Abstract
Cap-independent translation initiation on picornavirus mRNAs is mediated by an internal ribosomal entry site (IRES) in the 5′ untranslated region (5′ UTR) and requires both eukaryotic initiation factors (eIFs) and IRES-specific cellular trans-acting factors (ITAFs). We show here that the requirements for trans-acting factors differ between related picornavirus IRESs and can account for cell type-specific differences in IRES function. The neurovirulence of Theiler's murine encephalomyelitis virus (TMEV; GDVII strain) was completely attenuated by substituting its IRES by that of foot-and-mouth disease virus (FMDV). Reconstitution of initiation using fully fractionated translation components indicated that 48S complex formation on both IRESs requires eIF2, eIF3, eIF4A, eIF4B, eIF4F, and the pyrimidine tract-binding protein (PTB) but that the FMDV IRES additionally requires ITAF45, also known as murine proliferation-associated protein (Mpp1), a proliferation-dependent protein that is not expressed in murine brain cells. ITAF45 did not influence assembly of 48S complexes on the TMEV IRES. Specific binding sites for ITAF45, PTB, and a complex of the eIF4G and eIF4A subunits of eIF4F were mapped onto the FMDV IRES, and the cooperative function of PTB and ITAF45 in promoting stable binding of eIF4G/4A to the IRES was characterized by chemical and enzymatic footprinting. Our data indicate that PTB and ITAF45 act as RNA chaperones that control the functional state of a particular IRES and that their cell-specific distribution may constitute a basis for cell-specific translational control of certain mRNAs.
Keywords: IRES, ITAF45, picornavirus, RNA-binding proteins, translation initiation
The control of mRNA translation is an important component of the regulation of gene expression, and various cellular processes are regulated by enhancement or repression of translation initiation. Message-specific translational control relies on structural elements in either 5′ or 3′ nontranslated regions of mRNAs that may bind regulatory proteins, determine the affinity of binding of canonical translation initiation factors, or enable mRNAs to utilize variants of the canonical initiation mechanism (Standart and Jackson 1994).
Most examples of mRNA-specific translational control involve repression by RNA-binding proteins. Repression of initiation is exemplified by ferritin mRNA, one of a group of mRNAs that contain a cap-proximal 5′ UTR iron-responsive element (IRE; Hentze and Kühn 1996). Binding of the iron regulatory protein to the IRE sterically prevents recruitment by eIF4F of the 43S ribosomal complex to the cap-proximal region of the 5′ UTR before scanning to the initiation codon and thus represses translation (Muckenthaler et al. 1998). RNA protein interactions can also direct patterns of expression that are spatially restricted within cells or organisms, for example, during development or differentiation, and commonly involve repression of translation of specific mRNAs by regulatory 3′ UTR-binding proteins (Gray and Wickens 1998). Translational enhancement of specific mRNAs is less common. All mRNAs that are translated following conventional cap-mediated initiation require the full set of canonical eIFs, and variations in initiation factor levels have not been associated with translational enhancement of specific mRNAs that use this mechanism. Cell type-specific translational regulation is more likely to affect those mRNAs that require noncanonical protein factors for initiation in addition to the canonical set of eIFs.
Picornavirus 5′ NTRs contain a ∼ 400-nt-long internal ribosomal entry site (IRES) that mediates cap-independent translation initiation (Jackson and Kaminski 1995). These IRESs are divided into major groups on the basis of structural properties. One group includes poliovirus and rhinovirus and the other contains encephalomyocarditis virus (EMCV), foot-and-mouth disease virus (FMDV), and Theiler's murine encephalomyelitis virus (TMEV). There is significant genetic evidence that IRESs contain determinants of cell specificity. A mutant poliovirus in which the IRES had been substituted by the rhinovirus IRES replicated as well as wild-type poliovirus in HeLa cells, but replication of the mutant (but not wild-type) viruses was completely restricted in neuronal cells (Gromeier et al. 1996). Single substitutions in the IRESs of poliovirus vaccine strains are important determinants of their attenuated neurovirulence and impaired function in translation initiation, particularly in neural cells (Svitkin et al. 1985, 1988). Mutations in the IRES also attenuate TMEV neurovirulence in mice while having only minor effects on viral growth in cell culture (Pritchard et al. 1992; Pilipenko et al. 1995, 1999). Although such noncoding determinants of IRES function have been characterized in detail, the molecular basis for their cell-specific influence is not known.
IRES-mediated initiation may require both canonical initiation factors and message-specific cellular IRES trans-acting factors (ITAFs) that are not involved in cap-mediated initiation. EMCV, TMEV, and FMDV IRESs are all active in rabbit reticulocyte lysate (RRL), where translation mediated by poliovirus and rhinovirus IRESs is inefficient unless the lysate is supplemented by HeLa cell extracts (Brown and Ehrenfeld 1979; Dorner et al. 1984; Borman et al. 1993). Biochemical analyses led to the identification of the ITAFs present in these extracts as the pyrimidine tract-binding protein (PTB; Borman et al. 1993; Hellen et al. 1993), the La autoantigen (Meerovitch et al. 1993; Svitkin et al. 1994), the poly(rC)-binding protein 2 (PCBP2; Blyn et al. 1996, 1997), and unr and a unr-interacting protein (Hunt et al. 1999). All of these proteins except for the unr-interacting protein are RNA-binding proteins that bind specifically to structural elements in different IRESs and are thought to maintain these RNAs in an active conformation (Kaminski and Jackson 1998; Kolupaeva et al. 1998; Gamarnik and Andino 2000). A reasonable hypothesis is therefore that tissue-specific differences in the expression of an essential ITAF could determine those cells in which the IRES is active.
We report here the identification of a novel cellular mRNA-specific ITAF. We found that the activities of TMEV and FMDV IRESs were similar in RRL and in transfected BHK-21 cells but that replacement of the IRES of the neurotropic TMEV by the IRES of the epitheliotropic FMDV yielded viable but completely attenuated viruses that fail to replicate in mouse neurons. To identify the factor that is required for FMDV IRES function, we compared the factor requirements for initiation on TMEV and FMDV IRESs by biochemical reconstitution to the stage of 48S complex formation using fully fractionated components. Using this approach, we have previously found that initiation on the EMCV IRES up to the stage of 48S complex formation requires the canonical factors eIF2, eIF3, eIF4A, eIF4B, and eIF4F, and is enhanced less than twofold by PTB (Pestova et al. 1996a, 1996b). We also report that 48S complex formation on FMDV and TMEV IRESs requires the same set of canonical factors and that initiation on all three IRESs involves specific binding of a complex made up of the eIF4A and eIF4G components of eIF4F to the IRES. However, initiation on the TMEV IRES depended strongly on PTB, where initiation on the FMDV IRES required both PTB and a second protein, ITAF45, that is not required for TMEV initiation. ITAF45 bound specifically to a central domain of the FMDV IRES and acted synergistically with PTB to enhance binding of eIF4F to an adjacent domain. ITAF45 is a proliferation-dependent protein that is not detectable in murine brain cells and may thus function as a tissue-specific factor that controls translation of particular mRNAs.
Results
In vitro properties of TMEV GDVII/FMDV chimeras
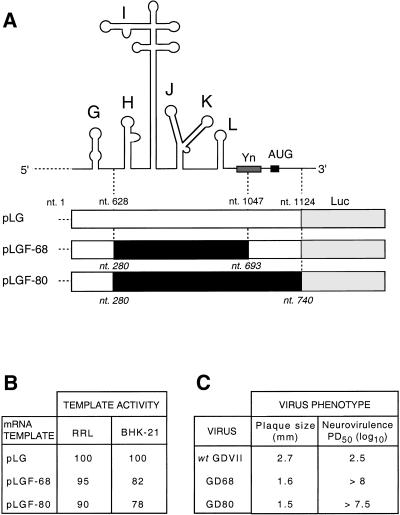
TMEV GDVII causes rapid fatal encephalitis in mice following intracerebral inoculation due to lytic infection of neurons (Theiler 1937). FMDV replicates in epithelial cells, causing an acute systemic infection, but does not replicate in the central nervous system (CNS; Bachrach 1968). We used FMDV and TMEV IRESs to investigate the molecular basis for cell-specific differences in IRES function. Chimeric GD68 and GD80 variants of TMEV GDVII were made by substituting its IRES by the FMDV IRES. IRES-mediated translation was assessed using constructs in which the luciferase coding region had been fused to TMEV nt 1–1124 in frame and downstream of the initiation codon AUG1068–70 (Fig. 1A). In the pLGF-68 and pLGF-80 chimeras, TMEV sequences from nt 628 (upstream of domain H) to either nt 1047 (the pyrimidine tract) or nt 1124 (past AUG1068–70) were replaced by equivalent FMDV sequences (nt 280–693 or nt 280–740, respectively; Forss et al. 1984). These substitutions had little effect on the efficiency of translation in RRL, and both caused a small reduction in IRES-mediated translation in BHK-21 cells (Fig. 1B). TMEV containing these substitutions was viable and genetically stable in BHK-21 cells, yielding plaques that were slightly reduced in diameter relative to wt TMEV GDVII (Fig. 1C).
Figure 1.
Structures of chimeric Theiler's murine encephalomyelitis virus (TMEV)/foot-and-mouth disease virus (FMDV) RNAs, template activities, and viral phenotypes. (A) Schematic representations of the TMEV IRES showing the positions of the oligopyrimidine tract (Yn) and the initiation codon AUG1068–70 and of constructs derived from the recombinant cDNA for TMEV GDVII (white) and FMDV 01K (black) fused to a luciferase reporter gene (grey). The sizes of cDNA fragments used in these constructs are as indicated; TMEV nucleotide positions (Law and Brown 1990) are shown above pLG, and FMDV nucleotide positions are shown below pLGF-68 and pLGF-80. (B) Template activities of pLG, pLGF-68, and pLGF-80 mRNA transcripts translated in RRL and in BHK-21 cells. (C) In vitro growth characteristics and neurovirulence of wt and chimeric TMEV GDVII viruses. Standard plaque assays were done using virus stocks produced by three accumulating passages posttransfection in BHK-21 cells, and plaques were measured 3 days postinfection. Neurovirulence was assayed by intracerebral inoculation of BALB/c mice with tenfold dilutions of virus. The paralytogenic activity of viruses was expressed as the TCD50 dose causing paralysis in 50% of animals (PD50).
