indicated times in unirradiated and UV-irradiated cultures after release from nocodazole, by direct visualization of nuclear division by propidium iodide staining. (E) Cell cultures were arrested in G1 with α-factor and then released at time zero in YEPD containing 200 mm HU. Aliquots of cells were collected at the indicated times and stained with antitubulin antibodies to score for the percentage of cells with elongated spindles by indirect immunofluorescence. FACS analysis of the DNA content and plating for cell survival were carried out concomitantly (see text for details).
The DDC2 gene is required for all known DNA integrity checkpoints
We tested whether the sensitivity of the ddc2Δ sml1Δ strain to genotoxic agents was related to checkpoint defects. As shown in Figure 1, ddc2Δ sml1Δ cells turned out to be defective in all known DNA integrity checkpoints. In fact, when ddc2Δ sml1Δ α-factor arrested cells were UV-irradiated in G1 and then released from the pheromone block, both entry into S phase (Fig. 1C) and budding kinetics (data not shown) were much faster than in wild-type and sml1Δ cell cultures under the same conditions. Cell survival after UV treatment was lower in ddc2Δ sml1Δ cells (5.2%) compared to wild-type and sml1Δ cells (94% and 98%, respectively). Furthermore, when α-factor-synchronized ddc2Δ sml1Δ cells were released from G1 arrest in the presence of MMS, they doubled their DNA content within 45 min, whereas MMS-treated wild-type and sml1Δ cell cultures progressed through S phase very slowly, completing DNA replication only after 150 min (Fig. 1C). The ddc2Δ sml1Δ cells progressively lost viability during MMS treatment (already down to 6.3% cell survival at 30 min), but viability of the MMS-treated wild-type and sml1Δ cells was substantially unaffected throughout the experiment. Finally, when cell cultures were released from G2 nocodazole arrest after UV irradiation (Fig. 1D), ddc2Δ sml1Δ cells divided nuclei much faster than wild-type and sml1Δ cells, which consistently delayed nuclear division compared to unirradiated cells. Moreover, cell survival of ddc2Δ sml1Δ double mutant cells after UV irradiation was much lower (8.3%) than that of wild-type (78%) and sml1Δ (90%) single mutants under the same conditions.
As shown in Figure 1E, ddc2Δ sml1Δ cells were also defective in slowing down the elongation of mitotic spindles in the presence of incompletely replicated DNA. In fact, when cells were released from α-factor arrest in the presence of 200 mm HU, although all cell cultures had arrested with approximately 1C DNA content (C = units of the haploid genome; data not shown), spindle elongation in ddc2Δ sml1Δ and mec1Δ sml1Δ cells took place with very similar kinetics along with aberrant chromosome segregation, whereas similarly treated wild-type and sml1Δ cells never showed elongated spindles throughout the experiment (Fig. 1E). The ddc2Δ sml1Δ and mec1Δ sml1Δ cells dramatically lost viability during HU treatment (already down to 5% and 6% cell survival at 30 min for ddc2Δ sml1Δ and mec1Δ sml1Δ cells, respectively), yet viability of HU-treated wild-type and sml1Δ cells was substantially unaffected throughout the experiment.
Thus, Ddc2 is required for all known DNA damage checkpoints and for response to incomplete DNA replication, and is thus a new component of the DNA-structure response-regulatory network.
DDC2 overexpression causes DNA damage checkpoint defects
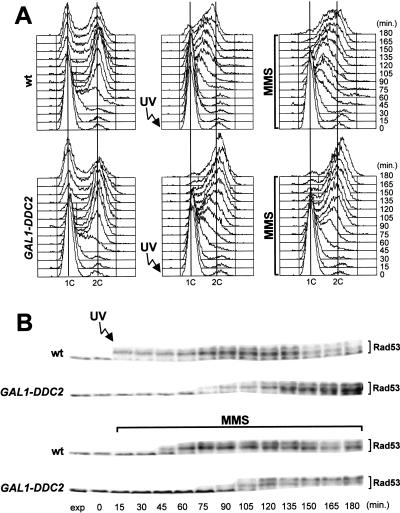
To study further the role of Ddc2 in the DNA damage checkpoint response, we analyzed the effect of overexpressing DDC2. To this purpose, a galactose-inducible GAL1–DDC2 gene fusion was integrated at the LEU2 locus of a DDC2 strain. As shown in Figure 2A, when GAL1–DDC2 cell cultures synchronized with α-factor were UV-irradiated prior to release from the G1 block or were released in the presence of MMS under galactose-induced conditions, they, respectively, entered or progressed through S phase much faster than similarly treated wild-type cells. Furthermore, they progressively lost viability during MMS treatment (9% survival at 120 min), whereas wild-type cell viability was unaffected. The inability of Ddc2 overproducing cells to respond properly to DNA damage correlated with defects in the induction of Rad53 phosphorylation. In fact, phosphorylation of Rad53 was delayed in UV-irradiated and MMS-treated GAL1–DDC2 cells under galactose-induced conditions, compared to similarly treated wild-type cells (Fig. 2B).
Figure 2.
Overexpression of DDC2 causes DNA damage checkpoint defects. Cultures of wild type (K699) and GAL1–DDC2 (YLL279.4) strains, logarithmically growing in YEP-raffinose, were synchronized with α-factor. Galactose to 2% was added 2.5 hr before α-factor addition. Release from α-factor block was performed by transferring cell cultures to YEP medium containing both raffinose and galactose, with or without 0.02% MMS. One-third of each α-factor-synchronized culture was UV-irradiated prior to the release in YEP–raf–gal. Time zero corresponds to cell samples withdrawn immediately before MMS addition or UV-irradiation and release from α-factor. The data presented in panels A and B all come from the same experiment. (A) Samples of untreated (left), UV-irradiated (middle), or MMS-treated (right) cells were taken at the indicated times after α-factor release and analyzed by FACS. (B) Extracts from UV- or MMS-treated cell cultures were analyzed by Western blot with anti-Rad53 antibodies. (exp) Exponentially growing cells.
