Abstract
Dictyostelium development is induced by starvation. The adenylyl cyclase gene ACA is one of the first genes expressed upon starvation. ACA produces extracellular cAMP that induces chemotaxis, aggregation, and differentiation in neighboring cells. Using insertional mutagenesis we have isolated a mutant that does not aggregate upon starvation but is rescued by adding extracellular cAMP. Sequencing of the mutated locus revealed a new gene, DdMYB2, whose product contains three Myb repeats, the DNA-binding motif of Myb-related transcription factors. Ddmyb2–null cells show undetectable levels of ACA transcript and no cAMP production. Ectopic expression of ACA from a constitutive promotor rescues differentiation and morphogenesis of Ddmyb2–null mutants. The results suggest that development in Dictyostelium starts by starvation-mediated DdMyb2 activation, which induces adenylyl cyclase activity producing the differentiation-inducing signal cAMP.
Keywords: myb-related gene, starvation response, REMI, adenylyl cyclase, cAMP oscillation, transcriptional regulation
Transcription factors and secreted signaling molecules play pivotal roles during development in multicellular organisms. The identification of mutants defective in those factors may provide powerful insight into the mechanisms of developmental regulation. Dictyostelium is an ideal model organism to investigate developmental regulation, because mutants that arrest at a certain stage of development are easy to isolate, and the secreted factor missing in such mutants can be provided by wild-type cells in simple mixing experiments (Loomis 1996; Parent and Devreotes 1996). Dictyostelium cells grow vegetatively as individual amebae feeding on bacteria or defined axenic media. Upon starvation they gather with neighboring cells forming aggregates consisting of up to 105 cells. Aggregating cells relay extracellular cAMP signals and move to the signaling center by chemotaxis to cAMP. After aggregation, cells behave as in more complex multicellular organisms; they differentiate to specific cell types, perform coordinated morphogenetic movement, and finally culminate into a fruiting body consisting of spores on top of a supporting stalk.
Development in Dictyostelium starts with the transcriptional activation triggered by amino acid depletion, inducing the essential molecules to produce and sense extracellular cAMP, such as an adenylyl cyclase, cell-surface cAMP receptors (cARs) and specific G protein α-subunits (Klein et al. 1988; Kumagai et al. 1988, 1989; Pitt et al. 1992). Intercellular signaling by secreted cAMP then induces the expression of another set of genes for the further stages of development (Kimmel and Firtel 1991). Adenylyl cyclase ACA is the key enzyme of this cAMP signaling and its mRNA shows an immediate sharp increase by starvation. Therefore the components that mediate the induction of adenylyl cyclase should have the central role in the growth/development transition in Dictyostelium. Several mutants that lack adenylyl cyclase activity or its activation have been identified. They are called Synag mutants, because they do not aggregate in their clonal populations, missing the cAMP signaling, but do aggregate and proceed to further development in mixed populations with wild-type cells (Theibert and Devreotes 1986).
Using plasmid insertional mutagenesis we have isolated a new Synag mutant with unique biochemical features. It shows poor chemotaxis to cAMP and no cAMP relay, but makes normal fruiting bodies after a few hours of stimulation with experimentally applied cAMP pulses, suggesting a defect in the initiation of cAMP signaling. By sequencing the insertion site we identified a novel myb-related gene, DdMYB2, as a key component that induces Dictyostelium development.
The vertebrate c-myb gene is essential for proliferation and differentiation of hematopoietic cells, and its truncated counterpart v-myb is responsible for the oncogenic transformation of myelomonocytic hematopoietic cells (Shen-Ong 1990). Both c-myb and v-myb encode transcriptional activators, but little is known about their target genes in the context of developmental control or the oncogenic transformation (Ness 1996). Proteins related to c-Myb have been identified and characterized as key regulators of differentiation and development in many eukaryotes, including vertebrates, fungi, insects, and plants (Lipsick 1996; Martin and Paz-Ares 1997). In Dictyostelium a Myb homolog, DdMyb1, also called as MybH, was identified previously (Stober-Grässer et al. 1992). The DNA-binding domain of DdMyb1 shows 65% amino acid identity to vertebrate c-Myb and is capable of binding to the same DNA sequence as c-Myb. However, target genes and function of DdMyb1 are not known.
In this report we demonstrate that the novel myb-related gene DdMYB2 is required for starvation-induced expression of the adenylyl cyclase ACA. Ectopic constitutive expression of ACA restores development of Ddmyb2-null cells, suggesting that DdMyb2 mediates the starvation signal and induces Dictyostelium development by direct or indirect activation of ACA gene expression.
Results
Isolation of Ddmyb2 mutants
To identify genes that regulate cAMP signaling in Dictyostelium development, we isolated a set of aggregateless mutants using restriction enzyme-mediated integration (REMI; Kuspa and Loomis 1992). REMI is a method of insertional mutagenesis, in which a linearized plasmid carrying a selection marker is integrated into the host genome with the help of a restriction enzyme. We screened 31 aggregateless mutants for cAMP-mediated activation of adenylyl and guanylyl cyclase, for chemotaxis to cAMP and folic acid, and for synergy with wild-type cells. Seven mutants were identified as Synags, as mixing them with wild-type cells restored development in these aggregateless mutants. Most Synags have a similar phenotype as aca–null cells, not producing extracellular cAMP but showing good chemotaxis to cAMP (Pitt et al. 1992). One Synag mutant, W67, showed a strongly reduced cAMP response and very weak chemotaxis to cAMP, suggesting a defect of a novel component that is required for both the secretion of cAMP and normal chemotaxis to cAMP.
A Southern blot of W67 genomic DNA probed with a part of the inserted plasmid revealed BglII restriction sites flanking the insertion site (see Fig. 1A). Plasmid pRW67 was recovered from W67 genomic DNA digested with BglII and was used to sequence the insertion–flanking region and to recapitulate the insertion event. For the latter, the plasmid was relinearized with BglII and introduced into wild-type cells by electroporation. About 70% of transformants showed an agg− phenotype. Southern analysis of four agg− and six agg+ clones revealed that all agg− clones had the expected homologous recombination, whereas all agg+ clones contained the wild-type DNA fragment (data not shown). Therefore, we conclude that the observed W67 phenotype results from the plasmid integration and not from potential secondary mutations.
Figure 1.