Attenuation of neurovirulence of TMEV GDVII/FMDV chimeric viruses
Several picornavirus IRESs have restricted activity in some cells and therefore contain determinants of the pattern and severity of host viral infection. We assayed the neurovirulence of the chimeric GD68 and GD80 viruses in mice. No clinical signs were observed 6 wk after intracerebral inoculation with up to 108 TCD50 of GD68 and up to 107.5 TCD50 of GD80 (Fig. 1C). For comparison, the PD50 of wt TMEV GDVII was 102.5 TCD50. Thus, GD68 and GD80 were completely attenuated as a consequence of replacing the TMEV IRES by that of FMDV. The sequences of these IRESs are ∼ 40% homologous (Kühn et al. 1990). Virus-specific PCR fragments were easily detected 1 day postinfection (p.i.) in all CNS samples from three mice inoculated with GD68 and from four mice inoculated with GD80. This result confirms that all mice were properly inoculated. Significantly, no virus-specific fragments were PCR-amplified from CNS samples prepared from the same number of mice 7 and 14 day p.i. Previously, we readily detected moderately attenuated TMEV mutant genomes up to 33 day p.i. (Pilipenko et al. 1995). TMEV GDVII replicates almost exclusively in neurons (Aubert and Brahic 1995). The results reported here imply that GD68 and GD80 did not replicate in the CNS and, more specifically, may be unable to interact functionally with the translational apparatus in neurons. These results indicated that cell-specific differences in the factors required for translation initiation could be analyzed using FMDV and TMEV IRESs.
Factor requirements for initiation on the TMEV GDVII IRES
IRES-mediated initiation involves recruitment of the 40S ribosomal subunit, Met-tRNAMeti, and eIFs to an internal site on an mRNA, leading to 48S complex formation at the initiation codon. We identified the factors required for 48S complex formation on the TMEV GDVII IRES by reconstituting this process in vitro, using purified translation components. The position of 48S complexes on this mRNA was determined by toeprinting, which involves cDNA synthesis by reverse transcriptase on a template RNA to which a ribosome or protein is bound. cDNA synthesis is arrested by the bound complex, yielding toeprints at its leading edge. Eukaryotic 48S complexes inhibit primer extension at positions that can vary from 14–21 nt 3′ to the A of the initiation codon depending on the mRNA on which the 48S complexes have assembled (Pestova et al. 1996a, 1998a,b). The positions of toeprints described here are shown on a structural model of an appropriate segment of the TMEV IRES (Fig. 2A).
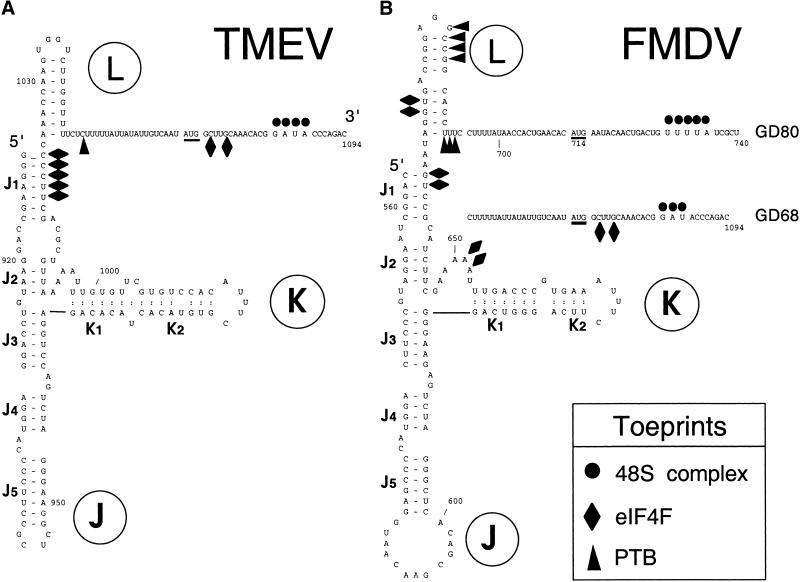
Figure 2.
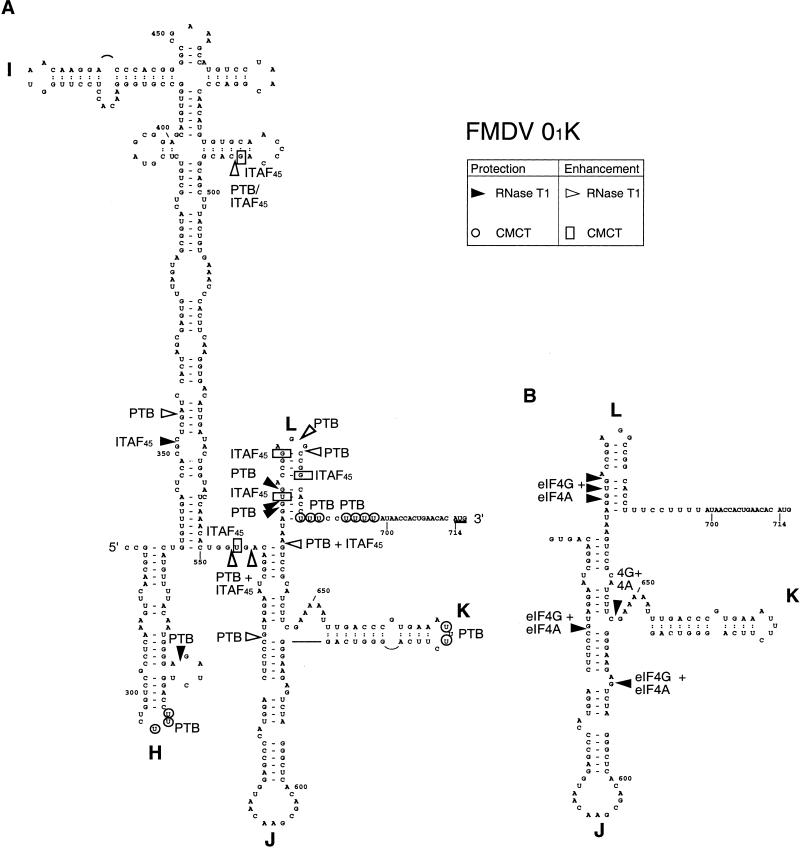
Secondary structures of segments of (A) Theiler's murine encephalomyelitis virus (TMEV) GDVII and (B) foot-and-mouth disease virus (FMDV) 01K internal ribosomal entry sites (IRESs; Pilipenko et al. 1989). The FMDV IRES was linked differently to downstream TMEV sequences in GD68 and GD80 as shown. FMDV and TMEV initiation codons are underlined. Prominent sites of RT arrest caused by eIF4F, PTB, and 48S complexes are indicated by symbols as described in the key at bottom right.
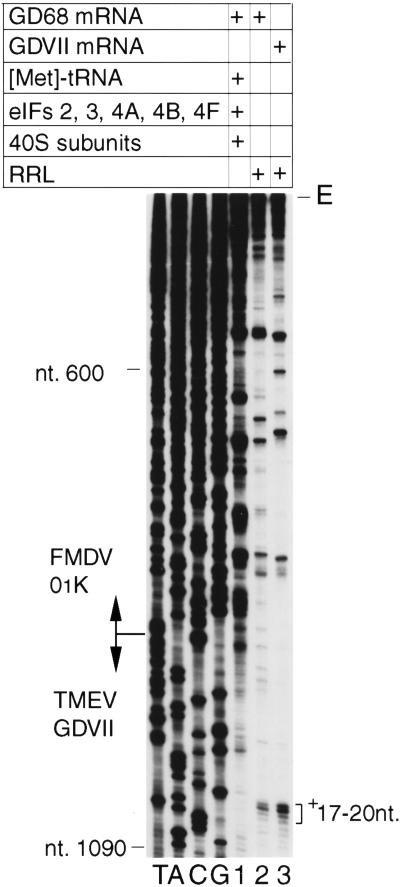
48S complexes accumulate when GMPPNP (a nonhydrolyzable GTP analog) is included with mRNA in RRL. Primer extension done on 48S complexes assembled on wt TMEV GDVII mRNA in RRL in the presence of 1mm GMPPNP yielded prominent toeprints 17 and 18 nt and weaker toeprints 19 and 20 nt 3′ to the A of AUG1068–70 (Fig. 3, lane 3).
Figure 3.
Toeprint analysis of 48S complexes assembled on Theiler's murine encephalomyelitis virus (TMEV) GDVII and foot-and-mouth disease virus (FMDV) 01K internal ribosomal entry sites in RRL. TMEV GDVII mRNA (lane 3) and chimeric GD68 mRNA containing the FMDV IRES (lanes 1,2) were incubated in RRL (lanes 2,3) or with translation components as indicated (lane 1) under standard conditions for 48S complex formation. A primer was annealed to TMEV nt 1195–1214 in the coding region of these mRNAs and extended with AMV-RT. The full-length cDNA extension product is marked E. cDNA products labeled + 17–20 nt terminated at these positions 3′ to the A of TMEV initiation codon AUG1068–70. Reference lanes T, A, C, and G depict the negative-strand sequence derived using the same primer and GD68 plasmid DNA. The junction between TMEV GDVII and FMDV 01K nucleotides is indicated.
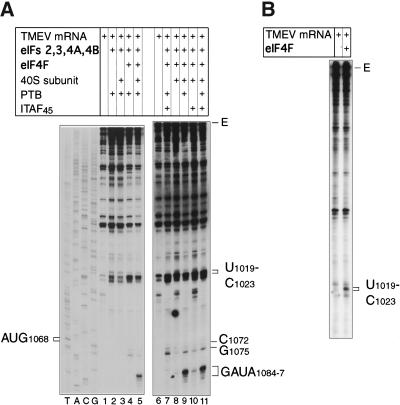
We next assayed formation of 48S complexes in vitro on this mRNA, using purified translation components. Incubation of TMEV GDVII mRNA with eIF2, eIF3, eIF4A, eIF4B, and eIF4F, PTB, 40S subunits, and Met-tRNAMeti resulted in formation of 48S complexes that yielded toeprints similar to those detected in RRL (Fig. 4A, lanes 5,9). Each translation component was then individually omitted. PTB enhanced 48S complex formation fivefold (average of six assays) (Fig. 4A, lanes 8,9). 48S complex formation was absolutely dependent on 40S subunits, Met-tRNAMeti, eIF2, eIF3, and eIF4F (Fig. 4A, lanes 3,4; data not shown). eIF4A was present as a subunit of eIF4F, and these experiments therefore did not enable us to determine the requirement for eIF4A. Neither eIF1 nor eIF1A were required for 48S complex formation (data not shown). This observation is consistent with reports that initiation on the TMEV IRES does not involve scanning (Pilipenko et al. 1994) for which these two factors are absolutely required (Pestova et al. 1998a). Taken together, these results indicate that eIF2, eIF3, eIF4F, and possibly eIF4A and eIF4B constitute a minimum set of factors that are sufficient for 48S complex formation on the TMEV IRES and that this process is strongly enhanced by PTB. We have previously found that eIF2, eIF3, eIF4A, eIF4B, and eIF4F constitute the minimum set of factors required for 48S complex formation on the EMCV IRES and that this process is enhanced less than twofold by PTB (Pestova et al. 1996a).
Figure 4.
