Abstract
Naturally occurring nitroalkene fatty acids (NAs) derived from oleic (NO2-OA) and linoleic (NO2-LA) acids mediate a variety of cellular responses. We examined the signaling pathways involved in NA activation of Nrf2/ARE-dependent versus PPARγ/PPRE-dependent transcription in human MCF7 breast cancer cells. Additionally, we compared the relative potencies of NO2-OA and NO2-LA in activating these two transcriptional programs. Here it is demonstrated that, in addition to the direct adduct formation of NA with the Nrf2 inhibitory protein, Keap1, shown by others, NA activation of Nrf2/ARE-mediated transcription results from increased nuclear Nrf2 levels and depends upon activation of the PI3K/AKT and PKC, but not ERK and JNK MAPK, signaling pathways. Examination of the relationship between NA stimulation of the Nrf2/ARE versus PPARγ/PPRE transcriptional programs revealed concentration-dependent activation of distinct signaling pathways that were readily distinguished by selective attenuation of Nrf2/ARE-dependent, but not PPARγ-dependent, transcription by inhibitors of PI3K and PKC. Moreover, measurable, statistically significant activation of PPARγ/PPRE-dependent transcription occurred at nanomolar concentrations of NAs—the 12-NO2 isomer of NO2-LA showing the most potent activity—whereas significant activation of Nrf2/ARE-dependent transcription occurred at much higher NA concentrations (≥ 3 micromolar) with the NO2-OA isomers the most potent. These findings have implications for the physiological roles of NAs suggesting that, at concentrations likely to be encountered in vivo, their direct activation of PPARγ transcription will dominate over their electrophilic activation of Nrf2 antioxidant/protective responses.
Nitroalkene fatty acids (NAs)1 are naturally occurring electrophilic derivatives of unsaturated fatty acids formed via •NO-dependent oxidative reactions (1). Due to the natural abundance of oleic and linoleic acids in the diet, circulation, and membranes, their nitrated derivatives, NO2-OA and NO2-LA, are quantitatively important species that mediate a variety of cellular responses including •NO-dependent vasodilation (2), anti-inflammatory processes (3, 4), heat shock responses (5), and others (1). Additionally, among the more striking specific properties of NAs are their abilities to activate PPARγ- and Nrf2-dependent transcription of genes containing, respectively, PPRE and ARE promoter elements (6–8). PPARγ, by governing the expression of a large network of genes, is important in the control of lipid and carbohydrate homeostasis, adipocyte differentiation, inflammation/anti-inflammation, and other functions (9–12). Nrf2, on the other hand, is crucial for mediating the expression of a family of genes associated with cellular defense and protection in response to oxidative and electrophilic stress (13, 14). NO2-LA and NO2-OA are among the most potent naturally occurring ligand agonists of PPARγ yet identified (6, 7, 15) and, more recently, have been shown to activate Nrf2/ARE-dependent transcription (8). What is not known is the relative potency of NAs and their physiological relevance, at NA levels likely to be achieved in vivo, towards activation of these alternative transcriptional programs.
It is well established that NO2-LA and NO2-OA activate PPARγ via direct interaction with the ligand-binding pocket of the transcription factor (15–18). Relatively less is known about the mechanism of NA-activation of Nrf2-dependent transcription. Under basal conditions, Nrf2 associates with its inhibitory partner, Keap1, which serves as an adaptor protein to bridge Nrf2 with a Cul3-based E3 ubiquitin ligase thereby targeting Nrf2 for ubiquitinylation and proteosomal degradation (19, 20). It is widely accepted that a major mechanism by which electrophiles activate Nrf2-dependent transcription is through disrupting Nrf2/Keap1 interactions resulting in Nrf2 stabilization, accumulation, and translocation to the nucleus where it can facilitate transcription of its target genes. This electrophile-mediated disruption of Nrf2/Keap1 interactions often occurs as a consequence of redox modification or direct adduction of key cysteine residues within Keap1 (21–24). Indeed, Kansanen et al. recently demonstrated formation of such adducts between Keap1 and the 9- and 10-E-nitro isomers of NO2-OA (25). While formation of these adducts is likely to contribute significantly to Nrf2 stabilization, other pathways are also implicated in the control of electrophile-stimulated Nrf2/ARE-dependent transcription—pathways that include PI3K/AKT, MAPK, and PKC signaling (26–34).
Accordingly, the studies described herein were designed to: 1) evaluate the roles of these kinase signaling pathways in NA-mediated activation of Nrf2/ARE-dependent transcription, 2) determine whether NA-mediated activation of the two alternative transcription programs (PPARγ- and Nrf2-driven) are independent of each other or if they share common signaling pathways or exhibit significant activation pathway cross-talk, and 3) quantitatively compare Nrf2/ARE versus PPARγ/PPRE transcriptional responses to NA over a range of NA concentrations in order to ascertain which transcription program will dominate at NA levels likely to be encountered in vivo.
Experimental Procedures
Materials
Nitroalkene fatty acids (NAs): total (E-9, 10, 12, and 13 nitro) nitrolinoleic acid (total NO2-LA) was synthesized (35) and the E-12-NO2 regioisomer (12-NO2-LA) was purified from total NO2-LA by preparative HPLC as described previously (15). E-9- and E-10-nitrooleic acids were synthesized regio- and stereoselectively using the previously reported procedure (36) and then combined as a 50:50 mixture (9/10-NO2-OA) for the experiments described herein. Chemicals, agonists and inhibitors: sulforaphane (SFN) was from LKT Laboratories (St. Paul, MN) and rosiglitazone (RSG) was from Cayman Chemicals (Ann Arbor, MI). Kinase inhibitors were from Cell Signaling Technologies, Danvers, MA (LY294002), Sigma-Aldrich, St. Louis, MO (staurosporine, Ro-31-8220), and Promega, Madison, WI (PD98059). Antibodies: antibodies directed against human Nrf2, topoisomerase I, β-actin, JNK, and phospho-JNK were from Santa Cruz Biotechnologies (Santa Cruz, CA) and against AKT, phospho-AKT, ERK, and phospho-ERK were from Cell Signaling.
Expression vectors and reporter genes: Use of the firefly luciferase reporter genes driven by the minimal TK promoter and three tandem PPREs, pPPREx3-tk-LUC (37), and the ARE derived from the murine gsta1 gene, pARE-TI-LUC (38); the control CMV-Renilla reporter gene, pGL4.75; and the human PPARγ1 expression vector, pcDNA3-PPARγ (39) have been described previously (35, 40). Additionally, the Renilla luciferase reporter vector driven by a minimal promoter and the murine gsta1 ARE, pARE-TI-REN, was constructed by replacing the luciferase cDNA in pARE-TI-LUC with the 1235 bp HindIII/BamHI fragment from pGL4.75 encoding the Renilla luciferase cDNA.
Cell culture, inductions, transfections, and assays
All cultures, inductions and transfections were accomplished with MCF7 breast cancer cells in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 10 μg/ml ciprofloxacin. Transfections: all were done using SuperFect Transfection Reagent (Qiagen, Valencia, CA) at a SuperFect:DNA ratio of 5:1 (μl:μg) according to the manufacturer’s recommendations. 4×104 cells were seeded in 12 well tissue culture plates. Twenty-four hours later cells were transfected with the following vector DNA combinations: Transfection scheme 1) For detection of ARE- or PPRE-dependent transcription in separate transfections, cells were co-transfected with, per well, 0.2–0.4 μg pARE-TI-LUC or pPPREx3-tk-LUC plus 0.1–0.2 μg pcDNA3-PPARγ and 20–40 pg pGL4.75 (control to correct for differences in transfection efficiencies). Transfection scheme 2) For simultaneous detection of ARE- and PPRE-dependent transcription, cells were co-transfected with 0.2 μg pARE-TI-REN and 0.2 μg pPPREx3-tk-LUC plus 0.1–0.2 μg pcDNA3-PPARγ. Approximately 17–20 hr later, cells were treated with nitroalkene fatty acid (NA), RSG, SFN, or vehicle control according to the induction protocol (below). Cells were harvested at various timed intervals thereafter for Dual Luciferase assay (Promega, Madison, WI). Inductions: all were accomplished by exposing cells to variable concentrations of NAs (9/10-NO2-OA, total NO2-LA, or 12-NO2-LA), SFN, RSG, or vehicle (dimethyl sulfoxide) control for 3 hr in complete medium supplemented with 10% fetal bovine serum. At the end of the 3 hr exposures, medium was replaced (minus inducing agent) and cells harvested at variably timed intervals thereafter for Western blot (40) or Dual Luciferase assay (Promega). For some experiments, kinase inhibitors were added 1 hr prior to induction and were maintained in the medium throughout the subsequent 3 hr exposure and beyond until cell harvesting.