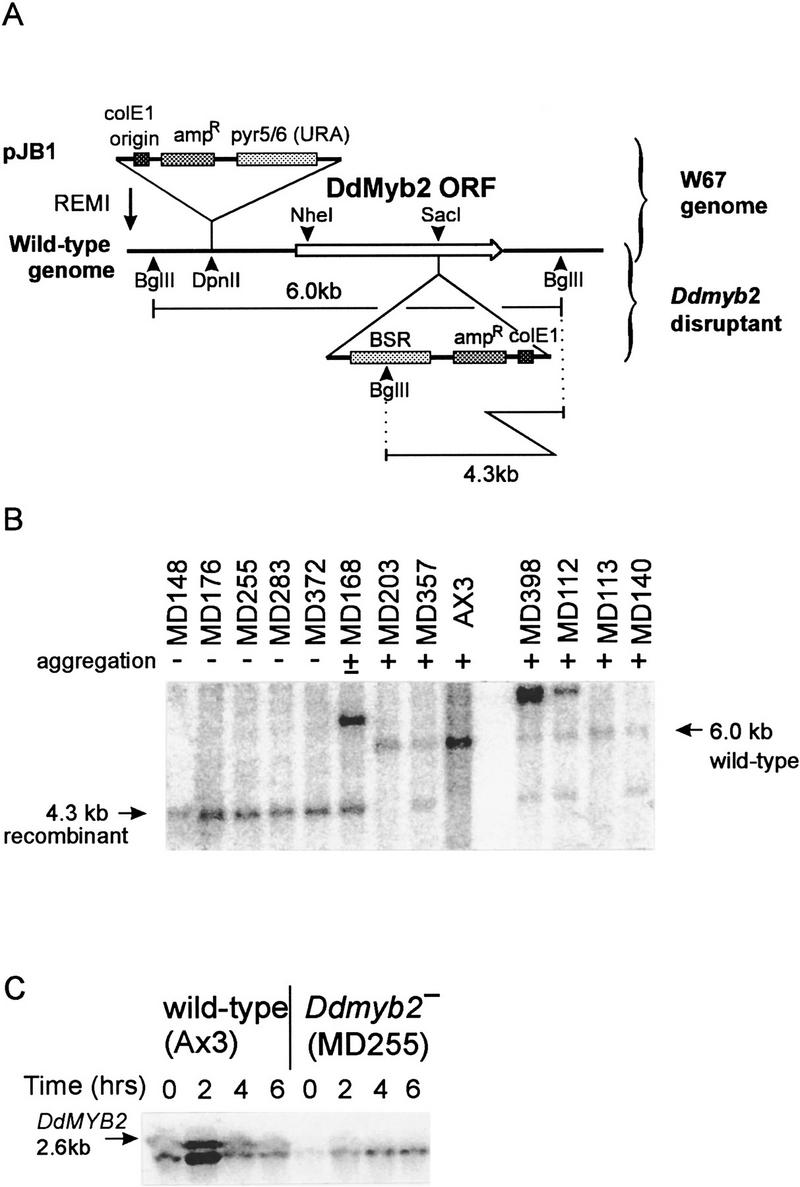
The DdMYB2 locus and isolation of a Ddmyb2–null mutant. (A) REMI and gene targeting at DdMYB2 locus. W67 mutant was made by REMI of the BamH1-linearized plasmid pJB1 into the genome of uracil–auxothroph wild-type DH1 with the restriction enzyme DpnII. The DdMYB2 locus was isolated from genomic DNA using the restriction enzyme BglII. The Ddmyb2-null mutant was made by homologous recombination in wild-type AX3 genome with pMKO1, which contains a BSR, and pBluescript at the SacI site of the NheI–BglII genomic DNA fragment. The expected sizes of BglII fragments from wild-type cells and Ddmyb2 disruptant are indicated. (B) Southern analysis of the transformants with pMKO1. DNA was isolated from randomly picked transformants, digested with BglII, and probed with the SacI–BglII genomic fragment containing 3′ portion of DdMYB2 gene. MD255 was used as Ddmyb2 disruptant for further analysis. (C) Expression of DdMYB2 in wild-type AX3 and in Ddmyb2–null mutant MD255. RNA was prepared from either growing cells or cells that had been starved in suspension for the time indicated. The blot was probed with partial cDNA for DdMYB2 RNA (nucleotides 1675–1974 in Fig. 2B).
pRW67 was analyzed by sequencing, revealing that the plasmid was not integrated into the coding region of a gene but upstream of an open reading frame (ORF). We did not find a possible coding sequence at the other side of the insertion, or in the other strand of the recovered genomic DNA. The deduced amino acid sequence of the ORF shows strong homology with c-myb; the new gene is addressed as DdMYB2, and its sequence has been deposited in the EMBL Nucleotide Sequence Database (accession no. AJ002383). The expression of DdMYB2 RNA was examined in wild-type and W67 cells. A Northern blot probed with the recovered fragment of genomic DNA revealed two messengers in wild-type cells, one of which was absent in mutant W67 (data not shown; result essentially the same as the Northern blot of MD255 shown in Fig. 2).
Figure 2.


A novel myb-related gene DdMYB2. (A) Nucleotide and deduced amino acid sequence of the DdMYB2 gene. Putative TATA-oligo(dT) and polyadenylation signals are underlined. (arrow) The putative start site of transcription, (*) the end of a cDNA isolate. The deduced amino acids sequence from 425 to 592 (boxed) consists of three Myb repeats. (B) Alignment of Myb domains from various Myb-related proteins. (D.d.) Dictyostelium discoideum Myb2 (this study), Myb1 (or MybH; Stober-Gräser et al. 1992); (H.s.) Homo sapiens c-Myb (Majello et al. 1986); (S.p.) Shizosacharomyces pombe Cdc5 (Ohi et al. 1994), (D.m.) Drosophila melanoguster c-Myb (Katzen et al. 1985); (Z.m.) Zea mays P (Grotewold et al. 1991). Boxes indicate amino acid identity between DdMyb2 and other proteins. Secondary structure prediction using PHD is shown for DdMyb2: (−) loop, (□) helix, (.) no prediction. The c-Myb helices determined by nuclear magnetic resonance (NMR) are indicated with shaded boxes (Ogata et al. 1994).
To prove that the W67 phenotype is caused by the reduced expression of DdMYB2, we performed gene targeting toward the coding region (Fig. 1A). The targeting construct pMKO1 contains the DdMYB2 coding region with an insertion of an Escherichia coli vector and a blasticidin cassette (Sutoh 1993). pMKO1 was linearized and introduced into wild-type cells. Among 296 clones randomly taken from a pool of transformants resistant to blasticidin, 270 clones were agg−, 13 clones were mutants making very small aggregates but no fruiting bodies (agg+/−), and 13 clones showed normal aggregation (agg+). Among these, 6 agg−, 1 agg+/−, and 6 agg+ clones were analyzed by Southern blotting (Fig. 1B). The blot was probed with the SacI–BglII fragment of genomic DNA containing the 3′ end of DdMYB2. By gene replacement, the 6.0-kb BglII band of wild-type is expected to shift to 4.3 kb. All agg− clones have only the 4.3-kb band and all agg+ clones still possess the 6.0-kb band. Some agg+ clones (e.g., MD203) have only the 6.0-kb band, as expected. Other agg+ clones (e.g., MD357) possess both 4.3- and 6.0-kb bands. We suppose that these agg+ clones were transformed to blasticidin-resistant clones with undigested plasmid by homologous recombination with a single crossover; then the fused gene consisting of the 5′ part of endogenous gene and the 3′ part of the targeting construct remains intact. We chose the Ddmyb2 disruptant clone MD255 for further analysis (Fig. 3 A and B shows aggregateless phenotype of MD255 and wild-type fruiting bodies). Ectopic expression of DdMYB2 in MD255 suppressed the phenotypic defects (data not shown). This confirms that the phenotypic defects in W67 and the Ddmyb2 disruptants result from the failed expression of DdMYB2.
