Abstract
Regulators of G protein signaling (RGS proteins) inhibit heterotrimeric G protein signaling by activating G protein GTPase activity. Many mammalian RGS proteins are expressed in the brain and can act in vitro on the neural G protein Go, but the biological purpose of this multiplicity of regulators is not clear. We have analyzed all 13 RGS genes in Caenorhabditis elegans and found that three of them influence the aspect of egg-laying behavior controlled by Go signaling. A previously studied RGS protein, EGL-10, affects egg laying under all conditions tested. The other two RGS proteins, RGS-1 and RGS-2, act as Go GTPase activators in vitro but, unlike EGL-10, they do not strongly affect egg laying when worms are allowed to feed constantly. However, rgs-1; rgs-2 double mutants fail to rapidly induce egg-laying behavior when refed after starvation. Thus EGL-10 sets baseline levels of signaling, while RGS-1 and RGS-2 appear to redundantly alter signaling to cause appropriate behavioral responses to food.
Keywords: RGS protein, heterotrimeric G protein, neurotransmission, C. elegans
Heterotrimeric G proteins mediate the effects of a vast array of hormones and neurotransmitters by acting as molecular switches that alternate between active and inactive forms (Hamm 1998). Signaling is initiated when an activated cell-surface receptor stimulates a G protein to bind GTP and is terminated when the G protein hydrolyzes GTP to return to the inactive GDP-bound form. Genetic experiments have shown that RGS proteins act as inhibitors of G protein signaling that can bind to G protein α subunits (De Vries et al. 1995; Dohlman et al. 1995; Druey et al. 1996; Koelle and Horvitz 1996). In vitro, RGS proteins accelerate the GTPase activity of G protein α-subunits, thus driving them to their inactive GDP-bound form (for review, see Berman and Gilman 1998). Mammals have ≥20 proteins containing the ∼120 amino acid RGS (regulators of G protein signaling) domain that defines RGS proteins. The RGS domain folds into a nine-helix structure that binds the Gα subunit to stimulate its GTPase activity (Tesmer et al. 1997). Although many RGS proteins consist of little more than an RGS domain, a subset of them also contain a large amino-terminal conserved region of unknown function, as well as a G gamma-like (GGL) domain that is able to bind a specific G protein beta subunit (Snow et al. 1998).
Despite many in vitro studies of RGS–G protein interactions, it remains largely unclear how cells benefit by using RGS proteins, rather than simply using G proteins with higher intrinsic GTPase activities to allow appropriate termination of signaling. The use of RGS proteins might be explained if they were themselves regulated to allow signaling to be altered. A current challenge is to understand whether RGS proteins are indeed regulated and, if so, for what purpose. A further puzzle concerns the G protein specificities of RGS proteins. Although a few RGS proteins have distinct G protein specificities in vitro, the majority of those studied behave similarly to each other, acting preferentially on members of the Gi/o subfamily of G proteins and, to a lesser extent, on Gq (for review, see Hepler 1999). If the in vivo specificities of RGS proteins are the same as those measured in vitro, it is unclear why cells need multiple regulators of the same G proteins. Analysis of RGS expression also raises this issue. For example, the G protein Go is expressed throughout the brain, and at least nine RGS genes are also expressed in the brain, some broadly distributed and others restricted to certain areas (Gold et al. 1997).
Genetic studies have the potential to clarify the roles of RGS proteins by revealing their genuine in vivo functions and G protein targets. The most detailed analysis to date is that of the yeast Sst2p RGS protein. This acts on the G protein Gpa1p, which mediates mating pheromone signaling (Apanovitch et al. 1998). sst2 mutations have two effects. First, they cause yeast to be supersensitive to the pheromone, such that they respond to a concentration that is ∼200-fold lower than that required for wild-type yeast (Chan and Otte 1982). Second, whereas wild-type yeast desensitize to mating pheromone after prolonged exposure, sst2 mutants fail to desensitize and thus cannot terminate the mating responses if mating fails. Desensitization is explained, at least in part, by the fact that pheromone signaling induces higher expression of Sst2p, which then feeds back to inhibit Gpa1p signaling (Dietzel and Kurjan 1987). Thus Sst2p is required both to set the baseline level of signaling sensitivity in pheromone-naive yeast and to adjust the level of sensitivity after pheromone exposure.
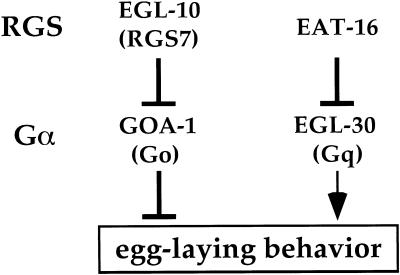
Two Caenorhabditis elegans RGS genes have been analyzed and shown to act on the homologs of the G proteins Go and Gq (known as GOA-1 and EGL-30, respectively). The RGS protein EGL-10 inhibits signaling by Go, which in turn inhibits egg-laying and locomotor behaviors (Mendel et al. 1995; Ségalat et al. 1995; Koelle and Horvitz 1996), whereas the RGS protein EAT-16 inhibits signaling by Gq, which has effects that are the opposite of those caused by Go (Fig. 1; Brundage et al. 1996; Hajdu-Cronin et al. 1999; Lackner et al. 1999; Miller et al. 1999). Studies of EGL-10 and EAT-16 have shown that these RGS proteins have roles in setting baseline levels of signaling, but have not provided evidence that they are regulated to adjust signaling levels.
Figure 1.
Model for RGS and G protein control of egg laying in C. elegans. Mammalian orthologs are indicated in parentheses below the names of the C. elegans proteins. No mammalian ortholog of EAT-16 has yet been identified. Genetic experiments show that Go and Gq signaling antagonize each other, and that Gq acts either downstream of or, as drawn here, parallel to Go (Hadju-Cronin et al. 1999).
The biological purpose of RGS control of Go and Gq in C. elegans remains obscure. Egg laying in C. elegans is strongly regulated, stopping when animals are starved and resuming when they are fed (Trent 1982). This allows worms to deposit their fertilized eggs where and when food is available for their progeny. Because the rate of egg laying is set by the balance between Go and Gq signaling, and because this balance is determined by RGS control, RGS proteins are ideally positioned to adjust signaling to alter egg laying. However, there is as yet no evidence that either EGL-10 or EAT-16 is regulated by starvation or feeding in a manner that could account for changes in egg-laying behavior. Another puzzle derives from the fact that many RGS genes other than egl-10 and eat-16 have been identified in the C. elegans genome sequence. Because in vitro studies of mammalian RGS proteins show that most can act on Go and Gq, the question arises as to whether the additional RGS proteins in C. elegans also regulate Go and Gq. If so, for what purpose?
This study was designed to identify the RGS proteins that regulate Go and Gq signaling in C. elegans and to understand the biological roles of these proteins. We take a functional-genomics approach, surveying all the RGS genes of C. elegans for effects on egg-laying behavior. We identify RGS-1 and RGS-2 as potential regulators of Go and use a recently developed gene knockout technology to delete the rgs-1 and rgs-2 genes. We find that these RGS genes redundantly adjust signaling when animals are fed to allow rapid induction of egg-laying behavior. Our results suggest that multiple RGS proteins control Go and Gq to set baseline and regulated levels of signaling.
Results
Overexpression of four of the 13 RGS genes of C. elegans affects Go/Gq-controlled egg-laying behavior
To identify RGS genes controlling Go and Gq in C. elegans, we generated transgenic strains that overexpressed each of the 13 RGS genes identified in the C. elegans genome sequence and analyzed these animals for defects in egg laying. This overexpression strategy is based on the observation, made in every previous genetic analysis of an RGS gene, that transgenic overexpression of the RGS gene induced phenotypic defects opposite to those caused by null mutations in the same RGS gene. This observation has been made in five cases: two from C. elegans (Koelle and Horvitz 1996; Hajdu-Cronin et al. 1999), two from yeast (Dohlman et al. 1995; Versele et al. 1999), and one from Aspergillus nidulans (Yu et al. 1996). These results suggest that RGS proteins are generally present at levels that partially inhibit their G protein targets, and that RGS overexpression can increase this inhibition.