Q-RT-PCR analysis of endogenous gene expression induced by NA
MCF7/PPARG1-4′, a cell line derived from parental MCF7 cells stably expressing PPARγ (15), cells were used to examine the induction of PPARγ/PPRE-responsive (FABP4/aP2) and Nrf2/ARE-responsive (NQO1) endogenous genes. Cells were treated with variable concentrations of 12-NO2-LA or vehicle for 3 hr, medium replaced, and cells harvested for total RNA (Trizol Reagent, Invitrogen) 9 hr later. From total RNA, cDNAs were generated by the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) and used for Q-RT-PCR with iQ Real-Time SYBR Green Supermix (Bio-Rad) and an ABI Prism 7700 thermocycler as described (41). From reference cDNA preparations, standard curves were generated for NQO1, FABP4/aP2, and GAPDH from which relative mRNA expression levels were determined for unknown samples. Levels of NQO1 and FABP4/aP2 mRNA were expressed in arbitrary units. Levels of GAPDH (housekeeping control) mRNA expression did not differ significantly between cells treated with 12-NO2-LA and untreated controls. Oligonucleotide pairs used for amplifications were: NQO1, TGCAGCGGCTTTGAAGAAGAAAGG and TCGGCAGGATACTGAAAGTTCGCA; FABP4/aP2, ACTGCAGCTTCCTTCTCACCTTGA and TGGCAAAGCCCACTCCTACTTCTT; and GAPDH, GAAGGTGAAGGTCGGAGTC and GAAGATGGTGATGGGATTTC.
Results
Time and concentration dependent activation of ARE-driven transcription by NAs in MCF7 cells
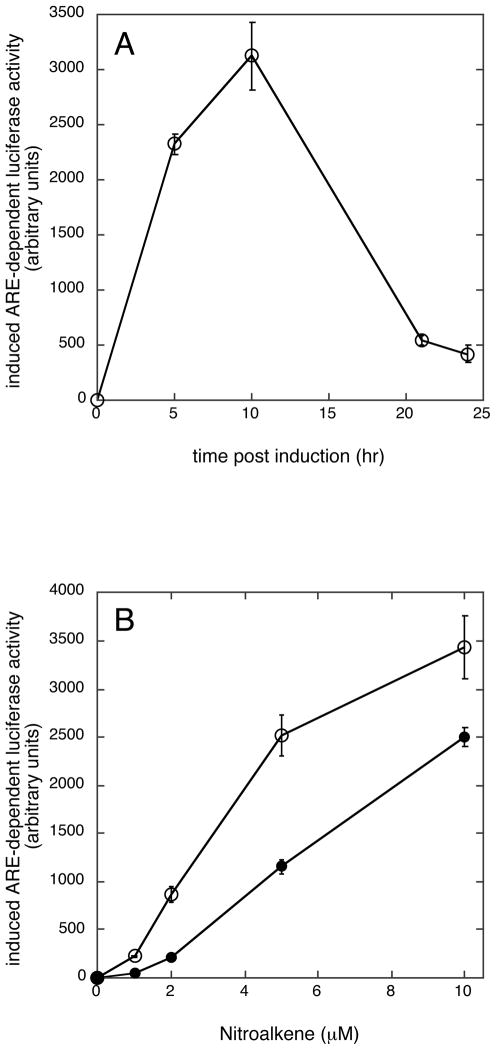
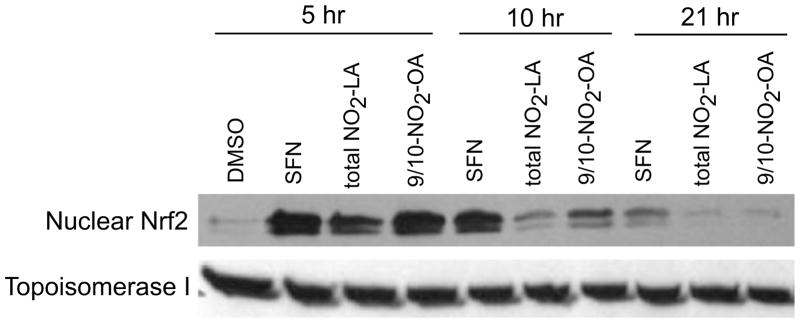
Treatment of MCF7 breast cancer cells with 9/10-NO2-OA results in a time-dependent activation of ARE driven transcription which reached a maximum at about 10 hr after the start of induction (Fig. 1A). Activation of ARE-driven transcription with nitrated derivatives of both oleic and linoleic acid is concentration dependent although 9/10-NO2-OA is somewhat more potent than total NO2-LA in this capacity (Fig. 1B). Induction of ARE-driven transcription is associated with a striking accumulation of nuclear Nrf2 in both NA-treated (total NO2-LA and 9/10-NO2-OA) and positive control (SFN-treated) cells (Fig. 2). For NAs, Nrf2 levels peak at 5 hr post-initiation of induction and decline thereafter. Peak reporter gene activity occurs later (Fig. 1A)—presumably reflecting a lag period during which mRNA and protein are accumulating in the cytosol.
Fig. 1. Induction of ARE/Nrf2-driven transcription by nitroalkene fatty acids (NAs) in MCF7 breast cancer cells is time and concentration dependent.
Cells transfected with the ARE-luciferase reporter according to transfection scheme 1 (Experimental Procedures) were exposed to 10 μM 9/10-NO2-OA (open circles), total NO2-LA (closed circles), or vehicle control for 3 hr. Medium was changed and incubations (minus NA) were continued for variable time intervals from the start of NA induction (panel A) or for 10 hr from the start of NA induction (panel B). Data was corrected for variations in transfection efficiencies using measurements of activity encoded by the co-transfected CMV-Renilla luciferase control, pGL4.75, and was expressed as induced luciferase activity in excess of vehicle controls (arbitrary units) ± 1 sd (n=3).
Fig. 2. Accumulation of nuclear Nrf2 in cells treated with NA.
Cells were treated for 3 hr with 10 μM total NO2-LA, 9/10-NO2-OA, or SFN (positive control). Cells were incubated for variable time periods after removing the inducing agent and harvested for preparation and Western blotting of nuclear protein 5, 10, and 21 hr after the start of NA or SFN induction (40) using antibodies directed against Nrf2 or topoisomerase I loading control.
Role of protein kinase signaling pathways in NA-induction of ARE/Nrf2-mediated transcription
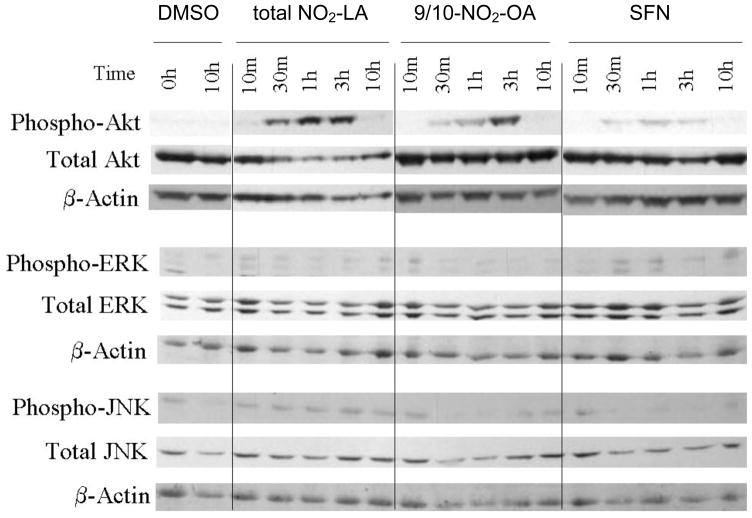
It has been recently shown that NO2-OA forms covalent adducts with key cysteine residues of Keap1 (25). These reactions presumably account, in part, for stabilization and accumulation of Nrf2 and its mediation of ARE-dependent transcription. To determine whether other signaling pathways are involved in Nrf2/ARE-transcription activation, we examined the roles of MAPK, PI3K and PKC signaling—pathways that have previously been implicated in Nrf2-dependent transcription in other systems (26–34). Indeed, PI3K/AKT signaling is activated by NAs (total NO2-LA and 9/10-NO2-OA) and, to a lesser extent, SFN as evidenced by a time-dependent phosphorylation of AKT (Fig. 3). That such activation has functional consequence in NA-induction of Nrf2/ARE-driven transcription is supported by the finding that PI3K inhibitors, wortmannin and LY294002, significantly attenuate NA (and SFN) induction of ARE-reporter gene activity (Fig. 4).