Figure 3.
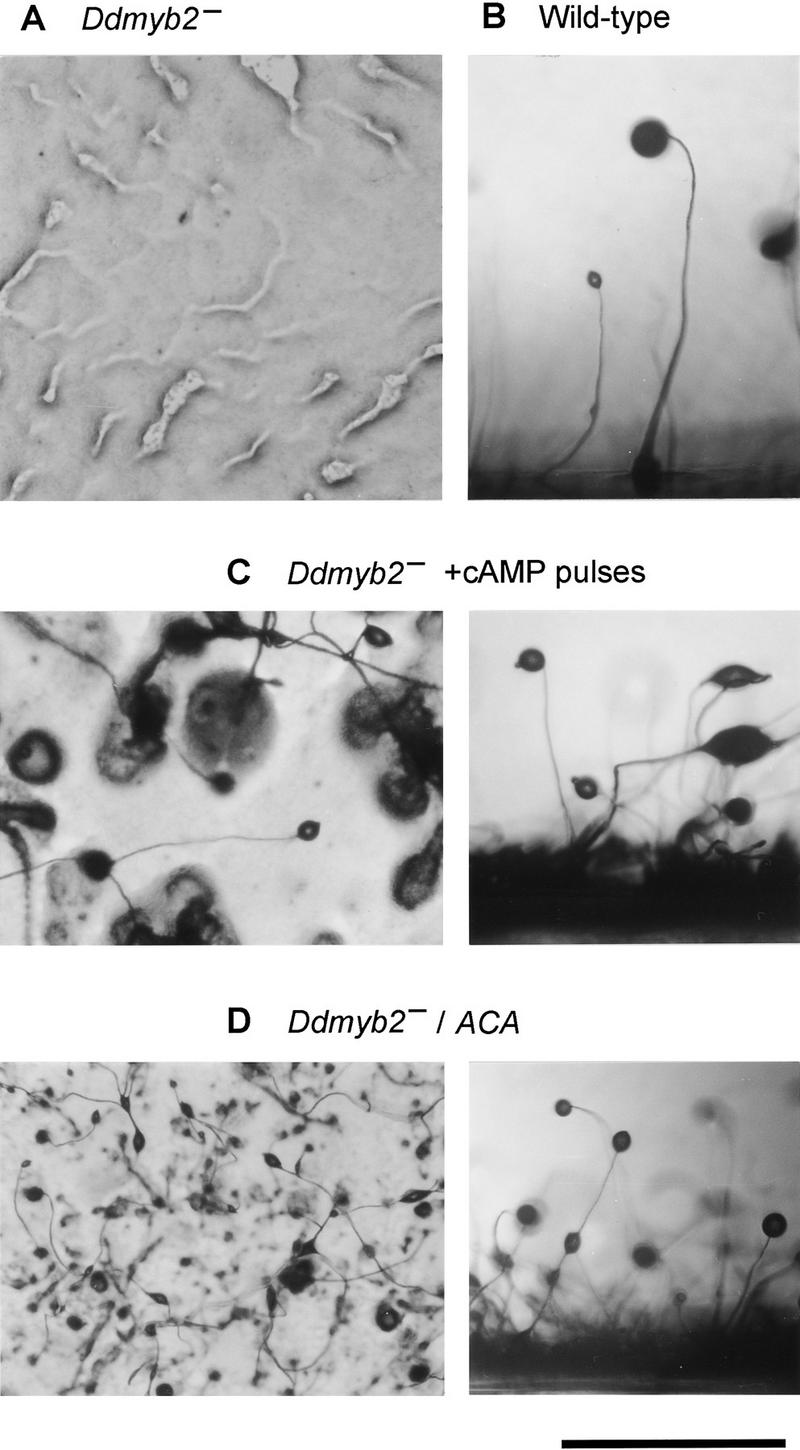
Phenotype of the Ddmyb2-null mutant and rescue by exogenous cAMP pulses or by ectopic expression of ACA. Cells starved in phosphate buffer for 5 hr were plated on nonnutrient agar at a density of 2 × 106/cm2. The photographs were taken after 30 hr; overhead views at left; side views at right. (A) No aggregation of Ddmyb2-null cells; (B) Wild-type fruiting bodies; development and fruiting body formation are restored by addition of exogenous cAMP pulses to Ddmyb2-null cells during 5 hr of starvation (C) or by constitutive expression of ACA (D). Scale bar is 1 mm.
Expression of DdMYB2
Using the SacI–BglII fragment of genomic DNA, a partial cDNA clone was isolated from a cDNA library made from 5-hr starved cells. The cDNA was used as a probe for Northern analysis to examine DdMYB2 expression in wild-type and Ddmyb2–null cells. Two transcripts with slightly different sizes ∼2.6-kb were detected in wild-type cells (Fig. 1C), whereas only the smaller transcript was visible in Ddmyb2–null cells. This identifies the larger one as the DdMYB2 transcript and suggests the existence of a gene closely related to DdMYB2. This second gene is not likely DdMYB1, which has low homology in nucleotide sequence with DdMYB2. A Northern blot of RNA from W67 cells also reveals the absence of the larger transcript (data not shown).
The Northern blot in Figure 1C shows that the DdMYB2 transcript is present during axenic growth. DdMYB2 expression increases about five-fold during the first 2 hr of starvation and decreases rapidly to 10% of the maximal level during the subsequent 4 hr of starvation.
A novel myb-related gene: DdMYB2
The isolated genomic fragment in pRW67 contains a single ORF of 699 codons (Fig. 2A). The sequence prior to the ORF is highly AT rich and contains a pair of TATAAA(T/A)A followed by oligo(dT), a usual Dictyostelium transcription start sequence (Kimmel and Firtel 1983). The ORF starts with the sequence AAAATG, which is a common translation initiation signal in Dictyostelium. The 3′ end of the transcript was determined by isolation of cDNA and contains several AATAAA sequences known as polyadenylation signal. Although the ORF is continuous (no frameshifts or stop codons), it contains a stretch of 87 nucleotides (nucleotide 955–1041 in Fig. 2A) that has the characteristics of a typical Dictyostelium intron consisting of oligo(dT) and oligo(dA) stretches flanked by consensus 5′ and 3′ splice sites GTAAGT and AG, respectively. Furthermore, no frameshift will occur when this region is spliced out. To examine the presence of an intron we performed PCR using primers flanking this region with genomic DNA and cDNA from a λgt11 cDNA library prepared from 5-hr starved cells as template. We observed products that were indistinguishable in size, suggesting that region 955–1041 was not spliced out in the mRNA used to prepare the cDNA library (data not shown).