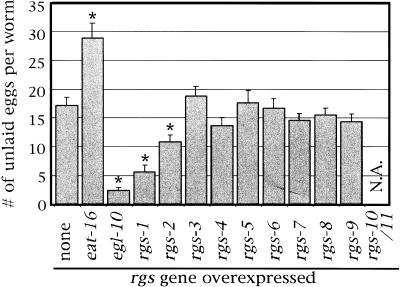
RGS genes were overexpressed by injecting genomic clones for each into C. elegans to produce multicopy extrachromosomal transgenic arrays of the injected DNA. The RGS genes were thus expressed from their own promoters, presumably in their normal temporal and spatial expression patterns, but overexpressed because of the high copy number of the transgene DNA (see below for analysis of overexpression levels). The transgenic strains were examined in assays for behavioral defects associated with changes in Go and Gq signaling. Rates of egg-laying behavior were measured using two assays that gave similar results for each strain tested. We quantitated the accumulation of unlaid eggs in adults and the developmental stages of the freshly laid eggs. The unlaid-egg assay is presented in Figure 2. Increased accumulation indicates reduced egg laying, presumably because of Gq inhibition, whereas reduced accumulation indicates a high rate of egg laying, presumably because of Go inhibition.
Figure 2.
Effects of transgenic overexpression of each of the 13 C. elegans RGS genes on egg-laying behavior. Each C. elegans RGS gene was overexpressed under its own promoter from multicopy extrachromosomal transgenes. Empty vector transgenes serve as the control (none). The number of unlaid eggs per adult worm is indicated as a mean ±95% confidence interval, determined for each gene from ≥50 animals (10 each from five independent transgenic strains). The adjacent, dicistronic genes rgs-10 and rgs-11 were overexpressed together. rgs-10/11 cooverexpression resulted in sick animals in which egg laying could not accurately be assessed. Asterisks indicate values that are statistically different from the control (P < 0.05). N.A., not applicable.
Although most transgenes had no significant effect on egg laying, we found that four RGS transgenes did cause changes relative to the control (Fig. 2). egl-10 overexpression resulted in hyperactive egg laying, whereas eat-16 overexpression resulted in reduced egg laying, which is consistent with the previously determined functions of these RGS proteins as inhibitors of Go and Gq, respectively. In addition, we identified the rgs-1 and rgs-2 genes as potential inhibitors of Go. Overexpression of either reduced the accumulation of unlaid eggs and thus appeared to cause hyperactive egg laying, similar to that seen in goa-1 null mutants. The effects of rgs-2 were weaker than those of either egl-10 or rgs-1. In addition to affecting egg-laying behavior, Go signaling also controls the rate of locomotion (Mendel et al. 1995; Ségalat et al. 1995). We noted strongly hyperactive locomotion in egl-10 and rgs-1 overexpressors, similar to that seen in goa-1 null mutants (data not shown). We did not observe any other behavioral defects in the egl-10, rgs-1, or rgs-2 overexpressors beyond those also seen in goa-1 mutants. The only defect that we noted in animals that overexpressed the other 10 RGS genes was decreased viability in strains that overexpressed the dicistronic gene rgs-10/rgs-11.
Overexpression of rgs-1 mimics the phenotype of mutants lacking the G protein Go, whereas rgs-2 overexpression shows similar but weaker effect
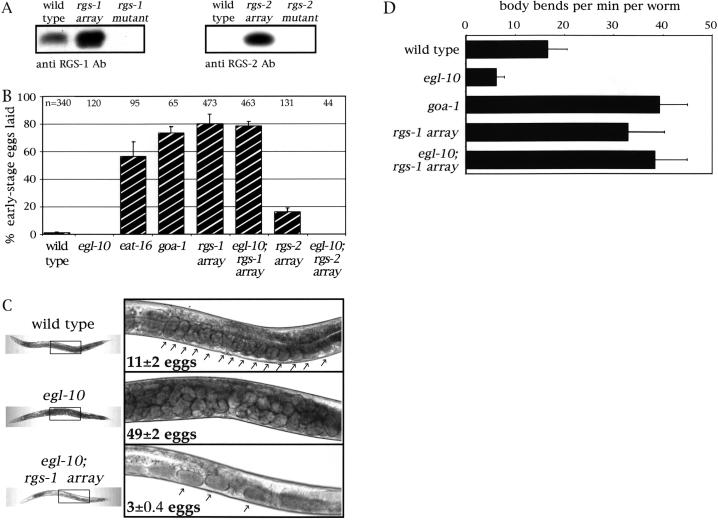
We investigated the effects of rgs-1 and rgs-2 overexpression in more detail by analyzing strains of C. elegans in which the unstable extrachromosomal transgenic arrays overexpressing these RGS genes were stably integrated into the chromosomes. To show that RGS proteins were indeed overexpressed in the stable transgenic strains, we generated polyclonal antibodies against recombinant RGS-1 and RGS-2 proteins and used them to probe Western blots of extracts from control and transgenic strains (Fig. 3A). The RGS-1 antibody detected a 25-kD protein that was overexpressed more than 10-fold in the stable rgs-1 transgenic strain and that was absent from the rgs-1 mutant. Although the RGS-2 protein was below our limit of detection in the wild-type strain, a 19-kD RGS-2 protein was detected in extracts of the rgs-2 transgenic strains.
Figure 3.
Analysis of transgenic strains stably overexpressing RGS-1 and RGS-2. (A) Immunoblots of protein extracts from C. elegans strains probed with anti-RGS-1 (left) or anti-RGS-2 (right) antibodies. Extracts were from the wild type, animals stably overexpressing the RGS genes, or knockout mutants (described below). The overexpressor lanes were loaded with 10-fold less total protein than were the wild-type or mutant lanes. The amount of total protein loaded in each lane was assessed by Coomassie blue staining of gels loaded with samples of the same extracts and by Ponceau S staining of the blots (not shown). (B) Egg-laying rates in C. elegans strains were assessed by counting the percentage of freshly laid eggs at early stages of development (≤ 4 cells). Each measurement was repeated at least four times, and the mean ± standard error is shown. Some values are zero, resulting in the absence of a visible bar. The total number of eggs scored for each strain is indicated at the top of the graph—some strains lay few eggs because of fertility or egg-laying defects, thus resulting in smaller samples. (C) Photographs of adult animals that were wild type (top), carrying an egl-10 null mutation (middle), or carrying both the egl-10 mutation and the stable rgs-1 transgene array (bottom). Thumbnail pictures of whole animals are shown at the left, and close-ups of the boxed regions are shown at the right. Numbers indicate the average unlaid eggs per strain ±95% confidence interval. Arrows point to individual unlaid eggs, except in the middle panel in which the body is fully packed and the eggs are too numerous to indicate individually. (D) Locomotion rates were assessed by counting body bends per min. For each strain, 30 worms were observed for 3 min each and the mean ±95% confidence interval is shown. The transgenes and mutations used for this Figure were: vsIs1 (“rgs-1 array”), vsIs4 (“rgs-2 array”), rgs-1(nr2017), rgs-2(vs17), egl-10(md176), eat-16(ad702), and goa-1(n363).
We also showed that the effects of the rgs-1 transgene were actually attributable to overexpression of the RGS-1 protein, as injection of an rgs-1 transgene modified by inserting a frame-shift mutation in the rgs-1 coding sequences failed to induce any of the phenotypic defects caused by the wild-type rgs-1 transgene (data not shown).