Factor dependence of 48S complex formation on the Theiler's murine encephalomyelitis virus (TMEV) internal ribosomal entry site. (A) Toeprinting analysis of RNP and 48S ribosomal complexes assembled on wt TMEV GDVII nt 1–1334 RNA under standard conditions as follows: with aminoacylated initiator tRNA and eIF2, eIF3, eIF4A, and eIF4B (lanes 2–11); with eIF4F (lanes 4–11); with 40S subunits (lanes 3,5,8–11); PTB (lanes 2–5,7,9,11); and ITAF45 (lanes 7,10,11). A primer was annealed within the TMEV coding sequence and was extended with AMV-RT. (B) Toeprinting analysis of the ribonucleoprotein complex assembled by incubating wt TMEV GDVII nt 1–1334 RNA with (lane 2) or without (lane 1) eIF4F. The full-length cDNA extension product is marked E. Other cDNA products terminated at the sites indicated on the right. Reference lanes T, A, C, and G depict the negative-strand TMEV sequence derived using the same primer.
Assembly reactions done using the TMEV IRES and all translation components also yielded prominent new toeprints at UUCCC1019–23 and weaker C1072 and G1075 toeprints (Fig. 4A, lanes 4,5,7–11). Their relative prominence increased on omission of 40S subunits (Fig. 4A, lanes 4,5), suggesting that they result from binding of factors to the IRES. Stable complexes were formed by the binding of eIF4F to the IRES and yielded the same pattern of toeprints at UUCCC1019–23 at the base of the J-K domain (Fig. 4B, lane 2). Mutations in the J-K domain that impair binding of eIF4F also strongly impair the ability of the IRES to support internal initiation and abrogate virus viability (T.V. Pestova and E.V. Pilipenko, unpubl.). The toeprints at C1072 and G1075 were seen only in the presence of eIF4F (Fig. 4A, cf. lanes 2 and 4), and they appear to be caused by the binding of a complex of its eIF4A and eIF4G subunits to the IRES because they were not observed in the presence of either eIF4G or eIF4A alone (E.V. Pilipenko and T.V. Pestova, unpubl.). These toeprints were strongly enhanced in reactions that contained PTB (Fig. 4A, cf. lanes 7,9,11 with lanes 8 and 10). This observation suggests that PTB binding facilitates formation of a specific ribonucleoprotein (RNP) complex on the GDVII IRES. Taken together, these results and previous reports (Pestova 1996a,b; Kolupaeva et al. 1998) suggest that the initiation processes on the related EMCV and TMEV IRESs differ in their requirement for PTB but that they otherwise occur by similar mechanisms that involve direct binding of eIF4F to the IRES upstream of the initiation codon and recruitment of a 43S complex.
TMEV and FMDV IRESs have different factor requirements for translation initiation
Primer extension done on 48S complexes assembled in RRL on the FMDV IRES yielded toeprints similar to those detected on the wt TMEV IRES (Fig. 3, lanes 2,3). The FMDV IRES therefore also promotes 48S complex assembly precisely at the initiation codon, and RRL contains all necessary activities for this process. Factors eIF2, eIF3, eIF4A, eIF4B, and eIF4F are sufficient for 48S complex formation on the EMCV IRES (Pestova et al. 1996a) and to a much lesser extent on the TMEV IRES (Fig. 4A, lane 8). This minimal set of five eIFs was not sufficient for 48S complex formation on the FMDV IRES (Fig. 3, lane 1; Fig. 5A, lane 3; Fig. 6A,B, lanes 3). PTB is a candidate for the missing essential activity because it binds specifically to the FMDV IRES and enhances its function (Luz and Beck 1990; Niepmann 1995; Kolupaeva et al. 1996). However, inclusion of PTB in reactions with the minimal set of factors did not lead to 48S complex formation on GD68 or GD80 mRNAs (Fig. 5C, lane 4; Fig. 6A,B, lanes 4). Internal initiation on the FMDV IRES therefore has different factor requirements than on either EMCV or TMEV IRESs, even though all three are related.
Figure 5.
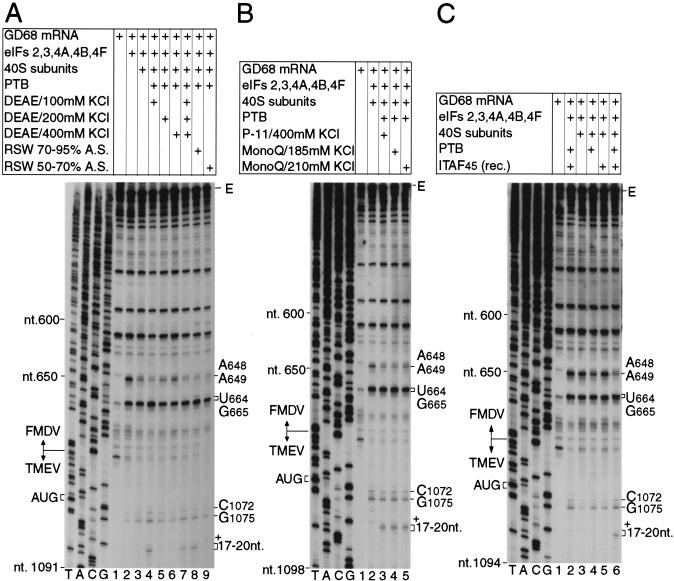
Purification of internal ribosomal entry site (IRES) trans-acting factors from rabbit reticulocyte lysate required for internal ribosomal entry on the foot-and-mouth disease virus (FMDV) 01K IRES, using toeprinting to assay 48S complex assembly. GD68 mRNA was incubated under standard reaction conditions for assembly of 48S complexes with translation components as indicated and (A) with the 70%–95% AS fraction (lane 8) or with the 50%–70% AS fraction (lane 9) and with the 100 mm KCl DEAE flow-though (lanes 4,7), the 250-mm KCl DEAE elution fraction (lanes 5,7), or the 400-mm DEAE elution fractions (lanes 6,7) of the RSW 70%–95% AS fraction; (B) with the 400-mm KCl phosphocellulose elution fraction (lane 3) or with 185-mm KCl (lane 4) or 210-mm KCl (lane 5) MonoQ elution fractions derived from this phosphocellulose elution fraction; and (C) with recombinant PTB (lanes 2,4,6) and recombinant ITAF45 (lanes 2,5,6). A primer was annealed to Theiler's murine encephalomyelitis virus (TMEV) nt 1195–1214 in the coding region of GD68 mRNA and was extended with AMV-RT. The full-length cDNA extension product is marked E. cDNA products labeled +17–+20 nt terminated at these positions 3′ to the A of TMEV initiation codon. Other cDNA products terminated at the sites indicated on the right. Reference lanes T, A, C, and G depict the negative-strand sequence derived using the same primer and GD68 plasmid DNA. The junction between TMEV GDVII and FMDV 01K nucleotides and the position of the initiation codon are indicated.
Figure 6.
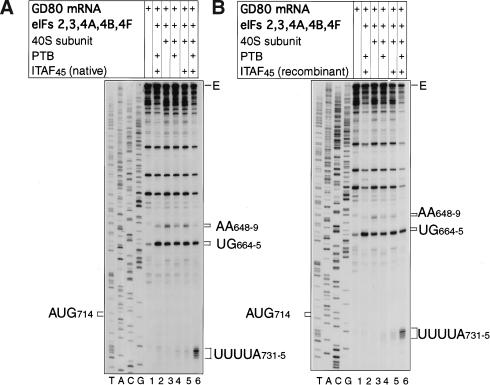
Identical activities of native and recombinant ITAF45 in foot-and-mouth disease virus internal-ribosomal-entry-site-mediated translation initiation. Toeprinting analysis of 48S and RNP complexes assembled on GD80 mRNA under standard conditions for assembly of 48S complexes as follows: with aminoacylated initiator tRNA and eIFs eIF2, eIF3, eIF4A, eIF4B, and eIF4F (lanes 2–6) with 40S subunits (lanes 3–6), recombinant PTB-1 (lanes 2,4,6) and (A) native ITAF45 (lanes 2,5,6) or (B) recombinant His6–ITAF45 (lanes 2,5,6). A primer was annealed to Theiler's murine encephalomyelitis virus nt 1195–1214 of GD80 mRNA and was extended with AMV-RT. The full-length cDNA extension product is marked E. Other cDNA products terminated at sites indicated on the right of each panel. Reference lanes T, A, C, and G depict the negative-strand sequence derived using the same primer and GD80 plasmid DNA. The position of the initiation codon is indicated.
ITAF45 is a novel trans-acting factor required for initiation on the FMDV IRES
RRL contains all factors required for FMDV IRES function (Fig. 1; Fig. 3, lane 2) and was therefore used as a source for purification of the additional factor(s) required together with the minimum set of eIFs for 48S complex formation. The 0.5m KCl ribosomal salt wash (RSW) was divided into 0%–40%, 40%–50%, 50%–70%, and 70%–95% ammonium sulfate (AS) fractions. Individually, none of these fractions promoted 48S complex formation on GD68 mRNA when included with 40S subunits and eIF2, eIF3, eIF4A, eIF4B, and eIF4F. However, a combination of 0%–40% and 70%–95% AS fractions together provided the additional activities necessary for efficient 48S complex formation on the FMDV IRES (data not shown).
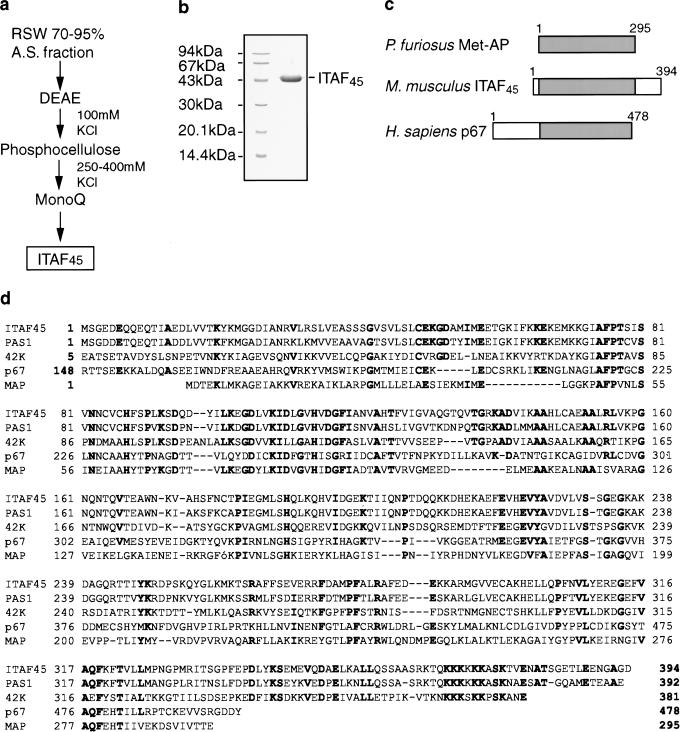
The active subfractions of the 0%–40% AS fraction were identical to those that contain PTB (Hellen et al. 1994). Recombinant PTB replaced the 0%–40% AS fraction without loss of activity in reactions that also contained the 70%–95% AS fraction (Fig. 5A, lane 8; data not shown). PTB is therefore essential for initiation on the FMDV IRES and was included in all subsequent reactions. The active component of the 70%–95% AS fraction was initially purified by DEAE-cellulose chromatography (Fig. 7A). The active 100 mm KCl flow-through fraction (Fig. 5A, lane 4) was applied to a phosphocellulose column, and proteins in it were separated by step-elution. The IRES trans-acting factor was in the 250–400-mm KCl fraction (Fig. 5B, lane 3), which was dialysed against 50 mm KCl buffer, applied to an FPLC MonoQ column and eluted with a 50–500 mm KCl gradient. Two well-resolved major peaks that eluted with 185 mm and 210 mm KCl both contained nearly homogenous 45-kD proteins (e.g., Fig. 7B). The former peak was half as large as the latter. The electrophoretic mobility and the activity of these proteins were identical (Fig. 5B, lanes 4,5). We named this protein IRES trans-acting factor 45 (ITAF45) on the basis of its function and apparent molecular weight. ITAF45 and the minimum set of eIFs without PTB were not sufficient to mediate 48S complex formation on GD68 mRNA (Fig. 5C, lane 5). Initiation on the FMDV IRES therefore requires both PTB and ITAF45. Separation of ITAF45 into two populations suggests that it may be modified posttranslationally.