The deduced amino acid sequence predicts a protein of 80.4 kD. Amino acids 425–561 show strong homology to a motif found in the Myb family of transcription factors. Myb-related proteins contain two or three imperfect repeats of ∼50 amino acids (Frampton et al. 1989). Each Myb repeat has a similar folding architecture, consisting of three well-defined helices. The second and the third helix form the helix–turn–helix structure. In vertebrate c-Myb, which contains three Myb repeats (R1, R2, R3), the third helices of R2 and R3 are involved in the specific base recognition for DNA binding (Ogata et al. 1994; Ness 1996).
The related domain of Dictyostelium Myb2 is located in the carboxy-terminal portion of the protein and contains three Myb repeats (Fig. 2B). The homology with the corresponding repeats in c-Myb is strongest for R1 (43% identity), whereas each of R2 and R3 shows 30% and 27% identity with the corresponding repeat in c-Myb, respectively. R3 in DdMyb2 shows a 12-amino-acid insertion between the second and the third helix. Secondary structure prediction using multisequence alignment (PHD: Profile fed neural network systems from HeiDelberg; Rost et al. 1994) predicts helix structures for DdMyb2 at the amino acids corresponding to helices of c-Myb, suggesting that the Myb repeats of DdMyb2 have a structure similar to that of c-Myb. A potential nuclear localization signal, 408-IKKKRERKR, is located just before the Myb domain (Garcia-Bustos et al. 1991). Beside the Myb domain we could not find any region with significant homology to other proteins in the database.
Exogenous cAMP pulses rescue morphogenesis and differentiation of Ddmyb2–null cells
Most Synag mutants show defects in cAMP signaling that can be at least partially restored by addition of cAMP pulses. Because the original mutant, W67, was identified as a Synag mutant, we examined the possibility that exogenous cAMP pulses can restore the development of MD255 Ddmyb2–null cells. They were stimulated with 100 nm cAMP at 5-min intervals for different lengths of time in suspension and plated on non-nutrient agar plates. Unpulsed cells or cells pulsed for up to 4.5 hr formed no fruiting bodies. However, after pulsing for 6 hr with cAMP, ∼80% of cells on the plates were recruited into aggregates, which developed to fruiting bodies (Fig. 3C). Spores in the fruiting bodies were morphologically indistinguishable from wild-type spores and resistant to treatment with detergent. They germinated upon food addition, and all colonies were aggregateless (data not shown). These findings led us to examine the cAMP signal transduction pathways in Ddmyb2–null cells starved with exogenous cAMP pulses comparing with those without pulses.
Signal transduction in Ddmyb2–null cells
cAMP binding
Secreted cAMP is detected by cARs. During early development, cAR1 represents most of cell-surface cAMP-binding activity and mediates many of the cAMP-induced responses, such as induction of gene expression, cAMP secretion, and chemotaxis (Sun and Devreotes 1991; Parent and Devereotes 1996). Expression of cAR1 is regulated transcriptionally and posttranscriptionally during Dictyostelium development. CAR1 mRNA is induced by starvation and by stimulation with extracellular cAMP (Louis et al. 1993). The binding of cAMP to wild-type cells increases to a maximal level at the 5.5-hr time point (Fig. 4A). In contrast, after an initial increase, Ddmyb2–null cells do not express a further increase of cAMP-binding activity upon starvation; at 5.5 hrs cAMP-binding activity is only 15% that of wild-type cells.
Figure 4.
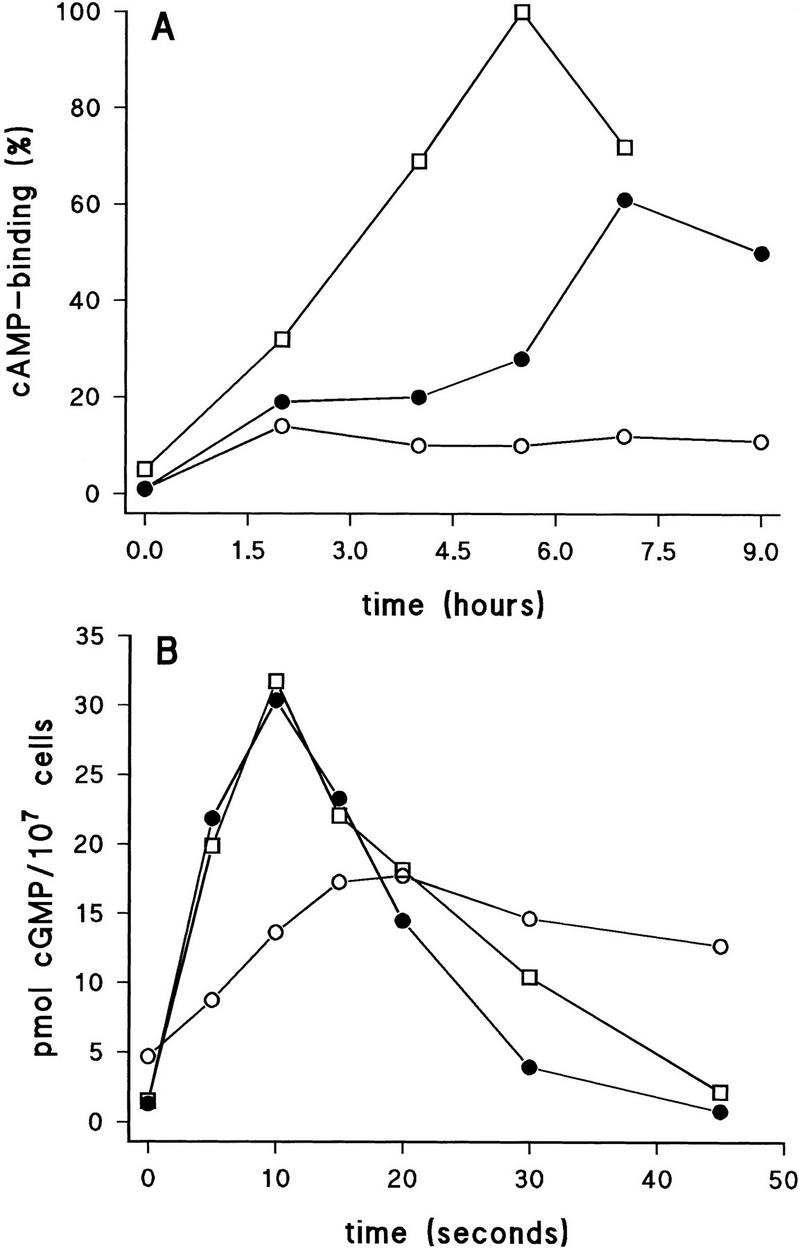
Defective signal transduction in Ddmyb2-null cells and its rescue by cAMP pulses. Cells were starved in suspension and pulsed with or without 100 nm cAMP added every 5 min. Wild-type Ax3 without pulses (□); Ddmyb2–null cells without (○) or with pulses (•). (A) Cells were starved with or without cAMP pulses for the indicated time and assayed for cAMP-binding activity. (B) cAMP-induced cGMP response, (C) cAMP relay response, and (D) adenylyl cyclase activity in wild-type AX3 cells starved for 5 hr or Ddmyb2–null cells starved for 7 hr. Adenylyl cyclase activity was measured in cell lysates with Mn2+–ATP as substrate.