Ddc2 and Mec1 are physically associated
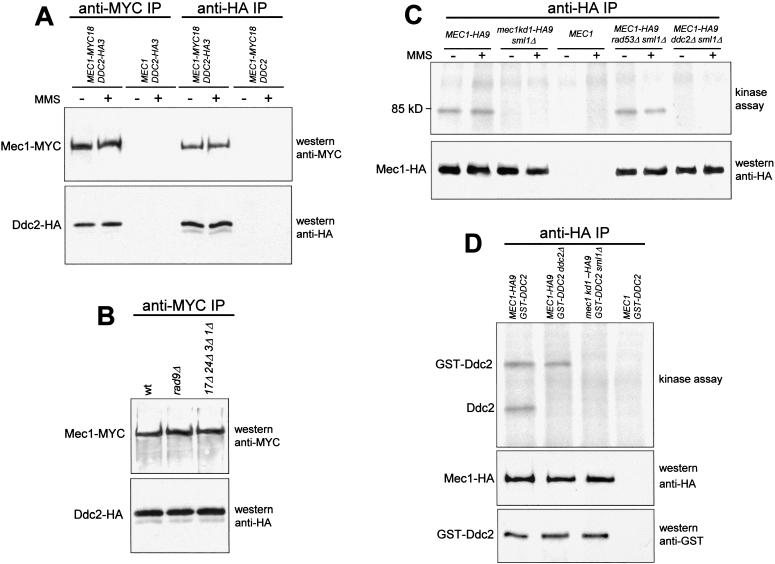
Based on the very similar behavior of ddc2Δ and mec1Δ strains, we analyzed a possible physical interaction between the proteins Ddc2 and Mec1. To this end, we generated a strain simultaneously expressing fully functional Mec1–MYC18 and Ddc2–HA3 tagged proteins from their own promoters. Western blots on crude extracts from this strain showed that anti-MYC and anti-HA antibodies specifically recognized Mec1–MYC18 and Ddc2–HA3, respectively (data not shown). These antibodies were then used to immunoprecipitate Mec1–MYC18 and Ddc2–HA3 independently from untreated and MMS-treated cells. As shown in Figure 3A, Ddc2–HA3 was specifically recognized by the anti-HA antibodies in Mec1–MYC18 immunoprecipitates, and anti-MYC antibodies detected Mec1–MYC18 in Ddc2–HA3 immunoprecipitates. Because we failed to detect Ddc2–HA3 in anti-MYC immunoprecipitates from cell extracts lacking the Mec1–MYC18 protein or to detect Mec1–MYC18 in anti-HA immunoprecipitates from cell extracts lacking the Ddc2–HA3 protein (Fig. 3A), it can be inferred that the observed Mec1–Ddc2 interaction was specific. Therefore, we conclude that Ddc2 physically interacts in vivo with Mec1. This interaction is not influenced by DNA insults, because equivalent amounts of Mec1 and Ddc2 could be coimmunoprecipitated both in untreated and in MMS-treated cells.
Figure 3.
Physical interaction between Ddc2 and Mec1 and in vitro kinase assay. (A) Immunoprecipitations with anti-MYC (anti-MYC IP) or anti-HA (anti-HA IP) antibodies were performed on extracts from exponentially growing untreated (−) or MMS-treated (+) (0.02% MMS for 1 hr) cells expressing Mec1–MYC18 (YLL447.32/1A) or Ddc2–HA3 (YLL683.8/3B) or both (DMP3084/3C), as indicated in the top part of the panel. Mec1 and Ddc2 were then detected by Western blot analysis of the immunoprecipitates probed with the antibodies indicated on the right side of the panel. (B) Western blot analysis of the anti-MYC immunoprecipitates of protein extracts from untreated wild-type (DMP3084/3C), rad9Δ (DMP3167/18A), and rad17Δ rad24Δ mec3Δ ddc1Δ (DMP3168/6D) cells concomitantly expressing Ddc2–HA3 and Mec1–MYC18. (C–D) Kinase assays were performed on anti-HA immunoprecipitates of protein extracts from exponentially growing untreated (−) or MMS-treated (+) cells with the genotypes indicated in the top parts of the panels. The same immunoprecipitates were also analyzed by Western blot using the antibodies indicated in the bottom parts of the right side of the panels. (C) Strains MEC1–HA9 (YLL476.34/2C), mec1kd1–HA9 sml1Δ (YLL593.1.3), MEC1 (K699), MEC1–HA9 rad53Δ sml1Δ (DMP2959/3A), MEC1–HA9 ddc2Δ sml1Δ (DMP3014/2A). (D) Strains MEC1–HA9 [pML103 GAL1–GST–DDC2] (YLL680), MEC1–HA9 ddc2Δ [pML103 GAL1–GST–DDC2] (YLL681), mec1kd1–HA9 sml1Δ [pML103 GAL1–GST–DDC2] (YLL682), and MEC1 [pML103 GAL1–GST–DDC2] (YLL678).
Furthermore, this interaction was unaffected both in a rad9Δ mutant and in a rad17Δ rad24Δ mec3Δ ddc1Δ quadruple mutant (Fig. 3B), as well as in a rad9Δ ddc1Δ double mutant (data not shown), indicating that none of the corresponding proteins is necessary for a stable Ddc2–Mec1 interaction.
The Mec1 protein has an associated kinase activity, which is required to phosphorylate Ddc2 in vitro
The Mec1 protein belongs to a PI-3 kinase motif family that includes S. pombe Rad3 as well as human ATM and ATR (Savitsky et al. 1995; Bentley et al. 1996; for review, see Carr 1997). The interaction between Ddc2 and Mec1, therefore, would also be consistent with a kinase–substrate relationship. Since evidence of Mec1-associated kinase activity has not yet been provided, we generated the “kinase-dead” mec1kd1 mutant allele, causing the amino acid change D2243E in the Mec1 putative kinase domain. The same amino acid change in the S. pombe Rad3 lipid kinase domain completely abolishes Rad3 function and reduces the Rad3-associated kinase activity (Bentley et al. 1996). Similarly to deletion of the MEC1 gene, the mec1kd1 allele caused cell lethality that was suppressed by deletion of SML1, and mec1kd1 sml1Δ strains were checkpoint defective (V. Paciotti et al., in prep.).
To gain insights into the role of the Mec1 conserved kinase domain and to look for putative Mec1 kinase substrates, we performed in vitro kinase assays on anti-HA immunoprecipitates from untreated and MMS-treated cells containing either HA-tagged Mec1 (Mec1–HA9) or Mec1kd1 (Mec1kd1–HA9). As shown in Figure 3C, incubation of the anti-HA immunoprecipitates with [γ-32P]ATP allowed detection of a DNA damage-independent protein kinase activity associated with Mec1–HA9. In fact, a phosphorylated protein of about 85 kD was specifically detected in both untreated and MMS-treated Mec1–HA9 immunoprecipitates (Fig. 3C). This cell-free phosphorylation was dependent on the integrity of the Mec1 kinase conserved domain, because we failed to detect the phosphorylated protein in kinase assays performed on anti-HA immunoprecipitates that either contained only Mec1kd1–HA9 or did not contain any HA-tagged proteins (Fig. 3C). Furthermore, the detected kinase activity was not caused by the possible presence of the Rad53 kinase, because we obtained identical results in kinase assays on immunoprecipitates from cell extracts lacking Rad53 (Fig. 3C). On the contrary, the phosphorylated protein, whose electrophoretic mobility was similar to that expected for Ddc2, was not detectable in kinase assays performed on immunoprecipitates from ddc2Δ sml1Δ cell extracts, indicating that either Ddc2 itself was the phosphorylated protein or Ddc2 was required for the Mec1-dependent phosphorylation event (Fig. 3C). We therefore generated wild-type and ddc2Δ strains ectopically expressing a fully functional GST–Ddc2 fusion protein along with a Mec1–HA9 or Mec1kd1–HA9 tagged protein. Anti-HA antibodies were then used to immunoprecipitate Mec1, with following kinase assays. As shown in Figure 3D, a protein species that co-migrated with GST–Ddc2 and was recognized by anti-GST antibodies was specifically phosphorylated in all Mec1–HA9 immunoprecipitates but not in Mec1kd1–HA9 immunoprecipitates, indicating that GST–Ddc2 phosphorylation was dependent on an intact Mec1 kinase domain, but was not influenced by the absence of a functional DDC2 chromosomal gene.