Fig. 3. Protein kinase activation in response to NA treatment.
Cells were treated with NAs or SFN for the indicated times up to 3 hr. For 10 hr incubations, medium was changed to remove NA or SFN and incubations continued an additional 7 hr. Cells were harvested at the indicated times after the start of NA or SFN treatment intervals. Total cellular protein was prepared for SDS-PAGE and Western blotting as described (40) using antibodies directed against the indicated total and phosphorylated signaling kinases or β-actin loading control.
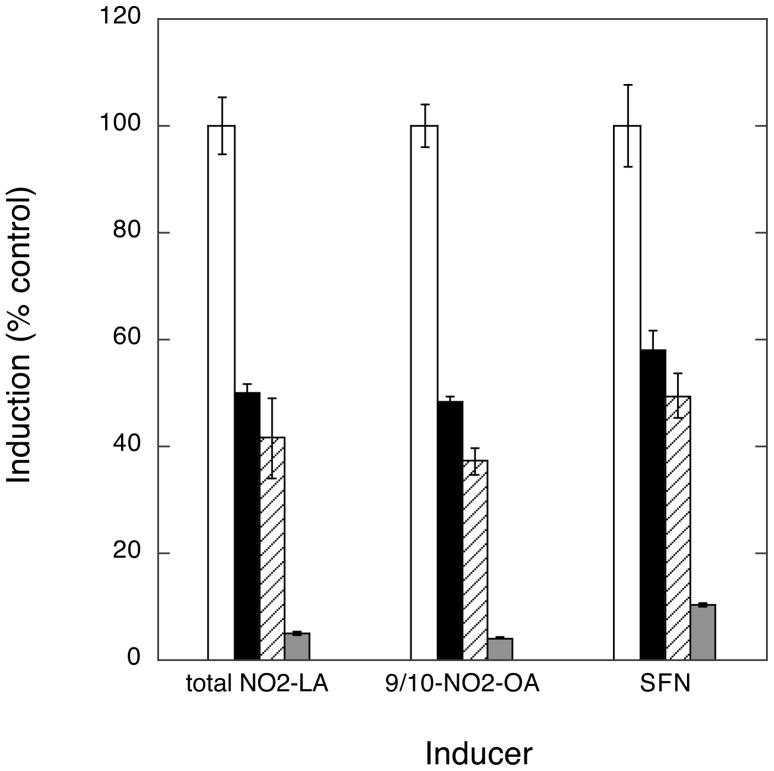
Fig. 4. PI3K activity is required for full activation of ARE/Nrf2-dependent transcription by NAs.
Cells were transfected with the ARE-luciferase reporter gene according to transfection scheme 1. One hr prior to the addition of 10 μM inducing agent (total NO2-LA, 9/10-NO2-OA, or SFN) or vehicle control, cells were treated with PI3K inhibitors 0.5 μM wortmannin (black bars) or 25 μM LY294002 (cross-hatched bars), or no inhibitor (open bars). Inhibitors remained in the medium throughout the subsequent 3 hr induction and until harvest 10 hr after the start of induction. Data was corrected for variations in transfection efficiencies and expressed as % maximum fold induction of control (minus inhibitors, open bars). Error bars: ± 1 sd (n=3).
Similarly, PKC is implicated in NA-induction of ARE-dependent transcription by experiments shown in Fig. 5: two different pan-PKC inhibitors, staurosporine and Ro-31-8220, significantly attenuate NA and SFN-induced transcription.
Fig. 5. PKC activity is required for optimal activation of ARE/Nrf2-dependent transcription by NAs.
These experiments were conducted exactly as described in Fig. 4 except that PKC inhibitors were used: staurosporine (15 nM, black bars; 50 nM cross-hatched bars), Ro-31-8220 (shaded bars). No inhibitor controls are shown as open bars. Data is expressed as % maximum fold induction ± 1 sd (n=3).
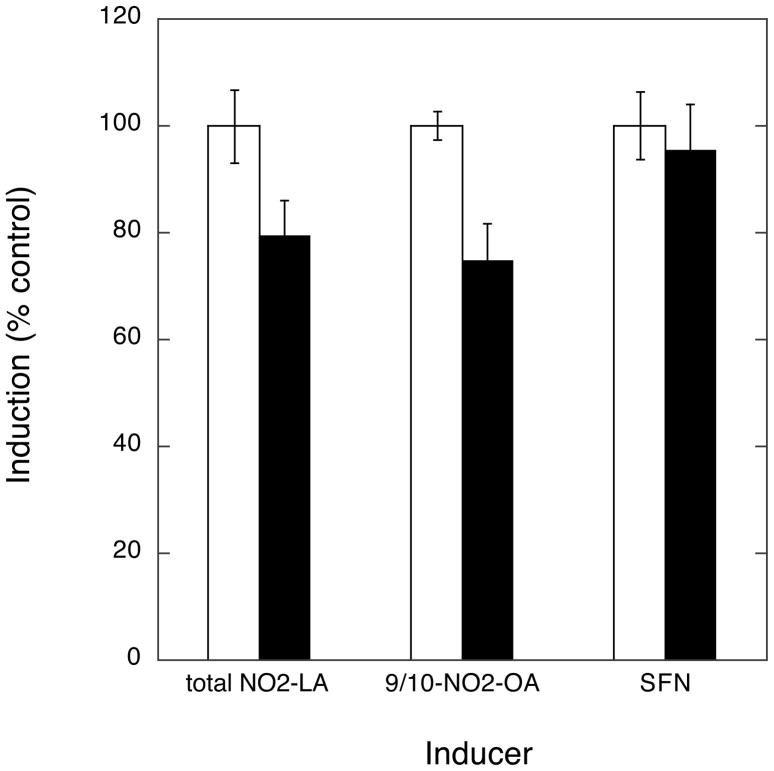
In contrast, no significant activation of the ERK or JNK pathways by NA or SFN was evident as shown by the absence of changes in phospho-ERK or phospho-JNK (Fig. 3). Consistent with the lack of ERK/MAPK involvement is the finding that inhibition of the upstream activator, MEK1, by PD98059 had minimal effect on NA- or SFN-induced ARE-reporter gene transcription (Fig. 6).
Fig. 6. Role of MAPK pathway in NA-induced ARE/Nrf2-dependent transcription.
The effect of the MEK1 inhibitor, PD98059 (20 μM, black bars), on induction of ARE/Nrf2-mediated transcription by 10 μM total NO2-LA, 9/10-NO2-OA, or SFN control was accomplished as described (Figs. 4 and 5). Data is expressed as a percent of induction observed in the absence of inhibitor (open bars). Error bars: ± 1 sd (n=3).