Figure 7.
Purification and structure of ITAF45. (A) Scheme for the purification of ITAF45 from the 0.5 m KCl RSW from RRL. (B) Resolution by SDS-PAGE of native rabbit ITAF45 (2 μg) after purification by MonoQ chromatography and elution with 210 mm KCl. The positions of molecular weight markers are indicated to the left and those of ITAF45 to the right. (C) Schematic illustration of ITAF45 with p67 and MAP to show regions of homology. (D) Deduced amino acid sequence of ITAF45 and alignment of it with sequences of the PAS1 protein from Fugu ribripes (PAS1; Gellner and Brenner 1999), p42 protein from Schizosaccharomyces pombe (p42; Yamada et al. 1994), and with type II methionine aminopeptidases from human (p67; Arfin et al. 1995) and from the hyperthermophile Pyrococcus furiosus (MAP; Tahirov et al. 1998). The sequences of these proteins and ITAF45 show 71%, 26%, 23%, and 35% amino acid identity and 80%, 44%, 43%, and 48% similarity, respectively. Residues in bold are present in the majority of sequences and have been introduced to facilitate visualization of alignments.
GD68 contains FMDV IRES sequences to the middle of the pyrimidine tract followed by TMEV nucleotides (Fig. 1). To confirm that initiation on the FMDV IRES requires both PTB and ITAF45 in addition to canonical eIFs, 48S complexes were also assembled on GD80 mRNA, which contains FMDV IRES sequences up to and including the initiation codon AUG714–6 and eight additional codons. PTB and ITAF45 were both required, along with the minimum set of eIFs for 48S complex assembly on this mRNA, yielding prominent toeprints at positions 18–22 nt 3′ of the A of AUG714 (Fig. 6A, lanes 3–6). 48S complex formation was absolutely dependent on Met-tRNAMeti, eIF2, eIF3, eIF4F, and 40S subunits but did not require eIF1 or eIF1A (data not shown).
Inclusion of ITAF45 in 48S complex assembly reactions on wt TMEV GDVII mRNA confirmed that ITAF45 could not substitute for PTB in this process and did not alter the pattern or intensity of toeprints caused by bound 48S complexes or by bound eIF4F (cf. Fig. 4A, lanes 9–11). These results confirm that initiation on the TMEV IRES requires PTB and show that the additional requirement for ITAF45 is specific for FMDV. This conclusion was supported by the observation that recombinant ITAF45 did not influence initiation on either the EMCV IRES or on capped native globin mRNA (data not shown).
Molecular cloning of ITAF45
The identity of ITAF45 was determined by amino acid sequence analysis. The amino terminus of ITAF45 was blocked (data not shown). The sequences of two well-resolved tryptic peptides from ITAF45 were LVKPGNQNTQVTEAWNK and RRFDAMPFTLR. They correspond exactly to amino acids 156–172 and 271–281 of murine proliferation-associated protein (Mpp1; Nakagawa et al. 1997) and PAG24, its human homolog (Lamartine et al. 1997), as well as to sequences of p38–2G4, a possible smaller (37kD) murine Mpp1 isoform (Radomski and Jost 1995).
To confirm that ITAF45 and Mpp1 are the same protein and that it plays a specific role in FMDV IRES-mediated initiation, cDNA for this protein was amplified by PCR, cloned, and sequenced. The single long ORF encodes a 394 amino acid protein with a calculated molecular mass of 43,669 Daltons. This value is in agreement with that estimated for ITAF45 by SDS-PAGE. The ITAF45 cDNA sequence differed from that of Mpp1 at two nucleotides, both of which resulted in coding changes (A279 and R311 in ITAF45 in place of T279 and K311 in Mpp1). As reported previously for Mpp1 and p38–2G4, homologous proteins of a similar size occur in many eukaryotes. These range from the closely related PAS1 protein from Fugu ribripes to a more distantly related DNA-binding protein from Schizosaccharomyces pombe (Yamada et al. 1994; Gellner and Brenner 1999). Significant sequence homologies also exist between these proteins, ITAF45, and type II methionine aminopeptidases from archaea and eukaryotes (Fig. 7C,D). These similarities extend over the entire ITAF45 coding sequence and include four of the five signature amino acid residues that coordinate two cobalt ions in these metallo-enzymes (Tahirov et al. 1998). This group of enzymes includes p67, a methionine aminopeptidase that regulates translation by interacting with the γ subunit of eIF2 and inhibiting phosphorylation of its α subunit (Ray et al. 1993; Arfin et al. 1995). ITAFs such as PTB, PCBP2, and unr are, respectively, RNP-motif, KH-domain, and cold shock domain proteins, but ITAF45 does not belong to any of these recognized families of RNA-binding proteins.
To confirm that the IRES-trans-activating activity of ITAF45 is not due to a contaminant, recombinant His6-ITAF45 was purified to homogeneity and its activity assayed in assembly reactions done on GD80 mRNA. Inclusion of PTB and His6-ITAF45 or native ITAF45 together with the minimum set of eIFs led to the formation of prominent 48S complexes that yielded identical toeprint patterns (Fig. 6A,B, lane 6). Significant assembly of 48S complexes on this mRNA did not occur unless PTB and ITAF45 were present together (Fig. 6A,B, lanes 3–6). Native and recombinant ITAF45 therefore have identical activities.
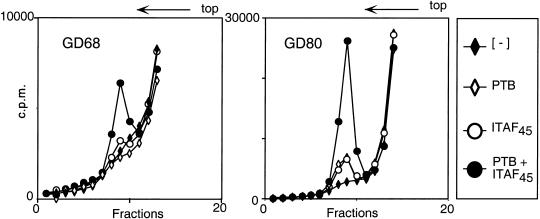
Ribosomal complexes assembled in these reactions were also resolved from RNP complexes by sucrose density gradient centrifugation to verify that they correspond to authentic 48S complexes and to confirm the factor requirements for their formation. 48S complexes were formed on GD68 mRNA in reactions that contained eIF2, eIF3, eIF4A, eIF4B, and eIF4F only if both PTB and His6–ITAF45 were present but not if either or both were omitted (Fig. 8). In parallel experiments done using GD80 mRNA, eIF2, eIF3, eIF4A, eIF4B, and eIF4F were not sufficient to mediate efficient 48S complex formation (Fig. 8). Inclusion of either ITAF45 or PTB individually in reactions containing these eIFs yielded small amounts of 48S complex. Inclusion of ITAF45 and PTB together in reactions had a very strong synergistic effect on 48S complex assembly (Fig. 8). These observations are exactly consistent with the results noted previously for assembly reactions on this mRNA assayed by toeprinting. Taken together, the results obtained using recombinant PTB and ITAF45 confirm that these two trans-acting factors both are necessary and are together sufficient for assembly of 48S complexes on the FMDV IRES in the presence of eIF2, eIF3, eIF4A, eIF4B, and eIF4F.
Figure 8.
Dependence of foot-and-mouth disease virus (FMDV) internal-ribosomal-entry-site (IRES)-mediated internal ribosomal entry on PTB and ITAF45, assayed by sucrose-density gradient centrifugation. Assays were carried out under standard conditions using GD68 or GD80 nt 1–1334 mRNA as indicated. Assembly reactions contained eIFs eIF2, eIF3, eIF4A, eIF4B, and eIF4F, and included recombinant PTB-1 and/or His6–ITAF45, as indicated. Sedimentation was from right to left. Fractions from the upper part of the sucrose gradient were omitted for clarity.
UV cross-linking of ITAF45
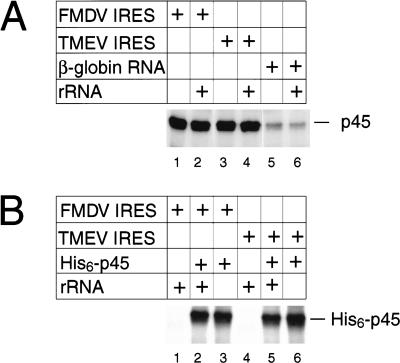
The role of ITAF45 in promoting initiation on the FMDV IRES could involve specific binding to it. A UV cross-linking assay was used to investigate whether ITAF45 is a specific RNA-binding protein (Fig. 9A). ITAF45 was labeled significantly more strongly after UV cross-linking to [32P]UTP-labeled nt 1–1334 GD80 mRNA (which contains the FMDV IRES) than to β-globin mRNA (Fig. 9A, lanes 1,5), both in the presence of a 50-fold molecular weight excess of unlabeled tRNA. Native ITAF45 was as strongly labeled after UV cross-linking to [32P]UTP-labeled wt TMEV nt 1–1334 RNA as to the equivalent chimeric GD80 RNA (Fig. 9A, lanes 1,3). The interaction of ITAF45 with wt TMEV and chimeric GD80 RNAs was specific since it was not significantly affected by the additional inclusion in cross-linking assays of a 15-fold molecular weight rRNA excess, where UV cross-linking of ITAF45 to β-globin RNA was slightly reduced by this competitor (Fig. 9A, cf. lanes 1,3,5 with lanes 2,4,6). Labeling of recombinant and native forms of ITAF45 after UV cross-linking to [32P]UTP-labeled wt TMEV and chimeric GD80 nt 1–1334 RNAs was comparable and was equally resistant to challenge by tRNA/rRNA competitors (Fig. 9B). In parallel experiments, recombinant and native ITAF45 also became strongly labeled after UV cross-linking to the EMCV IRES (data not shown). The interaction of ITAF45 with these RNAs is therefore specific and not caused by contaminants in preparations of the native protein.