Altogether, these data show that a Mec1-associated kinase activity is capable of phosphorylating Ddc2 in vitro, although they do not establish that Ddc2 is a direct target in vivo. The finding that phosphorylated Ddc2 is found in kinase assays on anti-HA immunoprecipitates from cells lacking Rad53 also indicates that Rad53 is required neither for Ddc2–Mec1 interaction nor for Ddc2 phosphorylation (Fig. 3C).
Ddc2 phosphorylation occurs periodically during the cell cycle and in response to DNA damage
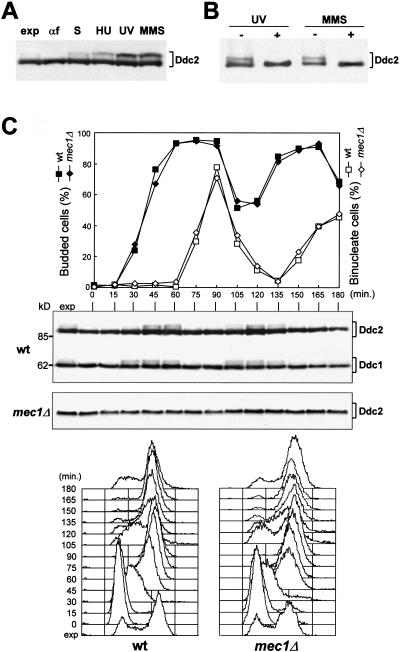
We further characterized the Ddc2 protein by analyzing strains carrying a DDC2–HA3 allele at the DDC2 chromosomal locus and expressing a fully functional 3HA-tagged Ddc2 protein. As shown in Figure 4A, when anti-HA antibodies were used in Western blots on crude extracts of exponentially growing cells, they specifically detected two major bands corresponding to Ddc2–HA3 that did not appear in extracts prepared from strains carrying the untagged DDC2 allele (data not shown). The slower migrating form was not present in α-factor-arrested cells, and its amount was increased when cells were collected during S phase or after HU treatment (Fig. 4A). MMS or UV treatment caused further increases in the amount of the slower migrating form of Ddc2 (Fig. 4A). The observed changes in Ddc2 electrophoretic mobility were owing to phosphorylation events. In fact, the slower migrating form of Ddc2 in MMS- and UV-treated cell extracts was converted to the fastest migrating protein species by treatment with bacteriophage λ-phosphatase (Fig. 4B).
Figure 4.
Ddc2 is phosphorylated during an unperturbed cell cycle and in response to DNA insults. (A–B) Protein extracts were prepared from strain YLL683.8/3B, expressing Ddc2–HA3 from the DDC2 promoter. (A) Western blot analysis with anti-HA antibodies of protein extracts from exponentially growing cultures untreated (exp) or treated with HU (50 mm; 1 hr), UV (40 J/m2), or MMS (0.02%; 1 hr). Protein extracts were also prepared from G1-arrested cells (αf) or from cells progressing through S phase 30 min after release from α-factor (S). (B) Protein extracts from UV- and MMS-treated exponentially growing cells were immunoprecipitated with anti-HA antibodies. Immunoprecipitates were then incubated at 30°C with (+) or without (−) λ-phosphatase, before electrophoresis and Western blot analysis using anti-HA antibodies. (C) Exponentially growing wild-type (DMP3198/1A) cells, concomitantly expressing Ddc2–HA3 and Ddc1–HA2 from the corresponding promoters, and mec1Δ sml1Δ (DMP3048/5B) cells, expressing Ddc2–HA3, were synchronized with α-factor and released into the cell cycle at time zero. Samples were collected at the indicated times after α-factor release to determine the percentage of budded and binucleate cells (top) and to perform Western blot analysis with anti-HA antibodies of protein extracts (middle) (molecular mass markers, kD, are indicated) and FACS analysis of the DNA content (bottom).
When α-factor-arrested cells were released from the pheromone block and Ddc2 was analyzed by Western blot during an unperturbed cell cycle, phosphorylated Ddc2 accumulated periodically, increasing in level when cells progressed through S and G2 phases and disappearing simultaneously with the appearance of binucleate cells (Fig. 4C). Among the DNA damage checkpoint proteins so far characterized, Ddc1 also undergoes cell-cycle-dependent phosphorylation (Longhese et al. 1997). As shown in Figure 4C, Ddc1 phosphorylation was already detected 30 min after α-factor release, when most cells initiated DNA replication, but Ddc2 phosphorylated forms appeared only after 45 min, when most cells were completing S phase or had reached G2. When a similar experiment was performed on a mec1Δ sml1Δ strain expressing Ddc2–HA3, we detected only the faster migrating Ddc2 band throughout the whole cell cycle, indicating that Mec1 is required for Ddc2 phosphorylation under unperturbed conditions (Fig 4C).
When G1-arrested DDC2–HA3 cell cultures were released in the presence of either HU or MMS or were UV-irradiated before α-factor release, the phosphorylated forms of Ddc2 became detectable only when cells progressed through S and G2 phases (Fig. 5A). Moreover, the amount of phosphorylated Ddc2 in HU-treated cells was indistinguishable from that accumulated during an unperturbed S phase, whereas it was increased in UV- or MMS-treated cells, suggesting that DNA damage in G1 or during S phase further stimulates Ddc2 phosphorylation, but only when cells reach S and G2.
Figure 5.
Ddc2 phosphorylation in response to DNA damage in different cell cycle phases or to replication block. The strain is YLL683.8/3B, expressing Ddc2–HA3 from the DDC2 promoter. (A) Cell cultures were synchronized with α-factor (αf) and released from the pheromone block in YEPD, YEPD containing 50 mm HU, YEPD containing 0.02% MMS, or were UV-irradiated (40 J/m2) prior to the release in YEPD. Cell samples collected at the indicated times after α-factor release were analyzed by FACS, and protein extracts were prepared and analyzed by Western blot with anti-HA antibodies. (B) Cell cultures were arrested with nocodazole (noc) and UV-irradiated (50 J/m2) prior to the release from the nocodazole arrest. Cell samples of unirradiated and UV-irradiated cultures collected at the indicated times after nocodazole release were analyzed for the percentage of nuclei division, and protein extracts were prepared and analyzed by Western blot using anti-HA antibodies.
Conversely, when nocodazole-arrested cells were UV-irradiated before release from the G2 block, phosphorylated forms of Ddc2 were detectable immediately after UV treatment, when most cells still contained undivided nuclei (Fig. 5B). This response to UV-induced DNA damage in G2 does not require cell cycle progression, since an identical extent of Ddc2 phosphorylation was observed in wild-type cells either released from nocodazole (Fig. 5B) or kept for 90 min in the presence of the drug after UV treatment in G2 (Fig. 6A).