Exogenously added cAMP pulses strongly induce cAMP-binding to Ddmyb2–null cells to about half the maximal level of wild-type cells (Fig. 4A). The induction of cAMP-binding sites on Ddmyb2–null cells by cAMP pulses was maximal between 5.5 and 7 hr of time point (after 4.5 and 6 hr of cAMP pulsation, respectively). A similar time course was observed in the rescue of fruiting body formation by cAMP pulsing, suggesting that the phenotypic defects of Ddmyb2–null cells can be suppressed by cAMP-induced gene expression mediated by the cell-surface receptor.
Chemotaxis
The chemotactic response of Ddmyb2–null cells to cAMP and folic acid was examined using the small population assay. The Ddmyb2–null cells starved for 5 hr showed weak chemotaxis to 10−6 m cAMP but do not respond to 10−8 or 10−7 m cAMP, whereas wild-type cells show clear responses to all these concentrations of cAMP. Upon pulsing Ddmyb2–null cells with 10−7 m cAMP for 5 hr, the mutant cells show normal chemotaxis to cAMP (data not shown). Chemotaxis to folic acid during the vegetative stage is identical in wild-type and mutant cells (data not shown). These observations confirm that the defect of Ddmyb2–null cells is specific to cAMP signaling that can be restored by cAMP pulses.
cAMP-mediated cGMP response
The cAMP binding to cAR1 induces production of intracellular signaling molecules, such as IP3, cAMP, and cGMP. In wild-type cells an extracellular cAMP stimulus induces cGMP accumulation with a peak at 10 sec; after the production by guanylyl cyclase, cGMP is degraded by intracellular phosphodiesterase. Synthesis of cGMP appears to be tightly linked to cAMP and folic acid chemotaxis (Parent and Devreotes 1996). Ddmyb2–null cells show significant accumulation of cGMP upon the cAMP stimulation (Fig. 4B); the response is ∼50% of that in wild-type cells. However if cells are pulsed for 6 hr with exogenous cAMP, the cGMP response is restored completely (Fig. 4B). Because unpulsed Ddmyb2–null cells have low levels of cAMP binding, this may explain the reduction of cGMP response. In the mutant, the basal cGMP level is about twofold higher than that of wild-type cells, and the rate of cGMP degradation is slower, which might result from low level of phosphodiesterase activity; these aberrations are also restored by treating the cells with cAMP pulses.
The folic acid-mediated cGMP response in Ddmyb2–null cells is normal (data not shown), which is consistent with the normal chemotaxis of these cells to folic acid.
cAMP relay response and adenylyl cyclase activity
Development and signal transduction in Ddmyb2–null cells are restored by cAMP pulses (Figure 3 and 4A,B). This suggests that mutant cells lack autonomous oscillations of cAMP, which are generated through synchronized cAMP production stimulated by extracellular cAMP (cAMP relay). This cAMP relay response is absent in Ddmyb2–null cells (Fig. 4C). Treatment of mutant cells with cAMP pulses for several hours partly restores the relay response to approximately half the level of the response in wild-type cells (Fig. 4C).
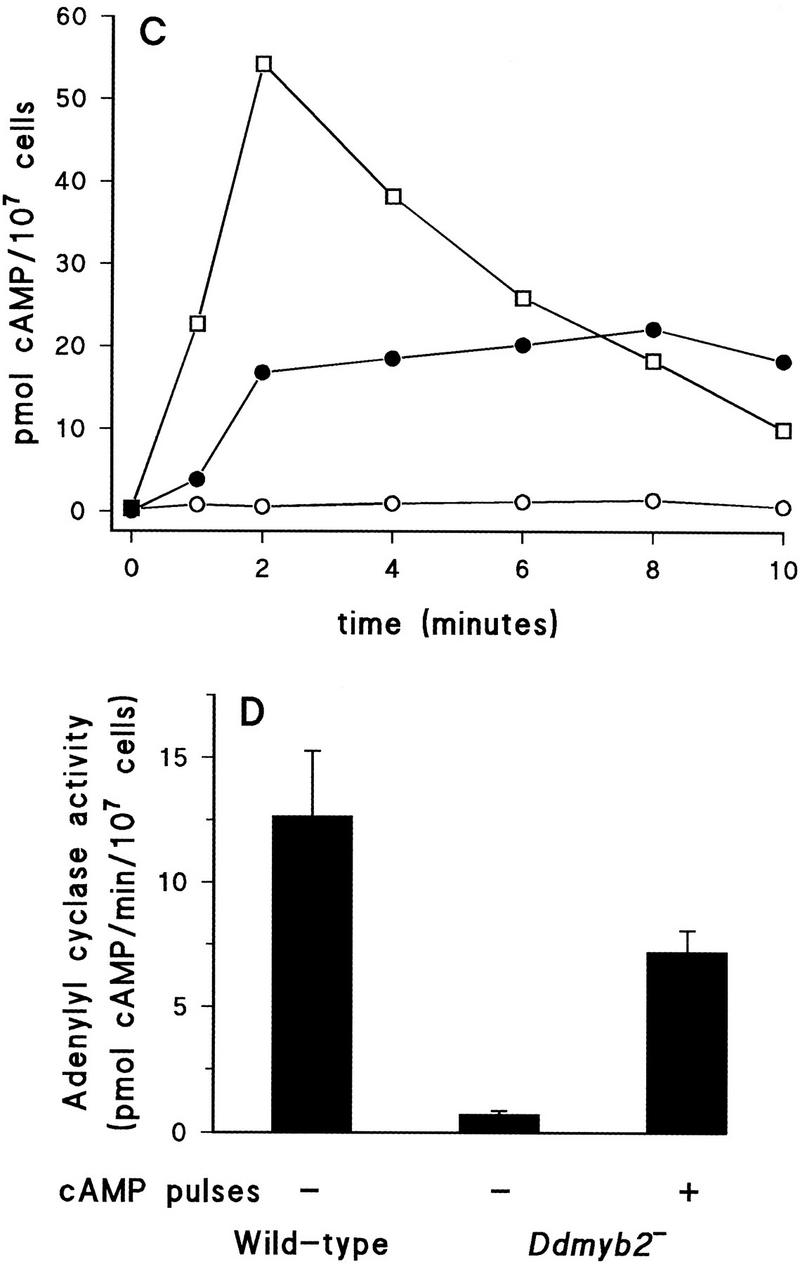
This severe defect in cAMP relay response in the mutant, combined with the moderate reduction of cAMP-binding and cGMP response, suggests another defect in cAMP production in addition to the reduced receptor expression (Fig. 4A–C). Adenylyl cyclase activity in cell lysates was measured in the presence of Mn2+, which bypasses the requirement for activation by G protein or receptor (Theibert and Devreotes 1986). The lysate from the mutant cells has very low activity of adenylyl cyclase, 5% of wild type; it increases to 56% of wild type by pulsing the cells with cAMP for 6 hr (Fig. 4D). This demonstrates that the reduced relay response in Ddmyb2–null cells is largely because of the very low activity of adenylyl cyclase.
