Abstract
Vg1 mRNA translocation to the vegetal cortex of Xenopus oocytes requires intact microtubules, and a 3′ UTR cis-acting element (termed VLE), which also mediates sequence-specific binding of several proteins. One protein, the 69-kD Vg1 RBP, associates Vg1 RNA to microtubules in vitro. Here we show that Vg1 RBP-binding sites correlate with vegetal localization. Purification and cloning of Vg1 RBP revealed five RNA-binding motifs: four KH and one RRM domains. Surprisingly, Vg1 RBP is highly homologous to the zipcode binding protein implicated in the microfilament-mediated localization of β actin mRNA in fibroblasts. These data support Vg1 RBP’s direct role in vegetal localization and suggest the existence of a general, evolutionarily conserved mechanism for mRNA targeting.
Keywords: RNA localization, Vg1 RBP, Xenopus, oocytes, microtubules, KH domain
Intracellular RNA localization leads to asymmetric protein synthesis, a necessary step in the process of pattern formation during early embryogenesis in many species (for review, see St. Johnston 1995; Gavis 1997). In Xenopus oocytes, morphological and molecular differences between the animal and vegetal hemispheres help define the primary axis around which subsequent development proceeds. Two temporally distinct pathways have been identified for vegetal RNA localization. The early pathway facilitates localization of several RNAs found to be localized during stages I–II of oogenesis (Xcat-2, Xlsirts, Xwnt-11), appears to be microtubule-independent, and is correlated with the migration of the mitochondrial cloud to a small region at the vegetal pole (Kloc et al. 1993; Mosquera et al. 1993; Forristall et al. 1995; Kloc and Etkin 1995; Zhou and King 1996a). The second pathway occurs during late stage III–early stage IV, requires intact microtubules, and targets RNA to a tight shell along the entire vegetal cortex (Melton 1987; Yisraeli and Melton 1988; Yisraeli et al. 1990). So far, only one mRNA, Vg1, is known to localize via the second pathway in vivo, although Xcat-2 RNA can employ this pathway when injected into stage III oocytes (Zhou and King 1996b). VegT/Xombi/Apod/Brat RNA is also localized to the vegetal cortex in a manner resembling the second pathway, but it is not yet known whether this localization requires intact microtubules; furthermore, it is released from the cortex in stage V/VI oocytes, significantly earlier than is Vg1 RNA (Lustig et al. 1996; Stennard et al. 1996; Zhang and King 1996). Cis-acting elements mediating one or the other pathway have been mapped, by deletion analysis, to extensive regions of the 3′ UTRs (Mowry and Melton 1992; Zhou and King 1996a,b; Gautreau et al. 1997). Trans-acting factors that bind these regions and may provide a link between the RNA and components of the cytoskeleton have been characterized only preliminarily, and the connection between these interactons and localization is just beginning to be defined (Schwartz et al. 1992; Mowry 1996; Deshler et al. 1997).
Vg1 RNA-binding protein (RBP) is a 69-kD oocyte protein that binds specifically to the vegetal localization element (VLE) of Vg1 RNA (Schwartz et al. 1992). Because it is enriched in the microtubule fraction of oocytes and mediates the association of VLE sequences to microtubules in vitro (Elisha et al. 1995), Vg1 RBP is a good candidate for a trans-acting factor involved in Vg1 RNA localization. In this paper we have mapped the Vg1 RBP-binding sites in the Vg1 VLE, and, using confocal microscopy to visualize injected wild-type and mutant VLEs in the same oocyte, we observe a direct correlation between the presence of two Vg1 RBP-binding sites and vegetal localization. The unexpected finding that the microtubule-associated protein Vg1 RBP is highly homologous to the microfilament-binding zipcode biding protein (ZBP-1) involved in β-actin mRNA localization suggests that microtubule- and microfilament-based systems of localization may be more related than originally anticipated.