NAs activate ARE/Nrf2- and PPRE/PPARγ-dependent transcription by distinct signaling pathways and with different potencies
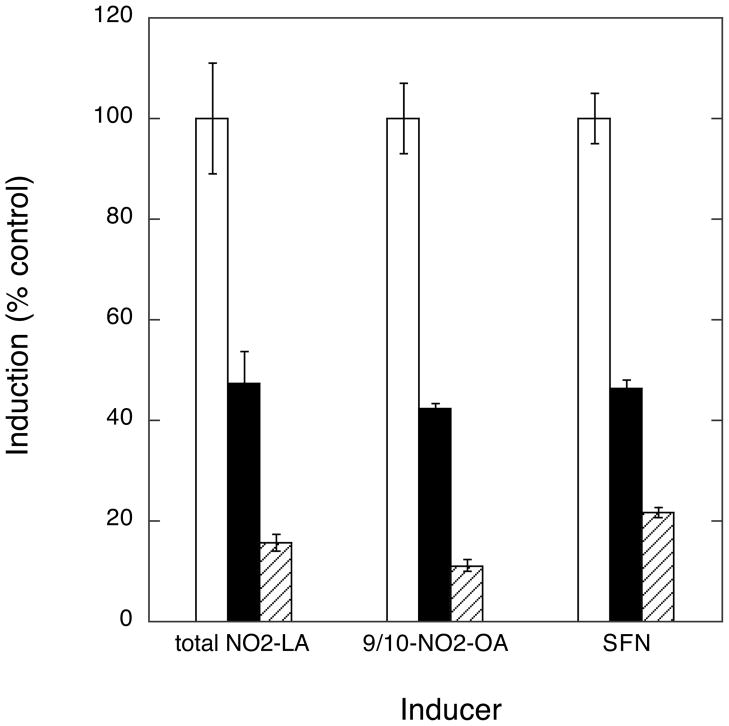
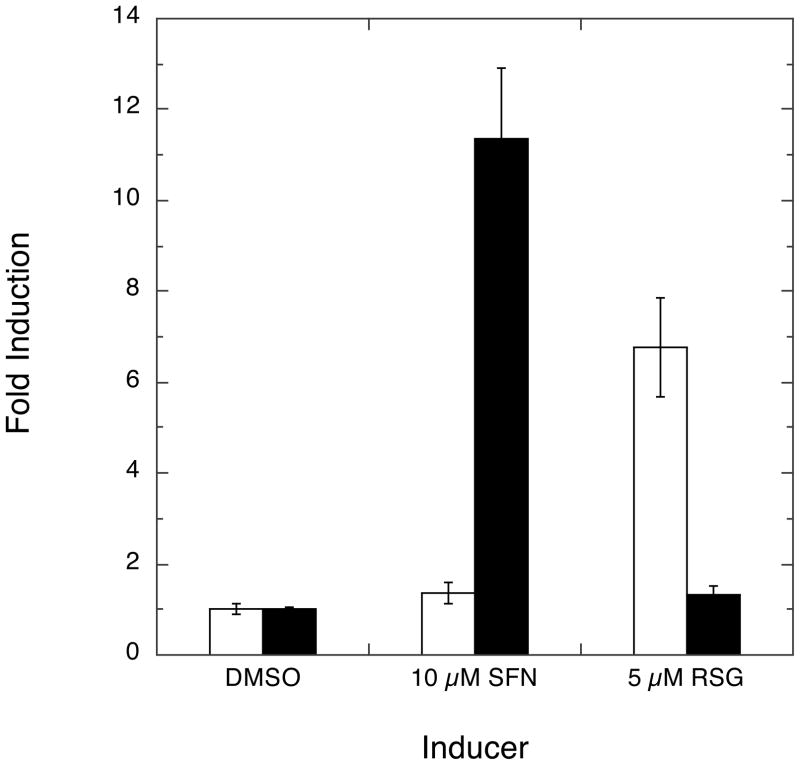
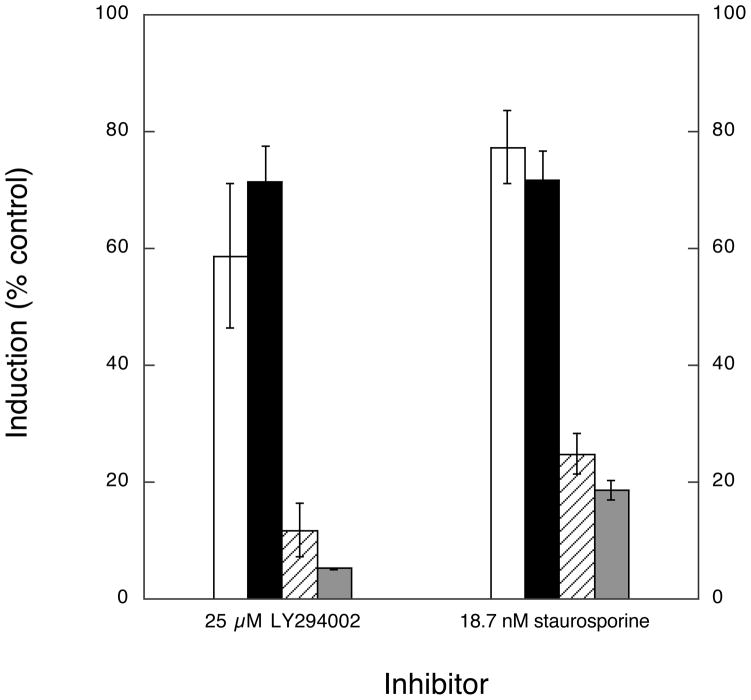
There is considerable opportunity for cross-talk between signaling pathways that have as their endpoints the activation of transcription from ARE- versus PPRE-containing genes. For example, recent results have linked Nrf2 and the induction of PPARγ (42). It was therefore important to establish if activation of these alternative transcriptional programs by NA could be dissociated or, instead, if these pathways were interdependent through the sharing of other common upstream effectors. First, in order to reveal any potential cross-talk between these transcriptional programs, we examined induction of transcription by the nutraceutical activator of ARE/Nrf2-directed transcription, SFN, and the relatively selective PPARγ agonist, RSG. As shown by experiments (Fig. 7) in which cells were co-transfected with PPRE-firefly luciferase and ARE-Renilla luciferase reporters, SFN resulted in selective induction of ARE/Nrf2-dependent transcription while RSG treatment exclusively induced PPRE/PPARγ-dependent transcription—suggesting that, at least in this system, there is little cross-talk between the transcriptional programs. Next, using the PI3K and PKC inhibitors, we asked whether the abilities of NAs to activate the alternative transcription programs could be dissociated. Indeed, as shown in Fig. 8, treatment with either inhibitor significantly attenuated induction of ARE-driven transcription by both NA (12-NO2-LA) and SFN. In contrast, these kinase inhibitors had very modest effects on activation of PPRE-driven transcription by either 12-NO2-LA or the control agonist, RSG. Hence, we conclude that the ARE/Nrf2 and PPRE/PPARγ activating properties of NAs involve distinct pathways that are distinguishable by preferential attenuation of ARE-driven transcription by PKC and PI3K inhibitors.
Fig. 7. Absence of cross-talk between ARE/Nrf2- and PPRE/PPARγ-directed transcription in response to treatment with SFN and rosiglitazone (RSG).
MCF7 cells were co-transfected with PPRE-driven firefly luciferase and ARE-driven Renilla luciferase reporters according to transfection scheme 2 (Experimental Procedures). Following induction with 10 μM SFN or 5 μM RSG and subsequent incubations, fold induction over vehicle (DMSO) control of PPRE/PPARγ-driven (open bars) and ARE/Nrf2-driven (black bars) was determined. Error bars: ± 1 sd (n=3).
Fig. 8. Resolution of NA-induced ARE/Nrf2 versus PPRE/PPARγ transcription programs by the preferential inhibition of ARE/Nrf2 activity with inhibitors of PI3K and PKC.
Cells were co-transfected with PPRE-driven and ARE-driven reporters according to transfection scheme 2 (Experimental Procedures). Transfected cells were treated with 10 μM NA (12-NO2-LA), 10 μM SFN, or 5 μM RSG ± inhibition with the PI3K or PKC selective inhibitors, LY294002 or staurosporine, as shown. Data is expressed as % maximum fold induction (minus inhibitor) ± 1 sd (n=3). PPRE/PPARγ-driven transcription: shown are cells treated with RSG (open bars) and 12-NO2-LA (black bars). ARE/Nrf2-driven transcription: shown are cells treated with SFN (cross-hatched bars) and 12-NO2-LA (shaded bars).