To further analyze changes in egg-laying behavior induced by RGS-1 and RGS-2 overexpression, we examined freshly laid eggs from the stable transgenic strains, an assay that measures hyperactive egg laying (Koelle and Horvitz 1996). The eggs of C. elegans are fertilized internally and normally accumulate in the uterus, where they sit for ∼2 hr and reach the ∼100-cell stage before being laid. In mutants that show hyperactive egg laying, however, eggs are laid shortly after fertilization resulting in freshly laid eggs that are often at early stages of development (≤4 cells). The frequency of egg laying can thus be indirectly assessed by examining freshly laid eggs and determining their developmental stages. Whereas wild-type animals or sluggish egg layers such as egl-10 mutants lay virtually no early-stage eggs, hyperactive egg layers such as eat-16 mutants or goa-1 mutants lay mostly early-stage eggs (Fig. 3B). Hyperactive egg laying induced by overexpression of rgs-1 is as strong as that seen in goa-1 null mutants, whereas overexpression of rgs-2 also induced hyperactive egg laying, but at a much lower level.
All the behavioral defects that have been described in goa-1 null mutants and egl-10 overexpressors (Mendel et al. 1995; Ségalat et al. 1995; Koelle and Horvitz 1996) also occur in RGS-1 overexpressors. In addition to hyperactive egg laying, these animals also display rapid foraging behavior, deep body bends, and abnormally fast locomotion (Fig. 3D).
These results indicate that rgs-1 and rgs-2 might act by inhibiting goa-1 signaling. To test this idea, we constructed animals carrying the rgs-1 overexpression transgene as well as a transgene expressing a GTPase-defective GOA-1(Q205L), which would be expected to be insensitive to RGS proteins (Berman et al. 1996). The rgs-1 transgene by itself causes hyperactivity in egg laying and locomotion (Fig. 3), whereas the GOA-1(Q205L) transgene alone causes sluggish locomotion and blocks egg laying (Mendel et al. 1995). In animals carrying both transgenes, the effects of rgs-1 overexpression were blocked. The animals moved sluggishly and did not lay early-stage eggs. These results suggest that the effects of rgs-1 overexpression occur mostly or entirely through inhibition of GOA-1 signaling.
Another RGS protein, EGL-10, was previously shown to be an inhibitor of the G protein GOA-1 (Koelle and Horvitz 1996). We therefore tested whether overexpression of RGS-1 or RGS-2 could correct the sluggish egg-laying in egl-10 null mutants. The accumulation of unlaid eggs (the Egl phenotype) that occurs in egl-10 mutants was completely corrected by overexpression of RGS-1 (Fig. 3C). In fact, overexpression of RGS-1 in an egl-10 mutant background still leads to hyperactive egg-laying behavior, as shown by the reduced accumulation of unlaid eggs (Fig. 3C) and by the high percentage of early-stage eggs laid (Fig. 3B). We also found that the sluggish locomotion seen in egl-10 null mutants could be rescued by RGS-1 overexpression (Fig. 3D and data not shown). The effects of RGS-2 overexpression were consistently weaker than those of RGS-1 overexpression. RGS-2 overexpression neither corrected the Egl phenotype of egl-10 mutants (data not shown) nor did it lead to hyperactive egg laying in the absence of EGL-10 (Fig. 3B).
In summary, we found that overexpression of RGS-1 mimics all the behavioral defects seen in goa-1 null mutants. Overexpression of EGL-10 also causes the same behavioral defects (Koelle and Horvitz 1996), and overexpression of RGS-1 rescues all the behavioral defects seen in egl-10 null mutants. These results suggest that RGS-1, like EGL-10, is able to inhibit signaling by GOA-1, and that overexpressed RGS-1 may substitute for EGL-10. RGS-2 overexpression causes weaker effects than overexpression of either EGL-10 or RGS-1 and does not substitute for EGL-10. RGS-2 may function differently than EGL-10, or the effects of RGS-2 overexpression may simply be too weak to overcome the absence of EGL-10. Although RGS-2 overexpression generates only weak effects on egg laying, its sequence similarity to RGS-1 and the overlapping expression patterns of RGS-1 and RGS-2 (see below) led us to continue to characterize both genes as potentially redundant regulators of GOA-1.
rgs-1 and rgs-2 expression overlaps with that of Go in the C. elegans nervous system
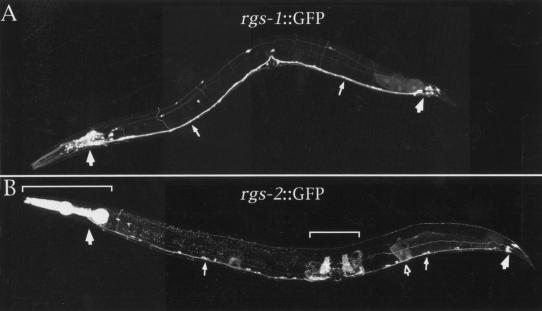
To examine the expression patterns of rgs-1 and rgs-2, we fused the promoters of these genes to the coding sequences for the green fluorescent protein (GFP) and examined transgenic animals carrying these constructs by confocal fluorescence microscopy. We saw rgs-1::GFP expression in most or all neurons (Fig. 4A) and rgs-2::GFP expression in a subset that included ventral cord and head- and tail-ganglia neurons (Fig. 4B). We did not observe rgs-2::GFP fluorescence in the hermaphrodite-specific neurons (HSNs), which also synapse onto the egg-laying muscles. We observed rgs-2::GFP fluorescence in some non-neuronal cells, including the pharyngeal muscles and uterine muscles, the latter of which are used for egg laying. The rgs-1 and rgs-2 expression patterns thus appear to overlap each other, and also to overlap the expression pattern of their potential target, the Go protein GOA-1, which is expressed in all neurons of the animal (Mendel et al. 1995; Ségalat et al. 1995).
Figure 4.
Expression patterns of rgs-1 and rgs-2. Transgenic animals carrying fusions of the rgs-1 or rgs-2 promoter regions to the coding sequences for the GFP were examined by fluorescence confocal microscopy. (A) rgs-1::GFP fluorescence is seen in most or all neurons. Large arrows indicate the head and tail ganglia at the left and right, respectively. Two small arrows point to the ventral nerve cord that runs the length of the animal. The only non-neuronal cells labeled are several posterior intestinal cells that are faintly fluorescent in some animals. (B) rgs2::GFP fluorescence is seen in a subset of neurons and in certain muscle cells. Large and small filled arrows indicate the same structures as in panel A. The bracket at the left lies over the pharynx; the bracket at the middle of the animal lies over the uterine muscles. The open arrow indicates faint fluorescence sometimes seen in the spermatheca.
RGS-1 and RGS-2 encode highly similar, small RGS proteins
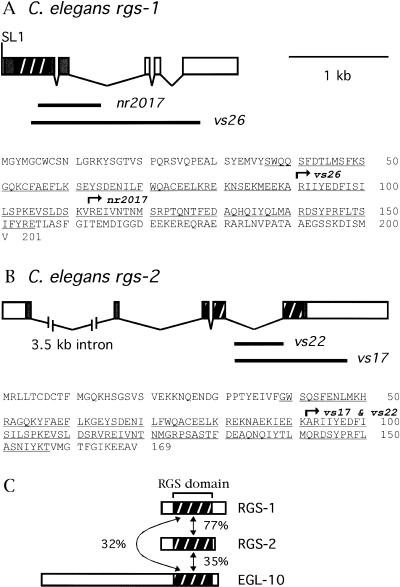
To express recombinant RGS-1 and RGS-2 for biochemical studies, we obtained cDNAs containing the complete coding sequences for these proteins. Figure 5 shows the RGS-1 and RGS-2 protein sequences encoded by the cDNAs, as well as the gene structures deduced by comparison with the C. elegans genome sequence.