Figure 6.
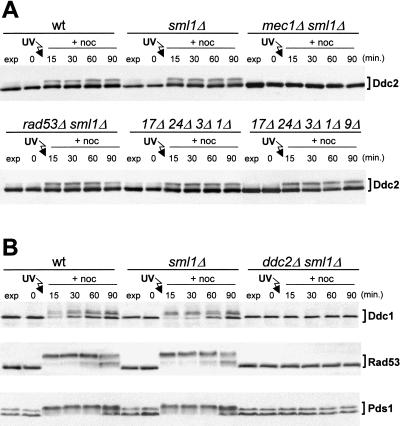
Interdependency of phosphorylation events in the checkpoint response. Cell cultures were arrested with nocodazole, UV-irradiated (50 J/m2), and resuspended in YEPD containing 15 μg/ml nocodazole (+noc). Time zero corresponds to cell samples taken immediately before UV irradiation. (A) Western blot analysis of protein extracts from wild-type (YLL683.8/3B), sml1Δ (DMP3048/8B), mec1Δ sml1Δ (DMP3048/5B), rad53Δ sml1Δ (DMP3080/7A), ddc1Δ rad24Δ rad17Δ mec3Δ (DMP3145/2D), and ddc1Δ rad24Δ rad17Δ mec3Δ rad9Δ (DMP3146/4B) strains, expressing Ddc2–HA3 from the DDC2 promoter. In all panels the Ddc2–HA3 protein was visualized with anti-HA antibodies on Western blots of protein extracts prepared at the indicated times. (B) The top part of the panel shows Western blot analysis with anti-HA monoclonal antibodies of protein extracts from wild-type (DMP3172/3A), sml1Δ (DMP3172/8A), and ddc2Δ sml1Δ (DMP3172/8C) cells, expressing Ddc1–HA2 from the DDC1 promoter. The middle and bottom parts of the panel show Western blot analysis with anti-Rad53 polyclonal antibodies and anti-MYC monoclonal antibodies, respectively, of protein extracts from wild-type (DMP3171/2A), sml1Δ (DMP3171/8B), and ddc2Δ sml1Δ (DMP3171/8A) cells, expressing Pds1–MYC18 from the PDS1 promoter.
Ddc2 phosphorylation depends on Mec1, but not on the other known DNA checkpoint proteins
DNA damage-induced Ddc2 phosphorylation after UV irradiation in G2 does not require any of the known DNA damage checkpoint proteins except Mec1. In fact, similar to what was observed during unperturbed conditions, Ddc2 phosphorylation did not occur in the absence of Mec1 after UV irradiation in G2 (Fig. 6A). Conversely, Ddc2 phosphorylation in rad53Δ sml1Δ double, ddc1Δ mec3 Δ rad17Δ rad24Δ quadruple, as well as in ddc1Δ mec3Δ rad17Δ rad24Δ rad9Δ quintuple mutant cells was indistinguishable from that observed in wild-type cells (Fig. 6A), and similar results were also obtained after UV irradiation in G1 (data not shown). Therefore, Rad17, Rad24, Mec3, Ddc1, Rad9, and Rad53 are not necessary for DNA damage-induced Ddc2 phosphorylation.
Because UV- and MMS-induced DNA damage leads to Mec1-dependent phosphorylation of Ddc1, Rad53, and the anaphase inhibitor Pds1 (Sanchez et al. 1996; Sun et al. 1996; Yamamoto et al. 1996; Cohen-Fix and Koshland 1997; Longhese et al. 1997; Paciotti et al. 1998), we asked whether Ddc2 was also required for any of these responses. As shown in Figure 6B, we failed to detect any mobility shift of these three proteins in ddc2Δ sml1Δ cells after UV irradiation in G2. Therefore, we conclude that Ddc2 is required to promote DNA damage-induced phosphorylation of Ddc1, Rad53, and Pds1.
Discussion
DNA is a reactive molecule and is prone to chemical and structural alterations, which may occur spontaneously or be induced by exogenous sources. These DNA insults induce many different forms of damage that subsequently undergo transformation and processing during the different cell cycle stages, thus challenging checkpoint and repair pathways (for review, see Paulovich et al. 1997b). One of the main problems still to be addressed is the definition of the signals leading to checkpoint activation. DNA intermediates and/or DNA–protein complexes, which can form during different stages of DNA repair, recombination, or replication, are good candidates for structures monitored by checkpoints. Whether the cell cycle phases at which any DNA alterations occur influence the chance to activate the checkpoint response is at present poorly understood. Characterization of the novel checkpoint gene DDC2 thus provides new insights into the mechanisms leading to checkpoint activation.
Ddc2 is a novel component of the DNA integrity checkpoints and undergoes periodic phosphorylation during an unperturbed cell cycle
The DDC2 gene is essential for cell viability, and its essential function is bypassed by deletion of SML1, suggesting that Ddc2, like Mec1 and Rad53, may regulate dNTP synthetic capacity (Desany et al. 1998; Zhao et al. 1998). Viable sml1Δ cells lacking DDC2 are defective in delaying cell cycle progression after DNA damage in G1, during S phase, and in G2, and fail to block spindle elongation when DNA synthesis is inhibited. The checkpoint defects and DNA damage hypersensitivity of ddc2Δ cells are indistinguishable from those of cells lacking Mec1, indicating that Ddc2 may also play a key role in the DNA integrity checkpoint response.
Moreover, cells overexpressing DDC2 are defective both in the DNA damage checkpoint response and in promoting Rad53 phosphorylation, indicating that, like Ddc2 loss of function, high levels of Ddc2 impair the ability of cells to react properly to DNA insults. The observed checkpoint defects are a peculiarity of DDC2 overexpression, because none of the other checkpoint genes analyzed so far causes checkpoint defects when overexpressed. One possibility that arises from the finding that Ddc2 physically interacts with Mec1, is that high levels of this protein may interfere with Mec1 activity or with its interaction with other regulatory subunits, implying that a functional checkpoint requires a threshold level of active Mec1. Yet, overproduction of Mec1 does not restore a proper DNA damage checkpoint response in cells overexpressing DDC2 (V. Paciotti et al., unpubl.), suggesting that other factors are involved in this phenomenon.
One important question is whether unperturbed DNA replication by itself is able to generate checkpoint signals. Indeed, although DNA replication is a remarkably accurate process, human cells are estimated to suffer DNA lesions every time their genome is replicated. Our finding that Ddc2 undergoes a Mec1-dependent phosphorylation during an unperturbed cell cycle suggests that this protein may be activated even in the absence of external DNA insults. Moreover, the finding that the Ddc2–Mec1 interaction also takes place in the absence of exogenous DNA damage suggests that these two proteins could be involved in a constant monitoring of the DNA structure integrity and in sensing possible alterations. The Ddc2 phosphorylated forms appear in the late S and G2 phases of the cell cycle and disappear at nuclear division. Among the checkpoint proteins so far identified, Ddc1 is also phosphorylated periodically during a normal cell cycle (Longhese et al. 1997). Although Ddc1 phosphorylated forms, after release from G1 block, appear as soon as cells initiate DNA replication, Ddc2 phosphorylation is detectable only when cells are completing DNA synthesis. This difference might reflect the ability of these two proteins to detect distinct sets of DNA structures.