Figure 9.
UV cross-linking of ITAF45 to wt Theiler's murine encephalomyelitis virus (TMEV) GDVII, chimeric foot-and-mouth disease virus (FMDV)/TMEV GD80 and β-globin mRNAs. (A) Native and (B) recombinant ITAF45 were UV cross-linked to [32P]UTP-labeled wt TMEV GDVII nt 1–1334, chimeric FMDV/TMEV GD80 nt 1–1334 or β-globin RNA nt 1–600 RNAs in the presence of a 50× molecular weight excess of tRNA and in the presence or absence a 15× molecular weight excess of ribosomal RNA, as indicated.
Interactions of initiation factors with the FMDV IRES assayed by toeprinting
IRES-mediated initiation involves specific interactions between the IRES and the components of the translation apparatus. We used toeprinting and footprinting to map the interactions of factors with the FMDV IRES. The presence of eIF2, eIF3, eIF4A, eIF4B, and eIF4F in assembly reactions on the FMDV IRES of GD68 mRNA with or without 40S subunits enhanced a toeprint at U664 and led to the appearance of prominent new toeprints at AA648–9 and G665 (e.g., Fig. 5C, lanes 1–3). Similar toeprints appeared on GD80 mRNA in the presence of these factors (Fig. 6A,B, lanes 1–3). UG664–5 and AA648–9 map to the base of the J1 and K1 helices of the J-K domain, respectively (Fig. 2B), and correspond to the binding site for the eIF4G subunit of eIF4F on EMCV and TMEV IRESs (Figs. 4B, 2A; Pestova et al. 1996a,b; Kolupaeva et al. 1998). The prominence of the AA648–9 toeprints was reduced on GD68 and more strongly on GD80 mRNA on inclusion of PTB in assembly reactions, and more strongly on inclusion of ITAF45 with PTB (Fig. 5C, lanes 3–6; Fig. 6A,B, lanes 3–6), implying some changes in the conformation of the IRES or in the character of its interaction with eIF4F as a result of binding PTB and ITAF45. Binding of eIFs to GD68 mRNA yielded weak toeprints at C1072 and G1075 (e.g., Fig. 5C, lane 3) that were previously detected on GDVII RNA (Fig. 4A). These toeprints resulting from binding of the eIF4A/eIF4G complex to the IRES are markedly enhanced in the presence of PTB or ITAF45 individually and are enhanced to an even greater extent on inclusion of PTB and ITAF45 together (Fig. 5C, lanes 2,6). We do not know whether these toeprints are indicative of conformational changes in the IRES or whether they represent additional contacts of the IRES with the eIF4G/4A complex. In any case, enhancement of these toeprints in the presence of PTB and ITAF45 indicates that these two proteins may facilitate binding of the eIF4G/4A complex to the IRES, which might in turn enhance 48S complex formation.
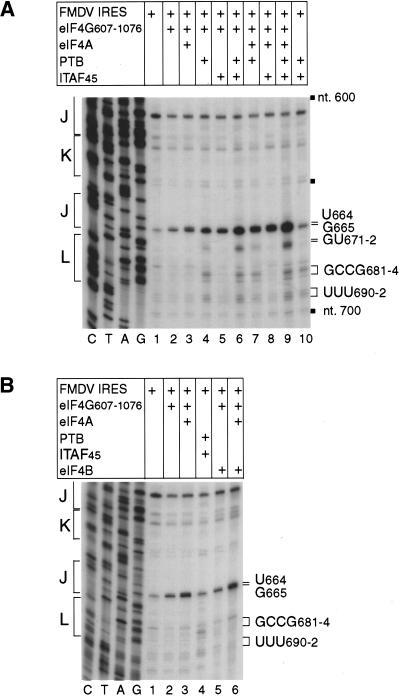
Toeprint analysis of RNP complexes formed with different combinations of factors indicated that eIF4F bound specifically to the FMDV IRES, yielding toeprints at UG664–5 in the J-K domain (data not shown). More detailed analysis indicated that the central domain of eIF4G (residues 607–1076) bound specifically but weakly to the FMDV IRES, yielding a toeprint at G665 (Fig. 10A, lanes 1,2). Inclusion of eIF4A with eIF4G607–1076 resulted in the appearance of an additional toeprint at U664 (Fig. 10A, lanes 2,3). eIF4A alone did not lead to the appearance of toeprints anywhere on the IRES (data not shown). The intensity of the G665 toeprint due to bound eIF4G607–1076 and of the UG664–5 toeprints due to the bound eIF4A/eIF4G607–1076 complex were enhanced by PTB and ITAF45 individually and together (Fig. 10A, lanes 1–9). Inclusion of PTB in these reactions led to the appearance of toeprints at GCCG681–4 and UUU690–2 that did not depend on the presence of other factors, including eIF4G, eIF4A, eIF4B, and ITAF45 (Fig. 2; Fig. 10A, lanes 1,4,6,7,9,10; Fig. 10B, lanes 1,4). The toeprints at UUU690–2 correspond to residues that are protected by PTB from chemical modification (Kolupaeva et al. 1996). Toeprints appeared at GU671–2 on inclusion of PTB and ITAF45 in binding reactions only in the presence of eIF4G607–1076 (cf. Fig. 10A, lane 6 and Fig. 10B, lane 4). Taken together, these observations indicate that the specific binding of eIF4G to the FMDV J-K domain is enhanced by eIF4A, PTB, and ITAF45.
Figure 10.
(A,B) Primer extension analysis of a ribonucleoprotein complex formed on the foot-and-mouth disease virus (FMDV) internal ribosomal entry site (IRES). GD80 mRNA was incubated under standard conditions with ITAF45, PTB, eIF4A, eIF4B, and eIF4G607–1076 individually or in combination as indicated. An oligonucleotide was annealed within the coding sequence and extended with AMV-RT. The cDNA products labeled U664, G665, GU671–2, GCCG681–4, and UUU690–2 terminated at these nucleotides. Reference lanes C, T, A, and G depict the negative strand FMDV sequence.
Localization of factor-binding sites on the FMDV IRES by footprinting
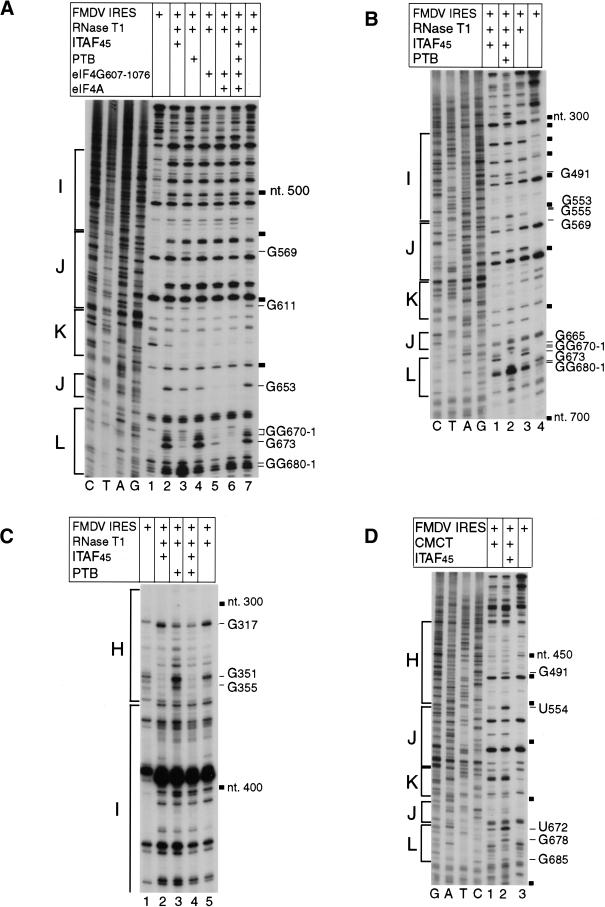
Footprinting was used to map binding sites for eIFs, PTB, and ITAF45 on the FMDV IRES and to identify structural changes in it caused by their binding. In these experiments we used RNase T1 (which cleaves RNA specifically after unpaired G residues) and 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluene sulphate (CMCT), which reacts with N-3 of unpaired uracil and N-1 of unpaired guanine residues (Ehresmann et al. 1987). Chemical modification or strand scission both arrest reverse transcriptase and can thus be analyzed by primer extension. CMCT modification causes primer extension arrest at the nucleotide immediately 3′ to the modified position, and enzymatic cleavage results in arrest of primer extension at the nucleotide on the 3′-side of the cleaved bond. Some residues in the FMDV IRES at which cleavage or modification patterns appear to be altered in the presence of factors such as G401 coincide with strong stops formed during primer extension on this highly structured RNA. Altered cleavage or modification of such residues is therefore equivocal and is not discussed below. Results of this analysis are summarized in Fig. 12 (see below).
Figure 12.
Summary of changes in modification by CMCT and in cleavage by RNAse T1 of the foot-and-mouth disease virus (FMDV) internal ribosomal entry site (IRES) caused by binding of ITAF45, PTB, eIF4A, and eIF4G607–1076 individually or in combination as indicated. These probes are indicated by symbols, as described in the key at top right. Results are displayed on a secondary structure model (Pilipenko et al. 1989). The initiation codon AUG714 is underlined.
Individually, neither eIF4A nor eIF4G607–1076 protected any part of the IRES from cleavage or modification (Fig. 11A, lanes 4,7; data not shown). However, together they specifically protected the IRES from cleavage at G569, G611, and G653 in the J-K domain and at GG670–1 and G673 in domain L (Fig. 11A, lane 5). These results are consistent with the results of toeprinting (Fig. 10A) and with the identification of eIF4G binding sites at identical locations in EMCV and TMEV IRESs.
Figure 11.
Chemical and enzymatic footprinting of the foot-and-mouth disease virus (FMDV) internal ribosomal entry site (IRES) in complexes formed with the IRES and ITAF45, PTB, eIF4A, eIF4B, and eIF4G607–1076 individually or in combination as indicated. Polyacrylamide–urea gel fractionation of cDNA products obtained after primer extension showing the sensitivity of the internal ribosomal entry site to RNase T1 cleavage (A, lanes 2–7; B, lanes 1–3; C, lanes 2–5) and to CMCT modification (D, lanes 1,2) either alone or complexed with factors as indicated. cDNA products derived from untreated RNA are shown in lane 1 of panels A and C, in lane 4 of panel B, and in lane 3 of panel D. A dideoxynucleotide sequence generated with the same primer was run in parallel on each gel and is shown in panels A, B, and D. The positions of protected residues or enhanced cleavage are indicated to the right of each panel. The positions of FMDV nucleotides at 50-nt intervals are indicated by black squares to the right of each panel. Individual subdomains are indicated to the left of each panel.