Results and Discussion
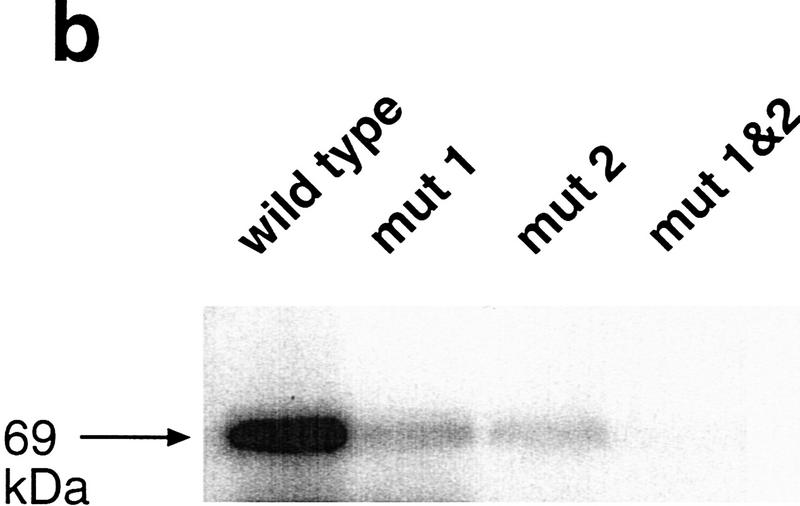
To determine whether Vg1 RBP binding to the VLE correlates with vegetal localization, we first used UV cross-linking to precisely map cis-acting Vg1 RBP-binding sites in the 366-nucleotide-long VLE. Initial mapping experiments indicated that two nonoverlapping regions at each end of the VLE can both bind Vg1 RBP (Fig. 1a). We chose to pinpoint the Vg1 RBP-binding sites using a linker–scan (LS) approach, which allowed us to compare the effects of relatively minor sequence changes (unaltered in length by deletions) to wild-type sequences. Twenty-nucleotide segments of VLE sequence were sequentially replaced with a 20-nucleotide linker sequence that does not bind Vg1 RBP (Figs. 1 and 2; data not shown). In each of the regions that binds Vg1 RBP independently, one 20-nucleotide element was found to be essential for Vg1 RBP binding. Although substitution of each of these elements individually (leaving the other intact) in the context of the full-length VLE partially reduces Vg1 RBP binding, loss of both elements abolishes binding almost completely (Fig. 1b). Thus, Vg1 RBP appears to bind independently at two distinct sites in the VLE. We note that a hexanucleotide repetitive sequence, UUUCUA, is found in two copies in the first element, once in the second element, and nowhere else in Vg1 RNA (Fig. 1c; see below).
Figure 1.
Cis-acting binding elements in the VLE and their role in localization. (a) The ability of different fragments of the VLE, as well as of the LS, substituted VLE fragments, to bind Vg1 RBP is schematically represented. The coordinates of the fragments and the substitution used for each binding assay (with the first nucleotide of the VLE numbered 1 and the substitution indicated as Ins) are listed at the left. The two regions identified as being important for Vg1 RBP binding are indicated by the boxes at the top. Domains protected by oocyte proteins from RNase degradation, mapped and labeled by Mowry (1996) as C, A/B, and D, respectively, are shown as lines beneath the full-length VLE. The degree of binding is indicated in the inset. (b) A representative UV cross-linking experiment (summarized in a) is shown for wild-type VLE, 1-366/ins1–20 (mut 1), 1-366/ins256–275 (mut 2), and 1–366/ins1-20&256–275 (mut 1&2) RNAs. (c) The nucleotide sequences of the first and second binding elements in the VLE are compared with a sequence from the 3′ UTR of Xcat-2 RNA from the region mapped as containing a late pathway localization element (Zhou and King 1996b). The hexanucleotide UUUCUA present in all these sequences is indicated in boldface type.
Figure 2.
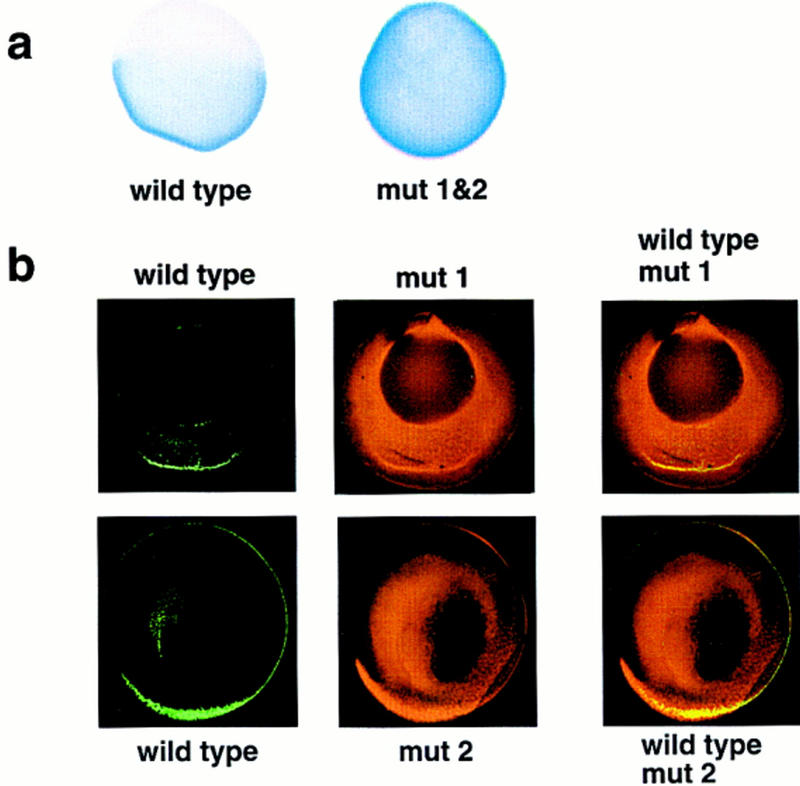
Localization of wild-type and mutant VLEs in oocytes. (a) The distribution of wild-type VLE RNA or double substituted VLE mut 1&2 RNA (1–366/ins1–20&256–275, see Fig. 1a for map) following injection into late stage III oocytes. mut 1&2 RNA appears to be fairly uniformly distributed throughout the oocyte; wild-type VLE shows strong cortical localization to the vegetal hemisphere alone. (b) Confocal micrographs of late stage III oocytes coinjected with wild-type VLE RNA (green channel) and either mut 1 RNA (1–366/ins1–20, see Fig. 1a) or mut 2 RNA (1–366/ins256–275) (red channel). Although the majority of both the mutant RNAs is not localized, some colocalization with the wild-type VLE RNA (yellow, third column) is observable.