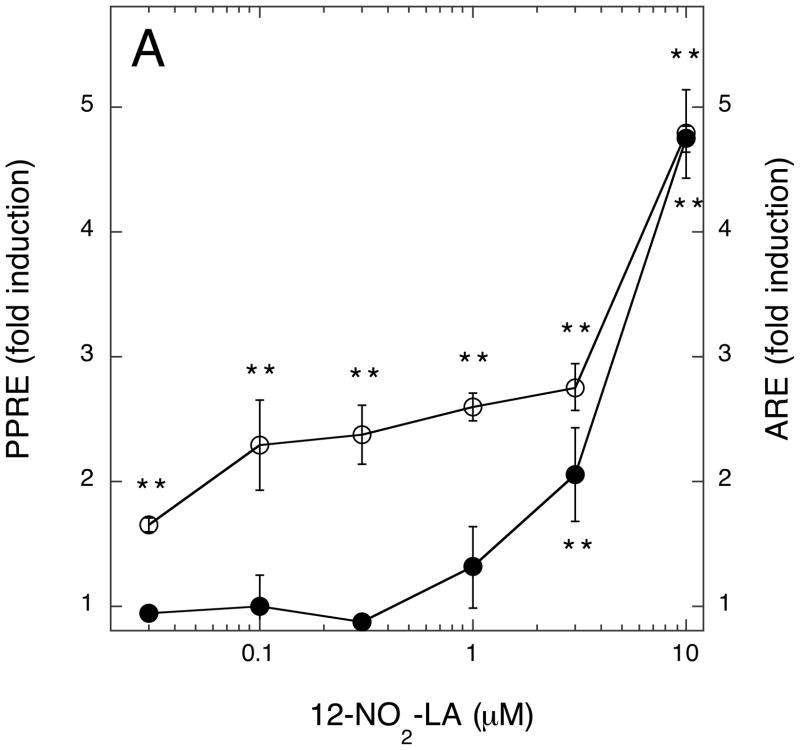
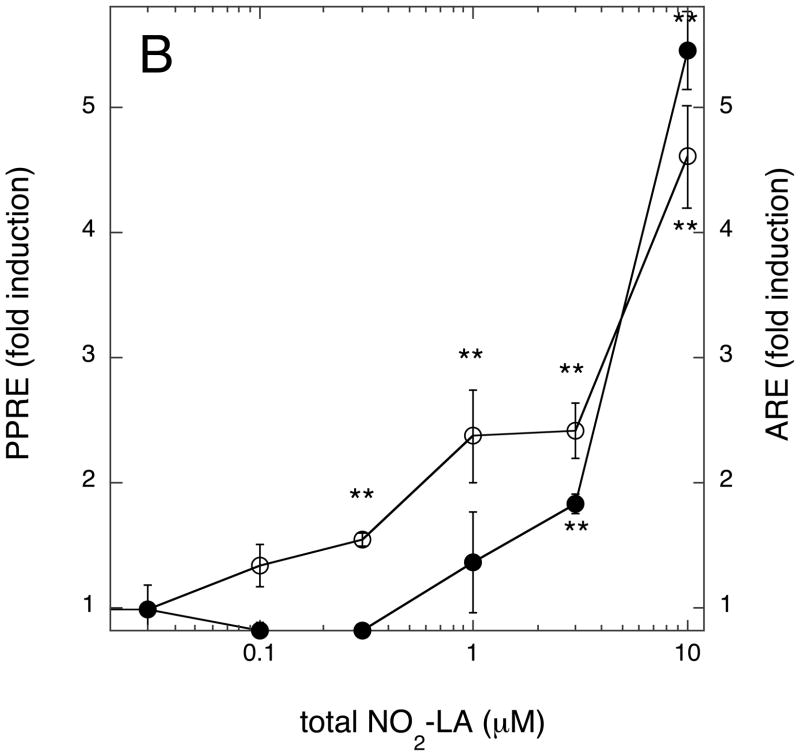
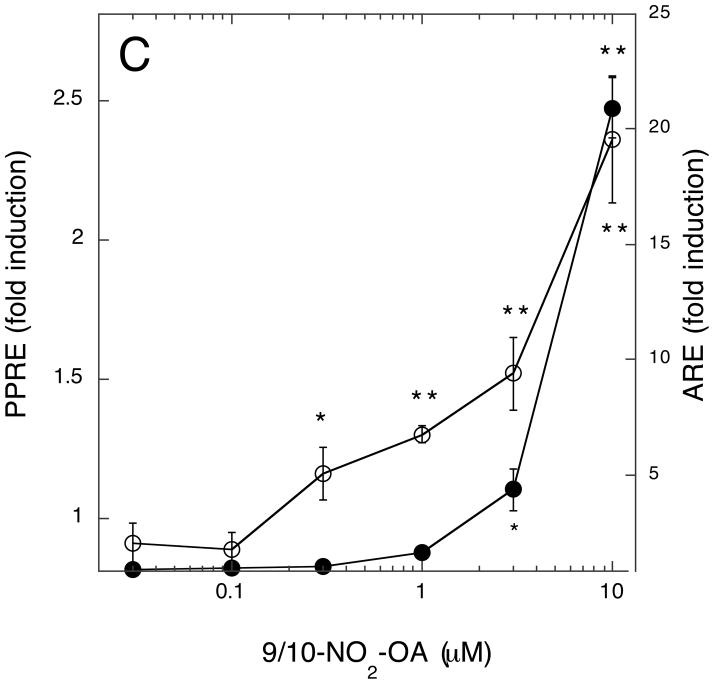
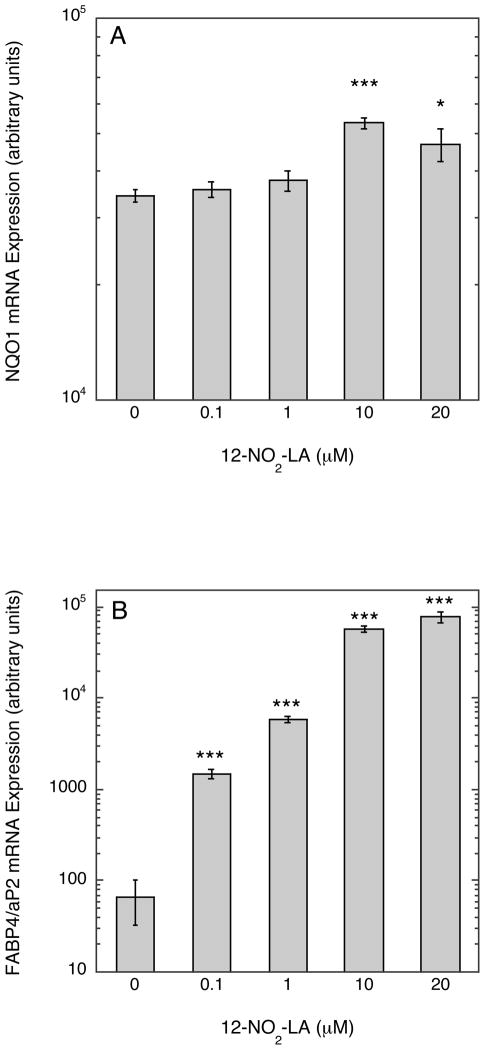
Lastly, we compared the dose-response profiles of NA-activated ARE/Nfr2- versus PPRE/PPARγ-driven transcription. This was important for two reasons: first to ascertain the relative strength of NA towards activation of these alternative transcription programs; and, secondly, to determine at what NA concentrations measurable effects on transcription could be seen and, hence, to determine which of the two transcription programs will dominate upon activation by NAs at concentrations likely to be encountered in vivo. Three NA preparations were used—9/10-NO2-OA, total NO2-LA, and 12-NO2-LA—the latter chosen because, of all NAs previously tested, this isomer has the greatest PPARγ agonist activity at submicromolar concentrations (15, 18). As shown for all three NAs (Fig. 9A-C), significant induction of PPRE/PPARγ-directed transcription occurs at much lower NA concentrations than for induction of ARE/Nrf2-directed transcription. For 12-NO2-LA (Fig. 9A), significant PPRE-dependent transcription was evident at the lowest concentration tested (30 nM) and, for the other NAs (Fig. 9B & C), at somewhat higher concentrations (≥ 300 nM). In contrast, significant activation of ARE/Nrf2-driven transcription was not observed until considerably higher concentrations, ≥ 3 μM, of NA were added. Finally, as also observed in Fig. 1B, induction of ARE-dependent transcription at higher NA concentrations is more robust for 9/10-NO2-OA (Fig. 9C) than for the NO2-LA isomers (Fig. 9A & B). Additionally, we examined the concentration dependent induction of Nrf2-responsive (NQO1) and PPARγ-responsive (FABP4/aP2) endogenous genes (14, 43) by 12-NO2-LA using the MCF7/PPARG1-4′ cell line—a cell line derived from MCF7 that stably expresses recombinant PPARγ (15). As shown in Fig. 10 and in agreement with reporter gene findings, induction of the Nrf2-responsive NQO1 mRNA (Fig. 10A) does not occur at low 12-NO2-LA concentrations (≤ 1 μM); rather, induction reaches significance only at much higher NA concentrations (10 and 20 μM). In contrast, robust induction of PPARγ-responsive FABP4/aP2 mRNA is observed at the lowest concentration, 0.1 μM, of 12-NO2-LA used (Fig. 10B).