PTB alone protected the FMDV IRES from cleavage at G317 in domain H (Fig. 11C, lane 3) and at GG670–1 and G673 in domain L (Fig. 11A, lane 3). The protected site in domain H is very close to residues in its apical loop that are protected by PTB from CMCT modification (Kolupaeva et al. 1996; Fig. 12). PTB alone also enhanced cleavage at GG680–1 at the apex of domain L and, weakly, at G569 at the junction of domains J and K (Fig. 11A, line 3). Binding of ITAF45 alone to the IRES protected it from cleavage at G351 in the basal part of domain I (Fig. 11C, lanes 2,5; Fig. 12) and enhanced CMCT modification at G491 in the apical half of domain I and at U554, U672, G678, and G685 near and within domain L (Fig. 11D, lane 2). Taken together, the data indicate that PTB and ITAF45 interact functionally with the IRES at several specific sites and may therefore act as RNA chaperones that foster correct IRES folding. Importantly, the footprint pattern caused by binding of PTB and ITAF45 together to the IRES differed somewhat from the summation of the footprints caused by binding of these two proteins individually. Thus, PTB alone enhanced RNase T1 cleavage at G355, near the base of domain I, and this enhanced cleavage was suppressed by ITAF45 (Fig. 11C, lanes 3,4). A combination of PTB and ITAF45 enhanced cleavage at G491, G553, and G555 and exposed a cleavage site at G665 (Fig. 11B, lane 2). These results are indicative of a cooperative chaperone-like action of PTB and ITAF45 in RNA recognition and folding, and are consistent with and can account for the synergistic enhancement by these two proteins of 48S complex formation on the FMDV IRES.
The strong protection of the FMDV IRES from cleavage at nt 670–673 caused by the binding of eIF4G607–1076, eIF4A, PTB, and ITAF45 is consistent with the appearance of additional toeprints at GU671–2 on the IRES on inclusion of both PTB and ITAF45 with either eIF4G607–1076 or, to a greater extent, with both eIF4G607–1076 and eIF4A (Fig. 10A, lanes 2,4–6,10).
The results of footprinting and toeprinting are thus wholly consistent. They indicate that ITAF45 binds at the base of domain I; that PTB makes multiple contacts with the IRES in domain H, domain K, and downstream of domain L; and that the eIF4G/eIF4A binary complex binds to the J-K domain. eIF4A does not bind specifically or stably to the IRES except in the presence of eIF4G. PTB and ITAF45 also increase the susceptibility of the IRES to cleavage at several positions by inducing conformational changes in it and synergistically enhance binding of the eIF4G/eIF4A complex in a manner that may account for the dependence of FMDV IRES function on PTB and ITAF45.
Discussion
We report here that we have identified a cellular RNA-binding protein, ITAF45, that mediates mRNA-selective translation initiation. We identified ITAF45 by reconstituting 48S complex formation in vitro on FMDV and TMEV IRESs, using fully fractionated translation components. Initiation on these IRESs and the related EMCV IRES required the same set of canonical eIFs, but they had strikingly different ITAF requirements. TMEV IRES function was enhanced fivefold by PTB, but initiation on the FMDV IRES was dependent on ITAF45 as well as on PTB. EMCV IRES function did not depend on either of these ITAFs.
The role of canonical initiation factors in internal ribosomal entry
The canonical factors eIF2, eIF3, eIF4A, eIF4B, and eIF4F are required for 48S complex formation on TMEV and FMDV IRESs just as for initiation on the EMCV IRES (Pestova et al. 1996a). Significantly, we found that eIF4F binds specifically to an analagous site in the J-K domains of each of these IRESs (Fig. 4; Pestova et al. 1996a,b; Kolupaeva et al. 1998). The specificity of binding to each of these IRESs is due to its eIF4G subunit. We noted that inclusion of eIF4A with eIF4G in toeprinting assays strongly enhanced its binding to the FMDV IRES and led to the appearance of a new toeprint at U664. Moreover, individually neither eIF4A nor eIF4G protected any part of the IRES from cleavage or chemical modification in footprinting assays. Taken together, these results indicate that the eIF4G/4A complex rather than eIF4G alone is responsible for recognition of the FMDV IRES. This conclusion is consistent with our finding that the affinity of eIF4G for the EMCV IRES is enhanced by up to two orders of magnitude by eIF4A (Lomakin et al. 2000). There is no eIF4E requirement for this interaction or for 48S complex formation. The IRES–eIF4G/4A interaction is essential for initiation; mutations in either the J-K domain of these IRESs or in the IRES-binding domain of eIF4G that abrogate this interaction also abrogate IRES function (Pestova et al. 1996a; T.V. Pestova and E.V. Pilipenko, unpubl.). The central domain of eIF4G contains binding sites for eIF4A and eIF3 (Morino et al. 2000) and is presumed to coordinate their activities. In the context of these picornavirus IRESs, the role of the eIF4G/4A complex may be to prepare the IRES for attachment of a 43S complex in the vicinity of the initiation codon, for example, by interaction with associated factors such as eIF3 and eIF4B or by causing conformational changes in the IRES/factor RNP complex.
The role of IRES trans-acting factors in internal ribosomal entry
The requirement for PTB in FMDV IRES-mediated initiation is consistent with previous reports that it binds specifically to this IRES (Luz and Beck 1990; Kolupaeva et al. 1996) and is involved in FMDV translation (Niepman 1995). However, PTB alone is not sufficient to enhance 48S complex formation above background levels on GD68 and GD80 mRNAs, and its activity is dependent on a second factor, ITAF45. ITAF45 and PTB bind specifically to nonoverlapping sites on the FMDV IRES and individually and together cause localized structural changes in adjacent regions of it (Figs. 10–12). These two ITAFs enhance binding of the eIF4G/4A complex to the IRES and likely promote their assembly into a stable RNP complex (Fig. 10). The binding sites for protein components of this RNP complex include all IRES domains except for the apical region of domain I and extend from the 5′ border of the IRES to within 15 nt of the initiation codon. The latter distance is only slightly greater than the number of residues upstream of the initiation codon of an mRNA that are covered by the 40S ribosomal subunit (Legon 1976; Kozak 1977). Ribosomes are recruited to the FMDV IRES at or immediately upstream of the initiation codon at the 3′ border of the IRES without scanning from an upstream position (Belsham 1992; Lopez de Quinto and Martinez-Salas 1999). The function of ITAFs on the FMDV IRES may therefore be to prepare an RNP complex that contains RNA and protein binding determinants for components of the 43S complex oriented in three dimensions in such a way that it can be recruited and loaded onto the mRNA precisely, placing the initiation codon at the ribosomal P site.
A robust model for IRES-mediated initiation should be able to account for the significant differences between closely related IRESs in their requirement for ITAFs. For example, ribosomal attachment to the initiation codon without scanning is also characteristic of EMCV and TMEV IRES (Kaminski et al. 1990; Pilipenko et al. 1994). There is significant conservation of sequence and structure between EMCV, TMEV, and FMDV IRESs (Pilipenko et al. 1989; Jackson and Kaminski 1995), and as described above, initiation on each of them requires the same set of canonical factors and involves the same interaction between the eIF4G/4A complex and the IRES J-K domain. EMCV, FMDV, and TMEV IRESs all bind PTB with high affinity (Jang and Wimmer 1990; Luz and Beck 1990; Witherell et al. 1993; Kaminski et al. 1995), and the location of PTB binding sites on them appears to be similar, although not identical (Kolupaeva et al. 1996). We have reported here that ITAF45 can be strongly UV cross-linked to both TMEV and FMDV IRESs (Fig. 9). Despite these many similarities, initiation on the EMCV IRES is only weakly dependent on PTB, initiation on the TMEV IRES is strongly dependent on PTB, and initiation on the FMDV IRES is strongly dependent both on PTB and on a second protein, ITAF45, that has no influence on EMCV or TMEV IRESs. Differences in the ITAF requirement for these three IRESs must therefore be related to the small sequence and structural differences between them.
One hypothesis is that ITAFs have a chaperone activity (cf. Herschlag 1995) that functions in IRES-mediated initiation to remodel aberrantly folded IRESs so that they achieve and maintain an active conformation. This hypothesis is supported by the finding that EMCV IRES-mediated translation (which does not require PTB) became strongly PTB dependent following insertion of a single extra residue into the A-rich loop between J and K domains (Kaminski and Jackson 1998). The presence of an additional A residue in this same loop in the IRES of the TMEV GDVII strain relative to the equivalent loop of the TMEV DA strain IRES may account for the apparent difference in their PTB-dependence for activity (Kaminski et al. 1995; this paper). This hypothesis is also consistent with the conclusion that it is the requirement for, rather than the binding of per se, PTB and ITAF45 to EMCV, FMDV, and TMEV IRESs that is altered as a consequence of nucleotide differences between them.
We suggest that the observation that FMDV IRES requires ITAF45 in addition to PTB, where the TMEV IRES requires only PTB for activity may reflect sequence differences between these IRESs that lead to differences between them in the mode of binding PTB. For example, cardiovirus (EMCV, TMEV) IRESs contain pyrimidine residues in the bulge between the two helices of domain K that are specifically bound by PTB (Kolupaeva et al. 1996), whereas analogous residues are not present in the FMDV IRES. We hypothesize that PTB alone cannot fold the FMDV IRES into a functionally competent structure because of such differences in PTB binding sites and that this IRES only achieves an active conformation in the presence of both RNA chaperones, PTB and ITAF45.
It is interesting to note that members of the second major group of picornavirus IRESs are also closely related yet also appear to have different ITAF requirements. Initiation on the poliovirus IRES is strongly dependent on PTB and PCBP2 and does not respond to unr, where in contrast to the poliovirus IRES, initiation on the rhinovirus IRES is strongly dependent on unr and is enhanced by PTB but is less responsive to PCBP2 (Blyn et al. 1997; Hunt and Jackson 1999; Hunt et al. 1999; Walter et al. 1999). Nevertheless, PTB, PCBP2, and unr all bind to both poliovirus and rhinovirus IRESs (Borman et al. 1993; Hellen et al. 1993, 1994; Hunt et al. 1999; Walter et al. 1999; T.V. Pestova, unpubl.).
ITAF45 is a proliferation-dependent protein that is distributed throughout the cytoplasm from metaphase through to telophase (Radomski and Jost 1995; Lamartine et al. 1997; Nakagawa et al. 1997). Its expression is up-regulated in response to mitogen stimulation, and it is not detectable in nonproliferating differentiated cells such as murine brain cells (Radomski and Jost 1995). The strict difference in factor requirements for initiation of translation on TMEV and FMDV IRESs is sufficient to limit the activity of the latter, and it is thus tempting to speculate that the requirement for ITAF45 may be a cell-specific determinant that limits replication of the chimeric GD68 and GD80 viruses as well as FMDV. An attractive hypothesis is that ITAF45 may function as a tissue-specific and cell-cycle-dependent factor that specifically binds and controls translation on a particular set of mRNAs during cell proliferation and/or specific phases of the cell cycle. Indeed, IRES-mediated translation of a number of cellular proteins, including ornithine decarboxylase, c-myc, the p58PITSLRE protein kinase, and platelet-derived growth factor B, is specifically activated in a cell-cycle or proliferation-dependent manner (Bernstein et al. 1997; Cornelis et al. 2000; Pyronnet et al. 2000).