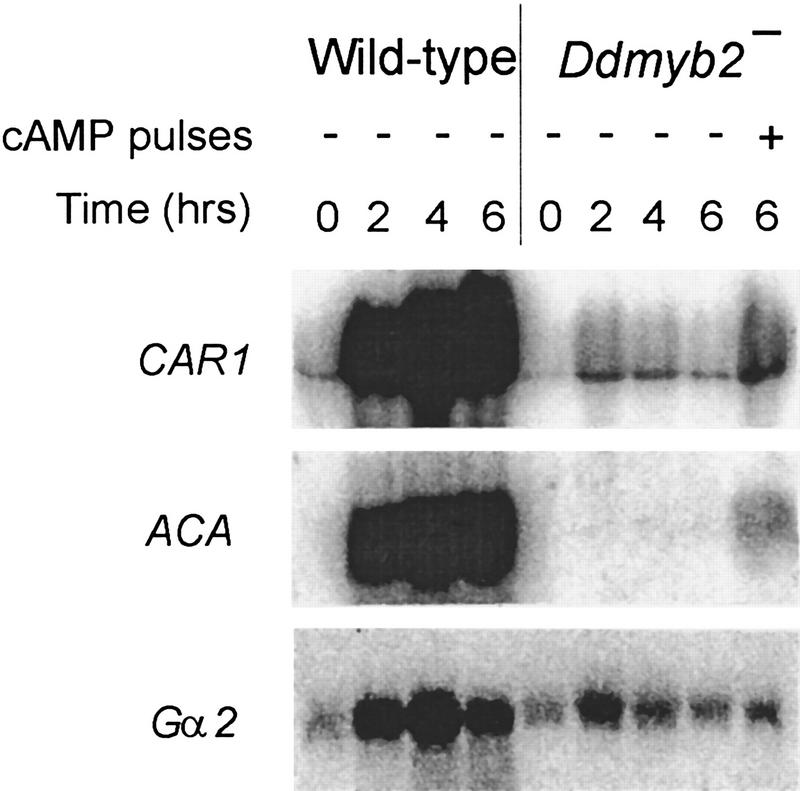
Expression of genes encoding cAMP-signaling components
Because DdMyb2 has a Myb motif, the defects in Ddmyb2–null cells may relate to transcriptional regulation. We compared the expression of the cAMP receptor gene CAR1 (Klein et al. 1988), the adenylyl cyclase gene ACA (Pitt et al. 1992), and the G protein α subunit gene Gα2 (Kumagai et al. 1988) in Ddmyb2–null and wild-type cells by Northern analysis (Fig. 5). CAR1 mRNA levels are strongly reduced in Ddmyb2–null cells. Although exogenous cAMP pulsing induces the expression of CAR1, they are still significantly lower than in wild-type cells, which is in accordance with biochemical data. It is notable that in the mutant, the expression of ACA is undetectable without cAMP pulses, but induced by cAMP pulses. The expression of Gα2 is slightly reduced in Ddmyb2–null cells. Though induction by cAMP pulses was not detected on this blot, upon longer electrophoresis two different transcripts (2.3 and 2.7 kb) were separated, revealing the severalfold induction of the 2.3-kb and reduction of the 2.7-kb transcript (data not shown).
Figure 5.
Gene expression of cAMP-signaling components in Ddmyb2-null cells. Wild-type AX3 and Ddmyb2–null cells were starved in suspension with or without cAMP pulses for the indicated time. Total mRNA was isolated and analyzed. The Northern blot shows the expression of CAR1, ACA, and Gα2.
The messengers for cAR1 and Gα2 are present at low levels in vegetative cells and have increased significantly during the first 2 hr of starvation. Whereas in wild-type cells the expression of these genes increases further between 2 and 6 hr, it slightly decreases in Ddmyb2–null cells. The presence of low but significant levels of CAR1 and Gα2 messenger in Ddmyb2–null cells explains the ability of these cells to show the cAMP-mediated cGMP responses and pulse-induced gene expression. The virtual absence of ACA messenger is consistent with the very low level of adenylyl cyclase activity and the lack of cAMP relay. It predicts that cells can not generate autonomous cAMP oscillations and therefore no autonomous pulse-induced gene expression. The low levels of CAR1 and Gα2 messenger in 6-hr starved Ddmyb2–null cells could be caused by the lack of this pulse-induced gene expression.
Ectopic expression of ACA but not CAR1 rescues morphogenesis in Ddmyb2–null cells
To confirm that the aggregateless phenotype of Ddmyb2–null cells results from the reduced expression of ACA and/or CAR1, we transformed Ddmyb2–null cells with plasmids carrying ACA or CAR1 under the transcriptional control of the constitutive actin-15 promoter (Knecht et al. 1986; Johnson et al. 1991). Increased expression of ACA or CAR1 was confirmed by adenylyl cyclase or cAMP-binding assays using growing cells. While the Ddmyb2−/CAR1 strain remained aggregateless at low or high cell density (data not shown), the Ddmyb2−/ACA strain made fruiting bodies of nearly normal size and shape at high cell density (Fig. 3D); at low cell density, fruiting bodies were small and a part of the cells did not participate in cell aggregation.
Starved wild-type cells show autonomous cAMP oscillations with a periodicity of ∼4–6 min (Fig. 6A). As expected from the very low level of adenylyl cyclase, constant low cAMP levels are detected in Ddmyb2–null cells. The results of Figure 6B reveal that ectopic expression of ACA in Ddmyb2–null cells is sufficient to induce cAMP oscillations, although the pulses are not as well separated as in wild-type cells; this could be caused by a disturbed balance between phosphodiesterase and adenylyl cyclase (Parent and Devreotes 1996). As we have shown that exogenous cAMP pulses restore development in Ddmyb2–null cells, the reappearance of autonomous oscillations and normal development in Ddmyb2−/ACA cells implies that DdMyb2 regulates starvation-mediated development mainly by induction of adenylyl cyclase activity.
Figure 6.