Fig. 9. Concentration-dependence of NA-mediated induction of ARE/Nrf2- versus PPRE/PPARγ-driven transcription programs.
Cells were transfected with ARE or PPRE reporter genes according to transcription scheme 1. Transfected cells were treated for 3 hr with the indicated concentrations of NAs—12-NO2-LA (panel A), total NO2-LA (panel B), or 9/10-NO2-OA (panel C). Medium was then replaced (minus NA) and cells harvested for luciferase determinations, corrected for variations in transfection efficiencies, 12 hr later. Data is expressed as fold induction (luciferase activity with NA ÷ luciferase activity without NA) for PPRE/PPARγ-dependent (open circles) or ARE/Nrf2-dependent (closed circles) reporter gene activity ± 1 sd (n=3–4). NA-induced values that are significantly different from un-induced controls are indicated: *, P<0.05; **, P<0.01 (unpaired, two-tailed t test with Welch correction, GraphPad InStat3).
Fig. 10. Induction of Nrf2- (NQO1) and PPARγ-responsive (FABP4/aP2) endogenous gene expression by 12-NO2-LA.
MCF7-derived cells stably expressing recombinant PPARγ (MCF7/PPARG1-4′) were exposed to variable concentrations of 12-NO2-LA or vehicle control for 3 hr and harvested 9 hr later (12 hr after the start of 12-NO2-LA exposures) for mRNA quantification by Q-RT-PCR as described in Experimental Procedures. Shown are levels of NQO1 (A) and FABP4/aP2 (B) mRNAs expressed in arbitrary units. Bars represent the means of replicate independent experiments (n=6) ± 1 sem. * P<0.05 and *** P<0.001 (t test).
Discussion
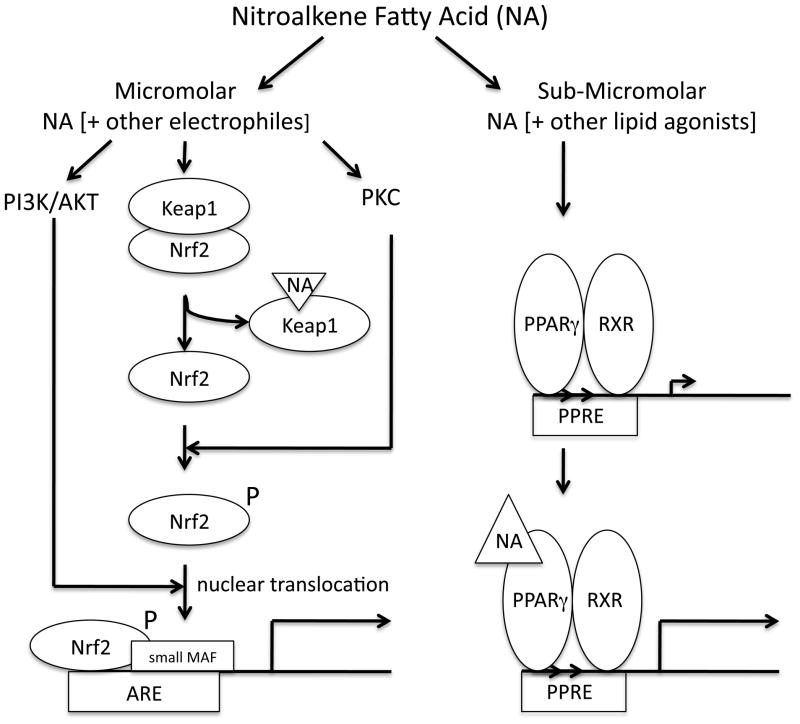
The studies described herein establish that NA-mediated inductions of ARE/Nrf2- and PPRE/PPARγ-dependent transcription occur by entirely different pathways in MCF7 breast cancer cells. The body of evidence presented in this work and the work of others are consistent with a mechanism of ARE/Nrf2-transcription activation that involves both direct modification of Keap1 by NA adduction at key cysteine residues (25) and redox activation of PKC plus PI3K/AKT signaling pathways by the NA electrophiles. As summarized in Fig. 11, PKC likely activates Nrf2-mediated transcription by phosphorylation of Nrf2—in particular, serine 40, as described by others (31–33). This modification, along with Keap1 adduct formation with NA, very likely disrupts Keap1-Nrf2 interactions facilitating dissociation, stabilization, and nuclear translocation of Nrf2. Our data would suggest that both processes—adduct formation and PKC activation—are required for full activation of Nrf2-mediated transcription by NAs. Our data would also suggest a crucial role for the PI3K/AKT pathway in NA induction of ARE/Nrf2-directed transcription. The mechanism whereby PI3K/AKT may stimulate this activity is less clear but could occur via indirect activation of PKC (44) or, alternatively, by facilitating the cytoplasmic-to-nuclear transport of Nrf2 as reported (28, 45).
Fig. 11.
Model showing the distinct pathways involved in nitroalkene fatty acid (NA) induction of ARE- and PPRE-dependent transcription programs.
In contrast, NA activation of PPRE/PPARγ-directed transcription is, in our system, independent of PKC, PI3K/AKT, or the MAPK pathways tested. Based upon previous studies from our laboratory and others showing potent activation of PPARγ (6, 7, 15, 35), avid binding to recombinant PPARγ (15, 18), and specific occupancy of the ligand-binding pocket of PPARγ (16, 17) by NAs, it is most likely that NAs induce PPARγ transcriptional activity directly via agonist-receptor interactions. Importantly, measurable and significant PPARγ activation occurs at much lower concentrations than those required for Nrf2-driven transcription activation. This is especially noteworthy for the naturally occurring 12-NO2-LA isomer that supports PPARγ activation at concentrations at least as low as 30 nM. This observation is particularly relevant physiologically because, in contrast to earlier estimates (46), under normal conditions, the levels of circulating NAs measured in vivo are considerably lower—at best, in the nanomolar range (47)—than originally thought. While it is true that NA concentrations in circulation or in local cellular compartments and membranes could be much higher under conditions of inflammation and oxidative/nitrosative stress, and it is true that much higher and biologically active levels can be achieved with direct infusion of NAs as pharmaceuticals (48, 49), it is unlikely NAs will exceed nanomolar levels under normal or common pathological conditions. Thus it would appear likely that under such conditions, NA activation of PPARγ and its associated biological effects—anti-inflammation, insulin sensitization, altered lipid and carbohydrate metabolism, and others—will dominate over NA effects upon ARE/Nrf2 associated responses. Therefore, by themselves, the contribution of endogenous NAs to activation of the ARE/Nrf2 cytoprotective response are estimated to be relatively minor; but, considered in combination with other endogenous and nutraceutical electrophiles, the NAs may contribute a meaningful additive effect to overall activation of the ARE/Nrf2 transcriptional program. A final note on NA concentrations: in our experiments, NAs were administered exogenously and in the presence of 10% fetal bovine serum. Given the known propensity of NAs to react with protein sulfhydryls, the actual local concentration of free NAs at their Keap1 and PPARγ targets is unknown and may well be less than the concentration added to the medium. Nevertheless, this possibility does not affect the conclusion that NAs, at low concentrations, are significantly more potent activators of PPARγ-dependent over Nrf2-dependent transcription.