Figure 5.
Gene structures, protein sequences, and knockout mutations of rgs-1 and rgs-2 . (A) Gene structure, protein sequence, and knockout mutations for rgs-1. Exons are boxed, protein coding sequences are filled and the RGS domain coding region is hatched. An SL1 trans-spliced leader found at the 5′ end of the cDNA sequence is indicated. Thick lines below the exon structure indicate the extents of the genomic regions deleted in two mutant alleles. The protein sequence deduced from the cDNA sequence is at the bottom, with the RGS domain underlined. Arrows indicate the regions of protein for which the coding sequences are removed by the indicated deletion mutations. (B) Gene structure, protein sequence, and knockout mutations for rgs-2. Features are shown in the same manner as in panel A. The vs22 mutation removes the beginning of exon five and thus shifts the reading frame for the remainder of the RGS domain coding sequences. (C) Schematic diagrams of the predicted RGS-1, RGS-2, and EGL-10 proteins. The RGS domains are hatched, and percent sequence identities between them are indicated.
The RGS-1 and RGS-2 proteins have 201 and 169 amino acid residues, respectively, and each sequence contains a 119–amino acid RGS domain, but no other features conserved with other proteins in the sequence database. There are no clear orthologs of RGS-1 and RGS-2 among the mammalian RGS proteins identified to date. The RGS domain sequences of the two proteins are 77% identical, making them much more similar to each other than either is to any other RGS domain sequence found in C. elegans (Fig. 5C).
RGS-1 and RGS-2 are GTPase activators of the C. elegans Go protein in vitro
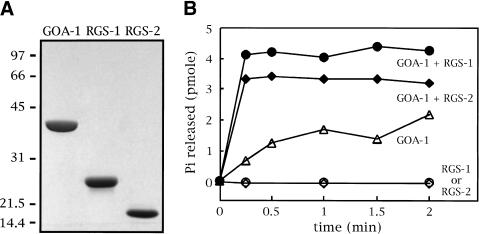
Because the genetic experiments and expression patterns described above suggested that RGS-1 and RGS-2 might act on the Go protein GOA-1, we tested whether RGS-1 and RGS-2 can act as GTPase activators of GOA-1. The proteins were expressed in Escherichia coli fused to glutathione S-transferase (GST), partially purified on glutathione agarose, cleaved by a protease to remove the GST tag, and finally obtained in a high state of purity after ion exchange chromatography. A Coomassie-stained gel of the purified proteins is shown in Figure 6A.
Figure 6.
Activation of the GTPase activity of GOA-1 by RGS-1 and RGS-2. (A) Coomassie-stained SDS-PAGE gel showing 4 μg each of the purified preparations of GOA-1, RGS-1, and RGS-2. The positions of molecular weight markers are indicated, with their sizes in kD. (B) Rates of GTP hydrolysis by the purified proteins were measured in a single-turnover assay. 0.6 μm GOA-1 was assayed either alone or in the presence of 0.2 μm RGS-1 or RGS-2. RGS-1 and RGS-2 were tested by themselves for GTPase activity (bottom curves) at 2 μm. The data shown represent the average of two experiments. The top two curves plateau at different levels because of differences in the amounts of GTP-loaded GOA-1 present at the outset of each assay.
We preloaded purified GOA-1 with radiolabeled GTP and monitored the GTP hydrolysis rate of GOA-1 by measuring the release of the radioactive γ phosphate as a function of time (Fig. 6B). GOA-1 by itself had a relatively slow rate of GTP hydrolysis, as has been described for other Gα proteins (Berman and Gilman 1998). In the presence of RGS-1 or RGS-2, however, GTP hydrolysis by GOA-1 was markedly enhanced, and the reactions went to completion before the first time point of the experiment. RGS-1 and RGS-2 by themselves had no detectable GTPase activity. These results suggest that the genetic results described above, in which overexpression of RGS-1 or RGS-2 mimicked the effects of GOA-1 loss-of-function mutations, could be explained by the direct action of RGS-1 and RGS-2 on the GOA-1 protein to inhibit its ability to signal.
Knockout mutants of rgs-1 and rgs-2
The experiments described above are not sufficient to determine the true physiological functions of the RGS-1 and RGS-2 proteins. Overexpression in living animals may cause these proteins to carry out abnormal activities. The in vitro studies show that RGS-1 and RGS-2 can act on GOA-1, but do not show that GOA-1 must be their true Gα-target in vivo, and do not reveal the biological purpose for which they might regulate GOA-1. Most previous studies of RGS proteins have used the same experimental approaches and suffer from the same limitations. To rigorously determine the in vivo functions of rgs-1 and rgs-2, we produced knockout mutants of C. elegans deleted for these genes. The high sequence similarity of RGS-1 and RGS-2, their overlapping expression patterns, similar overexpression phenotypes, and similar biochemical activities all suggested that these two proteins might function redundantly. Therefore, we decided to study both knockouts, individually and in combination. We used an adaptation of a recently developed gene knockout technology in which the progeny of several hundred thousand mutagenized animals are screened by PCR to identify rare individuals in which a gene of interest has suffered a deletion of ∼0.5–3 kb (Liu et al. 1999). We produced two independent deletion mutations for each RGS gene. The regions deleted are indicated in Figure 5. We present results obtained with the alleles rgs-1(nr2017) and rgs-2(vs17). Each experiment shown was repeated with the alleles rgs-1(vs26) and rgs-2(vs22). These repetitions yielded similar results (data not shown), verifying that the defects seen were indeed the result of deletion of the rgs-1 and rgs-2 genes and not attributable to any genetic background mutations that might exist. Because each deletion mutation removes a large portion of the coding sequences for the RGS domain (nr2017, vs26, and vs17) or shifts the reading frame in the middle of the RGS domain (vs22), each mutation is likely to be a null allele.
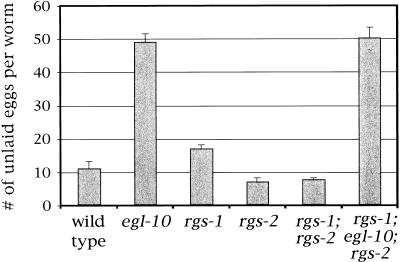
rgs-1and rgs-2 knockout mutations cause only weak egg-laying defects in well-fed worms
In the egg-laying assays presented in Figures 2 and 3, the animals were grown and examined in the presence of abundant food. Although egl-10 and eat-16 mutations have strong effects on egg laying under these conditions, rgs-1 and rgs-2 mutations cause only mild defects. Figure 7 shows that the numbers of unlaid eggs in well-fed rgs-1 and rgs-2 mutants are similar to those of the wild type, in comparison with the strong defect seen in egl-10 mutants. rgs-1; rgs-2 double mutants are also similar to the wild type under these conditions and rgs-1; egl-10; rgs-2 triple mutants appear similar to egl-10 single mutants. Because no strong synthetic behavioral defects are revealed in these double and triple mutants, rgs-1, rgs-2, and egl-10 do not have obvious redundant functions in controlling egg laying under well-fed conditions.
Figure 7.
Accumulation of unlaid eggs under well-fed conditions in RGS mutants. The number of unlaid eggs per worm was determined from 30 worms for each strain. Mean ±95% confidence interval is shown. The mutations used were egl-10(md176), rgs-1(nr2017), and rgs-2(vs17).