Materials and methods
Plasmids
Plasmids pET(His6–eIF4A), pET(His6–eIF4B) (Pestova et al. 1996a), pBS-(β-globin), and pE15 (Hellen et al. 1993) have been described. Mouse embryo cDNA (Clontech) corresponding to the ITAF45 coding region (Nakagawa et al. 1997) was amplified by PCR using the primers 5′-CGGGATCCGATGTCGGGCGAAGACGAGC-3′ and 5′CTCCGCTCGAGTTATCAGTCCCCAGCTCCATTCTC-3′, digested with BamH1 and XhoI and inserted into pET28b (Novagen) to yield pET(His6–ITAF45).
Chimeric TMDV/FMDV IRES-luciferase reporter constructs were derived from TMEV GDVII plasmids pUGD (Pilipenko et al. 1994), pGLV and pLG (Pilipenko et al. 1999), and the FMDV 01K plasmid pSP449 (Luz and Beck 1990). pLG contains TMEV GDVII nt 1–1124 (corresponding to the entire 5′ UTR, the initiation codon, and 18 codons) fused in-frame to the luciferase gene and downstream of a T7 promoter. pLGF-68 and pLGF-80 were derived from pLG by replacing TMEV nt 628–1047 with FMDV 01K nt 280–693 and TMEV nt 628–1124 with FMDV 01K nt 280–740, respectively. FMDV nucleotides are numbered starting after the poly(C) tract (Forss et al. 1984). Chimeric full-length TMEV clones GD68 and GD80 were derived from an infectious full-length wt TMEV GDVII clone GD18 by replacing TMEV nt 628–1047 with FMDV 01K nt 280–693 and TMEV nt 628–1124 with FMDV 01K nt 280–740, respectively.
Translation of luciferase expressing plasmids
Plasmids pLG, pLGF-68, and pLGF-80 were linearized with SalI, and mRNA was transcribed, purified, and translated in RRL at 30°C for 1 hr (Pilipenko et al. 1994). Template activity was quantified by measurement of [35S]methionine incorporation into the luciferase band separated by SDS-PAGE. For in vivo translation, 1-day-old quadruplicate cultures of BHK-21 cells grown in 50-mm plastic dishes were each transfected with 2 μg of mRNA using DEAE-dextran (Pilipenko et al. 1999). Cells were removed after 3 hr incubation at 37°C, and luciferase activity was determined using the Promega Luciferase Assay System according to the manufacturer's protocol. The signals were normalized to 106 cells by determining protein concentration in the lysates using the Lowry method. The signal variation in quadruplicate cultures did not exceed 15% of the average value for a given template. The saturating amount of RNA in this assay corresponded to 1μg/dish.
Preparation and characterization of chimeric viruses
Viruses were recovered from transcript RNA-transfected BHK-21 cells, accumulated, and characterized (Pilipenko et al. 1994, 1995). A region of the viral RNA (between nt 579 and 1214 of the wt TMEV GDVII sequence) was amplified by RT–PCR and sequenced as described (Pilipenko et al. 1995). No additional sequence changes were detected.
Neurovirulence assay and detection of replicating virus in mouse CNS
BALB/C mice (10–12 g) were inoculated intracerebrally with tenfold virus dilutions and monitored for clinical signs for 6 week p.i. Thirty mice were inoculated with 108 TCD50 (50% tissue culture infective dose) of GD68 virus, and 10 mice were each infected with 107 TCD50–103 TCD50 doses. For GD80 virus, 25 mice were inoculated with 107.5 TCD50, and 12 mice each with 106.5 TCD50 and 105.5 TCD50 doses. The paralytogenic activity was expressed as the viral dose causing paralysis in 50% of animals (PD50). The presence of the viral genome in the CNS was detected by RT–PCR amplification of the appropriate region of the viral RNA (nt 579–1214), as described above. Four mice for GD80 and three mice for GD68 were sacrificed at 1-, 7- and 14-day time-points p.i. Forty cycles of PCR amplification were performed with RNA samples extracted from CNS at days 7 and 14 p.i.
Purification of factors and 40S ribosomal subunits
Ribosomal subunits, eIF2, eIF3, and eIF4F were purified from RRL as described (Pestova et al. 1996a, 1998b). ITAF45 was purified from the ribosomal salt wash (RSW) 70%–95% ammonium sulfate (AS) precipitation fraction from RRL. This fraction was applied in buffer A (20 mm Tris at pH 7.5, 1 mm DTT, 0.1 mm EDTA, 10% glycerol) containing 100 mm KCl to a DEAE cellulose column. The flow-through fraction was applied to a phosphocellulose column and fractionated by step-elution using buffer A with 100 mm and then 250 mm, 400 mm, 600 mm and finally 800 mm KCl. The active 250–400-mm KCl elution fraction was dialyzed against buffer A with 50 mm KCl and applied in this buffer to an FPLC MonoQ HR 5/5 column. Fractions were collected across a 50–500 mm KCl gradient. Peaks of active, apparently homogenous ITAF45 eluted with 185 and 210 mm KCl. Recombinant PTB-1 and eIFs eIF1, eIF1A, eIF4A, and eIF4B were purified as described (Pestova et al. 1996a, 1998a). Recombinant ITAF45 was expressed in Escherichia coli BL21(DE3) and purified by Ni2+-NTA (Qiagen) and MonoQ HR 5/5 column chromatography.
Sequencing of ITAF45
Purified ITAF45 was resolved by gel electrophoresis. Sequencing of two well-resolved ITAF45 tryptic peptides was done as described (Pestova et al. 1998a,b).
Assembly and analysis of ribosomal and ribonucleoprotein complexes
For toeprinting analysis, ribosomal complexes were assembled essentially as described (Pestova et al. 1998a). For 5 min, 0.5 μg of GD18, GD68, or GD80 mRNA were incubated at 30°C in a 40-μl reaction volume that contained buffer B (2 mm DTT, 100 mm KAc, 20 mm Tris at pH 7.6, 2.5 mm MgAc, 1 mm ATP, 0.1 mm GMPPNP, 0.25 mm spermidine), eIF2 (3 μg), eIF3 (6 μg), eIF4A (2 μg), eIF4B (0.3 μg), eIF4F (1.5 μg), 6 pmole of 40S subunits, and 6 pmole of Met-tRNAMeti and with or without PTB (0.5 μg), ITAF45 (1 μg), eIF1 (0.3 μg), eIF1A (0.3 μg), or different RSW fractions (4–8 μg protein), as indicated. Incubation was continued for 3 min at 30°C following addition of 4 pmole of primer 5′-CCAGAAGACGTCATCGTCCA-3′ (complementary to TMEV nt 1195–1214). Ribosomal and RNP complexes were analyzed by primer extension (Pestova et al. 1996a) using avian myeloblastosis virus (AMV) reverse transcriptase (Promega) in the presence of [α-32P]dATP (∼ 6000 Ci/mmole; ICN Radiochemicals). cDNA products were compared with appropriate dideoxynucleotide sequence ladders.
For toeprinting analysis of 48S complexes assembled in RRL, 0.5 μg of RNA was incubated in 15 μl of RRL in the presence of 1 mm GMPPNP for 5 min at 30°C. The reaction mixture was diluted with buffer B to 40 μl final volume before the addition of 4 pmole of primer 5′-CCAGAAGACGTCATCGTCCA-3′ complementary to TMEV nt 1195–1214. Toeprint analysis was done as described above.
Ribonucleoprotein (RNP) complexes were formed by incubating eIFs and FMDV IRES mRNA for 5 min at 30°C in buffer (100 mm KAc, 2 mm MgAc, 2 mm Tris-HCl at pH 7.5, 1mm DTT). Reactions contained 1 μg of RNA (4 pmole), 1 μg of eIF4G607–1076 (10 pmole), 1 μg of eIF4A (22 pmole), 1.5 μg (21 pmole) of eIF4B, and 0.75 μg (12 pmole) of PTB and ATP (1mm), as indicated in the text, in a 20-μl volume. Toeprint analysis of FMDV mRNA/factor complexes was done essentially as described (Pestova et al. 1996a,b), using the primer 5′-CCAGAAGACGTCATCGTCCA-3′ (complementary to TMEV nt 1214–1195).
Ribosomal complexes were assembled for analysis by sucrose-density gradient centrifugation essentially as described above for toeprinting analysis, except that the reaction volume was increased to 100 μl and mRNAs were labeled during transcription with [32P]UTP (∼ 3000 Ci/mmole; ICN Radiochemicals) to a specific activity of 200,000 cpm/μg. Reaction mixtures containing translation components as indicated in the text were incubated for 5 min at 30°C. 48S and RNP complexes were resolved by centrifugation through 10%–30% sucrose-density gradients as described (Pestova et al. 1996a).
UV cross-linking of ITAF45 to RNA
The UV cross-linking reaction was done using a UV-Stratalinker (Stratagene) essentially as described (Pestova et al. 1996b). Purified wt TMEV RNA, chimeric GD80 RNA, or β-globin RNA (100 ng) that had been labeled with [32P]UTP during transcription with T7 polymerase were incubated for 10 min at 30°C in a 20 μl reaction volume containing buffer B and ITAF45 (3 μg) either with or without competitor RNAs, as indicated in the text. Samples were digested with cobra venom nuclease and RNAse A. Proteins were separated by 12% SDS-PAGE.
Chemical and enzymatic footprinting
RNP complexes were probed with RNase T1 and CMCT as described (Kolupaeva et al. 1996, 1998). Cleaved or modified sites were identified by primer extension, using AMV RT and the primers 5′-CCAGAAGACGTCATCGTCCA-3′ (complementary to TMEV nt 1214–1195), 5′-CAGTCAGTTGTATTCATG-3′ (complementary to FMDV nt 730–713), or 5′-CCAGTATCAATGTCACC-3′ (complementary to FMDV nt 539–523), as appropriate.
Acknowledgments
We thank E. Beck for the FMDV plasmid and L. Siconolfi-Baez for sequencing ITAF45. This study was supported by grant MCB-9726958 from the National Science Foundation to C.U.T.H., by a grant from the Russian Foundation of Basic Research to V.I.A., and by grants from the U.S. Civilian Research and Development Foundation (RB1-271) and the International Association for the promotion of cooperation with scientists from the former Soviet Union (INTAS-97-0501) to E.V.P.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tpestova@netmail.hscbklyn.edu; FAX (718) 270-2656.