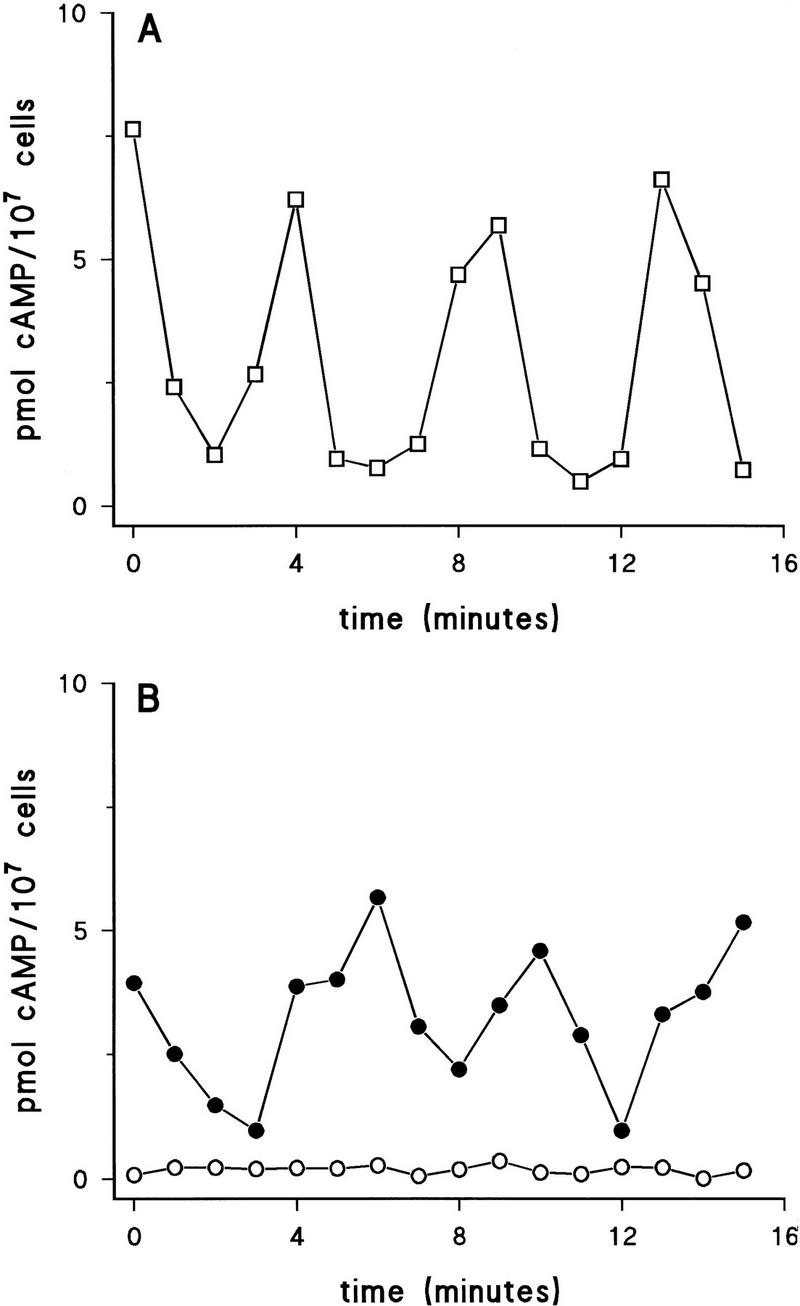
Restoration of cAMP oscillations in Ddmyb2-null cells by ectopic expression of ACA. Cells were starved for 5 hr, washed, and resuspended to a density of 108 cell/ml. Samples of the cell suspension were taken at 1-min intervals and used to determine the cAMP concentration. (A) Wild-type Ax3; (B) Ddmyb2–null cells (○), Ddmyb2−/ACA cells (•). The results shown are from a typical experiment reproduced two times.
Discussion
We have cloned a novel myb-related gene, DdMYB2, which encodes a key component regulating early development in Dictyostelium. DdMyb2 contains three Myb repeats, which is a sequence motif with a DNA-binding helix-turn-helix structure (Frampton et al. 1989; Ness 1996). Many Myb-related proteins from various eukaryotes have been shown to bind DNA in a sequence-specific manner and presumably regulate the expression of genes involved in growth control and differentiation (Lipsick 1996). This may suggest that the new Dictyostelium MYB2 gene encode a transcription factor. We have demonstrated that DdMYB2 is essential for starvation-induced transcription of a set of genes involved in early development. Furthermore, the deduced amino acid sequence of DdMyb2 contains, besides the Myb domain, a potential nuclear localization signal, IKKKRERKR, and glutamine-, proline-, and acidic amino acid-rich regions that are often found in transactivation domains (Latchman 1990).
DdMYB2 was identified by REMI mutagenesis. The integration event in the original W67 mutant occurred upstream of the DdMyb2 ORF. Nevertheless, W67 and a Ddmyb2–null mutant with a disruption of this ORF have essentially the same phenotype, including the absence of the DdMYB2 transcript. Apparently the insertion took place in the promotor, thereby disrupting DdMYB2 expression. Ddmyb2–null cells fail to aggregate. The expression of some early genes such as CAR1 and Gα2, and cellular responses such as cAMP-mediated chemotaxis and cGMP response are slightly but significantly increased by 2 hr of starvation. In contrast to wild-type cells in which these responses continue to increase during longer starvation, Ddmyb2–null cells stop development at this point. This suggests that DdMYB2 plays a key role in early development as has been observed for other myb-related genes in a variety of organisms. FlbD in Aspergillus nidulans regulates conidiophore development (Wieser and Adams 1995), stonewall in Drosophila is required for germ-cell development (Clark and McKearin 1996), and in Xenopus c-myb plays an important role in early mesodermal pattern formation (Amaravadi and King 1994). In addition, many plant myb-related genes are known to control pigment biosynthesis, and some genes such as GL1 in Arabidopsis are involved in the control of cell fate (Martin and Paz-Ares 1997). Developmental regulations by myb-related genes seem to be widely conserved in higher and lower eukaryotes.
How does DdMYB2 regulate early development in Dictyostelium? The most striking phenotype of the Ddmyb2–null mutant is the undetectably low level of ACA transcript resulting in very low levels of adenylyl cyclase activity and cAMP relay response. Ectopic expression of ACA under control of a constitutive actin promotor in Ddmyb2–null cells rescues differentiation and morphogenesis. This not only suggests that the absence of ACA is the main cause of the aggregateless phenotype of the Ddmyb2–null mutant, but also that DdMyb2 has no other essential function in Dictyostelium. This conclusion is supported by the observation that exogenously added cAMP pulses also restore development of Ddmyb2–null cells. Apparently, the lack of cAMP oscillations because of the absence of ACA is the only defect in Ddmyb2–null cells. All other components of the cAMP signaling pathway, including surface receptors, G proteins, and effector enzymes, are present in sufficient amounts to transmit cAMP signals. Once cAMP oscillations have started, either by exogenous cAMP pulses or by ectopic ACA expression, positive feedback loops enhance the expression of these components including cAR1 and Gα2 to enlarge the oscillations. It also explains the good synergy between wild-type and Ddmyb2–null cells: Wild-type cells provide cAMP oscillations to which Ddmyb2–null cells are fully responsive. This synergy is much better than that between wild-type and aca–null cells, probably because cAMP pulses do induce moderate levels of ACA expression, which is independent of its regulation by DdMyb2.