The slight increase in accumulated eggs in rgs-1 single mutants relative to the wild type (Fig. 7) is statistically significant (P < 0.0005). This effect is additive with the strong defects induced by egl-10 null mutations and it is suppressed by a goa-1 null mutation (data not shown). These results are consistent with RGS-1 causing a mild inhibition of GOA-1 signaling under well-fed conditions, whereas most of the inhibition of GOA-1 is attributable to EGL-10. The slight decrease in accumulated eggs in rgs-2 mutants is also reproducible and may be caused in part by a mild defect in egg production. Other mutants shown in Figure 7 had brood sizes close to that of the wild type (see Materials and Methods).
rgs-1 and rgs-2 redundantly regulate signaling to allow starved animals to rapidly induce egg laying when fed
Because analysis of egg laying in well-fed animals failed to identify strong functions for RSG-1 and RSG-2, we examined the mutants for defects in the regulation of egg laying caused by the removal and return of food. Worms lay eggs regularly on petri dish in the presence of a lawn of bacteria, the food used for laboratory culture of C. elegans. However, as shown below, egg-laying behavior virtually ceases if worms are starved by removing them from bacteria and resumes only if the animals are returned to bacteria. In this way, worms can lay their eggs when and where food is available for their progeny. Because RGS-1 and RGS-2 are potential regulators of GOA-1 based on their overexpression phenotypes, we reasoned that RGS-1 and/or RGS-2 might exist to alter GOA-1 signaling in response to starving or feeding, thereby altering egg laying under these conditions.
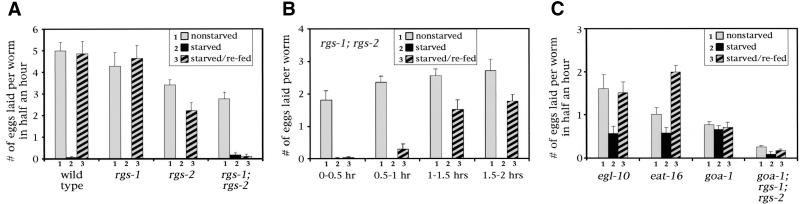
We found that RGS-1 and RGS-2 are redundantly required to induce egg laying when starved animals are fed. Figure 8A shows the rates of egg laying by wild-type or mutant animals that have been (1) maintained in constant food (nonstarved); (2) removed from food for 2 hr (starved); or (3) removed from food for 2 hr and then returned to food (starved/refed). Wild-type animals strongly regulate egg laying under these conditions, such that virtually no eggs are laid by starved animals, and refeeding starved animals returns egg laying to the same rate as that seen in nonstarved animals. rgs-1 single mutants also show strong regulation of egg laying. rgs-2 mutants show weak defects. Their baseline (nonstarved) level of egg laying is slightly reduced, probably because of the mild defect in egg production noted above, and they do not fully return to their baseline level of egg laying when starved and refed.
Figure 8.
Egg-laying behavior responses to starving and feeding in wild-type and mutant strains. (A) The strains indicated were preconditioned for 2 hr on plates either containing bacteria (to feed the animals) or not containing bacteria (to starve them). Animals were then moved to assay plates either with or without bacteria and the number of eggs laid in 30 min was counted. (Nonstarved) Both the preconditioning and assay plates had bacteria; (starved) both plates lacked bacteria; (starved/refed) the animals were preconditioned without bacteria and moved to assay plates with bacteria. Each value was determined by measuring 80 animals (8 experiments with 10 animals each) and is shown as a mean ± standard error. The mutations used were rgs-1(nr2017) and rgs-2(vs17). The mutations rgs-1(vs26) and rgs-2(vs22) were also tested and gave similar results (data not shown). The values for starved animals were often zero and thus did not result in visible bars in the graph. (B) rgs-1;rgs-2 double mutants were assayed as in panel A, except that animals were left on the assay plates for 2 hr, and the number of eggs laid during each 30-min period within the 2 hr was determined. (C) The mutants indicated were assayed as in panel A. The mutations used were egl-10(md176), eat-16(ad702), and goa-1(n363). The absolute levels of egg laying in these mutants are reduced relative to the wild type, either because of sluggish egg-laying behavior (egl-10) or because the animals have relatively few available eggs in their uteri (eat-16 and goa-1).
A dramatic defect, however, is seen in rgs-1; rgs-2 double mutants. These animals show a reduced baseline level of egg laying, just as seen in the rgs-2 single mutant, and they are able to stop laying eggs when starved. However, the double mutants fail to induce egg laying when refed after starvation (Fig. 8A). This result suggests a model in which feeding induces the activities of both RGS-1 and RGS-2, thus inhibiting signaling by the G protein GOA-1 and causing an increase in egg-laying behavior. Egg laying can be induced in wild-type animals by treatment with the neurotransmitter serotonin, which is normally released from the HSNs onto the egg-laying muscles to stimulate their contraction (Desai et al. 1988). Serotonin also stimulates rgs-1; rgs-2 double mutants to lay eggs, even after starvation (data not shown), suggesting that the rgs-1 and rgs-2 mutations may interfere with activity of the egg-laying neurons rather than with the responsiveness of the egg-laying muscles to serotonin. In addition to the synthetic egg-laying defect seen in rgs-1; rgs-2 double mutants, these animals also have smaller bodies than do the wild type or the single mutants (data not shown). RGS-1 and RGS-2 therefore have at least two redundant functions.
The blocked induction of egg laying that is seen in rgs-1; rgs-2 double mutants is temporary. Figure 8B shows the results of a time course in which the rate of egg laying was measured in 30-min periods after the return of starved animals to food. The block of egg laying is virtually complete in the first 30 min, but is largely gone after 2 hr. This suggests that a mechanism that is not dependent on RGS-1 or RGS-2 eventually allows the induction of egg laying when animals are fed, and that the RGS proteins are responsible mainly for the rapid phase of the normal response.
The RGS proteins EGL-10 and EAT-16 were identified based on their strong effects on egg laying under well-fed conditions (Koelle et al. 1996; Hadju-Cronin et al. 1999). egl-10 mutants lay eggs infrequently, whereas eat-16 mutants are hyperactive egg layers (Fig. 3; Hadju-Cronin et al. 1999). We tested these mutants in the starving and feeding assay (Fig. 8C). Unlike rgs-1; rgs-2 double mutants, the egl-10 and eat-16 mutants induce egg laying when refed after starvation. Both mutants also lay some eggs even when starved. In the case of egl-10, this is probably attributable to the fact that the animals are bloated with excess unlaid eggs and may be unable to retain any more. In the case of eat-16, hyperactive egg-laying behavior appears to continue even in the absence of food. Therefore, unlike rgs-1 and rgs-2, the RGS genes egl-10 and eat-16 are not required for the induction of egg laying after starved animals are refed.
If RGS-1 and RGS-2 cause changes in egg-laying behavior by regulating GOA-1, then a goa-1 null mutation should abolish these changes. In fact, goa-1 null mutants show no changes in rates of egg laying when starved or refed. Furthermore, goa-1; rgs-1; rgs-2 mutants behave like goa-1 mutants, with similar egg-laying rate under all conditions (Fig. 8C). However, interpretation of these results is complicated by the fact that goa-1 mutants are partially sterile (average brood size, 36; Ségalat et al. 1995) and have few eggs to lay. Thus egg production, rather than egg-laying behavior, may limit the number of eggs laid. The fertility of goa-1; rgs-1; rgs-2 mutants is even lower. Analysis of these mutants thus does not serve as a strong test of the idea that RGS-1 and RGS-2 act via regulation of GOA-1. However, the other data presented in this work do indicate that RGS-1 and RGS-2 act to inhibit GOA-1 and show that RGS-1 and RGS-2 are required to induce egg laying when starved animals are fed. These results suggest a model in which feeding after starving induces RGS-1 and RGS-2 activity, thus inhibiting GOA-1 signaling to induce egg laying.
Discussion
The discovery of a large family of RGS proteins, many of which can act as GTPase activators of the same G proteins in vitro, has raised the issue of whether one G protein might be regulated by several RGS proteins in vivo. If so, for what biological purpose? In this work, we have taken advantage of the completed genome sequence of C. elegans to survey the in vivo functions of all of its RGS proteins. Our results suggest that three RGS proteins, EGL-10, RGS-1, and RGS-2, all act on GOA-1, the C. elegans homolog of Go. The combined action of these regulators allows C. elegans to set the frequencies of egg-laying behavior in a manner appropriate for both constant and changing supplies of food.
Multiple RGS proteins control Go signaling in C. elegans
EGL-10 was previously shown to function in vivo by inhibiting GOA-1, the C. elegans homolog of Go (Koelle and Horvitz 1996). Several lines of evidence suggest that RGS-1 and RGS-2 also induce behavioral change by negatively regulating Go. First, overexpression of RGS-1 or RGS-2 in well-fed animals, like overexpression of EGL-10, mimics the behavioral defects of a Go null mutant, and these are the only behavioral defects we have noted (Fig. 3). RGS-1 or EGL-10 overexpression induces defects as strong as those seen in the Go null mutant, whereas RGS-2 overexpression causes weaker defects. Second, the effects of RGS-1 overexpression can be abolished by expression of GOA-1(Q205L), a GTPase-deficient mutant of GOA-1. Third, some neurons that express GOA-1 also express RGS-1 and RGS-2 (Fig. 4). Fourth, purified RGS-1 and RGS-2 proteins both stimulate the GTPase activity of purified GOA-1 (Fig. 5). Fifth, well-fed rgs-1 mutants show a modest but significant decrease in egg laying (Fig. 7), which is consistent with RGS-1 causing a modest inhibition of Go under these circumstances (compared with the much greater inhibition caused by EGL-10). Overexpression of RGS-1 through the increase of the copy number of its gene thus magnifies this modest effect into a large one, and can even compensate for the lack of EGL-10 in egl-10 mutants (Fig. 3). Defects in well-fed rgs-2 single mutants are more subtle than those of rgs-1 mutants. Sixth, the major role of RGS-1 and RGS-2 is to induce egg laying when starved animals are fed, a function that these two RGS proteins carry out redundantly (Fig. 8). The Go null mutation appears to abolish the effects of feeding and starving on egg laying, suggesting that the behavior may be normally regulated through this G protein. Finally, a genetic study of all the Gα-proteins encoded in the C. elegans genome has identified only one C. elegans G protein, namely Go, that inhibits egg laying, making Go the best candidate to be itself inhibited by RGS-1 and RGS-2 to cause the observed effects (Jansen et al. 1999).
C. elegans adjusts behavior in response to food by adjusting G protein signaling
In this article we show that worms demonstrate robust behavioral changes when they are removed from and returned to food. Egg-laying behavior is almost completely suppressed after animals are starved for 2 hr, but returns to its starting levels within 30 min after they are returned to food.
Previous work has shown that signaling through the Go protein GOA-1 stimulates egg laying, whereas signaling through the Gq protein EGL-30 inhibits this behavior (Fig. 1), and that the rate of egg laying is set by the balance between signaling by these two G proteins (Mendel et al. 1995; Ségalat et al. 1995; Brundage et al. 1996; Hadju-Cronin et al. 1999). Receptors for neurotransmitters that control egg laying, such as serotonin, acetylcholine, and certain neuropeptides, may directly stimulate signaling by GOA-1 and/or EGL-30 (Desai et al. 1988; Weinshenker et al. 1995; Waggoner et al. 1998, 2000). In response to feeding or starving, animals might alter release of these or other neurotransmitters to alter egg laying. Indeed, the release of the FLP-1 neuropeptide appears to be required for animals to properly halt egg laying when starved (Waggoner et al. 2000).
We have identified an additional mechanism by which animals alter signaling to control egg laying. As an alternative to altering the release of the neurotransmitters that couple to GOA-1 and/or EGL-30, animals might change the ability of these G proteins to signal in response to neurotransmitter release. Our work shows that this latter mechanism stimulates egg laying when starved animals are refed. A feeding-induced signal apparently stimulates RGS-1 and RGS-2 activity to reduce the efficiency of GOA-1 signaling, thus increasing egg-laying behavior.
Different RGS proteins control baseline and regulated levels of Go signaling
Our results begin to address why many RGS proteins with similar G protein specificities might exist in higher organisms such as C. elegans and mammals. We first consider yeast, which has only two RGS proteins, one specific for each of the two Gα-proteins in this organism (Apanovitch et al. 1998; Versele et al. 1999). The yeast RGS protein Sst2p, in particular, has been well studied (for review, see Dohlman et al. 1998). Sst2p inhibits Gpa1p, the G protein that mediates signaling by mating pheromone. In yeast that have never been exposed to pheromone, Sst2p partially inhibits Gpa1p signaling and thus sets the baseline level of sensitivity to pheromone. Once yeast are exposed to pheromone, Gpa1p signaling induces Sst2p expression, which feeds back to further inhibit Gpa1p and thus desensitizes yeast to pheromone. In this way Sst2p sets both baseline and regulated levels of signaling by Gpa1p.
In C. elegans, RGS proteins appear to set baseline and regulated levels of signaling by the G protein Go, but the roles played by just one RGS protein in yeast appear to be distributed among three different RGS proteins in C. elegans. The RGS protein EGL-10 has a very strong effect on egg laying under all circumstances, but feeding and starving still alter the residual egg laying seen in egl-10 mutants. These results suggest that the baseline levels of signaling by Go are set by EGL-10 activity, and that other factors besides EGL-10 are responsible for changing signaling to alter egg laying when worms are starved or fed. In contrast, the RGS proteins RGS-1 and RGS-2 have little effect on baseline levels of signaling, as indicated by their mild effects on egg laying in animals that are given constant food. However, RGS-1 and RGS-2 are redundantly required for the rapid induction of egg laying when animals are fed, suggesting that they are induced to alter signaling under these circumstances. We have not yet determined the molecular mechanism by which RGS-1 and RGS-2 activity might be induced upon feeding. Their expression could be controlled transcriptionally, as in the case of SST2 (Dietzel and Kurjan 1987) or their activity could be altered at other levels. Analysis with RGS-1 and RGS-2 antibodies and observation of rgs-1::GFP and rgs-2::GFP reporters has not yet revealed changes in animals that have been starved and fed (M.Q. Dong, unpubl.).
Our results suggest the logic that underlies the use of the many RGS proteins in mammals and in C. elegans. First, these proteins show some level of functional redundancy, as exemplified both by the redundant effects of RGS-1 and RGS-2 in inducing egg laying when starved animals are refed and by the partial redundancy between EGL-10 and RGS-1 in well-fed animals. RGS proteins thus appear to be typical of many other protein families, which often have more members in higher eukaryotes than in yeast, and which often show overlapping expression patterns and some redundancy in higher eukaryotes. In addition, it appears that many RGS proteins may exist in higher eukaryotes so that individual RGS proteins can be specialized to adjust signaling under particular circumstances. In the case of Go in C. elegans, EGL-10 appears to play the major role in setting the baseline level of signaling, whereas RGS-1 and RGS-2 appear to be specialized to modify signaling when animals are fed. It is possible that additional RGS proteins also regulate Go by altering signaling levels in particular cells and under circumstances that we have not examined. In general, the use of different RGS proteins to alter signaling in particular circumstances may explain the existence of the large number of these proteins.
Two classes of RGS proteins emerge from the genetic studies that have so far been performed on higher eukaryotes. EGL-10 and EAT-16 are members of the class of large RGS proteins that have an amino-terminal conserved region, as well as G gamma-like (GGL) and RGS domains. These proteins appear to set the baseline level of signaling by their respective Gα targets. The second class includes the small RGS proteins RGS-1 and RGS-2, which consist of little more than an RGS domain. These proteins mainly adjust signaling under particular circumstances and do not affect baseline signaling. The mammalian RGS proteins that are known to be transcriptionally regulated, and thus to have the potential to adjust signaling, are also members of the small RGS protein class (Druey et al. 1996; Pepperl et al. 1998; Miles et al. 2000). It remains to be seen if, in general, the large RGS proteins always function to set baseline signaling levels, whereas the small RGS proteins always function to adjust signaling under various circumstances. It is interesting to note that in this study we found that overexpressing the small RGS-1 protein can compensate for the loss of EGL-10 despite the fact that RGS-1 lacks the amino terminal and GGL domains found in EGL-10.