The Ddc2–Mec1 complex can respond to DNA perturbations independently of the other DNA damage checkpoint proteins at specific cell cycle stages
Like Ddc1, Rad9, and Rad53, Ddc2 is phosphorylated in response to DNA damage. Ddc1, Rad9, and Rad53 are phosphorylated immediately in response to DNA damage at any phase of the cell cycle (Sanchez et al. 1996; Sun et al. 1996; Longhese et al. 1997; Emili 1998; Paciotti et al. 1998; Sun et al. 1998; Vialard et al. 1998), while DNA damage-induced phosphorylation of Ddc2 occurs only in specific cell cycle phases. In fact, although UV irradiation of G2-arrested cells results in immediate Ddc2 phosphorylation independently of cell cycle progression, UV irradiation in G1 is able to induce Ddc2 phosphorylation only when cells reach the middle of S phase. Therefore, although DNA damage in G2 is probably capable of triggering immediate Ddc2 phosphorylation, the G1-induced DNA damage seems unable to promote Ddc2 phosphorylation until UV-induced lesions experience DNA replication. Replication of a damaged template might play an active role in generating structures that induce a checkpoint-mediated cell cycle arrest. On the other hand, the recombinational repair capacity involving replicated chromosomes may be directly involved in the activation of the checkpoint response in G2.
We have shown that Ddc2 and Mec1 physically interact and that their association does not depend on Rad17, Rad24, Mec3, Ddc1, Rad9, or Rad53. Like Mec1, Ddc2 is also required for Ddc1, Rad53, and Pds1 DNA damage-induced phosphorylation, indicating that Ddc2 and Mec1 share common functional properties. Although Mec1 has been implicated in the phosphorylation of Rad53, Rad9, and Ddc1, no evidence for an associated kinase activity had been provided so far. We have shown that a kinase activity is associated with Mec1 and depends on the integrity of its kinase domain. This kinase activity is capable of phosphorylating Ddc2 in vitro, suggesting that Ddc2 may be a direct target of Mec1 in vivo. The Mec1-dependent phosphorylation of Ddc2 in response to DNA damage is not affected by the absence of any of the other tested checkpoint proteins, indicating that the Ddc2–Mec1 complex responds to specific DNA structures independently of other checkpoint components. Moreover, the cell cycle dependent DNA damage-induced Ddc2 phosphorylation suggests that the Ddc2–Mec1 complex responds differently to the presence of aberrant DNA structures during specific cell cycle stages. From this perspective, it is reasonable to hypothesize that sensing and processing DNA damage in G1 may require more specialized sensor proteins than are required during S phase to activate the checkpoint-mediated cell cycle arrest. In fact, broken forks could arise during S phase as polymerase traverses pre-existing nicks, and DNA replication on a damaged template has an intrinsic probability to form double strand breaks (Haber 1999). Furthermore, recombination intermediates can be generated when stalling of the replication fork is caused by DNA polymerase encountering a lesion and DNA synthesis is resumed downstream from the damage (Neecke et al. 1999). Conversely, when DNA is damaged in G1, active processing of DNA damage might be necessary. This would explain the fact that although DNA damage-induced Rad53 phosphorylation requires Rad24, Rad17, Mec3, Ddc1, and Rad9 in G1, this requirement is greatly reduced in S phase cells (Navas et al. 1996; Pellicioli et al. 1999). Moreover, Rad9 and the Rad24 group of proteins appear unnecessary for checkpoint activation when chromosomes containing unrepaired damage experience DNA replication (Neecke et al. 1999). Therefore, replication of a damaged template could generate checkpoint signals capable of activating Mec1, either directly or through other S-phase-specific sensors; this, in turn, would allow Ddc2 phosphorylation.
The Ddc2 phosphorylation response to DNA damage in G2 also requires only Mec1 and is immediate, suggesting that the Mec1–Ddc2 sensing apparatus may be directly involved in recognizing or processing DNA lesions in G2. Indeed, homologous chromosome pairs are present in G2, and the repair capacity of cells that experience DNA damage in G2 is mainly represented by recombinational repair. Ddc2 works together with Mec1, and recent data have implicated Mec1 and its human homolog ATM in recombination mechanisms (Meyn 1993; Shafman et al. 1997; Chen et al. 1999; Cortez et al. 1999; Grushcow et al. 1999; Thompson and Stahl 1999; Bashkirov et al. 2000; Morrison et al. 2000). If Mec1 had a role in promoting the recombination pathways and in stabilizing and resolving the recombination intermediates, this could explain the reduced requirement of any other known checkpoint proteins for Ddc2 phosphorylation in response to DNA damage in G2.
Is Ddc2 the functional homolog of S. pombe Rad26?
Although the Ddc2 protein has no easily identifiable homologs in other species, it is interesting to note that search for homology with the S. pombe Rad26 protein sequence among the translational products of all S. cerevisiae genome sequences with different programs always picks up the Ddc2 protein. The best alignment between the two protein sequences shows only 13.6% identity and 33% similarity, but the data reported in this paper indicate that several properties are shared by Rad26 and Ddc2. In fact, Rad26 interacts with Rad3, the Mec1 S. pombe homolog, independently of other known checkpoint factors, and undergoes Rad3-dependent DNA damage-induced phosphorylation that does not require any other known checkpoint proteins (Edwards et al. 1999). Moreover, the ability of cells to promote Rad26 phosphorylation is influenced by the cell cycle stages at which DNA alterations occur. The two proteins Rad26 and Ddc2 show differences as well as similarities in properties. In fact, unlike Ddc2, Rad26 seems not to be phosphorylated during an unperturbed cell cycle and after HU treatment. If Ddc2 phosphorylation in HU-treated cells reflects its cell-cycle-dependent phosphorylation, the absence of Rad26 phosphorylation in the unperturbed condition may reflect differences in DNA metabolism between the two yeasts, implying additional roles for the budding yeast checkpoint proteins during unperturbed conditions. This is also the case for S. pombe Rad9, the Ddc1 functional homolog, which does not show any modification during an unperturbed cell cycle.
Altogether, our data strongly suggest that Ddc2 is the functional equivalent of S. pombe Rad26. This phenomenological similarity underscores not only a structural conservation of the checkpoint response between the two yeasts but also provides further evidence that the mechanisms leading to checkpoint activation are shared by evolutionarily distant organisms.
Materials and methods
Cloning and disruption of the DDC2 gene
The ddc2-1 mutation was previously identified as pip6-1 (Longhese et al. 1996). To clone the gene identified by this mutation, strain DMP635/1B (see Table 1; square brackets indicate plasmids), derived from the original mutant, was transformed with a yeast genomic DNA library constructed in the pUN100 LEU2 centromeric plasmid (Jansen et al. 1993). Transformants were screened for the presence of recombinant plasmids that are capable of restoring a Sect+ 5-FOA+ phenotype and therefore possibly complement the pri1-2 ddc2-1 synthetic lethality (Longhese et al. 1996). Sequencing of both ends of the smallest yeast DNA insert identified by this screening and search through the yeast genome database revealed that the cloned fragment was located on S. cerevisiae chromosome IV, between positions 1445133 and 1454619. Further analysis allowed us to establish that a 3681-bp NsiI–NsiI DNA fragment was sufficient to complement the pri1-2 ddc2-1 synthetic lethal phenotype and that it contained the YDR499W ORF we call DDC2.
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain
|
Genotype
|
Source
|
|---|---|---|
| K699 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 | Longhese et al. 1997 |
| YLL244 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ddc1Δ::KanMX4 | Longhese et al. 1997 |
| YLL275 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ddc2Δ::KanMX4 [pML94 CEN4 URA3 DDC2] | This study |
| YLL279.4 | MATa ade2-1 can1-100 his3-11,15 trp1-1 ura3 leu2-3,112::GAL1-DDC2::LEU2 | This study |
| YLL447.32/1A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 MEC1–MYC18::LEU2::mec1 | This study |
| YLL476.34/2C | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 MEC1–HA9::LEU2::mec1 | This study |
| YLL488 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 sml1Δ::KanMX4 | Longhese et al. 2000 |
| YLL490 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 mec1 Δ::HIS3 sml1Δ::KanMX4 | Longhese et al. 2000 |
| YLL593.1.3 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 mec1D2243E–HA9::LEU2::mec1 sml1Δ::KanMX4 | This study |
| YLL678 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 [pML103 2μ URA3 GAL1–GST–DDC2] | This study |
| YLL680 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 MEC1–HA9::LEU2::mec1 [pML103 2μ URA3 GAL1–GST–DDC2] | This study |
| YLL681 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 MEC1–HA9::LEU2::mec1 ddc2Δ::KanMX4 [pML103 2μ URA3 GAL1–GST–DDC2] | This study |
| YLL682 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 mec1D2243E–HA9::LEU2::mec1 sml1Δ::KanMX4 [pML103 2μ URA3 GAL1–GST–DDC2] | This study |
| YLL683.8/3B | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2–HA3::URA3 | This study |
| DMP635/1B | MATα ade2-1 ade3 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 pri1-2 pip6-1 [pML9 ADE3 URA3 PRI1] | This study |
| DMP2959/3A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 MEC1–HA9::LEU2::mec1 rad53Δ::HIS3 sml1Δ::KanMX4 | This study |
| DMP2995/1B | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 sml1Δ::KanMX4 ddc2Δ::KanMX4 | This study |
| DMP2995/7A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 sml1Δ::KanMX4 ddc2Δ::KanMX4 [pML94 CEN4 URA3 DDC2] | This study |
| DMP3014/2A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 MEC1–HA9::LEU2::mec1 ddc2Δ::KanMX4 sml1Δ::KanMX4 | This study |
| DMP3048/5B | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2–HA3::URA3 mec1Δ::HIS3 sml1Δ::KanMX4 | This study |
| DMP3048/8B | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2–HA3::URA3 sml1Δ::KanMX4 | This study |
| DMP3080/7A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2–HA3::URA3 rad53Δ::HIS3 sml1Δ::KanMX4 | This study |
| DMP3084/3C | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 MEC1–MYC18::LEU2::mec1 DDC2–HA3::URA3 | This study |
| DMP3145/2D | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2–HA3::URA3 rad17Δ::LEU2 rad24Δ::TRP1 mec3Δ::TRP1 ddc1Δ::KanMX4 | This study |
| DMP3146/4B | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2–HA3::URA3 rad17Δ::LEU2 rad24Δ::TRP1 mec3Δ::TRP1 ddc1Δ::KanMX4 rad9Δ::URA3 | This study |
| DMP3167/18A | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 MEC1–MYC18::LEU2::mec1 DDC2–HA3::URA3 rad9Δ::URA3 | This study |
| DMP3168/6D | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 MEC1–MYC18::LEU2::mec1 DDC2–HA3::URA3 rad17Δ::LEU2 rad24Δ::TRP1 mec3Δ::TRP1 ddc1Δ::KanMX4 | This study |
| DMP3171/2A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 PDS1–MYC18::LEU2::pds1 | This study |
| DMP3171/8A | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 PDS1–MYC18::LEU2::pds1 sml1Δ::KanMX4 ddc2Δ::KanMX4 | This study |
| DMP3171/8B | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 PDS1–MYC18::LEU2::pds1 sml1Δ::KanMX4 | This study |
| DMP3172/3A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC1–HA2::LEU2::ddc1 | This study |
| DMP3172/8A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC1–HA2::LEU2::ddc1 sml1Δ::KanMX4 | This study |
| DMP3172/8C | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC1–HA2::LEU2::ddc1 sml1Δ::KanMX4 ddc2Δ::KanMX4 | This study |
| DMP3198/1A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2-HA3::URA3 DDC1-HA2::LEU2::ddc1 | This study |
To construct a DDC2 chromosomal deletion, the ddc2Δ:: KanMX4 cassette was constructed by PCR using plasmid pFA6a–kanMX4 (Wach et al. 1994) as a template and oligonucleotides PRP23 (3′-GGAATTAGGCACCAGGCCTCCAAGGTTTACTCAAATACCGCCATCCGTACGCTGCAGGTCGAC-5′) and PRP24 (3′-CCTTTATCGTCAGATCATGCAAACTATCTACAAGGTGTCGATGATCTCTCATCGATGAATTCGAGCTCG-5′) as primers. One-step replacement of 1879 bp of the DDC2 coding region with the KanMX4 cassette was carried out by transforming the diploid strain W303. G418-resistant transformants were shown by PCR analysis to be heterozygous for the replacement. Analysis of meiotic tetrads derived from the DDC2/ddc2Δ heterozygous strain showed that only two spores were viable in each tetrad, and each carried the wild-type DDC2 allele, whereas no viable spores containing the ddc2Δ::KanMX4 allele were found. The same one-step replacement was carried out by transforming a diploid W303 derivative strain heterozygous for the deletion of the SML1 gene, thus generating SML1/sml1Δ DDC2/ddc2Δ strains. When such a double heterozygous strain was allowed to sporulate, several tetrads contained more than 2 viable spores. Viable ddc2Δ sml1Δ spores were present with the frequency expected for spores simultaneously carrying the two unlinked ddc2Δ and sml1Δ alleles, indicating that deletion of SML1 suppresses the cell lethality caused by the absence of Ddc2. The identity of the cloned DDC2 sequence with that of the gene identified by the ddc2-1 mutation was demonstrated by constructing a ddc2Δ/ddc2-1 SML1/sml1Δ diploid strain. This diploid strain was as hypersensitive to UV, MMS, and HU as the parental strains (data not shown). Furthermore, although spore viability was affected, we could test 145 viable meiotic segregants from 66 tetrads of this diploid strain, which were all sensitive to UV, MMS, and HU, confirming that the ddc2Δ and ddc2-1 mutations are allelic.
Plasmids
Plasmid pML81 is the original pUN100 derivative plasmid (Jansen et al. 1993) carrying the S. cerevisiae chromosome IV fragment between positions 1445133 and 1454619. Plasmid pML94 was obtained by cloning the DDC2 NsiI–NsiI fragment from plasmid pML81 into the PstI site of plasmid YCplac33 (Gietz and Sugino 1988). To construct plasmid pML105, in which the 2722-bp fragment spanning from the DDC2 ATG to the XbaI site and containing the entire DDC2 coding region is fused to the GAL1 promoter, a DDC2 fragment spanning from position +1 to position +621 from the translation initiation codon was amplified by PCR using plasmid pML94 as a template and oligonucleotides PRP36 (5′-CGCGGATCCATTATGAGACGAGAAACGGTGGGTGAAT-3′) and PRP37 (5′-CTGAGGTTCTACATGCAAATTCAT-3′) as primers and was then cloned into the pML95 BamHI–XbaI sites (Longhese et al. 1997), to give rise to plasmid pML100. The 2105-bp XbaI fragment from plasmid pML81 was then cloned into the XbaI site of plasmid pML100, creating plasmid pML105.
To construct plasmid pML103, in which the 2724-bp fragment spanning from the DDC2 ATG to the XbaI site and containing the entire DDC2 coding region is fused to GAL1–GST, a DDC2 fragment spanning from position +1 to position +621 from the translation initiation codon was amplified by PCR using plasmid pML94 as a template and oligonucleotides PRP36 and PRP37 as primers and was then cloned into the BamHI–XbaI sites of plasmid pEG(KT) (Mitchell et al. 1993), to give rise to plasmid pML99. The 2105-bp XbaI fragment from plasmid pML81 was then inserted into the XbaI site of plasmid pML99, creating plasmid pML103.
Plasmids pML191.17 and pML205.5, used to generate the MEC1–MYC18 and MEC1–HA9 alleles, respectively, were originated by inserting the 1103-bp EcoRI–BstXI MEC1 fragment into the EcoRI–SmaI sites in the YIplac128 polylinker region (Gietz and Sugino 1988). Sequences encoding 18 tandem MYC epitopes or 9 tandem HA epitopes were then inserted into a NotI restriction site introduced by PCR at the MEC1 codon 53 in plasmids pML191.17 and pML205.5, respectively.
Plasmid pML224, used to generate the mec1kd1 allele, was originated by inserting the 1243-bp KpnI–BamHI MEC1 fragment from plasmid pML79 (Longhese et al. 1997) into the KpnI–BamHI sites in the YIplac211 polylinker region (Gietz and Sugino 1988). Plasmid pML228.1, carrying the carboxy-terminal region of MEC1 and containing the D2243E amino acid change, was generated by site-directed mutagenesis PCR using PRP154 (3′-CGGGTAAAGTTCTTCATGTAGAATTCGACTGTTTATTTGAGAAAG-5′) and PRP155 (3′-CTTTCTCAAATAAACAGTCGAATTCTACATGAAGAACTTTACCCG-5′) as primers and pML224 as a template.
Yeast strains and media
The genotypes of all the yeast strains used in this study are listed in Table 1. All yeast strains were derivatives of, or were back-crossed to, W303 (MATa or MATα, ade2-1, trp1-1, leu2-3,112, his3-11,15, ura3). Strain YLL275 was constructed by transforming a K699 strain containing plasmid pML94 with the ddc2Δ::KanMX4 cassette. Strain YLL279.4, carrying three copies of the GAL1–DDC2 fusion integrated at the LEU2 locus, was obtained by transforming strain K699 with ClaI-digested plasmid pML105. Strain YLL683.8/3B, carrying the DDC2–HA3 allele at the DDC2 chromosomal locus, was generated by the PCR one-step tagging method (Knop et al. 1999) using plasmid 3748, kindly provided by K. Nasmyth (IMP, Vienna), as template and oligonucleotides PRP179 (5′-CTTGAGTCAAAATCATTCGATCTAACCACACTAGAGGAGGCCGATTCATTATATATCTCAATGGGACTGTCCGGTTCTGCTGCTAG-3′) and PRP180 (3′-ATATAGTTAATATTAAGCATTACAAGGTTTCTATAAAGCGTTGACATTTTCCCCTTTTGATTGTTGCCCCTCGAGGCCAGAAGAC-5′) as primers. Strains YLL447.32/1A and YLL476.34/2C were generated by integration of StuI-digested plasmids pML191.17 and pML205.5 in W303, respectively, followed by tetrad dissection. Strain YLL593.1.3, carrying the mec1D2243E–HA9 allele at the MEC1 chromosomal locus was constructed by integration of XhoI-digested plasmid pML228.1 into a MEC1–HA9 sml1Δ strain followed by excision of the URA3 marker.
The MEC1–MYC18, MEC1–HA9, and DDC2–HA3 alleles are fully functional, because strains K699, YLL447.32/1A, YLL476.34/2C, and YLL683.8/3B were indistinguishable from one another with respect to viability, growth rates at any temperature, and sensitivity to UV, MMS, and HU. Strains YLL678, YLL680, and YLL682 were obtained by transforming, respectively, strains K699, YLL476.34/2C, and YLL593.1.3 with pML103 plasmid DNA. Strain YLL681 has been obtained by transforming a ddc2Δ/DDC2 MEC1–HA9/MEC1 diploid strain with pML103 plasmid DNA, followed by tetrad dissection. The accuracy of all gene replacements and integrations was verified by Southern blot analysis or PCR. All the other listed strains were obtained by crosses followed by sporulation and tetrad dissection.
Standard yeast genetic techniques and media were according to Rose et al. (1990). Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone, 50 mg/l adenine) supplemented with 2% glucose (YEPD), 2% raffinose (YEP–raf), or with 2% raffinose and 2% galactose (YEP–raf–gal). Transformants carrying the KanMX4 cassette were selected on YEPD plates containing 400 μg/ml G418 (US Biological).
Synchronization experiments
Cell synchronization in G1 was obtained by treatment of exponentially growing cell cultures with 2 μg/ml of α-factor. Yeast cells were synchronized in G2 by treating exponentially growing YEPD cell cultures with 5 μg/ml of nocodazole in 1% dimethylsulfoxide (DMSO). UV, MMS, and HU treatments were performed as previously described (Longhese et al. 1997). Flow cytometric DNA quantitation was determined on a Becton-Dickinson FACScan. In situ immunofluorescence was performed according to Nasmyth et al. (1990). All the experiments were performed at 26°C.
Western blot analysis, immunoprecipitation, and phosphatase treatment
For Western blot analysis, protein extracts were prepared by TCA precipitation as previously described (Longhese et al. 1997). Protein extracts were resolved by electrophoresis on 10% SDS–polyacrylamide gels and on 7.5% SDS–polyacrylamide gels to detect Mec1–MYC18, Mec1–HA9, and Ddc2–HA3 proteins. Proteins were transferred to Protran membranes (Schleicher and Schuell), which were incubated for 2 hr with the anti-HA monoclonal antibodies 12CA5 or with the anti-MYC monoclonal antibodies 9E10. Rad53 was detected using anti-Rad53 polyclonal antibodies kindly provided by C. Santocanale (Pharmacia-Upjohn, Italy). Secondary antibodies were purchased from Amersham, and proteins were visualized by an enhanced chemiluminescence system according to the manufacturer. Immunoprecipitation and phosphatase treatments were performed as described in Longhese et al. (1997).
Kinase assay
Protein extracts were prepared as described in Longhese et al. (1997). After addition of 1:1 volume acid-washed glass beads, 2.5 mg of clarified protein extracts was incubated for 1 hr at 4°C with 75 μl of a 50% (vol/vol) protein A–Sepharose, covalently linked to 12CA5 monoclonal antibodies. Immunoprecipitates were then washed twice with 1 ml of phosphate-buffered saline, resuspended in 60 μl of 40 mm Hepes (pH 7.4), 10 mm MnCl2, 10 mm MgCl2, 1 μm ATP, 1 mm dithiothreitol, 100 μm sodium orthovanadate, and protease inhibitor cocktail, and were incubated at 30°C for 30 min with 2 μl of [γ-32P]ATP. Twenty-five μl of SDS-gel loading buffer was added to the resins; bound proteins were resolved by electrophoresis on a 7.5% SDS–polyacrylamide gel and visualized after exposure of the gels to autoradiography films.
Acknowledgments
We are grateful to R. Fraschini for help in the cloning experiments and to Luca De Gioia for computer analysis. We wish to thank C. Santocanale, R. Deschenes, K. Nasmyth, and M. Shirayama for providing antibodies, plasmids, and strains; S. Piatti for critical reading of the manuscript; and all the members of our laboratory for useful discussions and criticisms. This work was supported by grants from Associazione Italiana Ricerca sul Cancro and Cofinanziamento 1999 MURST-Università di Milano-Bicocca to G. Lucchini and by CNR Target Project on Biotechnology Grant CT.97.01180.PF49(F). V. Paciotti was supported by a fellowship from Fondazione Italiana per la Ricerca sul Cancro.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mariapia.longhese@unimib.it; FAX 39-02-64483565.
References
- Bashkirov VI, King JS, Bashkirova EV, Schmuckli-Maurer J, Heyer WD. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol Cell Biol. 2000;20:4393–4404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, Demaggio A, Ford JC, Hoekstra M, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Carr AM. Control of cell cycle arrest by the Mec1sc/Rad3sp DNA structure checkpoint pathway. Curr Opin Genet Dev. 1997;7:93–98. doi: 10.1016/s0959-437x(97)80115-3. [DOI] [PubMed] [Google Scholar]
- Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: A PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;74:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Yuan SF, Liu W, Xu Y, Trujillo K, Song B, Cong F, Goff SP, Wu Y, Arlinghaus R, et al. Radiation-induced assembly of Rad51 and Rad42 recombination complex requires ATM and c-Abl. J Biol Chem. 1999;274:12748–12752. doi: 10.1074/jbc.274.18.12748. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Koshland D. The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway. Proc Natl Acad Sci. 1997;94:14361–14366. doi: 10.1073/pnas.94.26.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Quin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of Brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- de la Torre-Ruiz M, Green CM, Lowndes NF. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 1998;17:2687–2698. doi: 10.1093/emboj/17.9.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes & Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RJ, Bentley RJ, Carr AM. A Rad3–Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nature Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- Emili A. MEC1-Dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base-pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF. A novel Rad24 checkpoint complex closely related to Replication factor C. Curr Biol. 1999;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- Griffiths DJ, Barbet NC, McCready S, Lehmann AR, Carr AM. Fission yeast RAD17: A homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grushcow JM, Holzen TM, Park KJ, Weinert T, Lichten M, Bishop DK. Saccharomyces cerevisiae checkpoint genes MEC, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics. 1999;153:607–620. doi: 10.1093/genetics/153.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. DNA recombination: The replication connection. TIBS. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- Jansen R, Tollervey D, Hurt EC. A U3 snoRNP protein with homology to splicing factor PRP4 and Gβ domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kondo T, Matsumoto K, Sugimoto K. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol Cell Biol. 1999;19:1136–1143. doi: 10.1128/mcb.19.2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Foiani M, Muzi Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Fraschini R, Plevani P, Lucchini G. Yeast pip3/mec3 mutants fail to delay entry into S phase and to slow DNA replication in response to DNA damage, and they define a functional link between Mec3 and DNA primase. Mol Cell Biol. 1996;16:3235–3244. doi: 10.1128/mcb.16.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Paciotti V, Fraschini R, Zaccarini R, Plevani P, Lucchini G. The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 1997;16:5216–5226. doi: 10.1093/emboj/16.17.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese, M.P., Paciotti, V., Neecke, H., and Lucchini, G. 2000. Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics (In press). [DOI] [PMC free article] [PubMed]
- Lowndes NF, Murguia JR. Sensing and responding to DNA damage. Curr Opin Gen Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- Lydall D, Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: Further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- Meyn MS. High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science. 1993;260:1327–1330. doi: 10.1126/science.8493577. [DOI] [PubMed] [Google Scholar]
- Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J. 2000;19:463–471. doi: 10.1093/emboj/19.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Adolf G, Lydall D, Seddon A. The identification of a second cell cycle control in the HO promoter in yeast: Cell cycle regulation of SWI5 nuclear entry. Cell. 1990;62:631–647. doi: 10.1016/0092-8674(90)90110-z. [DOI] [PubMed] [Google Scholar]
- Navas TA, Sanchez Y, Elledge SJ. RAD9 and DNA polymerase ɛ form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes & Dev. 1996;10:2632–2643. doi: 10.1101/gad.10.20.2632. [DOI] [PubMed] [Google Scholar]
- Neecke H, Lucchini G, Longhese MP. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1- and Rad53-dependent checkpoint in budding yeast. EMBO J. 1999;18:4485–4497. doi: 10.1093/emboj/18.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V, Lucchini G, Plevani P, Longhese MP. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:101–111. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- Paulovich AG, Margulies RU, Garvik BM, Hartwell LH. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997a;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Toczyski DP, Hartwell LH. When checkpoints fail. Cell. 1997b;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore P, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;286:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Shafman T, Khanna KK, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D, et al. Interaction between ATM protein and c-Abl in response to DNA damage. Nature. 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- Siede W, Friedberg AS, Friedberg EC. RAD9-Dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc Natl Acad Sci. 1993;90:7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Fay DS, Marini F, Foiani M, Stern DF. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes & Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- Sun Z, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- Thelen MP, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- Thompson DA, Stahl FW. Genetic control of recombination partner preference in yeast meiosis: Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics. 1999;153:621–641. doi: 10.1093/genetics/153.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialard JE, Gilbert CS, Green CM, Lowndes NF. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruption in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Weinert T. DNA damage and checkpoint pathways: Molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes & Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EGD, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]