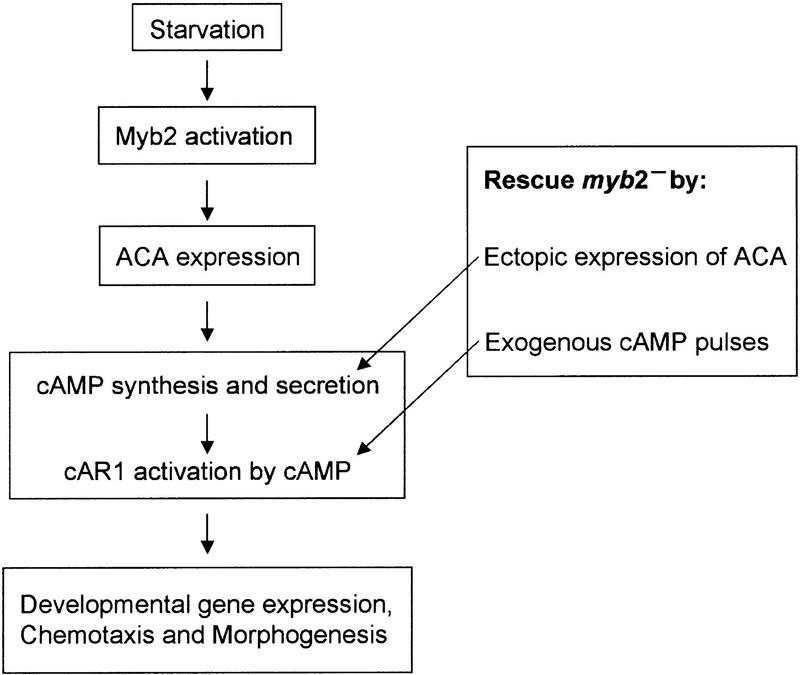
Expression of ACA is one of the earliest responses of cells to starvation. Therefore the factor that induces ACA expression might be activated by the starvation signal and directly enhance the transcription of ACA. It is possible that the ACA promotor is the target of the starvation-activated transcription factor DdMyb2. A model for the onset of Dictyostelium development is shown in Figure 7. Starvation induces the activation of DdMyb2 thereby inducing the expression of ACA. This leads to cAMP oscillations that induce the expression of a set of genes required for chemotaxis, cell aggregation, and further development. Deletion of DdMYB2 can be rescued either by ectopic expression of ACA leading to autonomous cAMP oscillations, or by adding exogenous cAMP pulses that mimic these oscillations. Two pertinent questions require further investigation: How is DdMyb2 activated by starvation, and how does activated DdMyb2 induce the expression of ACA? Studies using mutants with altered PKA activity suggest that this protein kinase might be involved in starvation-mediated ACA expression in Dictyostelium. A mutant with a deletion of the catalytic subunit of PKA is unable to express ACA (Mann et al. 1997). Since DdMyb2 contains two potential PKA phosphorylation sites, KRSYS193 in the amino-terminal part and KRVLS475 in the third helix of Myb repeat R1, DdMyb2 activity could be regulated by PKA directly. PKA may also indirectly regulate DdMyb2 via phosphorylase kinase that may phosphorylate the serine in IKKKRERKRES418I; the consensus phosphorylation site overlaps with the putative nuclear localization signal (underlined), suggesting another possible mechanism of DdMyb2 regulation at the level of translocation to the nucleus. Site-directed mutagenesis of DdMyb2 along with PKA mutants might answer how DdMyb2 is activated. Possible interactions with other regulatory proteins should not be ignored, as accumulating evidence suggests that c-Myb and several Myb-related proteins alter gene expression in cooperation with other transcription factors or bridging proteins (Lipsick 1996). In this context it is interesting to note that in DdMyb2, amino acid sequence is less well conserved in the second and third helix of Myb repeats R2 and R3, which are supposed to contact DNA, than in the rest of the Myb domain. This might imply that Myb domains, besides their ability to bind DNA, have other important conserved functions such as protein–protein interactions.
Figure 7.
Model of the regulation of early development in Dictyostelium by Myb2.
Materials and methods
Cell culture and development
Dictyostelium discoideum axenic strains DH1 and Ax3 were used as wild-type strains. DH1 (ura−) was grown in HG5 medium supplemented with uracil at 100 μg/ml, and derived URA+ transformants were selected and maintained in FM medium (GIBCO-BRL) lacking uracil. Ax3 was used for transformation using BSR or neo selection markers (Witke et al. 1987; Sutoh 1993); axenic medium was supplemented with 10 μg/ml of blasticidin S or geneticin to select and maintain transformants.
Vegetative cells were obtained from axenic cultures at a cell density of 3 × 106 cells/ml. For the biochemical assays and Northern blotting, cells were starved in 10 mm phosphate buffer (pH 6.5) at 107 cells/ml for the indicated time. When indicated, cells were pulsed with 10−7 m cAMP at 5-min intervals.
Morphogenesis was observed on nonnutrient agar plates; cells were plated at low (105 cells/cm2) or high cell density (2 × 106 cells/cm2), from the latter photographs were taken after 30 hr of incubation. For quantification of morphogenesis, cells were put on nonnutrient agar plates at 4 × 105 cells/cm2 after the indicated length of starvation with or without cAMP pulses; the number of cells not included in fruiting bodies were estimated after 30 hr of incubation.
For synergy experiments, vegetative mutant and wild-type cells were mixed at ratios of 1:1 and 9:1. Small droplets (0.1 μl) of the mixtures were deposited on a hydrophobic agar plate. The resulting cell density was ∼105 cells/cm2. The presence of fruiting bodies was examined after 40 hr of incubation.
REMI mutagenesis
REMI was carried out as described by Kuspa and Loomis (1992) with the following modification. The plasmid pJB1, which carries the pyr5-6 (URA+) gene in Bluescript, was digested with BamHI and introduced into DH1 cells along with restriction enzyme DpnII by electroporation. Transformants were selected in FM medium lacking uracil and plated clonally onto LP agar plates in association with E. coli B/r. Aggregateless mutants were picked and characterized by biochemical assays.
pRW67 was recovered from W67 cells as follows: 20 μg of genomic DNA isolated from W67 was digested with BglII. The digested DNA was purified by phenol/chloroform extraction and ethanol precipitation. To promote recircularization, the DNA was ligated at a concentration of 50 μg/ml. After ethanol precipitation 0.2 μg of the ligated product was used for transformation of SURE electroporation-competent E. coli cells (Stratagene). