Abstract
The nonsense-mediated mRNA decay pathway is an example of an evolutionarily conserved surveillance pathway that rids the cell of transcripts that contain nonsense mutations. The product of the UPF1 gene is a necessary component of the putative surveillance complex that recognizes and degrades aberrant mRNAs. Recent results indicate that the Upf1p also enhances translation termination at a nonsense codon. The results presented here demonstrate that the yeast and human forms of the Upf1p interact with both eukaryotic translation termination factors eRF1 and eRF3. Consistent with Upf1p interacting with the eRFs, the Upf1p is found in the prion-like aggregates that contain eRF1 and eRF3 observed in yeast [PSI+] strains. These results suggest that interaction of the Upf1p with the peptidyl release factors may be a key event in the assembly of the putative surveillance complex that enhances translation termination, monitors whether termination has occurred prematurely, and promotes degradation of aberrant transcripts.
Keywords: mRNA decay, translation termination, release factors, nonsense mutation, ribosome, mRNA surveillance
Recent studies have demonstrated that cells have evolved elaborate mechanisms to rid themselves of aberrant proteins and transcripts that can dominantly interfere with their normal functioning (He et al. 1993; Pulak and Anderson 1993; for review, see Maquat 1995; Jacobson and Peltz 1996; Ruiz-Echevarria et al. 1996; Gottesman et al. 1997; Suzuki et al. 1997; Weng et al. 1997). Such pathways can be viewed both as regulators of gene expression and as sensors for inappropriate polypeptide synthesis. The nonsense-mediated mRNA decay (NMD) pathway is an example of a translation termination surveillance pathway, because it eliminates aberrant mRNAs that contain nonsense mutations within the protein coding region (He et al. 1993; Pulak and Anderson 1993; Maquat 1995; Caponigro and Parker 1996; Jacobson and Peltz 1996; Ruiz-Echevarria et al. 1996; Gottesman et al. 1997; Suzuki et al. 1997; Weng et al. 1997). The NMD pathway has been observed to function in all eukaryotic systems examined so far and appears to have evolved to ensure that termination of translation occurs at the appropriate codon within the transcript. Transcripts containing premature nonsense codons are rapidly degraded, thus preventing synthesis of incomplete and potentially deleterious proteins. There are well over 200 genetic disorders that can result from premature translation termination (McKusick 1994). Conceivably, understanding how this process affects translation termination and mRNA degradation can lead to rational approaches for the treatment of a subset of these disorders (Howard et al. 1996).
The proteins involved in promoting NMD have been investigated in Caenorhabditis elegans, mammalian cells, and in the yeast Saccharomyces cerevisiae. Three factors involved in NMD have been identified in yeast. Mutations in the UPF1, UPF2, and UPF3 genes were shown to selectively stabilize mRNAs containing early nonsense mutations without affecting the decay rate of most wild-type mRNAs (Leeds et al. 1991, 1992; Cui et al. 1995; He and Jacobson 1995; Lee and Culbertson 1995). Recent results indicate that the Upf1p, Upf2p, and Upf3p interact and form a complex (He and Jacobson 1995; Weng et al. 1996b; He et al. 1997). Therefore, we define a putative surveillance complex consisting of at least Upf1p, Upf2p, and Upf3p. In C. elegans, seven smg alleles have been identified that result in an increased abundance of nonsense-containing transcripts (Pulak and Anderson 1993). A human homolog of the UPF1 gene, called RENT1 or HUPF1, has been identified, indicating that NMD is an evolutionarily conserved pathway (Perlick et al. 1996; Applequist et al. 1997).
Although the cellular compartment in which NMD occurs in mammalian cells is controversial (Maquat 1995; Weng et al. 1997; Zhang and Maquat 1997), it appears that in yeast, however, NMD occurs in the cytoplasm when the transcript is associated with ribosomes. The following results support this conclusion. (1) Nonsense-containing and intron-containing RNAs that are substrates of the NMD pathway in yeast become polysome associated and are stabilized in the presence of the translation elongation inhibitor cycloheximide (Zhang and Maquat 1997). The polysome-associated RNAs, however, regain their normal rapid decay kinetics when the drug is washed out of the growth medium and translation resumes (Zhang and Maquat 1997). (2) Upf1p, Upf2p, and Upf3p have been shown to be associated with polysomes (Peltz et al. 1993b, 1994; Atkin et al. 1995, 1997). (3) As revealed by fluorescent in situ hybridization analysis, the cytoplasmic abundance of an intron-containing lacZ reporter RNA containing mutations in the 5′ splice site or branch point was dramatically reduced in UPF1+ strain but increased in cytoplasmic abundance in upf1Δ cells (Long et al. 1995). (4) NMD can be prevented by nonsense-suppressing tRNAs (Losson and Lacroute 1979; Gozalbo and Hohmann 1990; Belgrader et al. 1993). (5) The NMD pathway is functional only after at least one translation initiation/termination cycle has been completed (Ruiz-Echevarria and Peltz 1996; Zhang and Maquat 1997; Ruiz-Echevarria et al. 1998). Furthermore, a translation reinitiation event can prevent activation of the NMD pathway (Ruiz-Echevarria and Peltz 1996; Zhang and Maquat 1997; Ruiz-Echevarria et al. 1998). Taken together, these results indicate that the NMD pathway in yeast is a cytoplasmic and translation-dependent event. The observation that the rent1/hupf1 protein is predominantly cytoplasmic (Applequist et al. 1997) also strongly indicates a cytoplasmic function for this protein.
On the basis of studies from a number of laboratories investigating the sequences and factors involved in NMD, the following model for how NMD occurs has been proposed (for review, see Jacobson and Peltz 1996; Ruiz-Echevarria et al. 1996; Weng et al. 1997). Following transport of the mRNA from the nucleus to the cytoplasm, ribosomes begin translating the mRNA. The ribosome encounters a nonsense codon, which causes termination of translation. The decision whether translation termination has occurred prematurely is made by a putative surveillance complex, which consists of, at a minimum, the Upfps (Upf proteins). The surveillance complex scans 3′ of the termination codon and searches for a sequence element called the downstream element (DSE; Peltz et al. 1993a; Zhang et al. 1995; Ruiz-Echevarria and Peltz 1996; Ruiz-Echevarria et al. 1998). The DSE is a degenerate sequence that is present multiple times in virtually all mRNAs (Zhang et al. 1995). We hypothesize that the surveillance complex interacts with the DSE or associated factors and signals that premature termination has occurred. This interaction leads to rapid decapping and 5′→3′ decay of the aberrant mRNA.
The yeast UPF1 gene and its protein product have been the most extensively investigated factor of the putative surveillance complex (Altamura et al. 1992; Koonin 1992; Leeds et al. 1992; Atkin et al. 1995, 1997; Czaplinski et al. 1995; Cui et al. 1996; Weng et al. 1996a,b; 1998). The Upf1p contains a cysteine- and histidine-rich region near its amino terminus and all the motifs required to be a member of the superfamily group I helicases. The yeast Upf1p has been purified and demonstrates RNA-binding and RNA-dependent ATPase and RNA helicase activities (Czaplinski et al. 1995; Weng et al. 1996a,b). Disruption of the UPF1 gene results in stabilization of nonsense-containing mRNAs and suppression of certain nonsense alleles (Leeds et al. 1991; Cui et al. 1995; Czaplinski et al. 1995; Weng et al. 1996a,b).
In addition to its role in accelerating the decay of nonsense-containing transcripts, recent evidence suggests that the Upf1p enhances translation termination at a nonsense codon (Leeds et al. 1992; Weng et al. 1996a,b, 1998). Nonsense suppression results when a near cognate tRNA successfully competes with the termination factors at a nonsense mutation so that amino acid incorporation into the peptide chain occurs rather than prematurely terminating translation. Sufficient levels of nonsense suppression allow production of completed polypeptide protein which can support growth. The initial result suggesting that the Upf1p is involved in translation termination was the finding that a upf1Δ strain demonstrated a nonsense suppression phenotype (Leeds et al. 1992; Cui et al. 1995; Weng et al. 1996a,b). More significantly, a set of mutations in the UPF1 gene separated its mRNA decay function from its activity in modulating translation termination at a nonsense codon (Weng et al. 1996a,b). A subset of mutations in the helicase region of the UPF1 gene inactivate the decay activity of the Upf1p while still retaining its ability to enhance translation termination at a nonsense codon and prevent nonsense suppression (Weng et al. 1996a). Conversely, another subset of mutations in the cysteine- and histidine-rich amino-terminal region reduce the ability of the Upf1p to enhance translation termination at a nonsense codon, allowing suppression of certain nonsense alleles (Weng et al. 1996b). These alleles, however, do not affect the activity of the Upf1p in promoting decay of nonsense-containing mRNAs (Weng et al. 1996b). On the basis of these results, we hypothesized that the Upf1p is a factor that in addition to its role in promoting NMD, modulates the efficiency of translation termination at a nonsense codon.
As a consequence of the results described above, we hypothesize that the Upf1p enhances translation termination by interacting with the peptidyl release factors, eukaryotic release factor 1 (eRF1) and 3 (eRF3), to augment their activity. Both eRF1 and eRF3 are conserved proteins that interact and promote peptidyl release in eukaryotic cells (Frolova et al. 1994; Stansfield et al. 1995; Zhouravleva et al. 1995). In yeast, eRF1 and eRF3 are encoded by the SUP45 and SUP35 genes, respectively (Frolova et al. 1994; Zhouravleva et al. 1995). Sup45p and Sup35p have been shown to interact (Stansfield et al. 1995; Paushkin et al. 1997b). eRF1 contains intrinsic peptide hydrolysis activity, whereas eRF3, which has homology to the translation elongation factor EF1α (Didichenko et al. 1991), demonstrates GTPase activity (Frolova et al. 1996), and enhances the termination activity of eRF1 (Zhouravleva et al. 1995). The results presented in this report demonstrate a biochemical interaction between the human and yeast Upf1p and the peptidyl release factors eRF1 and eRF3. On the basis of these results, a model will be presented describing how the Upf1p, as part of a putative surveillance complex, assembles at the termination event, enhances translation termination, and degrades aberrant mRNAs.
Results
Upf1p interacts with the peptidyl release factors eRF1 and eRF3
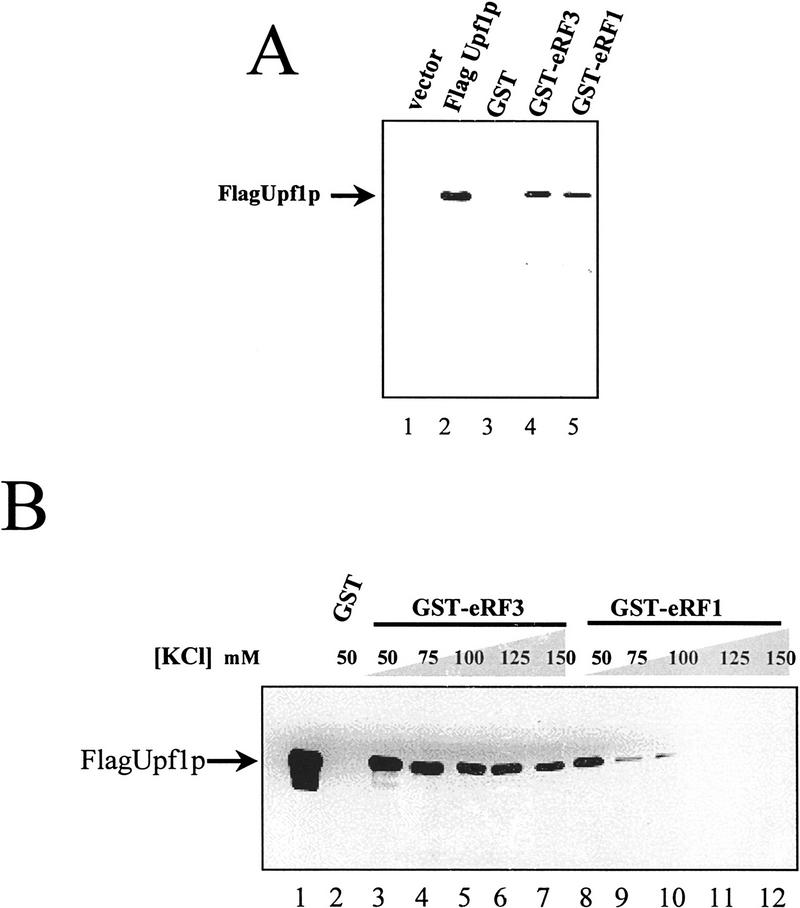
As described above, previous genetic results indicate that the Upf1p enhances translation termination at a nonsense codon (Weng et al. 1996a,b). We hypothesized that Upf1p modulates translation termination by interacting with the peptidyl release factors eRF1 and eRF3. To test this prediction, eRF1 and eRF3 were individually expressed in Escherichia coli as GST fusion proteins and purified by use of glutathione–Sepharose beads. The purified GST–RF (release factor) fusion proteins associated with the glutathione–Sepharose beads were added to a yeast cytoplasmic extract containing a Flag epitope-tagged Upf1p (Czaplinski et al. 1995; Weng et al. 1996a,b). Following incubation, the GST–RFs and associated proteins were purified by affinity chromatography and subjected to SDS-PAGE. Immunoblotting was performed and the presence of the Upf1p was assayed by use of an antibody against the Flag epitope. The anti-Flag antibody recognized only the 109-kD Upf1p in cytoplasmic extracts from cells transformed with plasmid expressing the Flag–Upf1p (Fig. 1A, cf. lanes 2 and 1). This analysis also demonstrated that the Upf1p specifically copurified with either eRF1 (Fig. 1A, lane 5) or eRF3 (Fig. 1A, lane 4). Upf1p did not copurify with GST protein that was not fused to another protein (Fig. 1A, lane 3) or a GST–JIP protein, in which a Jak2 interacting protein fused to GST was used to monitor the specificity of the reaction (data not shown).
Figure 1.
The yeast Upf1 protein interacts specifically with the peptidyl release factors. (A) GST–eRF1 or GST–eRF3 fusion proteins bind specifically to Upf1p in a yeast extract. Cytoplasmic extracts from a yeast strain BJ3505 transformed with either pG-1 (vector) or pG-1FLAGUPF1 (Flag-Upf1p) were prepared in IBTB and incubated with 30 μl of GST, GST–eRF1, or GST–eRF3 Sepharose–protein complexes. The Sepharose–protein complexes were washed twice in IBTB (see Materials and Methods), resuspended in SDS–polyacrylamide loading buffer, separated on an 8% SDS-polyacrylamide gel, and immunoblotted by use of anti-Flag antibody. (B) Upf1p interacts directly with both eRF1 and eRF3. Upf1p was purified as described previously (Czaplinski et al. 1995). Upf1p (200 ng) was added to 10 μl of GST, GST–eRF1, or GST–eRF3 Sepharose–protein complexes in a total reaction volume of 200 μl in IBTB supplemented with KCl to the final concentration indicated above each lane. After 1 hr at 4°C, Sepharose–protein complexes were washed for 3 min with 1 ml of IBTB supplemented with KCl to the final concentration indicated above each lane. The purified Sepharose–protein complexes were resuspended in SDS-polyacrylamide loading buffer and separated on a 7.5% SDS-polyacrylamide gel and immunoblotted as in A.
The interaction of purified Upf1p with either eRF1 or eRF3 was also monitored. The purification for epitope-tagged Upf1p (Flag–Upf1p) has been described previously (Czaplinski et al. 1995). Purified Flag–Upf1p was incubated with the GST–RF fusion proteins in the presence of increasing salt concentrations and the interactions of these proteins were monitored as described above. The results demonstrated that the purified Flag–Upf1p interacted with either eRF1 or eRF3 [Fig. 1B, lanes 8–12 (eRF1) and lanes 3–7 (eRF3)]. The Upf1p–eRF3 complex was less sensitive to increasing salt concentrations than the Upf1–eRF1 complex (Fig. 1B). The interactions were specific, because the purified Upf1p did not interact with the GST protein (Fig. 1B, lane 2) or GST–JIP (data not shown). Interaction of Upf1p with either eRF1 or eRF3 was shown to be dose dependent (data not shown).
The Upf1p is associated with the aggregates of eRF3 in [PSI+] strains
The biochemical results described above suggest that the Upf1p could enhance translation termination at a nonsense codon by interacting with the peptidyl release factors and enhancing their activity. We sought to confirm and extend this observation by asking whether the interaction of the Upf1p with the release factors can be observed in yeast cells. Recent results have shown that the nonsense suppressor phenotype observed in strains carrying the cytoplasmically-inherited determinant [PSI+] is a consequence of a specific alternative protein conformational state of the yeast eRF3 (Sup35p). In a [PSI+] state, eRF3 forms high-molecular-weight aggregates, or an amyloid-like fiber, which inhibit eRF3 activity, leading to increased readthrough of translation termination codons by ribosomes (Wickner 1994; Patino et al. 1996; Paushkin et al. 1996; Glover et al. 1997). It was also suggested that this specific alternative conformation of eRF3 is capable of self-propagation by an autocatalytic mechanism, analogous to that of mammalian prions (Wickner 1994; Glover et al. 1997; Paushkin et al. 1997a). Thus, the alternative protein conformational state of the eRF3, and not a mutation in the SUP35 gene, allows self-propagation of the [PSI+] phenotype. Yeast eRF1 (Sup45p) interacts with eRF3 and was also found in the aggregates present in [PSI+] cells (Paushkin et al. 1997b).
Because of the interaction of Upf1p with eRF1 and eRF3, we reasoned that Upf1p may be associated with the eRF3 aggregates in [PSI+] cells. To test this possibility, the presence of the Upf1p in the eRF3 and eRF1 aggregates found in [PSI+] cells was monitored. Previous results demonstrated that the eRF1/eRF3 aggregates sedimented through a sucrose pad in extracts prepared from [PSI+] cells. Cytoplasmic extracts from isogenic [psi−] and [PSI+] cells were prepared and centrifuged through a sucrose cushion and the presence of Upf1p, eRF1, and eRF3 was monitored in different fractions by Western blotting analysis. The results demonstrated that Upf1p, eRF1, and eRF3 were present in the pellet fraction in extracts from [PSI+] cells but were not found in the pellet fraction in a [psi−] extract (Fig. 2, cf. lanes 3 and 6). This result provides evidence that the Upf1p interacts with the translation termination factors in yeast cells.
Figure 2.
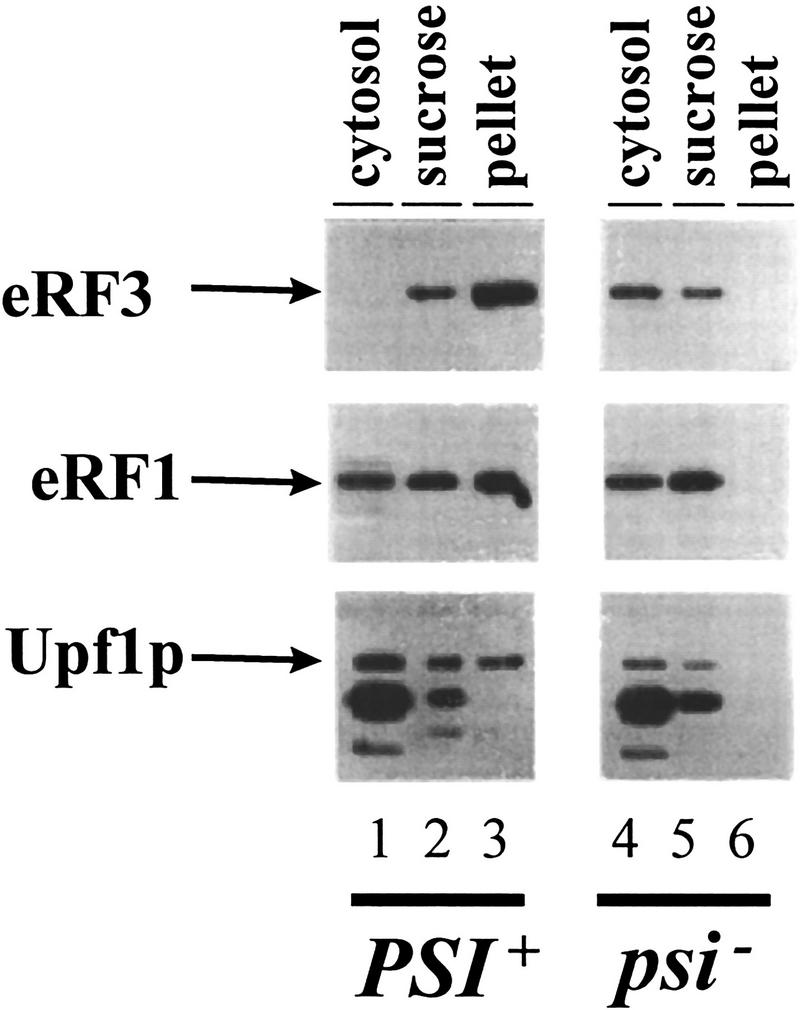
The Upf1p is associated with eRF3 [PSI+] aggregates. Cytoplasmic extracts from isogenic [PSI+] and [psi−] variants of strain 7G-H66 upf1Δ and containing FLAG–UPF1 inserted into a centromere plasmid were fractionated by centrifugation through a sucrose cushion as described previously (Paushkin et al. 1997b). Supernatant (cytosol), sucrose pad (sucrose), and pellet fractions were subjected to SDS–PAGE, and the distribution of eRF1, eRF3, and Upf1p within these fractions was determined by immunoblotting with polyclonal antibody against eRF1 and eRF3 and a monoclonal antibody against the Flag epitope. A 95-kD protein cross-reacts with anti-flag antibody in strain 7G-H66 and has the same distribution in [PSI+] and [psi−] cells. This 95-kD protein is not present in extracts prepared from strain BJ3505 (see Fig. 1).
eRF3 and RNA compete for interaction with Upf1p
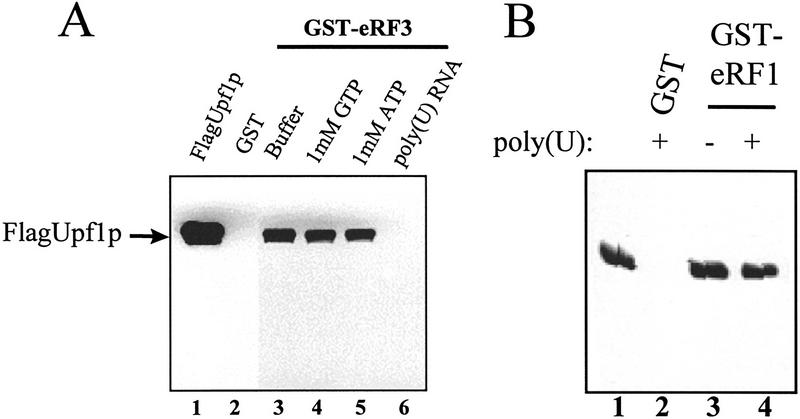
We also asked whether GTP, which binds to eRF3 (Frolova et al. 1996), or RNA, which binds to Upf1p, could affect the interaction between the release factors and the Upf1p. Reaction mixtures were prepared containing purified Flag–Upf1p and either purified GST–eRF1 or GST–eRF3 and containing either GTP or poly(U) RNA. Following incubation, the sepharose–GST–RF fusion complexes were washed with the same buffer containing either GTP or poly(U) RNA. The remaining bound proteins were subjected to SDS-PAGE followed by immunoblotting by use of an antibody against the Flag epitope. The results demonstrated that the interaction between Upf1p and eRF3 was not affected by GTP (Fig. 3A, cf. lanes 3 and 4; data not shown). A similar experiment showed that ATP did not affect the interaction between eRF3 and Upf1p (Fig. 3A, cf. lanes 3–5). Although poly(U) RNA did not affect the Upf1p–eRF1 interaction (Fig. 3B), the Upf1p–eRF3 interaction was dramatically reduced in reactions containing poly(U) RNA (Fig. 3A, cf. lanes 3–6).
Figure 3.
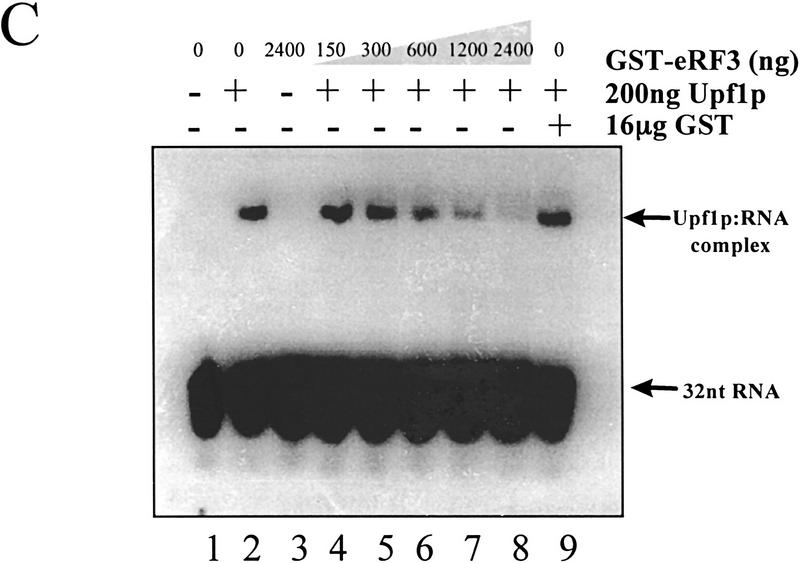
eRF3 and RNA compete for binding to Upf1p. (A) Poly(U) RNA prevents Upf1p from binding to eRF3. Reaction mixtures were prepared as described in Fig. 1B, except that binding was performed in TBSTB (TBST with 100 μg/ml BSA) and reaction mixtures contained 1 mm ATP, 1 mm GTP, or 100 μg/ml poly(U) RNA as indicated above each lane. The reaction mixtures were mixed for 1 hr at 4°C. Following mixing, the complexes were washed as in Fig. 1B with TBSTB containing 1 mm ATP, 1 mm GTP, or 100 μg/ml poly(U) RNA as indicated above each lane. (B) Poly(U) RNA does not prevent Upf1 and eRF1 interaction. Reaction mixtures were prepared as in Fig. 1B, in the presence or absence of 100 μg/ml poly(U) RNA as indicated above each lane. (C) eRF3 inhibits Upf1p RNA binding. A uniformly labeled 32-nucleotide RNA was synthesized by SP6 transcription of SstI-digested pGEM5Zf(+). The indicated amounts of GST–eRF3 were incubated with 200 ng of Upf1p for 15 min at 4°C. RNA substrate (50 fmoles) was added and incubated for 5 min. Stop solution was added, and reactions electrophoresed in a 4.5% native polyacrylamide gel (0.5× TBE, 30:0.5 acrylamide/bisacrylamide, with 5% glycerol).
The results described above indicated that RNA and eRF3 may be actually competing for binding to Upf1p. To test this hypothesis further, the effect of eRF3 on the ability of Upf1p to complex with RNA was monitored. Reaction mixtures containing Upf1p and RNA, and either lacking or containing increasing concentrations of eRF3, were prepared and the formation of the Upf1p:RNA complex was monitored by an RNA gel shift assay (Czaplinski et al. 1995; Weng et al. 1996a,b, 1998). Although Upf1p–RNA complexes formed in the absence of eRF3 (Fig. 3C, lane 2), increasing concentrations of eRF3 in the reaction mixtures reduced the amount of the Upf1p–RNA complex that formed (Fig. 3C, lanes 4–8). Inhibition was specific to eRF3, because the GST protein had no effect on Upf1–RNA complex formation (Fig. 3C, lane 9). eRF3–RNA complexes did not form (Fig. 3C, lane 3), indicating that the observed complexes were the result of binding to the Upf1p. Taken together, these results suggest that RNA and eRF3 bind competitively to Upf1p.
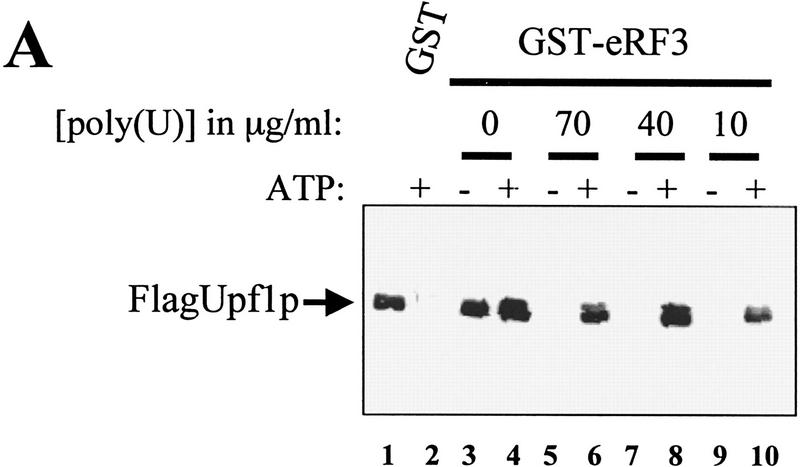
Previous results have shown that ATP bound to the Upf1p reduces its affinity for RNA (Weng et al. 1997a, 1998). This observation suggests that the ATP-bound form of Upf1p would favor interacting with eRF3 under conditions in which RNA and eRF3 are competing for interaction with Upf1p. This hypothesis was tested by incubating purified Flag–Upf1p with poly(U) RNA in the presence or absence of ATP. Following incubation, GST–eRF3 was added to the reaction mixtures and the Upf1–eRF3 interaction was monitored by immunoblotting analysis as before. The results demonstrated that when poly(U) and ATP were both present in the reaction mixture, the Upf1p interacted with eRF3 with the same affinity as in reactions lacking poly(U) RNA (Fig. 4A, lanes 6, 8, 10). Control experiments demonstrated that ATP did not prevent association of Upf1p with eRF3 (Fig. 4A, lane 4), and poly(U) RNA completely inhibited the interaction (Fig. 4A, lanes 5, 7, 9). These results are consistent with the notion that ATP binding to Upf1p functionally enhances interaction of Upf1 with eRF3, by preventing binding of competing RNAs.
Figure 4.
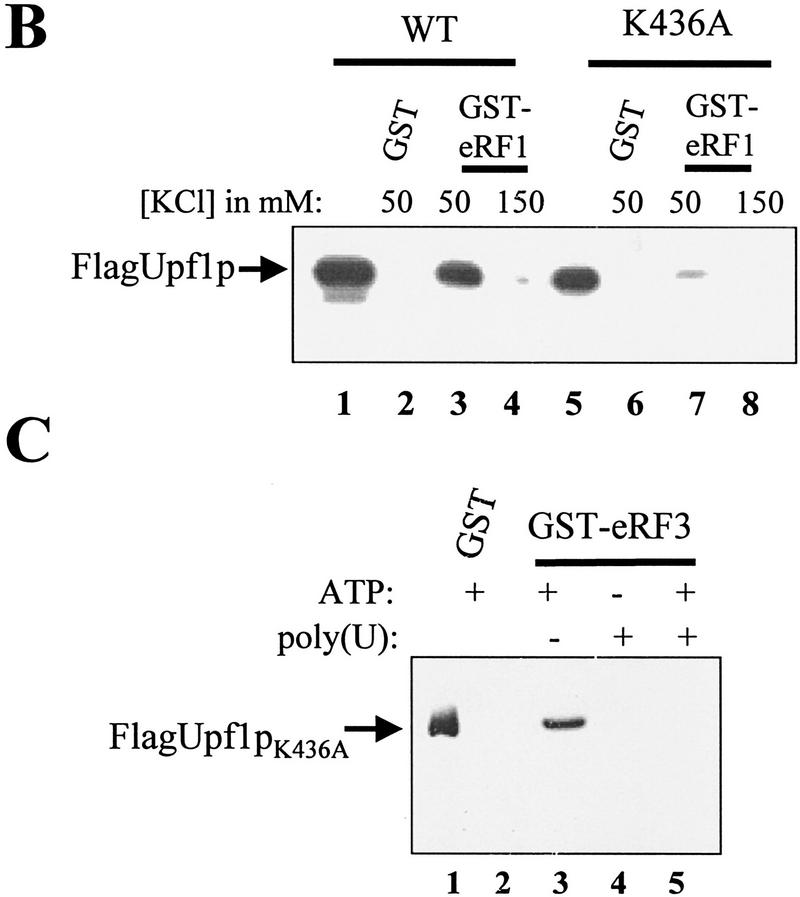
ATP prevents RNA from competing with eRF3 for binding to Upf1p. (A) Reaction mixtures were prepared as described in Fig. 3A, except that binding was performed in IBTB, and reaction mixtures contained 1 mm ATP or poly(U) RNA at the concentrations indicated above each lane. The reaction mixtures were mixed for 1 hr at 4°C. Following mixing, the complexes were washed with IBTB containing 1 mm ATP or poly(U) RNA at concentrations as indicated above each lane. (B) Upf1pK436A interacts weakly with eRF1. Reaction mixtures were prepared as in Fig. 1B, substituting Upf1pK436A for the wild-type protein (lanes 5–8). (C) A mutant Upf1p is unable to interact with eRF3 in the presence of RNA. Reaction mixtures were prepared as in A, substituting purified Upf1pK436A for the wild-type protein. Reactions contained 1 mm ATP or 40 μg/ml poly(U) RNA as indicated above each lane.
The K436A form of the Upf1p demonstrates altered interactions with the translation termination release factors
Next, we wanted to determine whether a mutation in the UPF1 gene that inactivated its mRNA turnover and translation termination activities affected the ability of the Upf1p to interact with the translation termination release factors. Previous results have shown that strains harboring mutations in the conserved lysine residue in position 436 of the Upf1p (K436) result in stabilization of nonsense containing mRNAs and a nonsense suppression phenotype (Weng et al. 1996a). By use of a purified K436A form of the Upf1p (Weng et al. 1996a, 1998), we asked whether this mutation affected the ability of the Upf1p to interact with the eRF1. Reaction mixtures containing the K436A form of Upf1p, GST–eRF1, and various KCl concentrations were prepared and their interaction was monitored as described above. The results demonstrated that the K436A mutation dramatically reduced the interaction of Upf1pK436A with eRF1 at least four- to sixfold relative to the interaction of wild-type Upf1 with eRF1 (Fig. 4B, cf. lanes 3 and 4 with lanes 7 and 8; data not shown).
We also monitored the ability of the K436A Upf1p to interact with eRF3. A reaction mixture containing the K436A Upf1p and GST–eRF3 was prepared and the Upf1p–eRF3 interaction was monitored as described above. The result demonstrated that the mutant form of Upf1p was capable of interacting with eRF3 with an equivalent affinity as the wild-type Upf1p (Fig. 4C, lane 3; data not shown).
Next, we determined whether the K436A mutation affected the ability of the Upf1p to preferentially interact with eRF3 versus RNA when ATP is present in the reaction mixture. The K436A mutation has been shown to reduce the affinity of the Upf1p for ATP (Weng et al. 1996a, 1998). However, although the K436A form of the Upf1p is still capable of binding RNA, unlike the wild-type Upf1p, ATP is unable to dissociate the RNA:Upf1pK436A complex (Weng et al. 1996a, 1998). Therefore, we monitored the ability of the Upf1pK436A to interact with eRF3 in the presence of ATP and RNA. Reaction mixtures containing the mutant Upf1p and either ATP, poly(U) RNA, or ATP and poly(U) RNA were prepared and interaction of the Upf1p with eRF3 was monitored as described above. The results demonstrated that, analogous to the wild-type Upf1p, poly(U) RNA prevented the interaction of Upf1pK436A with eRF3 (Fig. 4C, lane 4). However, unlike the wild-type Upf1p, ATP was unable to restore interaction of Upf1pK436A with eRF3 in the presence of poly(U) RNA (Fig. 4C, lane 5). This result indicates that the Upf1pK436A will not favor the Upf1p–eRF3 complex over the Upf1p-RNA complex when ATP is present in the reaction. Taken together, these results suggest that strains harboring the K436A upf1 allele, which no longer degrades aberrant mRNAs and displays a nonsense suppression phenotype, demonstrate altered interactions with the translation termination release factors. The altered Upf1pK436A:eRF interactions observed in the in vitro reactions correlate well with the in vivo mRNA decay and nonsense suppression phenotypes of this mutant upf1 allele.
eRF1 and eRF3 inhibit Upf1p ATPase activity
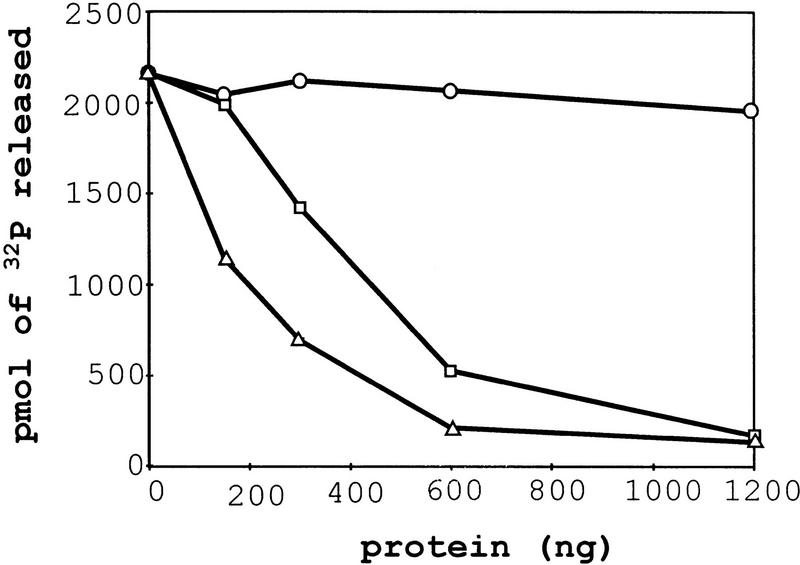
The genetic and biochemical data indicated that the ATPase/helicase activities were not required for enhancing translation termination, but were necessary to degrade nonsense-containing transcripts (Weng et al. 1996a,b, 1997). On the basis of these results, we predicted that interaction of the Upf1p with the eRFs would inhibit its ATPase/helicase activity, thus allowing the Upf1p to enhance translation termination. Therefore, we also examined whether the interaction of Upf1p with either eRF1 or eRF3 would affect the RNA-dependent ATPase activity of Upf1p. Reaction mixtures were prepared containing radiolabeled [γ32P]ATP and (1) Upf1p; (2) Upf1p and RNA; (3) Upf1p, RNA, and GST; (4) Upf1p, RNA, and GST–eRF1; or (5) Upf1p, RNA, and GST–eRF3. The ATPase activity in these reactions was monitored by use of a charcoal assay as described previously (Czaplinski et al. 1995; Weng et al. 1996a,b). The results demonstrated that reactions containing only Upf1p had no detectable ATPase activity, whereas reactions containing Upf1p and poly(U) RNA demonstrated maximal ATPase activity (data not shown). Addition of either eRF1 or eRF3 inhibited RNA-dependent ATPase activity of the Upf1p in a dose-dependent manner (Fig. 5; GST–eRF1 and GST-eRF3). Addition of the GST protein to the reaction mixtures had no effect on the RNA-dependent ATPase activity of the Upf1p (Fig. 5; GST). Neither eRF1 nor eRF3 demonstrated any intrinsic ATPase activity or stimulated the Upf1p ATPase activity in reactions lacking RNA (data not shown). The inhibition of the Upf1p ATPase activity by eRF1 was not simply a consequence of inhibiting its RNA-binding activity, because eRF1 does not inhibit this function of Upf1p (data not shown). Taken together, these results demonstrate that the ATPase activity of the Upf1p can be modulated by its interaction with the translation termination factors.
Figure 5.
eRF1 and eRF3 inhibit Upf1p RNA-dependent ATPase activity. Upf1p RNA-dependent ATPase activity was determined in the presence of GST–RF fusions by a charcoal assay with 1 μg/ml poly(U) RNA and 100 μg/ml BSA. The results are plotted as picomoles of 32P released vs. the amount of the indicated protein. (○) GST; (□) GST–eRF3; (▵) GST–eRF1.
The yeast/human UPF1 allele functions to modulate translation termination
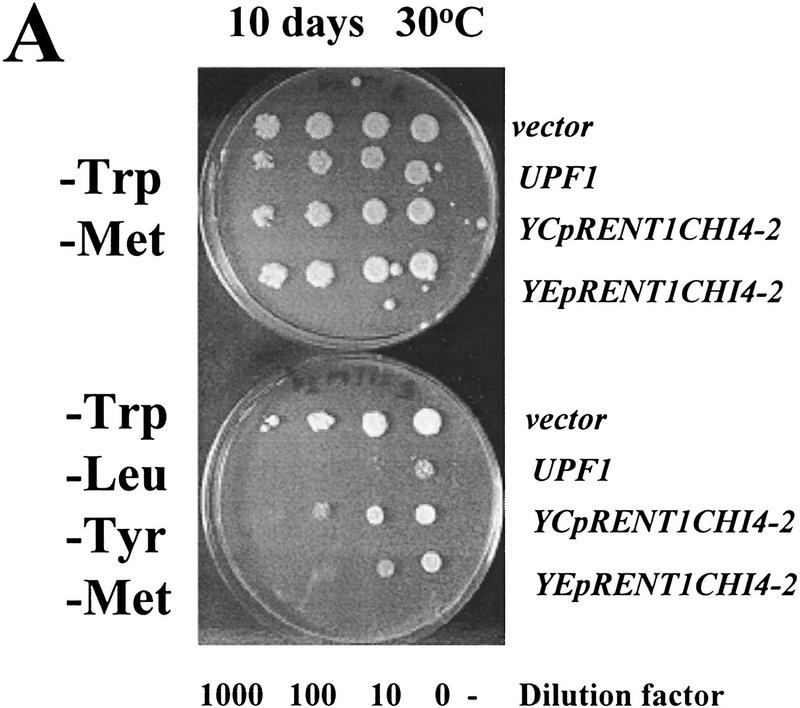
We wanted to determine whether the human homolog of the yeast Upf1p, called rent1 or hupf1, also modulated translation termination and mRNA turnover, suggesting a conserved role for this protein throughout evolution. We were unable to express the rent1/hupf1 in yeast cells (Perlick et al. 1996; data not shown). Therefore, we asked whether expression of a yeast/human UPF1 hybrid gene would prevent nonsense suppression in a upf1Δ strain and promote decay of aberrant transcripts. Although the amino- and carboxy-terminal ends of the human and yeast Upf1p are divergent, the rent1/hupf1 contains both the cysteine/histidine-rich region and helicase motifs found in the yeast UPF1 gene and displays 60% identity and 90% similarity over this region (Perlick et al. 1996; Applequist et al. 1997). The hybrid construct used in these experiments consisted of the conserved domains from the human protein sandwiched between the amino and carboxyl termini from the yeast UPF1 gene (Perlick et al. 1996). This hybrid gene was shown previously to complement a upf1Δ strain in a frameshift allosuppression assay (Perlick et al. 1996). We initially asked whether expression of the hybrid gene would function to prevent nonsense suppression. To test this possibility, a upf1Δ strain harboring leu2-2 and tyr7-1 nonsense alleles was transformed with plasmids harboring (1) the vector alone, (2) the wild-type yeast UPF1 gene, or (3) the yeast/human hybrid gene expressed from a MET25 promoter inserted into either a centromere (YCpRENT1CHI4-2) or a high copy plasmid (YEpRENT1CHI4-2). Methionine was omitted from the media to increase the expression of the hybrid gene (Perlick et al. 1996). Suppression of the leu2-2 and tyr7-1 nonsense alleles was monitored by plating cells on −Trp −Met −Leu −Tyr medium. As a control, these cells were plated on −Trp −Met medium. The results demonstrated that the upf1Δ cells harboring the vector grew on both types of medium (Fig. 6A), indicating suppression of these nonsense alleles. Cells harboring the yeast UPF1 gene were unable to grow on −Trp −Met −Leu −Tyr medium, demonstrating that the presence of the yeast UPF1 gene prevented suppression of these nonsense alleles (Fig. 6A). Similarly, expression of the hybrid yeast/human UPF1 gene prevented growth of these cells on −Trp −Met −Leu −Tyr medium, demonstrating the ability of this protein to substitute for the yeast Upf1p in preventing suppression of the leu2-2 and tyr7-1 alleles (Fig. 6A). The hybrid gene functioned better when expressed from a multicopy plasmid (Fig. 6A). The expression of the chimeric protein had no effect on normal cell growth, because cells harboring these plasmids grew as well as wild-type on the −Trp −Met medium (Fig. 6A).
Figure 6.
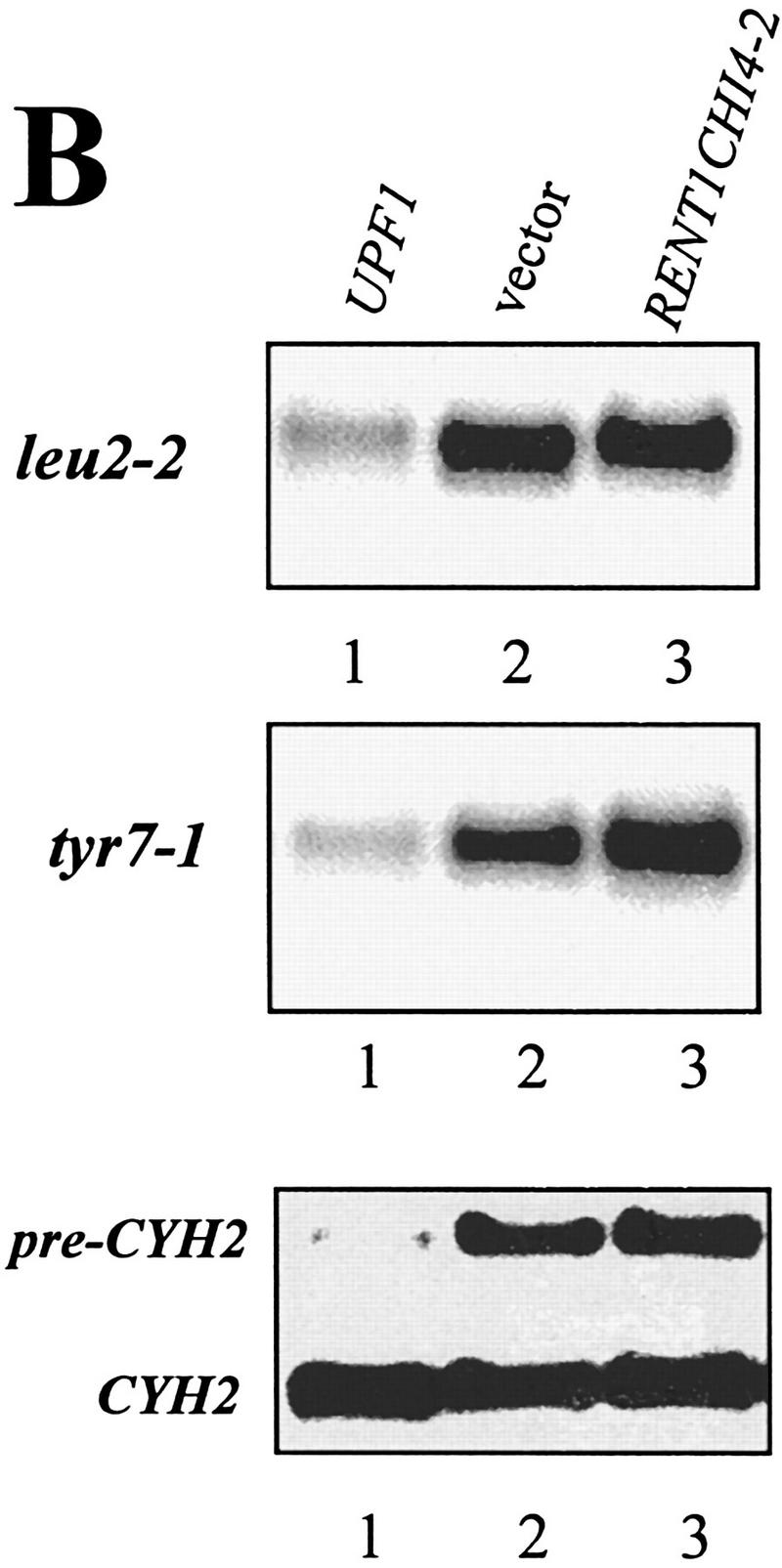
A RENT1/HUPF1 chimeric allele functions in translation termination. (A) A RENT1/HUPF1 chimeric allele prevents nonsense suppression in a upf1Δ strain. Strain PLY146 (MATα ura3-52 trp1Δ upf1::URA3 leu2-2 tyr7-1) was transformed with YCplac22 (vector), YCpUPF1 (UPF1), YCpRent1CHI4-2, or YEpRent1CHI4-2, and cells were grown to OD600 = 0.5 in -Trp -Met medium. Dilutions of 1/10, 1/100, and 1/1000 were prepared in -Trp -Met medium and 5 μl of these dilutions was plated simultaneously on -Trp -Met (top plate) or -Trp -Met -Leu -Tyr (bottom plate) media. Cells were monitored for growth at 30°C. (B) A RENT1/HUPF1 chimeric allele does not promote decay of nonsense containing mRNAs. Total RNA was isolated from cells at OD600 = 0.8 from the strains described in A. RNA (40 μg) from strains PLY146 transformed with YCplac22 (vector), YCpUPF1 (UPF1), or YEpRent1CHI4-2 (YEpRENT1CHI4-2)(10) was subjected to Northern blotting analysis and probed with either the LEU2, TYR7, or CYH2 probes.
Next, we asked whether the yeast/human UPF1 gene promotes decay of nonsense-containing transcripts in yeast cells. To test this, the abundance of the tyr7-1 and leu2-2 nonsense-containing transcripts was determined in a upf1Δ strain harboring either the vector plasmid, the yeast UPF1 gene, or the human/yeast hybrid UPF1 allele in a high copy plasmid. Total RNAs from these cells were isolated and the abundances of the tyr7 and leu2 transcripts were analyzed by RNA-blotting analysis, probing the blots with radiolabeled DNA probes encoding the TYR7 and LEU2 genes (Weng et al. 1996a,b). The results demonstrated that the leu2-2 and tyr7-1 mRNAs were low in abundance in a UPF1+ cell but were abundant in both a upf1Δ strain and a upf1Δ containing the yeast/human hybrid allele (Fig. 6B). Similarly, the CYH2 precursor, which is an endogenous substrate for NMD (He et al. 1993), was abundant in cells expressing the yeast/human hybrid allele, whereas the CYH2 mRNA levels were similar in all three strains (Fig. 6B). Taken together, these results indicated that the product of the yeast/human UPF1 hybrid gene functions in translation termination, but does not activate the NMD pathway in yeast cells.
The human Upf1p interacts with the peptidyl release factors eRF1 and eRF3
The results described above suggest that the human homolog of the UPF1 gene may also function in modulating the translation termination activity of the peptidyl release factors. Therefore, we asked whether the full-length rent1/hupf1 would interact with eRF1 and eRF3. To test this possibility, radiolabeled rent1/hupf1 protein was synthesized in a coupled in vitro transcription/translation system. In vitro synthesis of the rent1/hupf1 produced a band of ∼130 kD (Fig. 7, lane 1), consistent with the reported size of rent1/hupf1 (Applequist et al. 1997). The luciferase protein was also synthesized as described above and was used as a control protein for specificity of the interaction. Synthesis of the luciferase protein produced a 68-kD protein (Fig. 7, lane 5). The rent1/hupf1 or the luciferase protein was incubated with either GST, GST–eRF1, or GST–eRF3 as described above and the interactions of rent1/hupf1 or luciferase with these proteins were monitored by SDS-PAGE followed by autoradiography. The results demonstrated that the rent1/hupf1 interacted with both the GST–eRF1 or GST–eRF3 (Fig. 7, lanes 3 and 4). The interaction was specific, because rent1/hupf1 did not form a complex with GST protein (Fig. 7, lane 2). Further, the in vitro synthesized luciferase protein did not interact with GST, GST–eRF1, or GST–eRF3 (Fig. 7, lanes 6–8). Furthermore, poly(U) RNA prevented the interaction of hupf1/rent1 with eRF3 (data not shown). Taken together, these results indicate that the rent1/hupf1 also interacts with the peptidyl release factors eRF1 and eRF3, suggesting that the role of the Upf1p in the surveillance complex in modulating translation termination is conserved throughout evolution.
Figure 7.
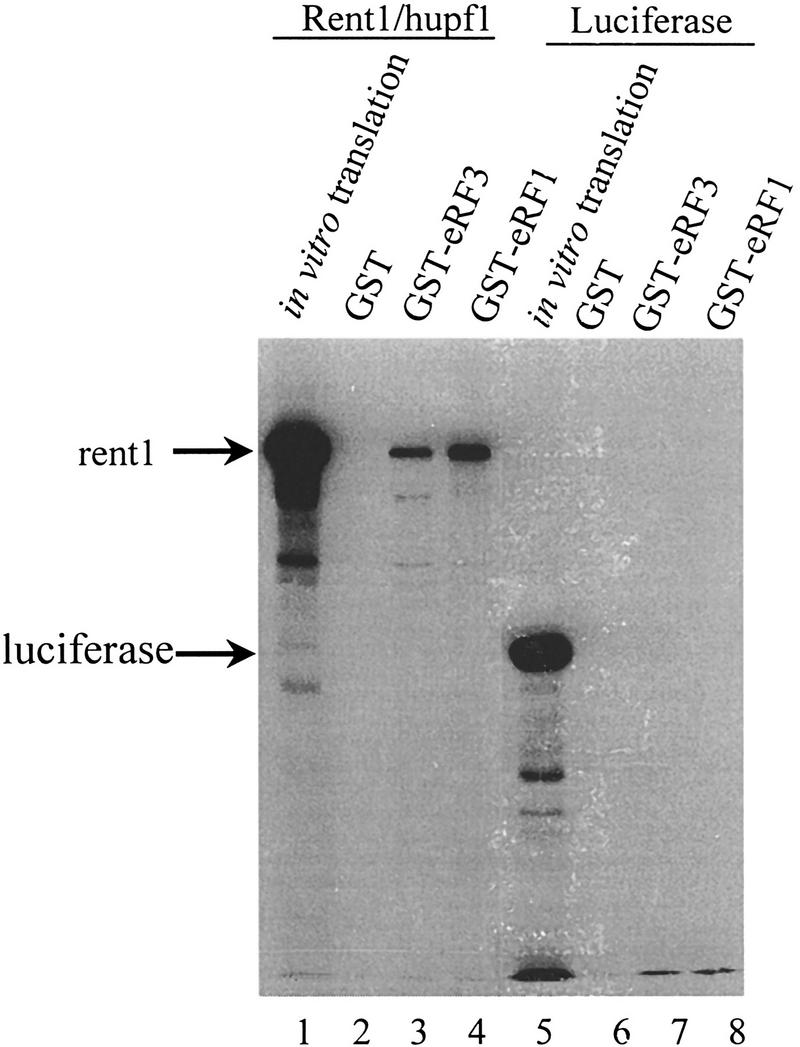
Rent1/hupf1 interacts with eRF1 and eRF3. NotI linearized pT7RENT1 (lanes 1–4) or luciferase template (lanes 5–8) was used in the TNT-coupled reticulocyte in vitro transcription translation as per manufacturer’s directions (Promega). Completed translation reactions (2 μl) were electrophoresed in lanes 1 and 5. Completed reactions (5 μl) were incubated in 200 μl of IBTB with 10 μl of GST, GST–eRF1, or GST–eRF3 Sepharose–protein complexes as indicated above each lane. Following mixing for 1 hr at 4°C, the Sepharose–protein complexes were washed as in Fig. 1A, and the bound proteins were subjected to SDS-PAGE in an 8% gel. Following electrophoresis, gels were fixed for 30 min in 50% methanol, 10% acetic acid, and then treated with 1 m salicylic acid for 1 hr. Gels were dried and subjected to autoradiography.
Discussion
Previous results indicated that the Upf1p is a multifunctional protein involved in enhancing translation termination at nonsense codons and in promoting decay of nonsense-containing transcripts (Weng et al. 1996a,b, 1998). The results presented here begin to elucidate how the Upf1p functions in enhancing translation termination. We demonstrated that both the yeast and human forms of the Upf1p probably affect translation termination by interacting with the peptidyl release factors eRF1 and eRF3 and modulating their activity (Fig. 1). These results were substantiated by demonstrating that the Upf1p was also observed as part of the peptidyl release factor aggregates, or fibers, observed in [PSI+] yeast cells, and a mutant form of Upf1 has altered interactions with the release factors. These observations will be discussed below and, on the basis of these observations, a model for how the surveillance complex assembles at a termination codon to enhance translation termination and promote NMD will be described.
The interaction of the Upf1p with the peptidyl release factors suggests that the Upf1p enhances the activity of these factors
Previous genetic data suggested that the Upf1p modulates the translation termination activity in a cell (Weng et al. 1996a,b, 1998). Consistent with this notion, we have demonstrated that the Upf1p interacted with eRF1 and eRF3, the two major components known to be involved in translation termination (Stansfield et al. 1995; Zhouravleva et al. 1995). Furthermore, the finding that the Upf1p is also associated with the eRF3 aggregates found in [PSI+] cells is consistent with this protein interacting with the translation termination release factors in vivo (Fig. 2). This result suggests that a portion of the Upf1p that is normally utilized by the cell to enhance translation termination is depleted from the cellular pool in yeast [PSI+] cells. At present, the effect of removing this portion of the Upf1p on NMD is not known. It will be interesting to determine whether under- or overexpression of the Upf1p, or any of the other Upfps, affects the efficiency of the formation of the [PSI+] phenotype. In any event, the results presented here identify Upf1p as a component of the [PSI+] complexes, suggesting that it may play a role in aggregate formation or maintenance.
The precise mechanism of how eRF1 and eRF3 promote termination when the A site of the ribosome is occupied by a termination codon has not been fully elucidated (for review, see Buckingham et al. 1997). One suggestion is that eRF1 may structurally mimic a stem of a tRNA, whereas eRF3 may mimic the function of EF-1α (Didichenko et al. 1991). The interaction of these two proteins at the ribosomal A site promotes cleavage of the peptide associated with the tRNA in the P site (Zhouravleva et al. 1995). There are several steps in the termination process in which interaction of the release factors with Upf1p could be envisioned to enhance its translation termination efficiency. These include (1) increasing the efficiency in which the eRFs compete with near cognate tRNAs and productively interact with the ribosome to promote termination, (2) the efficiency of the eRFs to promote peptidyl hydrolysis, or (3) increasing the recycling of the eRFs so that there is a larger free pool of these factors that can promote termination. Future experiments will be required and assays need to be developed to determine how the Upf1p functions to enhance translation.
The role of the Upf1p in enhancing translation termination may be conserved throughout evolution
The human homolog of the yeast UPF1 gene has been recently isolated (Perlick et al. 1996; Applequist et al. 1997). Although the human gene contained amino- and carboxy-terminal domains that were not present in the yeast UPF1 gene, the human gene contained the cysteine/histidine-rich region and the helicase motifs found in the yeast homolog (Perlick et al. 1996; Applequist et al. 1997). Further, expression of a yeast/human hybrid of the UPF1 genes functioned in a frameshift suppression assay when expressed in a upf1Δ strain (Perlick et al. 1996). The results presented here demonstrate that, analogous to the Upf1p, expression of the yeast/human UPF1 allele prevented the nonsense suppression phenotype observed in a upf1Δ strain harboring the nonsense-containing leu2-2 and tyr7-1 alleles (Fig. 6). Although the yeast/human hybrid was able to complement the translation termination phenotype of the yeast Upf1p, it did not promote rapid decay of nonsense-containing mRNAs (Fig. 6). Furthermore, consistent with a role in translation termination, the human rent1/hupf1 protein also interacted with the translation termination factors eRF1 and eRF3 (Fig. 6). These results, as well as the predominantly cytoplasmic localization of both the yeast Upf1p and rent1/hupf1 (for review, see Jacobson and Peltz 1996; Applequist et al. 1997), are consistent with a role of this protein in modulating translation termination. Taken together, these results suggest that the role of the Upf1p in translation termination is likely to be conserved throughout evolution.
Interaction with the release factors modulates the biochemical activities of the Upf1p
Our results demonstrate that interaction of Upf1p with the release factors inhibited its ATPase activity and prevented Upf1p from binding to RNA (Figs. 3 and 5). These results are consistent with our previous biochemical and genetic results demonstrating that the Upf1p ATPase/helicase and RNA-binding activities were required to promote NMD, but were dispensable for its translation termination activity (Weng et al. 1996a,b, 1998). We have also shown that RNA and eRF3 compete for binding to Upf1p (Fig. 3). This result suggests that factors that reduce the Upf1p affinity for RNA would consequently favor binding to the release factors. We have demonstrated previously that binding of ATP to Upf1p reduces its affinity for RNA (Weng et al. 1996a, 1998). The results shown here demonstrated that ATP causes Upf1 to favor interaction with eRF3 over RNA (Figs. 4C and 8A). On the basis of these results, we hypothesize that ATP is a cofactor of the Upf1p that allows it to switch between its translation termination and NMD activities. The results from our genetic and biochemical analysis of the Upf1p are consistent with this hypothesis (Weng et al. 1996a,b, 1998). For example, a mutant form of the Upf1p that lacked ATPase activity, but still bound ATP, was still functional in preventing translation termination (Weng et al. 1996a, 1998). Significantly, the binding of ATP to this mutant form of the Upf1p still modulated its RNA-binding affinity (Weng et al. 1998). Furthermore, a mutant Upf1pK436A, whose RNA-binding activity could not be modulated by ATP, did not function in enhancing translation termination at a nonsense codon (Weng et al. 1996a, 1998). This Upf1pK436A also demonstrated a dramatically reduced interaction with eRF1 (Fig. 4B), and did not interact with eRF3 in the presence of RNA and ATP (Fig. 4C).
Figure 8.
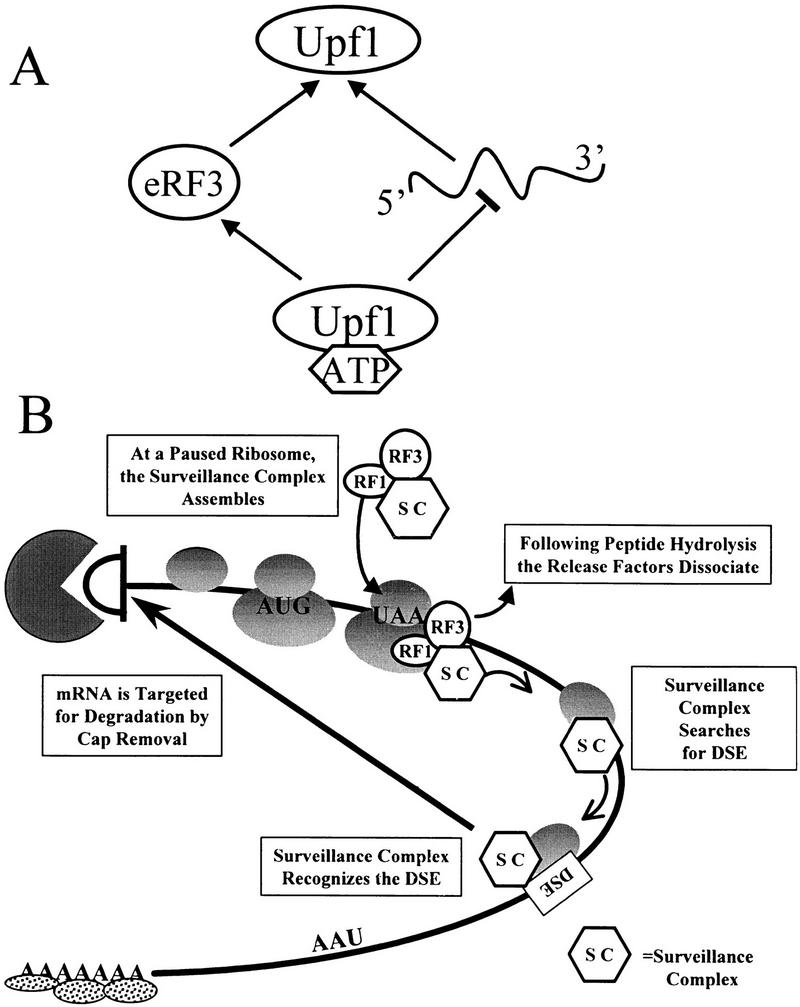
Model for Upf1 function in mRNA surveillance. (A) Modulation of RNA binding enhances interaction of Upf1 with peptidyl release factors. ATP binding to Upf1p decreases the affinity of Upf1 for RNA. Because RNA and eRF3 compete for binding to Upf1, interaction with eRF3 is favored. (B) A model for mRNA surveillance. Interaction of Upf1p with peptidyl release factors assembles an mRNA surveillance complex at a termination event. This interaction prevents Upf1 from binding RNA and hydrolyzing ATP, and enhances translation termination. Following peptide hydrolysis, the release factors dissociate from the ribosome, activating the Upf1p helicase activity. The surveillance complex then scans 3′ of the termination codon for a DSE. Interaction of the surveillance complex with the DSE signals that premature translation termination has occurred and the mRNA is then decapped and degraded by the Dcp1p and Xrn1p exoribonuclease, respectively.
A model for how the complex that promotes NMD assembles and determines whether aberrant termination has occurred
On the basis of the results described here, we suggest the following model for how the NMD pathway functions to enhance translation termination and subsequently recognize and degrade a nonsense-containing transcript (Fig. 8B). A termination codon in the A site of a translating ribosome causes the ribosome to pause (step 1). The translation termination factors eRF1 and eRF3 interact at the A site and promote assembly of the surveillance complex by interacting with Upf1p, which is most likely complexed with other factors (step 2). The interaction of Upf1p with the release factors inhibits its ATPase and RNA-binding activities. This inhibition may be necessary for the Upf1p to enhance the activity of the termination factors and ensure that the Upfp complex does not prematurely disassociate from release factors and search for a DSE. Peptide hydrolysis occurs while the release factors are associated with the surveillance complex. Following GTP hydrolysis by eRF3 and completion of termination, the eRFs disassociate from the ribosome (step 3). We hypothesize that disassociation of the release factors activates the RNA-binding and ATPase activities of the Upf1p and triggers the Upfp complex to scan 3′ of the termination codon in search of a DSE (step 4). If the complex becomes associated with the DSE or DSE-associated factors, an RNP complex forms such that the RNA is a substrate for rapid decapping by Dcp1p (step 5; Muhlrad and Parker 1994). We hypothesize that the RNP complex that forms as a consequence of the surveillance complex interacting with the DSE prevents the normal interaction between the 3′ poly(A)–PABP complex and the 5′ cap structure (Jacobson 1996; Tarun et al. 1997). The uncapped mRNA is subsequently degraded by the Xrn1p exoribonuclease (step 6; Muhlrad and Parker 1994; Hagan et al. 1995).
An important point that this model suggests is that the interaction of the peptidyl release factors with a ribosome paused at a termination codon is a key event in the assembly of the complex that monitors translation termination. Because the concentration of either release factors or Upfps in cells is less than the concentration of ribosomes (Didichenko et al. 1991; Stansfield et al. 1992; Atkin et al. 1997), not every ribosome will be associated with a surveillance complex. Thus, it is conceivable that a paused ribosome signals assembly of this complex. Interaction of the release factors with the Upf1p enhances translation termination and also ensures that the RNA-dependent ATPase and/or helicase activities of Upf1p function at the appropriate time in this process. The goal of future experiments will be to determine how the Upf1p enhances translation termination and to identify other factors that are part of this surveillance complex and determine how they function to modulate translation termination and degrade aberrant mRNAs.
On the basis of the model described above, we speculate that the termination event is a key point in the assembly of the surveillance complex and leads to enhanced translation termination and degradation of nonsense-containing transcripts. Further, we hypothesize that translation termination may also be an important event in regulating the stability or translation efficiency of wild-type transcripts. The 3′-untranslated regions of many transcripts encode regulatory elements that modulate the translation efficiency and/or stability of their respective mRNAs (for review, see Beelman and Parker 1995; Ross 1995; Jacobson 1996; Jacobson and Peltz 1996; Wickens et al. 1997). It is conceivable that the termination event is also the cue for the assembly of complexes that subsequently interact with the elements in the 3′ UTR that modulate their stability and/or translation efficiency. Interestingly, one subunit of the protein phosphatase 2A (PP2A) is the translation termination factor eRF1 (Andjelkovic et al. 1996). It is possible that one role of eRF1 is to bring the PP2A phosphatase into the ribosome at the termination event. The PP2A may be then positioned in the appropriate location to modulate the activity of factors that regulate the translation efficiency or stability of the given transcripts. Interestingly, this scenario is very similar to how we perceive the NMD pathway function. The basic premise for both wild type and NMD is that termination is a rate-limiting event that pauses the ribosome and signals the assembly of complexes that regulate subsequent events in the life span of a given transcript. Interestingly, although the role of PP2A in translation has not been investigated, mutations in the SAL6 gene that encodes a phosphatase have been shown to promote suppression of nonsense mutations (Vincent et al. 1994). Clearly, further experimentation is required to test this hypothesis.
Materials and methods
General yeast methods
Yeast media was prepared as described (Rose et al. 1990). Yeast transformations were performed by the lithium acetate method (Scheistl and Geitz 1989). RNA isolation, blotting, and hybridization was as described (Hagan et al. 1995; Weng et al. 1996a).
Plasmids
Plasmid YCp and YEp RENTCHI4-2 were created by ligating a 4.5-kb SstI–Asp718 fragment from pMET25CHIMERA (Perlick et al. 1996) harboring the chimeric gene under the MET25 promoter into YCplac22 and YEplac112 (Ferguson et al. 1981), respectively. YCpFLAGUPF1 and YEpFLAGUPF1 were described previously (Weng et al. 1996a). GST–RF fusion plasmids, pGEX2T, pGEX2T–SUP35, and pGEX2T–SUP45 were described previously (Paushkin et al. 1997b).
Preparation of glutathione Sepharose–RF fusion complexes
Strain BL21(DE3) pLysS transformed with pGEX2T, pGEX2T–SUP35, or pGEX2T–SUP45 (Paushkin et al. 1997b) were grown at 24°C in LB with 50 μg/ml ampicillin and 30 μg/ml chloramphenicol to OD600 = 0.6. IPTG (0.3 mm) was added and cells grown overnight. Cells were collected and washed once with cold TBST (50 mm Tris at pH 7.4, 150 mm NaCl, 0.1% Triton X-100) with 0.5 mm PMSF. Cells were resuspended in 50 μl of TBST with 0.5 mm PMSF per milliliter of culture and lysed by sonication. Triton X-100 was added to a final concentration of 1% and lysates mixed for 20 min at 4°C. Cell debris was removed by centrifugation at 30,000g for 30 min. Eighty microliters of a 50% slurry of glutathione–Sepharose (Pharmacia) equilibrated in TBST was added per milliter of extract and incubated at 4°C with mixing for 30 min. Sepharose beads were collected at 500g for 3 min, washed for 3 min with TBST supplemented with NaCl to 500 mm, and collected as before for a total of two times. The Sepharose–protein complexes were then washed and collected as before with IBTB (25 mm Tris-HCl at pH 7.5, 50 mm KCl, 10 mm MgCl2, 2% glycerol, 0.1% Triton X-100, 100 μg/ml BSA) for a total of two times, and resuspended in IBTB to yield a 2:1 ratio of buffer to packed bead volume. One microliter of GST–RF complexes typically contained 0.9 μg of GST–eRF1 or 1.5 μg of GST–eRF3, whereas GST complexes typically contained 4.5 μg of GST per microliter of resin.
Preparation of cytoplasmic extracts
BJ3505 (MATα pep4::HIS3 prb- Δ1.6R HIS3 lys2-208 trp1- Δ10 ura3-52 gal2 can1) cells were grown to an OD600 = 1.0 and washed in 5 ml of cold buffer IB (IBTB lacking BSA) with 0.5 mm PMSF. Cells were repelleted and suspended in 1.3 ml of cold IB with 0.5 mm PMSF and protease inhibitors (PI, 1 μg/ml each leupeptin, aprotinin, and pepstatin A) per gram of cell weight. An approximately equal volume of glass beads was added and lysis was achieved by vortexing six times for 20 sec, with 1 min cooling on ice in between vortexing. The lysate was removed, and the beads washed two times with an equal volume of IB with 0.5 mm PMSF and 1 μg/ml each leupeptin, aprotinin, and pepstatin A. The washes were combined with the lysate and the cell debris was removed by centrifugation at 30,000g for 20 min.
Preparation of [PSI+] upf1Δ strains
UPF1 was deleted from [PSI+] strain 7G-H66 (MATa ade2-1 SUQ5 trp1-289 leu2-3,112 ura3-52 [PSI+]) as described (Cui et al. 1995). The deletion was confirmed by Southern blot analysis (data not shown). To cure the [PSI+] determinant, 7G-H66 upf1Δ was grown in medium containing 3 mm GuHCl (Ter-Avanesyan et al. 1994). Disruption of UPF1 resulted in suppression of ade2-1, which is used to monitor the suppressor phenotype of [PSI+], therefore the [psi−] status of clones obtained after growth on GuHCl medium was identified in crosses with the 1A-H19 [psi−] tester strain (MATα ade2-1 lys2-1 his3-11,15 leu2-3,112 SUQ5 [psi−]) (Ter-Avanesyan et al. 1994). The suppressor phenotype of the upf1Δ allele is a recessive trait, whereas the [PSI+] determinant is dominant. Therefore, the nonsuppressor phenotype of the diploids indicated [psi−] state of the clones. The [PSI+] and [psi−] isolates of strain 7G-H66 upf1Δ were then transformed with the centromeric-based plasmid YCplac22FLAGUPF1 (Weng et al. 1996a,b).
Preparation of lysates for [PSI+] aggregate cocentrifugation
7G-H66 upf1Δ cells transformed with YCplac22 or YCpFLAGUPF1 were grown in medium lacking tryptophan to OD600 = 1.5, washed in water, and lysed by mixing with glass beads in buffer A (25 mm Tris-HCl at pH 7.5, 50 mm KCl, 10 mm MgCl2, 1 mm EDTA, 2% glycerol) containing 1 mm PMSF and PI (2 μg/ml aprotinin, 1 μg/ml pepstatin A, 0.5 μg/ml leupeptin, 2.5 μg/ml anti-pain, 0.5 μg/ml TLCK, 0.5 μg/ml TPCK, 0.1 mm benzamidine, 0.1 mm sodium metabisulfite). Lysates were centrifuged at 15,000g for 20 min, then treated with RNase A (400 μg/ml) to disrupt polyribosomes. Extracts were then subjected to centrifugation through a sucrose cushion as described previously (Paushkin et al. 1997b). Ribosomes migrate primarily to the sucrose fraction and because eRF1, eRF3, and Upf1p are all ribosome associated, they are present in this fraction in [psi−] extracts.
Preparation of purified GST–RF fusion proteins
Extracts from 400-ml cultures of strain BL21(DE3) pLysS transformed with pGEX2T, pGEX2T–SUP35, or pGEX2T–SUP45 were prepared as described above for preparation of GST–RF fusion complexes. Eight hundred microliters of a 50% slurry of glutathione–Sepharose was added and incubated with mixing for 30 min. Sepharose beads were collected and washed twice for 3 min with TBST supplemented with NaCl to 500 mm, and collected by centrifugation at 500g for 3 min. The Sepharose beads were then washed in TBST and collected for a total of two times. GST fusion proteins were eluted by resuspending the washed Sepharose beads in 400 μl glutathione elution buffer (50 mm Tris-HCl at pH 8.0, 10 mm glutathione) and incubating at room temperature for 10 min with mixing. Sepharose beads were collected and the supernatant removed. Elution was repeated as before for a total of three times, and the elution fractions combined. Concentration of proteins was determined by the Bradford assay.
Immunodetection of Upf1, eRF1, and eRF3
Upf1 was detected by use of the M2 mouse monoclonal antibody against the Flag epitope as described previously (Czaplinski et al. 1995; Weng et al. 1996a,b). eRF3 was detected as described in Didichenko et al. (1991). eRF1 was detected as described in Stansfield et al. (1992).
ATPase assays
Upf1p RNA-dependent ATPase activity was determined by use of 20 ng Upf1p in the presence of GST–RF fusion proteins by a charcoal assay as described previously (Czaplinski et al. 1995) with 1 μg/ml poly(U) RNA and 100 μg/ml BSA. The results are plotted as picomoles of 32P released versus the concentration of the indicated protein.
RNA-binding assay
A uniformly labeled 32-nucleotide RNA was synthesized by SP6 transcription of SstI digested pGEM5Zf(+) as described previously (Czaplinski et al. 1995). RNA-binding buffer was as described previously (Czaplinski et al. 1995), with the exception that 100 μg/ml BSA was included in all reactions. The indicated amounts of GST–eRF3 (28) were incubated with 200 ng of Upf1p for 15 min at 4°C. Fifty femtomoles of the RNA substrate was added and incubated for 5 min. Stop solution was added, and reactions electrophoresed in a 4.5% native polyacrylamide gel (0.5× TBE, 30:0.5 acrylamide/bisacrylamide, with 5% glycerol).
Acknowledgments
We thank M.F. Tuite for the gift of anti-eRF1 antibody and I.A. Valouev for performing the knockout of the UPF1 gene in the strain 7G-H66. This work was partially supported by the Russian Foundation for Basic Research to M.D.T-A. and S.V.P.; and was supported by grants from the National Institutes of Health (GM48631-01) and an Established Investigator Award given to S.W.P. M.J.R-E. and K.C. acknowledge the New Jersey American Heart Association for their support. We thank Carlos Gonzalez, Thomas Thisted, Terri Kinzy, and Jon Dinman for critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Peltz@RWJA.UMDNJ.EDU; FAX (732) 235-5223.
References
- Altamura N, Groudinsky O, Dujardin G, Slonimski PP. NAM7 nuclear gene encodes a novel member of a family of helicases with a Z1-6n-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol. 1992;224:575–587. doi: 10.1016/0022-2836(92)90545-u. [DOI] [PubMed] [Google Scholar]
- Andjelkovic N, Zolnierowicz S, Van Hoof C, Goris J, Hemmings BA. The catalytic subunit of protein phosphatase 2A associates with the translation termination factor eRF1. EMBO J. 1996;15:7156–7167. [PMC free article] [PubMed] [Google Scholar]
- Applequist SE, Selg M, Roman C, Jack HM. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin AL, Altamura N, Leeds P, Culbertson MR. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin AL, Schenkman LR, Eastham M, Dahlseid JN, Lelivelt MJ, Culbertson MR. Relationship between yeast polyribosomes and Upf proteins required for nonsense mediated mRNA decay. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- Beelman CA, Parker R. mRNA degradation in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- Belgrader P, Cheng J, Maquat LE. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci. 1993;90:482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham R, Grentzmann G, Kisselev L. Polypeptide chain release factors. Mol Microbiol. 1997;24:449–456. doi: 10.1046/j.1365-2958.1997.3711734.x. [DOI] [PubMed] [Google Scholar]
- Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes & Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- Cui Y, Dinman JD, Peltz SW. mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and -1 ribosomal frameshifting efficiency. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Weng Y, Hagan KW, Peltz SW. Purification and characterization of the Upf1p: A factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- Didichenko SA, Ter-Avanesyan MD, Smirnov VN. EF-1α-like ribosome-bound protein of yeast Saccharomyces cerevisiae. Eur J Biochem. 1991;198:705–711. doi: 10.1111/j.1432-1033.1991.tb16070.x. [DOI] [PubMed] [Google Scholar]
- Ferguson JJ, Groppe JC, Reed SI. Construction and characterization of three yeast-Escherichia coli shuttle vectors designed for rapid subcloning of yeast genes on small DNA fragments. Gene. 1981;16:191–197. doi: 10.1016/0378-1119(81)90075-5. [DOI] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis LE, Phillippe M, Justesen J, Kisselev L. A highly conserved eukaryotic protein family possessing properties of a polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;4:334–341. [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Wickner R, Maurizi MR. Protein quality control: Triage by chaperones and proteases. Genes & Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- Gozalbo D, Hohmann S. Nonsense suppressors partially revert the decrease of the mRNA levels of a nonsense mutant allele in yeast. Curr Genet. 1990;17:77–79. doi: 10.1007/BF00313252. [DOI] [PubMed] [Google Scholar]
- Hagan KW, Ruiz-Echevarria MJ, Quan Y, Peltz SW. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol Cell Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway using an interacting protein screen. Genes & Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1-mutant. Proc Natl Acad Sci. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M, Frizzell RA, Bedwell DM. Aminoglycoside antibiotics restore CFTR function by overcoming premature stop mutations. Nature Med. 1996;2:467–469. doi: 10.1038/nm0496-467. [DOI] [PubMed] [Google Scholar]
- Jacobson A. Poly(A) metabolism and translation: The closed loop model. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Koonin EV. A new group of putative RNA helicases. Trends Biochem Sci. 1992;17:495–497. doi: 10.1016/0968-0004(92)90338-a. [DOI] [PubMed] [Google Scholar]
- Lee BS, Culbertson MR. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes & Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Elliot DJ, Stutz F, Rosbash M, Singer RH. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. When cells stop making sense: Effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- McKusick VA (with the assistance of C.A. Francomano, S.E. Antonarakis, and P.L. Pearson). Mendelian inheritance in man: A catalog of human genes and genetic disorders. Baltimore, MD: Johns Hopkins University Press; 1994. . (Web site http://www.ncbi.nlm.nih.gov/Omim/). [Google Scholar]
- Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist L. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [PSI+] determinants mediated by oligomerization of the Sup35-encoded polypeptide chain-release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- ————— In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 1997a;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- ————— Interaction between yeast Sup45p(eRF1) and Sup35p(eRF3) polypeptide chain release factors: Implications for prion-dependent regulation. Mol Cell Biol. 1997b;17:2798–2805. doi: 10.1128/mcb.17.5.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on three mRNA sequence elements and at least one trans-acting factor. Genes & Dev. 1993a;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- Peltz SW, Trotta C, Feng H, Brown AH, Donahue JL, Welch EW, Jacobson A. Identification of the cis-acting sequences and trans-acting factors involved in nonsense-mediated mRNA decay. In: Tuite M, McCarthy J, Brown A, Sherman F, editors. Protein synthesis and targeting in yeast. New York, NY: Springer-Verlag; 1993b. pp. 1–10. [Google Scholar]
- Peltz SW, Feng H, Welch EW, Jacobson A. Nonsense-mediated mRNA decay in yeast. Prog Nucleic Acid Res Mol Biol. 1994;47:271–298. doi: 10.1016/s0079-6603(08)60254-8. [DOI] [PubMed] [Google Scholar]
- Perlick HA, Medghalchi SM, Spencer FA, Kendzior Jr RJ, Dietz HC. Mammalian orthologues of a yeast regulator of nonsense-transcript stability. Proc Natl Acad Sci. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes & Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston DF, Hieter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, Peltz SW. Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J. 1996;15:2810–2819. [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, Czaplinski K, Peltz SW. Making sense of nonsense in yeast. Trends Biochem Sci. 1996;21:433–438. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, Gonzalez CI, Peltz SW. Identifying the right stop: Determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheistl RH, Geitz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Stansfield I, Grant CM, Akhmaloka, Tuite MF. Ribosomal association of the yeast SAL4(SUP45) gene product: Implications for its role in translation fidelity and termination. Mol Microbiol. 1992;6:3469–3478. doi: 10.1111/j.1365-2958.1992.tb01782.x. [DOI] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagakesamanskaya AR, Poznyakov AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45(eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki CK, Rep M, van Dijl JM, Suda K, Grivell LA, Schatz G. ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem Sci. 1997;22:118–123. doi: 10.1016/s0968-0004(97)01020-7. [DOI] [PubMed] [Google Scholar]
- Tarun SZ, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor is involved in the maintenance of the non-Mendelian determinant [PSI+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A, Newmam G, Liebman SW. The yeast translational allosuppressor, SAL6: A new member of the PP1-like phosphatase family with a long serine-rich N-terminal extension. Genetics. 1994;138:597–607. doi: 10.1093/genetics/138.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of the mutations in the ATPase and helicase regions of Upf1 Protein. Mol Cell Biol. 1996a;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex, but not mRNA turnover. Mol Cell Biol. 1996b;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Ruiz-Echevarria MJ, Zhang S, Cui Y, Czaplinski K, Dinman JD, Peltz SW. Characterization of the nonsense-mediated mRNA decay pathway and its effect on modulating translation termination and programmed frameshifting. mRNA metabolism and post-transcriptional gene regulation. Mod Cell Biol. 1997;17:241–263. [Google Scholar]
- Wickens M, Anderson P, Jackson RJ. Life and death in the cytoplasm: Messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Zhang J, Maquat LE. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 1997;16:826–833. doi: 10.1093/emboj/16.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Ruiz-Echevarria MJ, Quan Y, Peltz SW. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol Cell Biol. 1995;15:2231–2244. doi: 10.1128/mcb.15.4.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Welch EW, Hagan KW, Brown AH, Peltz SW, Jacobson A. Polysome associated mRNAs are substrates for the nonsense mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA. 1997;3:234–244. [PMC free article] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, LeGoff X, LeGuellec R, Inge-Vechtomov S, Kisselev L, Phillippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]