References
- Arfin SM, Kendall RL, Hall L, Weaver LH, Stewart AE, Matthews BW, Bradshaw RA. Eukaryotic methionyl aminopeptidases: two classes of cobalt-dependent enzymes. Proc Natl Acad Sci. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert C, Brahic M. Early infection of the central nervous system by the GDVII and DA strains of Theiler's virus. J Virol. 1995;69:3197–3200. doi: 10.1128/jvi.69.5.3197-3200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrach HL. Foot-and-mouth disease. Annu Rev Microbiol. 1968;22:201–244. doi: 10.1146/annurev.mi.22.100168.001221. [DOI] [PubMed] [Google Scholar]
- Belsham GJ. Dual initiation sites of protein synthesis on foot-and-mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J. 1992;11:1105–1110. doi: 10.1002/j.1460-2075.1992.tb05150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J, Sella O, Le SY, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- Blyn LB, Swiderek KM, Richards O, Stahl DC, Semler BL, Ehrenfeld E. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: Identification by automated liquid chromatography-tandem mass spectrometry. Proc Natl Acad Sci. 1996;93:11115–11120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn LB, Towner JS, Semler BL, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman A, Howell MT, Patton J, Jackson RJ. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J Gen Virol. 1993;74:1775–1788. doi: 10.1099/0022-1317-74-9-1775. [DOI] [PubMed] [Google Scholar]
- Brown BA, Ehrenfeld E. Translation of poliovirus RNA in vitro: Changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology. 1979;97:396–405. doi: 10.1016/0042-6822(79)90350-7. [DOI] [PubMed] [Google Scholar]
- Cornelis S, Bruynooghe Y, Denecker G, van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- Dorner AJ, Semler BL, Jackson RJ, Hanecak R, Duprey E, Wimmer E. In vitro translation of poliovirus RNA: Utilization of internal initiation sites in reticulocyte lysate. J Virol. 1984;50:507–514. doi: 10.1128/jvi.50.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNA in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forss SK, Strebel K, Beck E, Schaller H. Nucleotide sequence and genome organization of foot-and-mouth disease virus. Nucleic Acids Res. 1984;12:6587–6601. doi: 10.1093/nar/12.16.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Interactions of viral protein 3CD and Poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J Virol. 2000;74:2219–2226. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellner K, Brenner S. Analysis of 148 kb of genomic DNA around the wnt1 locus of Fugu rubripes. Genome Res. 1999;9:251–258. [PMC free article] [PubMed] [Google Scholar]
- Gray NK, Wickens M. Control of translation initiation in animals. Ann Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- Gromeier M, Alexander L, Wimmer E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc Natl Acad Sci. 1996;93:2370–2375. doi: 10.1073/pnas.93.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CUT, Pestova TV, Litterst M, Wimmer E. The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5′ nontranslated region. J Virol. 1994;68:941–950. doi: 10.1128/jvi.68.2.941-950.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CUT, Witherell GW, Schmidt M, Shin SH, Pestova TV, Gil A, Wimmer E. A cytoplasmic 57kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Kühn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Hunt SL, Hsuan JJ, Totty N, Jackson RJ. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes & Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SL, Jackson RJ. Polypyrimidine tract-binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: The picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- Jang SK, Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: Structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes & Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- Kaminski A, Howell MT, Jackson RJ. Initiation of encephalomyocarditis virus RNA translation: The authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A, Hunt SL, Patton JG, Jackson RJ. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- Kaminski A, Jackson RJ. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA. 1998;4:626–638. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Hellen CUT, Shatsky IN. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalo-myocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1179–1188. [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CUT, Shatsky IN. Translation initiation factor eIF4G recognizes a specific structural element within the internal ribosomal entry site of encephalomyocarditis virus RNA. J Biol Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- Kozak M. Nucleotide sequences of 5′-terminal ribosome-protected initiation regions from two reovirus messages. Nature. 1977;269:390–394. doi: 10.1038/269390a0. [DOI] [PubMed] [Google Scholar]
- Kühn R, Luz N, Beck E. Functional analysis of the internal translation initiation site of foot-and-mouth disease virus. J Virol. 1990;64:4625–4631. doi: 10.1128/jvi.64.10.4625-4631.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamartine J, Seri M, Cinti R, Heitzmann F, Creaven M, Radomski N, Jost E, Lenoir GM, Romeo G, Sylla BS. Molecular cloning and mapping of a human cDNA (PA2G4) that encodes a protein highly homologous to the mouse cell cycle protein p38–2G4. Cytogen Cell Genet. 1997;78:31–35. doi: 10.1159/000134621. [DOI] [PubMed] [Google Scholar]
- Law KM, Brown TDK. The complete nucleotide sequence of the GDVII strain of Theiler's murine encephalomyelitis virus (TMEV) Nucleic Acids Res. 1990;18:6707–6708. doi: 10.1093/nar/18.22.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legon S. Characterization of the ribosome-protected regions of 125I-labelled rabbit globin messenger RNA. J Mol Biol. 1976;106:37–53. doi: 10.1016/0022-2836(76)90299-0. [DOI] [PubMed] [Google Scholar]
- Lomakin IB, Hellen CUT, Pestova TV. Physical association of eIF4G with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto S, Martinez-Salas E. Involvement of the aphthovirus RNA region located between the two functional AUGs in start codon selection. Virology. 1999;255:324–336. doi: 10.1006/viro.1999.9598. [DOI] [PubMed] [Google Scholar]
- Luz N, Beck E. A cellular 57kDa protein binds to two regions of the internal translation initiation site of foot-and-mouth disease virus. FEBS Lett. 1990;269:311–314. doi: 10.1016/0014-5793(90)81182-n. [DOI] [PubMed] [Google Scholar]
- Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan E K, Agol VI, Keene JD, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino S, Imataka H, Svitkin YV, Pestova TV, Sonenberg N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol. 2000;20:468–477. doi: 10.1128/mcb.20.2.468-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler M, Gray NK, Hentze MW. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol Cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Watanabe S, Akiyama K, Sarker AH, Tsutsui K, Inoue H, Seki S. cDNA cloning, sequence analysis and expression of a mouse 44-kDa nuclear protein copurified with DNA repair factors for acid-depurinated DNA. Acta Med Okayama. 1997;51:195–206. doi: 10.18926/AMO/30763. [DOI] [PubMed] [Google Scholar]
- Niepmann M. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth disease virus. FEBS Lett. 1995;388:39–42. doi: 10.1016/0014-5793(96)00509-1. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996a;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Hellen CUT. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996b;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998a;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT. A prokaryotic-like mode of binding of cytoplasmic eukaryotic ribosomes to the initiation codon during internal initiation of translation of Hepatitis C and classical swine fever virus RNAs. Genes & Dev. 1998b;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko EV, Blinov VM, Chernov BK, Dmitrieva TM, Agol VI. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 1989;17:5701–5711. doi: 10.1093/nar/17.14.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko EV, Gmyl AP, Maslova SV, Belov GA, Sinyakov AN, Huang M, Brown TDK, Agol VI. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J Mol Biol. 1994;241:398–414. doi: 10.1006/jmbi.1994.1516. [DOI] [PubMed] [Google Scholar]
- Pilipenko EV, Gmyl AP, Maslova SV, Khitrina EV, Agol VI. Attenuation of Theiler's murine encephalomyelitis virus by modifications of the oligopyrimidine/AUG tandem, a host-dependent translational cis element. J Virol. 1995;69:864–870. doi: 10.1128/jvi.69.2.864-870.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko EV, Viktorova EG, Khitrina EV, Maslova SV, Jarousse N, Brahic M, Agol VI. Distinct attenuation phenotypes caused by mutations in the translational starting window of Theiler's murine encephalomyelitis virus. J Virol. 1999;73:3190–3196. doi: 10.1128/jvi.73.4.3190-3196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard AE, Calenoff MA, Simpson S, Jensen K, Lipton HL. A single base deletion in the 5′ noncoding region of Theiler's virus attenuates neurovirulence. J Virol. 1992;66:1951–1958. doi: 10.1128/jvi.66.4.1951-1958.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle–dependent internal ribosome entry site. Mol Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- Radomski N, Jost E. Molecular cloning of a murine cDNA encoding a novel protein, p38–2G4, which varies with the cell cycle. Exp Cell Res. 1995;220:434–445. doi: 10.1006/excr.1995.1335. [DOI] [PubMed] [Google Scholar]
- Ray MK, Chakraborty A, Datta B, Chattopadhyay A, Satta D, Bose A, Kinzy TG, Wu S, Hileman RE, Merrick WC, Gupta NK. Characteristics of the eukaryotic initiation factor 2 associated 67-kDa polypeptide. Biochemistry. 1993;32:5151–5159. doi: 10.1021/bi00070a026. [DOI] [PubMed] [Google Scholar]
- Standart N, Jackson RJ. Regulation of translation by specific protein/mRNA interactions. Biochimie. 1994;76:867–879. doi: 10.1016/0300-9084(94)90189-9. [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Maslova SV, Agol VI. The genomes of attenuated and virulent poliovirus strains differ in their in vivo translation efficiences. Virology. 1985;147:243–252. doi: 10.1016/0042-6822(85)90127-8. [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Meerovitch K, Lee HS, Dholakia JN, Kenan DJ, Agol V I, Sonenberg N. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J Virol. 1994;68:1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Pestova TV, Maslova SV, Agol VI. Point mutations modify the response of poliovirus RNA to a translation initiation factor: a comparison of neurovirulent and attenuated strains. Virology. 1988;166:394–404. doi: 10.1016/0042-6822(88)90510-7. [DOI] [PubMed] [Google Scholar]
- Tahirov TH, Oki H, Tsukihara T, Ogasahara K, Yutani K, Ogata K, Izu Y, Tsunasawa S, Kato I. Crystal structure of methionine aminopeptidase from hyperthermophile, Pyrococcus furiosus. J Mol Biol. 1998;284:101–124. doi: 10.1006/jmbi.1998.2146. [DOI] [PubMed] [Google Scholar]
- Theiler M. Spontaneous encephalomyelitis of mice, a new virus disease. J Exp Med. 1937;65:705–719. doi: 10.1084/jem.65.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter BL, Nguyen JHC, Ehrenfeld E, Semler BL. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570–1585. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherell GW, Gil A, Wimmer E. Interaction of polypyrimidine tract binding protein with the encephalomyocarditis virus mRNA internal ribosomal entry site. Biochemistry. 1993;32:8268–8275. doi: 10.1021/bi00083a030. [DOI] [PubMed] [Google Scholar]
- Yamada H, Mori H, Momoi H, Nakagawa Y, Ueguchi C, Mizuno T. A fission yeast gene encoding a protein that preferentially associates with curved DNA. Yeast. 1994;10:883–894. doi: 10.1002/yea.320100704. [DOI] [PubMed] [Google Scholar]