Materials and methods
Detailed protocols for many of the methods used in this work are available electronically at http://info.med.yale.edu/mbb/koelle/.
Identification and transgenic overexpression of C. elegans RGS genes
Analysis of the C. elegans genome sequence identified 13 predicted genes with RGS domain similarity, including the previously studied egl-10 and eat-16 genes. The remaining genes were named as follows (designations from the genome sequence are given in parentheses): rgs-1 (C05B5.7), rgs-2 (F16H9.1a), rgs-3 (C29H12.3), rgs-4 (Y38E10A.v), rgs-5 (B0336.4), rgs-6 (C41G11.3), rgs-7 (F56B6.2), rgs-8 (F52D2.2), rgs-9 (ZC53.7), rgs-10 (F45B8.2), rgs-11 (F45B8.1). Mammalian RGS genes are given the names RGS-1, RGS-2, and so on, but the RGS numbering schemes in mammals and worms do not indicate the complex evolutionary relationships between these gene sets. The rgs-3 gene encodes a protein with two highly similar RGS domains. For each gene, genomic DNA containing the coding sequences and approximately 5 kb of flanking DNA on either side (presumed to contain the promoters and regulatory sequences) were subcloned into the plasmid pBluescript (Stratagene). The source of genomic DNA was either cosmid clones obtained from the C. elegans genome project, or (for rgs-4) a PCR product amplified from total genomic DNA. rgs-10 and rgs-11 are highly similar, adjacent genes predicted to be transcribed from the same promoter to form a dicistronic transcript. They were subcloned and overexpressed together as a pair. Transgenic strains were generated by injecting RGS subclones (or pBluescript as a control) at 80 ng/μl into lin-15(n765ts) animals along with a lin-15 rescuing plasmid as a coinjection marker. Extrachromosomal transgenes were chromosomally integrated by irradiating transgenic animals with γ-rays and the resulting strains were outcrossed at least four times to provide a clean genetic background. The integrated rgs-1 transgene array used in this work has the allele designation vsIs1. The integrated rgs-2 transgene array is vsIs4. The integrated transgene expressing GOA-1(Q205L) used was syIs9 (Mendel et al. 1995).
Behavioral assay
Measurement of accumulation of unlaid eggs, staging of freshly laid eggs, and measurement of locomotion rates were performed as described previously (Koelle and Horvitz 1996). We generally used accumulation of unlaid eggs to quantify the failure to lay eggs, and laying of early-stage eggs (≤4 cells) to quantify hyperactive egg laying. The use of both assays, along with measurements of brood size, ensures that changes in egg-laying behavior can be differentiated from changes in egg production (Koelle and Horvitz 1996). Brood sizes for strains shown in Figure 7 were: Wild type, 247 ± 12; egl-10, 226 ± 37; rgs-1, 253 ± 34; rgs-2, 220 ± 35; and rgs-1; rgs-2, 167 ± 41. All assays were performed on animals selected as late L4 larvae and aged at 20°C to produce precisely staged adults. The post-L4 aging times used were 36 hr (Fig. 2) and 30 hr (Figs. 3,7,8).
To measure responses to starving and feeding, staged adult animals were allowed to feed or starve for 2 hr at 20° C by transferring them to NGM plates either with or without a lawn of OP50 bacteria as food (Wood 1988). Ten of the starved worms were then transferred to a bacteria-free plate, and another 10 starved worms were transferred to a plate with a lawn of OP50. As a control, 10 nonstarved worms were put onto a plate with a lawn of OP50. The three assay plates were each ringed by a circle of 4m fructose solution as an osmotic barrier to prevent worms from leaving the agar surface. All three groups of worms were allowed to lay eggs for 30 min and the number of eggs laid was counted. For time course experiments the animals were allowed to continue to lay eggs and the accumulation of eggs was counted every 30 min. Each determination was repeated eight times and the results were averaged.
Antibodies
RGS-1 and RGS-2 were expressed in E. coli as glutathione S-transferase fusion proteins, purified, and injected into rabbits to raise polyclonal antisera. RGS-1 and RGS-2 were also expressed in E. coli, fused to the maltose binding protein, and these fusion proteins were used to affinity purify the antisera as described by Koelle and Horvitz (1996). For Western blots, the RGS proteins were visualized using horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) and chemiluminescence detection reagents (Pierce).
GFP transgenes
GFP reporter constructs were constructed by inserting genomic DNA fragments from rgs-1 or rgs-2 into the vectors pPD95.69 and pPD95.67 (provided by Dr. Andrew Fire, Carnegie Institute of Washington). These constructs contained the promoter regions and 5′ coding sequences of the RGS genes, such that a coding exon for each gene was fused in frame to the coding sequence for GFP. The rgs-1 transgene contained sequences from −2399 to +3 of the rgs-1 gene (coordinates relative to the translation start site of rgs-1). The rgs-2 transgene contained sequences from −4770 to +3592 (relative to the rgs-2 translation start), and thus included the large first intron of rgs-2.
cDNA sequences of rgs-1 and rgs-2
A 1131-bp rgs-1 cDNA clone was obtained from the library of Barstead and Waterston (1989) and extended at its 5′ end by an additional 179 bp using the 5′-RACE technique (Frohman et al. 1988). The resulting cDNA sequence appears to be full length as it has an SL1 trans-spliced leader at its 5′ end (Krause and Hirsh 1987), a poly(A) tail at its 3′ end, and it matches the length of the single 1.3-kb message seen on Northern blots. An rgs-2 expressed sequence tag identified an rgs-2 cDNA clone (yk78b6), which we obtained from Yuji Kohara. The 1562-bp sequence of this clone contains a complete open reading frame and is similar in length to the two rgs-2 transcripts of 1.4 and 1.6 kb seen on Northern blots. The structural differences between the two transcripts have not been investigated. The GenBank accession numbers for the rgs-1 and rgs-2 sequences are AF220159 and AF220265, respectively.
Protein purification and GTPase assays
RGS-1, RGS-2, and GOA-1 were expressed and purified using the same strategy. The proteins were expressed in E. coli as fusion proteins consisting of glutathione S-transferase, followed by a His(6) tag, followed by a cleavage site for the tobacco etch virus (TEV) protease (Parks et al. 1994), followed by the protein of interest. The fusion proteins were partially purified on glutathione agarose, cleaved by a His(6)-tagged recombinant TEV protease to remove the GST–His(6) tag, passed through Ni–NTA resin to absorb the released GST–His(6) tag as well as the His(6)–TEV protease and any uncleaved fusion protein, and further purified by anion exchange chromatography. Single-turnover GTPase assays were performed as described by Berman et al. (1996).
C. elegans gene knockouts
A library of frozen mutant worms derived from 460,000 trimethlypsoralen/UV mutagenized animals was screened by PCR for deletions in the rgs-1 and rgs-2 genes. Library construction and screening methods were adapted from Liu et al. (1999) and are described in detail at http://info.med.yale.edu/mbb/koelle/. The rgs-1(nr2017) mutation came from a similarly prepared library of ethylnitrosourea-mutagenized animals. Deletion mutants were outcrossed at least four times to the wild type to produce clean genetic backgrounds. rgs-1(nr2017) is a 638-bp deletion of sequences whose limits are CGAGAAATTGTCAACACTAAC…GTTTGGAATGGTTTATCAGTT. The deleted material is replaced by the following 35-bp insertion: TATGTTTAAGTTAAGTTTATAGTTTAAGTTTAAAG. rgs-1(vs26) is a 1409-bp deletion of sequences with limits GTATCATCTATGAAGATTTCA…GTGTAACTAATATGCAAAAGT. rgs-2(vs17) is a 1136-bp deletion of sequences with limits ATATATATATCTCATTACTGG…AATCAAGTGTAACACTAATAT. rgs-2(vs22) is a 495-bp deletion with limits ATATATATATATCTCATTACT…TTTCAGGCTCGCATTATCTAC.