Footnotes
This work was supported by grants from the NIH (CA 70338) and the Department of Defense (W81XWH-10-1-0439)
Abbreviations: ARE, antioxidant response element; ERK, extracellular signal-regulated kinase; FABP4/aP2, fatty acid-binding protein 4; GAPDH, glyceraldehye 3-phosphate dehydrogenase; JNK, c-Jun-NH2-kinase; Keap1, Kelch-like ECH-associated protein 1; MAPK, mitogen-activated protein kinase; NA, nitroalkene fatty acid; NO2-LA and NO2-OA, nitrolinoleic and nitro-oleic acids; NQO1, NAD(P)H quinone oxidoreductase 1; Nrf2, nuclear factor erythroid 2-related factor; PI3K, phosphatidylinositol 3-kinase; PKC, protein kinase C; PPARγ, peroxisomal proliferator-activated receptor gamma; PPRE, PPAR responsive element; RSG, rosiglitazone; SFN, sulforaphane
References
- 1.Freeman BA, Baker PR, Schopfer FJ, Woodcock SR, Napolitano A, d’Ischia M. Nitro-fatty acid formation and signaling. J Biol Chem. 2008;283:15515–15519. doi: 10.1074/jbc.R800004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim DG, Sweeney S, Bloodsworth A, White CR, Chumley PH, Krishna NR, Schopfer F, O’Donnell VB, Eiserich JP, Freeman BA. Nitrolinoleate, a nitric oxide-derived mediator of cell function: Synthesis, characterization, and vasomotor activity. Proc Natl Acad Sci (USA) 2002;99:15941–15946. doi: 10.1073/pnas.232409599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coles B, Bloodsworth A, Clark SR, Lewis MJ, Cross AR, Freeman BA, O’Donnell VB. Nitrolinoleate Inhibits Superoxide Generation, Degranulation, and Integrin Expression by Human Neutrophils: Novel Antiinflammatory Properties of Nitric Oxide-Derived Reactive Species in Vascular Cells. Circ Res. 2002;91:375–381. doi: 10.1161/01.res.0000032114.68919.ef. [DOI] [PubMed] [Google Scholar]
- 4.Cui T, Schopfer FJ, Zhang J, Chen K, Ichikawa T, Baker PRS, Batthyany C, Chacko BK, Feng X, Patel RP, Agarwal A, Freeman BA, Chen YE. Nitrated Fatty Acids: Endogenous Anti-inflammatory Signaling Mediators. J Biol Chem. 2006;281:35686–35698. doi: 10.1074/jbc.M603357200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kansanen E, Jyrkkannen HK, Volger OL, Leinonen H, Kivela AM, Hakkinen SK, Woodcock SR, Schopfer FJ, Horrevoets AJ, Yla-Herttuala S, Freeman BA, Levonen AL. Nrf2-dependent and -independent Responses to Nitro-fatty Acids in Human Endothelial Cells. J Biol Chem. 2009;284:33233–33241. doi: 10.1074/jbc.M109.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker PRS, Lin Y, Schopfer FJ, Woodcock SR, Groeger AL, Batthyany C, Sweeney S, Long MH, Iles KE, Baker LMS, Branchaud BP, Chen YE, Freeman BA. Fatty Acid Transduction of Nitric Oxide Signaling: Multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor γ ligands. J Biol Chem. 2005;280:42464–42475. doi: 10.1074/jbc.M504212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schopfer FJ, Lin Y, Baker PRS, Cui T, Garcia-Barrio M, Zhang J, Chen K, Chen YE, Freeman BA. Nitrolinoleic acid: An endogenous peroxisome proliferator-activated receptor γ ligand. Proc Natl Acad Sci (USA) 2005;102:2340–2345. doi: 10.1073/pnas.0408384102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villacorta L, Zhang J, Garcia-Barrio MT, Chen X-l, Freeman BA, Chen YE, Cui T. Nitro-linoleic acid inhibits vascular smooth muscle cell proliferation via the Keap1/Nrf2 signaling pathway. Am J Physiol Heart Circ Physiol. 2007;293:H770–776. doi: 10.1152/ajpheart.00261.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear Receptors and Lipid Physiology: Opening the X-Files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 10.Szanto A, Nagy L. The many faces of PPARγ: anti-inflammatory by any means? Immunobiology. 2008;213:789–803. doi: 10.1016/j.imbio.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Evans R, Barish G, Wang Y. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 12.Kintscher U, Law RE. PPARγ-mediated insulin sensitization: the importance of fat versus muscle. Am J Physiol Endocrinol Metab. 2005;288:E287–291. doi: 10.1152/ajpendo.00440.2004. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen T, Nioi P, Pickett CB. The Nrf2-Antioxidant Response Element Signaling Pathway and Its Activation by Oxidative Stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayes J, McMahon M, Chowdhry S, Dinkova-Kostova A. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal. 2010;13:1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 15.Alexander R, Wright M, Gorczynski M, Smitherman P, Akiyama T, Wood H, Berger J, King S, Morrow C. Differential potencies of naturally occurring regioisomers of nitrolinoleic acid in PPARγ activation. Biochemistry. 2009;48:492–498. doi: 10.1021/bi8016747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schopfer FJ, Cole MP, Groeger AL, Chen C-S, Khoo NKH, Woodcock SR, Golin-Bisello F, Motanya UN, Li Y, Zhang J, Garcia-Barrio MT, Rudolph TK, Rudolph V, Bonacci G, Baker PRS, Xu HE, Batthyany CI, Chen YE, Hallis TM, Freeman BA. Covalent Peroxisome Proliferator-activated Receptor γ Adduction by Nitro-fatty Acids. J Biol Chem. 285:12321–12333. doi: 10.1074/jbc.M109.091512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Zhang J, Schopfer FJ, Martynowski D, Garcia-Barrio MT, Kovach A, Suino-Powell K, Baker PR, Freeman BA, Chen YE, Xu HE. Molecular recognition of nitrated fatty acids by PPARγ. Nat Struct Mol Biol. 2008;15:865–867. doi: 10.1038/nsmb.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorczynski M, Smitherman P, Akiyama T, Wood H, Berger J, King S, Morrow C. Activation of PPAR by nitroalkene fatty acids: importance of nitration position and degree of unsaturation. J Med Chem. 2009;52:4631–4639. doi: 10.1021/jm900326c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB Protein Is an Adaptor That Bridges Nrf2 to a Cul3-Based E3 Ligase: Oxidative Stress Sensing by a Cul3-Keap1 Ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent Proteasomal Degradation of Transcription Factor Nrf2 Contributes to the Negative Regulation of Antioxidant Response Element-driven Gene Expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 21.Hong F, Freeman ML, Liebler DC. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem Res Toxicol. 2005;18:1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- 22.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci (USA) 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, Yamamoto M. Physiological Significance of Reactive Cysteine Residues of Keap1 in Determining Nrf2 Activity. Mol Cell Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang DD, Hannink M. Distinct Cysteine Residues in Keap1 Are Required for Keap1-Dependent Ubiquitination of Nrf2 and for Stabilization of Nrf2 by Chemopreventive Agents and Oxidative Stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kansanen E, Bonacci G, Schopfer FJ, Linna S, Tong KI, Leinonen H, Woodcock SR, Yamamoto M, Carlberg C, Yla-Herttuala S, Freeman BA, Levonen A-L. Electrophilic nitro-fatty acids activate Nrf2 by a Keap1 cysteine 151-independent mechanism. J Biol Chem. 286:14019–14027. doi: 10.1074/jbc.M110.190710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Chen Y, Sternberg P, Cai J. Essential Roles of the PI3 Kinase/Akt Pathway in Regulating Nrf2-Dependent Antioxidant Functions in the RPE. Invest Ophthalmol Vis Sci. 2008;49:1671–1678. doi: 10.1167/iovs.07-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JM, Hanson JM, Chu WA, Johnson JA. Phosphatidylinositol 3-Kinase, Not Extracellular Signal-regulated Kinase, Regulates Activation of the Antioxidant-Responsive Element in IMR-32 Human Neuroblastoma Cells. J Biol Chem. 2001;276:20011–20016. doi: 10.1074/jbc.M100734200. [DOI] [PubMed] [Google Scholar]
- 28.Kang KW, Lee SJ, Park JW, Kim SG. Phosphatidylinositol 3-Kinase Regulates Nuclear Translocation of NF-E2-Related Factor 2 through Actin Rearrangement in Response to Oxidative Stress. Mol Pharm. 2002;62:1001–1010. doi: 10.1124/mol.62.5.1001. [DOI] [PubMed] [Google Scholar]
- 29.Zipper LM, Mulcahy RT. Erk Activation Is Required for Nrf2 Nuclear Localization during Pyrrolidine Dithiocarbamate Induction of Glutamate Cysteine Ligase Modulatory Gene Expression in HepG2 Cells. Toxicol Sci. 2003;73:124–134. doi: 10.1093/toxsci/kfg083. [DOI] [PubMed] [Google Scholar]
- 30.Xu C, Yuan X, Pan Z, Shen G, Kim JH, Yu S, Khor TO, Li W, Ma J, Kong ANT. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol Cancer Ther. 2006;5:1918–1926. doi: 10.1158/1535-7163.MCT-05-0497. [DOI] [PubMed] [Google Scholar]
- 31.Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser40 by Protein Kinase C in Response to Antioxidants Leads to the Release of Nrf2 from INrf2, but Is Not Required for Nrf2 Stabilization/Accumulation in the Nucleus and Transcriptional Activation of Antioxidant Response Element-mediated NAD(P)H:Quinone Oxidoreductase-1 Gene Expression. J Biol Chem. 2003;278:44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- 32.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by Protein Kinase C Regulates Antioxidant Response Element-mediated Transcription. J Biol Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 33.Niture SK, Jain AK, Jaiswal AK. Antioxidant-induced modification of INrf2 cysteine 151 and PKC-{delta}-mediated phosphorylation of Nrf2 serine 40 are both required for stabilization and nuclear translocation of Nrf2 and increased drug resistance. J Cell Sci. 2009;122:4452–4464. doi: 10.1242/jcs.058537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Numazawa S, Ishikawa M, Yoshida A, Tanaka S, Yoshida T. Atypical protein kinase C mediates activation of NF-E2-related factor 2 in response to oxidative stress. Am J Physiol Cell Physiol. 2003;285:C334–342. doi: 10.1152/ajpcell.00043.2003. [DOI] [PubMed] [Google Scholar]
- 35.Alexander RL, Bates DJ, Wright MW, King SB, Morrow CS. Modulation of nitrated lipid signaling by multidrug resistance protein 1 (MRP1): glutathione conjugation and MRP1-mediated efflux inhibit nitrolinoleic acid-induced, PPARgamma-dependent transcription activation. Biochemistry. 2006;45:7889–7896. doi: 10.1021/bi0605639. [DOI] [PubMed] [Google Scholar]
- 36.Gorczynski M, Huang J, King S. Regio- and stereospecific syntheses and nitric oxide donor properties of (E)-9- and (E)-10-nitrooctadec-9-enoic acids. Org Lett. 2006;8:2305–2308. doi: 10.1021/ol060548w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 38.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci (USA) 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurnell M, Wentworth JM, Agostini M, Adams M, Collingwood TN, Provenzano C, Browne PO, Rajanayagam O, Burris TP, Schwabe JW, Lazar MA, Chatterjee VKK. A Dominant-negative Peroxisome Proliferator-activated Receptor γ (PPARγ) Mutant Is a Constitutive Repressor and Inhibits PPARγ-mediated Adipogenesis. J Biol Chem. 2000;275:5754–5759. doi: 10.1074/jbc.275.8.5754. [DOI] [PubMed] [Google Scholar]
- 40.Sibhatu MB, Smitherman PK, Townsend AJ, Morrow CS. Expression of MRP1 and GSTP1-1 modulate the acute cellular response to treatment with the chemopreventive isothiocyanate, sulforaphane. Carcinogenesis. 2008;29:807–815. doi: 10.1093/carcin/bgn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu M, Miller MS. Determination of murine fetal Cyp1a1 and 1b1 expression by real-time fluorescence reverse transcription-polymerase chain reaction. Toxicol Appl Pharmacol. 2004;201:295–302. doi: 10.1016/j.taap.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Cho H-Y, Gladwell W, Wang X, Chorley B, Bell D, Reddy SP, Kleeberger SR. Nrf2-regulated PPARγ Expression Is Critical to Protection against Acute Lung Injury in Mice. Am J Respir Crit Care Med. 182:170–182. doi: 10.1164/rccm.200907-1047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rival Y, Stennevin A, Puech L, Rouquette A, Cathala C, Lestienne F, Dupont-Passelaigue E, Patoiseau JFO, Wurch T, Junquéro D. Human Adipocyte Fatty Acid-Binding Protein (aP2) Gene Promoter-Driven Reporter Assay Discriminates Nonlipogenic Peroxisome Proliferator-Activated Receptor γ Ligands. J Pharm Exp Ther. 2004;311:467–475. doi: 10.1124/jpet.104.068254. [DOI] [PubMed] [Google Scholar]
- 44.Kim S, Kim S. PKC downstream of PI3-kinase regulates peroxynitrite formation for Nrf2-mediated gsta2 induction. Arch Pharm Res. 2004;27:757–762. doi: 10.1007/BF02980145. [DOI] [PubMed] [Google Scholar]
- 45.Nakaso K, Yano H, Fukuhara Y, Takeshima T, Wada-Isoe K, Nakashima K. PI3K is a key molecule in the Nrf2-mediated regulation of antioxidative proteins by hemin in human neuroblastoma cells. FEBS Lett. 2003;546:181–184. doi: 10.1016/s0014-5793(03)00517-9. [DOI] [PubMed] [Google Scholar]
- 46.Baker PRS, Schopfer FJ, Sweeney S, Freeman BA. Red cell membrane and plasma linoleic acid nitration products: Synthesis, clinical identification, and quantitation. Proc Natl Acad Sci (USA) 2004;101:11577–11582. doi: 10.1073/pnas.0402587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsikas D, Zoerner A, Mitschke A, Homsi Y, Gutzki FM, Jordan J. Specific GC-MS/MS stable-isotope dilution methodology for free 9- and 10-nitro-oleic acid in human plasma challenges previous LC-MS/MS reports. J Chromatography B. 2009;877:2895–2908. doi: 10.1016/j.jchromb.2008.12.062. [DOI] [PubMed] [Google Scholar]
- 48.Cole MP, Rudolph TK, Khoo NK, Motanya UN, Golin-Bisello F, Wertz JW, Schopfer FJ, Rudolph V, Woodcock SR, Bolisetty S, Ali MS, Zhang J, Chen YE, Agarwal A, Freeman BA, Bauer PM. Nitro-fatty acid inhibition of neointima formation after endoluminal vessel injury. Circ Res. 2009;105:965–972. doi: 10.1161/CIRCRESAHA.109.199075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Villacorta L, Chang L, Fan Z, Hamblin M, Zhu T, Chen CS, Cole MP, Schopfer FJ, Deng CX, Garcia-Barrio MT, Feng Y-H, Freeman BA, Chen YE. Nitro-Oleic Acid Inhibits Angiotensin II-Induced Hypertension. Circ Res. 107:540–548. doi: 10.1161/CIRCRESAHA.110.218404. [DOI] [PMC free article] [PubMed] [Google Scholar]