Localization of VLE RNAs with substitutions in either one or both of the Vg1 RBP-binding elements was compared with the localization of wild-type VLE using the oocyte culture system described previously (Yisraeli and Melton 1988). Microinjected VLE RNA with both sites substituted was homogeneously distributed throughout the oocyte after 5 days in culture, in contrast to wild-type VLE, which shows clearly defined vegetal localization (Fig. 2a). The effects of substituting either site alone were assayed by simultaneously injecting wild-type VLE RNA, labeled with digoxygenin–UTP, and substituted VLE RNA, labeled with fluorescein–UTP; their localization was visualized by confocal microscopy using rhodamine and Cy-5-labeled antibodies, respectively (Fig. 2b). Substitution of either site alone significantly impairs localization, as indicated by the presence of the substituted RNA throughout most of the oocyte (red channel). Significantly, a low level of cortical localization of the singly substituted VLEs can be detected colocalizing (in yellow) with the wild-type VLE RNA (green channel) (Fig. 2b), but this colocalization was not detected with the double substitutions (data not shown). This technique allows us to quantify the degree of partial localization of the substituted RNAs with respect to the internal wild-type VLE controls. For both singly substituted RNAs, we estimate this degree of partial localization to be on the order of 20%.
Initial characterization of the VLE performed by Mowry and Melton (1992) showed that deletions removing either 5′ or 3′ sequences from the VLE significantly impair localization; these deletions included, respectively, either the upstream or downstream elements we have pinpointed here. More recently, Mowry (1996) identified, by RNase footprinting, three distinct regions of the VLE protected by Xenopus oocyte proteins; two of these regions fall precisely at the sites identified here for Vg1 RBP binding (see Fig. 1a). As mentioned above, a 6-nucleotide sequence present in two copies in the first element is also present in one copy in the second element (Fig. 1c). This sequence is also present in the region of Xcat-2 RNA, which is important for its localization via the late pathway (Fig. 1c) (Zhou and King 1996b). In UV cross-linking assays, Vg1 RBP binds Xcat-2 RNA (Z. Elisha, pers. comm.). An oocyte protein, termed Vera, reported to be associated with the ER and of a slightly larger size than Vg1 RBP, has been recently described as also binding the VLE (Deshler et al. 1997). Deletion of two 9-nucleotide repeats from the first 20 nucleotides of the VLE (containing the hexanucleotide sequence mentioned above) also impairs localization significantly, as well as reducing Vera binding. (These investigators also found that additional combinations of deletions can affect localization and protein binding.) A very recent set of experiments by Gautreau et al. (1997) further emphasizes the importance of the Vg1 RBP-binding sites we have mapped here. They identified two subelements, at the 5′ and 3′ ends of the VLE, whose deletion severely impairs localization (the 5′ subelement is 1–135 and contains the first Vg1 RBP-binding site, and the 3′ subelement is 201–340 and contains the second Vg1 RBP-binding site; see Fig. 1, a and c). Each of these subelements is incapable, on its own, of vegetal localization, but a duplicated 5′ subelement localizes well. This first subelement contains two copies of the hexanucleotide repeat, and mutations in this repeat eliminate vegetal localization. These results complement those presented here, in which we have preserved the basic structure and length of the VLE. Taken together, these data strongly suggest that the two Vg1 RBP-binding sites represent a cis-acting element required for localization via the late pathway and that cooperative Vg1 RBP binding is required for proper localization.
To clone Vg1 RBP, we first purified it from a Xenopus oocyte S100 lysate by sequential column chromatography. As shown in Figure 3a, Vg1 RBP, assayed by UV cross-linking, elutes off a heparin–Sepharose column in the range of 250–500 mm KCl. Then, these fractions were passed over an RNA affinity column, consisting of in vitro-transcribed VLE RNA attached to CNBr-activated Sepharose (Kaminski et al. 1995). Here, Vg1 RBP activity, which elutes from this column between 500 and 700 mm KCl, coincides with a principal silver-stained band migrating at 69 kD (Fig. 3b). The purified protein does not bind VLE sequences with both binding elements substituted, indicating that it has the same RNA-binding specificity as the oocyte extract protein (data not shown). Microsequencing of the gel-purified polypeptide generated five peptide sequences of 9–15 residues each. Subsequent RT–PCR of total Xenopus oocyte RNA using degenerate primers made to two “back-translated” peptides (40 and 41; see Materials and Methods) yielded a product that encompassed the sequence of a third peptide (35). Screening of a Xenopus oocyte cDNA library with this PCR fragment resulted in a 2.4-kb cDNA, containing an ORF of 594 amino acids. All five peptides obtained from the purified Vg1 RBP were found in the conceptually translated cDNA protein sequence (see Fig. 4). By Northern blot analysis, Vg1 RBP mRNA appears to be ∼2.8–3.0 kb long, implying that this clone is missing some noncoding region sequences (data not shown).
Figure 3.
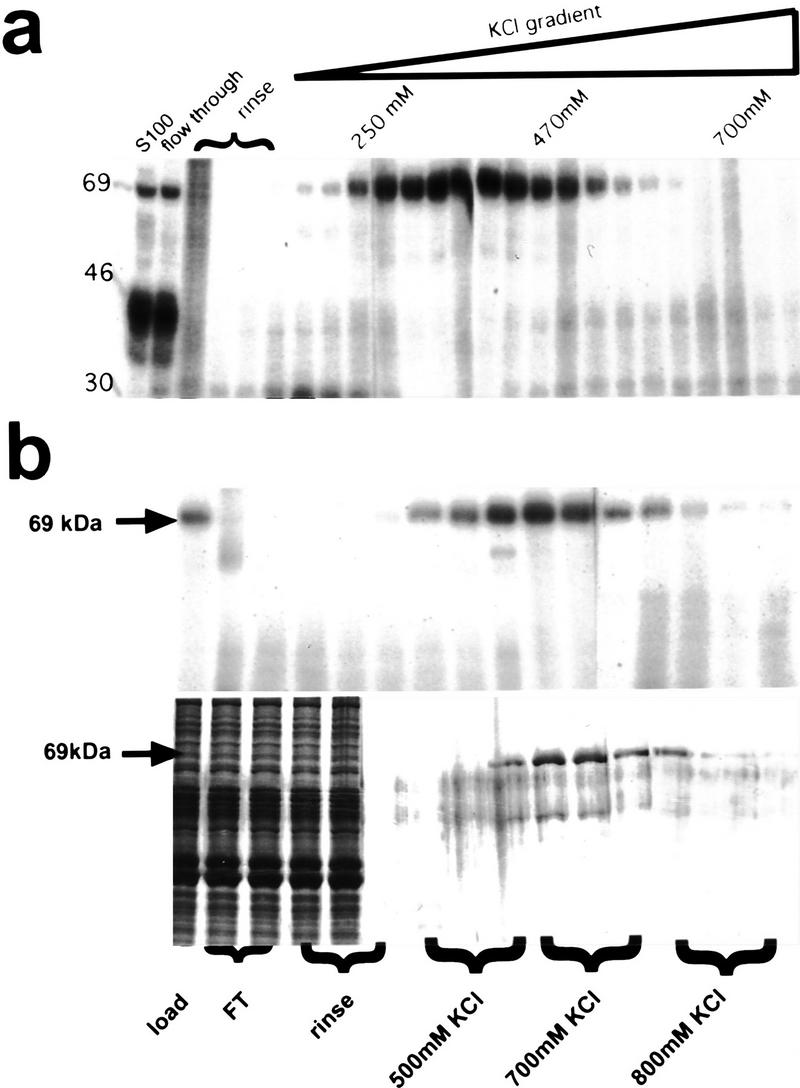
Biochemical purification Vg1 RBP. (a) Fractions of the heparin–Sepharose column were assayed for Vg1 RBP cross-linking activity. KCl concentrations (as determined by conductivity) of selected fractions are indicated. (b) Equivalent fractions of the RNA affinity column were assayed for Vg1 RBP cross-linking activity (autoradiograph, top) and presence of protein (silver-stained gel, bottom). (Arrows) The location of Vg1 RBP.
Figure 4.
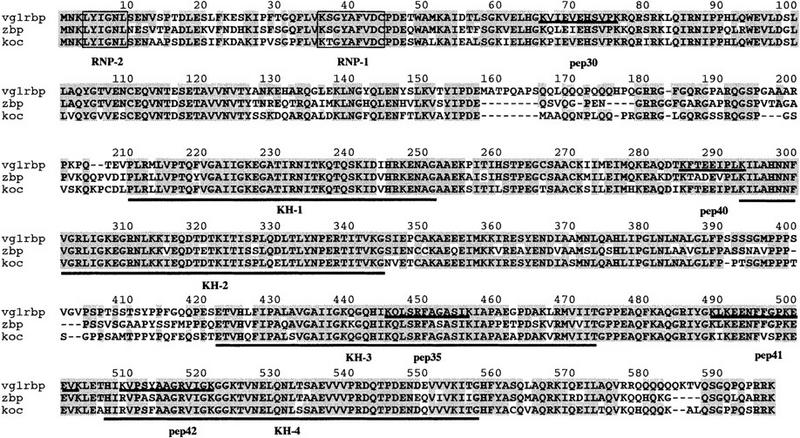
Vg1 RBP protein sequence and its homology to ZBP-1 and KOC. The protein sequence for Vg1 RBP is shown with the four KH (underlined) and one RRM (boxed) domains indicated. The five peptides that were obtained from the microsequencing of the purified Vg1 RBP are underlined and labeled. ZBP-1 and KOC sequences are also shown, aligned to Vg1 RBP, and identical amino acids are shaded. Percent identities/homologies between the proteins are Vg1 RBP/ZBP-1, 78%/84%; Vg1 RBP/KOC, 83%/87%; ZBP-1/KOC, 76%/80%.
Database and literature searches with the Vg1 RBP peptides and, subsequently, the complete ORF, revealed a striking sequence conservation with an RBP implicated in mRNA localization in polarized somatic cells. Localization of β-actin mRNA to the leading edge of chick fibroblasts requires the presence of conserved elements in its 3′ UTR, included in a 54-nucleotide sequence termed the zipcode (Kislauskis et al. 1993, 1994). The 68-kD ZBP-1 that binds to wild-type, but not to mutant zipcode sequences inactive in RNA localization, recently has been cloned (Ross et al. 1997). Vg1 RBP and ZBP-1 share 78% identical amino acids (Fig. 4). The searches also revealed that a human protein of ∼65 kD, of unknown function, called KOC [KH-domain-containing protein overexpressed in cancer (Mueller-Pillasch et al. 1997)] is a very close relative of the localization element-binding proteins. Alignment of all three proteins, shown in Figure 4, indicates the high sequence conservation between Vg1 RBP, ZBP-1, and KOC, which extends throughout their length. A prominent feature of all these polypeptides is their multiple predicted RNA-binding motifs, including one RNA recognition motif (RRM) domain and four tandem KH domains. The RRM, characterized by its RNP2 and RNP1 motifs, and the KH (hnRNP K homology) domain are responsible for the RNA-binding ability of a variety of snRNA, pre-mRNA, mRNA, and rRNA-binding proteins, including some that play an important role in early development (Siomi and Dreyfuss 1997). The RRM and KH domains are the most highly conserved regions of the three proteins. We note that a REV-like nuclear export sequence is also present and conserved in all three proteins (325–333 in Vg1 RBP), but the functional importance, if any, of this motif is not clear. Our database searches failed to identify any obvious homologs of Vg1 RBP in yeast, Drosophila, or Caenorhabditis elegans. On the other hand, we did find other vertebrate counterparts as partial EST clones in mouse (accession nos. aa073514 and aa117282) and zebrafish (aa495472).
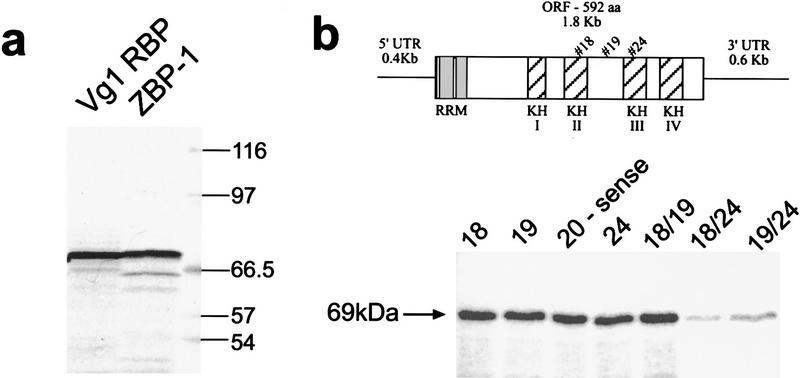
Two independent lines of evidence suggest that the cloned gene is indeed Vg1 RBP. First, the in vitro translation product made from the cloned gene is the same size as Vg1 RBP in S100 extracts or purified by RNA affinity chromatography; in vitro-translated ZBP-1 comigrates with this band as well (Fig. 5a). Second, to address the question of whether the cloned gene encodes the binding activity defined in S100 extracts as Vg1 RBP, antisense oligodeoxynucleotides directed against coding sequences were injected into stage III oocytes. The ability of the oligonucleotides, following overnight incubation, to cause a specific loss of binding activity as a result of RNase H-directed degradation of Vg1 RBP mRNA was monitored by extracting protein and then by performing UV cross-linking (Fig. 5b). Although none of the oligonucleotides causes a loss of Vg1 RBP-binding activity when injected singly, a combination of two oligonucleotides directs a clear drop in binding activity (70% loss of control activity). This effect is very specific, as neither the sense oligonucleotide nor another combination of two antisense oligonucleotides influences Vg1 RBP binding. Northern blot analysis of RNAs extracted in parallel from the injected oocytes, probed with a Vg1 RBP DNA probe, shows precisely the same effect (data not shown). These results strongly suggest that the cloned gene encodes Vg1 RBP.
Figure 5.
The cloned gene encodes Vg1 RBP. (a) In vitro translation of Vg1 RBP and ZBP-1 RNA. Both Vg1 RBP and ZBP-1 translation products migrate close to 69 kD, according to the molecular mass markers (right). (b) UV cross-linking of S100 extracts from oocytes injected with the indicated oligonucleotides was performed. Using a PhosphorImager to quantify the band intensities, we observed a 70% reduction in cross-linking activity (as compared to the control sense oligonucleotide 20) when oligonucleotides 18 and 24 are injected simultaneously, and a 50% reduction when 19 and 24 are injected together. The third pair of oligonucleotides (18 and 19) had no effect on Vg1 RBP-binding activity. The schematic drawing indicates the position of the oligonucleotides relative to the RNA-binding domains.
The identification of Vg1 RBP, shown here to mediate Vg1 mRNA localization in Xenopus oocytes, as a close homolog of ZBP-1, a protein implicated in targeting β-actin mRNA in fibroblasts, is startling. It clearly suggests that this specific 3′ UTR–RBP is a common component in localization pathways hitherto considered to be individual for mRNA and cell type, and raises several interesting questions. Despite the high degree of sequence similarity, significant functional differences exist between Vg1 RBP and ZBP-1. The zipcode motif that ZBP-1 binds includes a 6-nucleotide tandem repeat, ACACCC, which, when mutated, abrogates ZBP-1 binding (Kislauskis et al. 1993, 1994; Ross et al. 1997). The Vg1 RBP-binding sites described here contain a different hexanucleotide repeat, UUUCUA, and are AU rich (see Fig. 1c). The source of the different binding specificities of these proteins remains to be determined; certainly, as in the case of the fragile X FMR-1 gene, single-amino-acid substitutions in a KH domain can have profound affects (Siomi et al. 1994). An additional intriguing difference between the proteins lies in their cytoskeletal associations. β-Actin mRNA localization requires intact microfilaments (and not microtubules), and ZBP-1 coelutes from the RNA affinity column with other proteins including actin (Sundell and Singer 1991; Ross et al. 1997). Vg1 RBP, however, associates with microtubules, both in vivo and in vitro, and can mediate the specific association of Vg1 RNA to microtubules (Elisha et al. 1995). Furthermore, Vg1 mRNA localization requires intact microtubules in oocytes (Yisraeli et al. 1990). Importantly, localized β-actin mRNA in neurons appears to be complexed in large granules that are associated with microtubules rather than microfilaments (Bassell et al. 1998). Interestingly, several reports have suggested that microtubule- and microfilament-based localization mechanisms may be capable of interacting. For example, in squid axons, vesicles can move along either microtubules or microfilaments (Kuznetsov et al. 1992). In Drosophila oocytes, oskar mRNA localization to the posterior pole, which is sensitive to microtubule inhibitors and is mediated by Staufen protein (which can bind to microtubules), is disrupted by mutants in microfilament-binding cytoplasmic tropomyosin II (Erdelyi et al. 1995). In fact, microtubule-interacting sites and actin-binding domains may be topologically very similar, as suggested by a comparison of kinesin and myosin 3-D structures (Woehlke et al. 1997) and by the observation that in yeast, a kinesin-related protein can suppress some aspects of a myosin mutant phenotype (Lillie and Brown 1992). The presence of a highly conserved protein involved in the movement of both microtubule- and microfilament-based localized mRNAs suggests that the mechanism for RNA localization may be a very fundamental one that has been adapted by different cells for specific needs. It will be interesting to see what provides the cytoskeletal specificity for Vg1 RBP and ZBP-1 and how additional, cell-specific proteins might mediate these interactions.
Materials and methods
UV cross-linking
For the LS and antisense experiments, UV cross-linking was performed using either 3 or 4 μg of an oocyte S100 extract, respectively, as described previously (Elisha et al. 1995). UV cross-linking was also used to assay for the presence of Vg1 RBP activity in the column fractions; following normalization of the salt concentrations (as measured by conductivity), equivalent aliquots of each fraction were tested. Full-length VLE RNA was used as a radioactive probe for the column and antisense assays. Templates to synthesize the probes for the LS analysis were constructed by PCR (Schwartz 1996), with the 5′ primer containing a T3 RNA polymerase promoter and the linker, which replaced successive 20-nucleotide segments of the VLE, consisting of the sequence GGGAACAAAAGCTTGCATGC. Substitutions were verified by sequencing. Probes for the LS analysis were synthesized in the presence of cap analog to prevent increased degradation as a result of the absence of polylinker sequence at the 5′ end of the RNA (Schwartz 1996). Although bromo-UTP was used initially for synthesizing probes for the LS analysis, unsubstituted UTP was found to give identical results and used for subsequent experiments. In the LS and antisense experiments, binding was quantitated using a PhosphorImager, relative to internal controls.
Oocyte injections
Late stage III oocytes were injected with 1 ng of RNA (localization experiments) or oligodeoxynucleotides (antisense experiments) in 10 nl of water. For localization experiments, oocytes were cultured for 5 days, as described (Yisraeli and Melton 1988). For the whole mount immunohistochemical visualization of RNA, exogenous digoxygenin-labeled RNA was detected using alkaline phosphatase-conjugated anti-digoxygenin antibody and BM-Purple substrate (Boehringer Mannheim). The coinjection experiments (performed with wild-type oocytes to allow for orientation) were visualized by confocal microscopy: Wild-type VLE RNA, labeled with digoxygenin–UTP, was detected by a rhodamine-labeled anti-digoxygenin antibody (green channel), and the substituted VLE, labeled with fluorescein–UTP, was detected by a mouse anti-fluorescein first antibody and a donkey Cy5 anti-mouse second antibody (red channel). No fluorescence was detected with the fluorescein–UTP-labeled RNA alone, and autofluorescence of the yolk was not detectable under the conditions used (data not shown). In Figure 2, the degree of partial localization of the mutant RNA, relative to the wild-type RNA, was determined by calculating the fraction of all the mutant RNA fluorescent signal that was localized to the cortex and comparing that to the fraction of all the wild-type RNA that was localized to the cortex.
Antisense oligodeoxynucleotides consisted of the following sequences: (18) 5′-AAGTCCTGTAGTGGAGATAT; (19) 5′-ACTCCAACAGAAGGTGGTGG; and (24) 5′-TGCTTTCCAATAATCGCTCC. The sense oligodeoxynucleotide control (20) was the complementary sequence to number 18. All of the oligonucleotides were synthesized with the first four and last four nucleotides consisting of phosphorothioate links, to minimize degradation. When combinations of oligonucleotides were injected, 0.5 ng of each was used. Oocytes injected with oligonucleotides were incubated overnight at 19°C in 1× modified Barth’s saline, and S100 protein extracts were prepared the following morning. Oocyte equivalents (1.2) of S100 extract (∼3 μg) were used for UV cross-linking.
Purification and cloning of Vg1 RBP
S100 oocyte extract (200 mg) was applied to a heparin–Sepharose column and eluted with a salt gradient. The fractions containing the 69-kD Vg1 RBP-binding activity were pooled and concentrated using a Centricon filter. Six micrograms of the concentrated protein was loaded onto a CNBr-activated Sepharose column bound with 300 μg of VLE RNA synthesized in vitro (Kaminski et al. 1995). After allowing the sample to flow through the column, the column was washed with 100 mm KCl and eluted with a salt gradient of 250 mm–1 m KCl. UV cross-linking was performed on aliquots from representative fractions. Pooled fractions containing Vg1 RBP were electrophoresed on a preparative gel. The Coomassie blue-stained Vg1 RBP band (of the same size) was excised from the gel, degraded with the endoprotease LysC, and the peptide products were then eluted and subjected to HPLC. Five clearly separated peaks were microsequenced by Edman degradation (see Fig. 3).
One microgram of total Xenopus-mixed oocyte RNA was reverse transcribed into single-stranded cDNA using the First Strand cDNA Synthesis Kit (Boehringer Mannheim) and random primers. An aliquot of the RT reaction was amplified by cloned Pfu DNA polymerase (Stratagene) with degenerate oligonucleotides (5′-AARTTYACTGARGARATHCC-3′ based on peptide 40 KFTEEIPLK and 5′-CCRAARAARTTYTCYTCYTT-3′ based on peptide 41 KLKEENFFGPKEEVK). The 600-bp PCR product was gel-purified and, following verification by automated sequencing, labeled by random priming. Screening of a Xenopus oocyte cDNA library in Bluescript pBRN3 (generous gift of Nigel Garrett and John Gurdon, Wellcome/CRC Institute, Cambridge, UK) resulted in five clones that represented two allelic varients of Vg1 RBP (97% identical). In this work we used a full-length clone of allele D, containing an ORF of 594 amino acids. The first AUG encountered in this cDNA was predicted to serve as the initiator codon due to the 4 in-frame stop codons in the unusually long 378-nucleotides 5′ UTR.
In vitro translation
In vitro-transcribed capped mRNAs of 100 ng (Vg1-RBP) or 400 ng (ZBP-1) were translated in 10 μl of the reticulocyte cell-free lysate (Promega) in the presence of [35S]Met, and labeled products were separated by 15% SDS-PAGE. Loading volumes were adjusted to give similar intensity of products, to compensate for the lower efficiency of translation of ZBP-1 mRNA compared to that encoding Vg1 RBP.
Acknowledgments
We thank M. Tarshish for help and expertise in confocal microscopy; R. Honikman for help in finding conditions for the double labeling experiments; I. Marianovsky for excellent technical assistance with the heparin–Sepharose chromatography; N. Garret and J. Gurdon for the Xenopus oocyte cDNA library; R. Singer for the ZBP-1 clone; and Z. Paroush, H. Cedar, and D. St. Johnston for critical reading of the manuscript. This work was funded in part by the UK–Israel Science and Technology Research Fund (N.S. and J.K.Y.), the Israel Science Foundation (J.K.Y.), and the Israel Cancer Research Fund (J.K.Y). A.G. is the recipient of an Overseas Research Students Award.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
Two genes highly related to Vg1 RBP have appeared in GenBank while this paper was in press. B23 (accession no. AF042353) is a transcriptional activator of the TFIIIα gene in stage I Xenopus oocytes and is identical to an allelic variant of Vg1 RBP isolated during this cloning process (see Materials and Methods). Vera (accession no. AF055923) is a Vg1 RBP that has been implicated in Vg1 RNA localization. It appears to be identical to Vg1 RBP, although its recognition site is reported to be different than that of Vg1 RBP (Deshler et al. 1998, Curr. Biol. 8: 489–496). The functional significance of these proteins remains to be determined.
Footnotes
E-MAIL yisraeli@yam-suff.cc.huji.ac.il; FAX 972-2-675745.
References
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of β-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler JO, Highett MI, Schnapp BJ. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- Elisha Z, Havin L, Ringel I, Yisraeli JK. Vg1 RNA binding protein mediates the association of Vg1 RNA with microtubules in Xenopus oocytes. EMBO J. 1995;14:5109–5114. doi: 10.1002/j.1460-2075.1995.tb00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdelyi M, Michon AM, Guichet A, Glotzer JB, Ephrussi A. Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature. 1995;377:524–527. doi: 10.1038/377524a0. [DOI] [PubMed] [Google Scholar]
- Forristall C, Pondel M, Chen L, King ML. Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development. 1995;121:201–208. doi: 10.1242/dev.121.1.201. [DOI] [PubMed] [Google Scholar]
- Gautreau D, Cote CA, Mowry KL. Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocytes. Development. 1997;124:5013–5020. doi: 10.1242/dev.124.24.5013. [DOI] [PubMed] [Google Scholar]
- Gavis ER. Expeditions to the pole: RNA localization in Xenopus and Drosophila. Trends Cell Biol. 1997;7:485–492. doi: 10.1016/S0962-8924(97)01162-8. [DOI] [PubMed] [Google Scholar]
- Kaminski A, Hunt SL, Patton JG, Jackson RJ. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Li Z, Singer RH, Taneja KL. Isoform-specific 3′-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development. 1995;121:287–297. doi: 10.1242/dev.121.2.287. [DOI] [PubMed] [Google Scholar]
- Kloc M, Spohr G, Etkin LD. Translocation of repetitive RNA sequences with the germ plasm in Xenopus oocytes. Science. 1993;262:1712–1714. doi: 10.1126/science.7505061. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SA, Langford GM, Weiss DG. Actin-dependent organelle movement in squid axoplasm. Nature. 1992;356:722–725. doi: 10.1038/356722a0. [DOI] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Suppression of a myosin defect by a kinesin-related gene. Nature. 1992;356:358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- Lustig KD, Kroll KL, Sun EE, Kirschner MW. Expression cloning of a Xenopus T-related gene (Xombi) involved in mesodermal patterning and blastopore lip formation. Development. 1996;122:4001–4012. doi: 10.1242/dev.122.12.4001. [DOI] [PubMed] [Google Scholar]
- Melton DA. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature. 1987;328:80–82. doi: 10.1038/328080a0. [DOI] [PubMed] [Google Scholar]
- Mosquera L, Forristall C, Zhou Y, King ML. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with nanos-like zinc finger domain. Development. 1993;117:377–386. doi: 10.1242/dev.117.1.377. [DOI] [PubMed] [Google Scholar]
- Mowry KL. Complex formation between stage-specific oocyte factors and a Xenopus mRNA localization element. Proc Natl Acad Sci. 1996;93:14608–14613. doi: 10.1073/pnas.93.25.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry KL, Melton DA. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- Mueller-Pillasch F, Lacher U, Wallrapp C, Micha A, Zimmerhackl F, Hameister H, Varga G, Friess H, Buchler M, Beger HG, Vila MR, Adler G, Gress TM. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. Oncogene. 1997;14:2729–2733. doi: 10.1038/sj.onc.1201110. [DOI] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SP. “Localization of Vg1—A maternal mRNA”, Ph.D. thesis. Jerusalem, Israel: Hebrew University; 1996. [Google Scholar]
- Schwartz SP, Aisenthal L, Elisha Z, Oberman F, Yisraeli JK. A 69 kDa RNA binding protein from Xenopus oocytes recognizes a common motif in two vegetally localized maternal mRNAs. Proc Natl Acad Sci. 1992;89:11895–11899. doi: 10.1073/pnas.89.24.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G. Essential role for KH domains in RNA binding: Impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- St. Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- Stennard F, Carnac G, Gurdon JB. The Xenopus T-box gene, Antipodean, encodes a vegetally localised maternal mRNA and can trigger mesoderm formation. Development. 1996;122:4179–4188. doi: 10.1242/dev.122.12.4179. [DOI] [PubMed] [Google Scholar]
- Sundell CL, Singer RH. Requirement of microfilaments in sorting of actin messenger RNA. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom Booher N, Vale RD. Microtubule interaction site of the kinesin motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- Yisraeli JK, Melton DA. The maternal mRNA Vg1 is correctly localized following injection into Xenopus oocytes. Nature. 1988;336:592–595. doi: 10.1038/336592a0. [DOI] [PubMed] [Google Scholar]
- Yisraeli JK, Sokol S, Melton DA. A two-step model for the localization of a maternal mRNA in Xenopus oocytes: Involvement of microtubules and microfilaments in translocation and anchoring of Vg1 mRNA. Development. 1990;108:289–298. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]
- Zhang J, King ML. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development. 1996;122:4119–4129. doi: 10.1242/dev.122.12.4119. [DOI] [PubMed] [Google Scholar]
- Zhou Y, King ML. Localization of Xcat-2 RNA, a putative germ plasm component, to the mitochondrial cloud in Xenopus stage I oocytes. Development. 1996a;122:2947–2953. doi: 10.1242/dev.122.9.2947. [DOI] [PubMed] [Google Scholar]
- ————— RNA transport to the vegetal cortex of Xenopus oocytes. Dev Biol. 1996b;179:173–183. doi: 10.1006/dbio.1996.0249. [DOI] [PubMed] [Google Scholar]