Acknowledgments
We thank the NemaPharm Group of Axys Pharmaceuticals for isolating the rgs-1(nr2017) mutant, Rajesh Ranganathan for help isolating rgs-1(vs26), and David Shechner for help with the rgs-2 knockouts. This work was supported in part by grants from the Pew Scholars Program and the Merck Genome Research Institute. M.R.K. is a Leukemia Society of America Scholar.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Michael.Koelle@Yale.edu; FAX (203) 785-6404.
References
- Apanovitch DM, Slep KC, Sigler PB, Dohlman HG. Sst2 is a GTPase-activating protein for Gpa1: Purification and characterization of a cognate RGS-Gα protein pair in yeast. Biochemistry. 1998;37:4815–4822. doi: 10.1021/bi9729965. [DOI] [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. The basal component of the nematode dense-body is vinculin. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- Berman DM, Gilman AG. Mammalian RGS proteins: Barbarians at the gate. J Biol Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein α subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- Brundage L, Avery L, Katz A, Kim U, Mendel JE, Sternberg PW, Simon MI. Mutation in a C. elegans Gqα gene disrupts movement, egg laying, and viability. Neuron. 1996;16:999–1009. doi: 10.1016/s0896-6273(00)80123-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RK, Otte CA. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by α factor and α factor pheromones. Mol Cell Biol. 1982;2:21–29. doi: 10.1128/mcb.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries L, Mousli M, Wurmser A, Farquhar MG. GAIP, a protein that specifically interacts with the trimeric G protein Gαi3, is a member of a protein family with a highly conserved core domain. Proc Natl Acad Sci. 1995;92:11916–11920. doi: 10.1073/pnas.92.25.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C, Garriga G, McIntire SL, Horvitz HR. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature. 1988;336:638–646. doi: 10.1038/336638a0. [DOI] [PubMed] [Google Scholar]
- Dietzel C, Kurjan J. Pheromonal regulation and sequence of the Saccharomyces cerevisiae SST2 gene: A model for desensitization to pheromone. Mol Cell Biol. 1987;7:4169–4177. doi: 10.1128/mcb.7.12.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Apaniesk D, Chen Y, Song J, Nusskern D. Inhibition of G-protein signaling by dominant gain-of-function mutations in Sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:3635–3643. doi: 10.1128/mcb.15.7.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Song J, Apanovitch DM, DiBello PR, Gillen KM. Regulation of G protein signalling in yeast. Semin Cell Dev Biol. 1998;9:135–141. doi: 10.1006/scdb.1998.0218. [DOI] [PubMed] [Google Scholar]
- Druey KM, Blumer KJ, Kang VH, Kehrl JH. Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature. 1996;379:742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: Region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu-Cronin YM, Chen WJ, Patikoglou G, Koelle MR, Sternberg PW. Antagonism between Goα and Gqα in Caenorhabditis elegans: The RGS protein EAT-16 is necessary for Goα signaling and regulates Gqα activity. Genes & Dev. 1999;13:1780–1793. doi: 10.1101/gad.13.14.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm HE. The many faces of G protein signaling. J Biol Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- Hepler JR. Emerging roles for RGS proteins in cell signalling. Trends Pharmacol Sci. 1999;20:376–382. doi: 10.1016/s0165-6147(99)01369-3. [DOI] [PubMed] [Google Scholar]
- Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, Plasterk RH. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet. 1999;21:414–419. doi: 10.1038/7753. [DOI] [PubMed] [Google Scholar]
- Koelle MR, Horvitz HR. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner MR, Nurrish SJ, Kaplan JM. Facilitation of synaptic transmission by EGL-30 Gqα and EGL-8 PLCβ: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron. 1999;24:335–346. doi: 10.1016/s0896-6273(00)80848-x. [DOI] [PubMed] [Google Scholar]
- Liu LX, Spoerke JM, Mulligan EL, Chen J, Reardon B, Westlund B, Sun L, Abel K, Armstrong B, Hardiman G, et al. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 1999;9:859–867. doi: 10.1101/gr.9.9.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel JE, Korswagen HC, Liu KS, Hajdu-Cronin YM, Simon MI, Plasterk RH, Sternberg PW. Participation of the protein Go in multiple aspects of behavior in C. elegans. Science. 1995;267:1652–1655. doi: 10.1126/science.7886455. [DOI] [PubMed] [Google Scholar]
- Miles RR, Sluka JP, Santerre RF, Hale LV, Bloem L, Boguslawski G, Thirunavukkarasu K, Hock JM, Onyia JE. Dynamic regulation of RGS2 in bone: Potential new insights into parathyroid hormone signaling mechanisms. Endocrinology. 2000;141:28–36. doi: 10.1210/endo.141.1.7229. [DOI] [PubMed] [Google Scholar]
- Miller KG, Emerson MD, Rand JB. Goα and diacylglycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron. 1999;24:323–333. doi: 10.1016/s0896-6273(00)80847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks TD, Leuther KK, Howard ED, Johston SA, Dougherty WG. Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Anal Biochem. 1994;216:413–417. doi: 10.1006/abio.1994.1060. [DOI] [PubMed] [Google Scholar]
- Pepperl DJ, Shah-Basu S, VanLeeuwen D, Granneman JG, MacKenzie RG. Regulation of RGS mRNAs by cAMP in PC12 cells. Biochem Biophys Res Commun. 1998;243:52–55. doi: 10.1006/bbrc.1997.8056. [DOI] [PubMed] [Google Scholar]
- Ségalat L, Elkes DA, Kaplan JM. Modulation of serotonin-controlled behaviors by Go in Caenorhabditis elegans. Science. 1995;267:1648–1651. doi: 10.1126/science.7886454. [DOI] [PubMed] [Google Scholar]
- Snow BE, Krumins AM, Brothers GM, Lee SF, Wall MA, Chung S, Mangion J, Arya S, Gilman AG, Siderovski DP. A G protein gamma subunit-like domain shared between RGS11 and other RGS proteins specifies binding to Gβ5 subunits. Proc Natl Acad Sci. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4-activated Giα1: Stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- Trent C. “Genetic and behavioral studies of the egg-laying system of Caenorhabditis elegans.” Ph.D. thesis. Cambridge, MA: Massachusetts Institute of Technology; 1982. [Google Scholar]
- Versele M, de Winde JH, Thevelein JM. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 1999;18:5577–5591. doi: 10.1093/emboj/18.20.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner LE, Zhou GT, Schafer RW, Schafer WR. Control of alternative behavioral states by serotonin in Caenorhabditis elegans. Neuron. 1998;21:203–214. doi: 10.1016/s0896-6273(00)80527-9. [DOI] [PubMed] [Google Scholar]
- Waggoner LE, Hardaker LA, Golik S, Schafer WR. Effect of a neuropeptide gene on behavioral states in Caenorhabditis elegans egg-laying. Genetics. 2000;154:1181–1192. doi: 10.1093/genetics/154.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshenker D, Garriga G, Thomas JH. Genetic and pharmacological analysis of neurotransmitters controlling egg laying in C. elegans. J Neurosci. 1995;15:6975–6985. doi: 10.1523/JNEUROSCI.15-10-06975.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Yu JH, Wieser J, Adams TH. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 1996;15:5184–5190. [PMC free article] [PubMed] [Google Scholar]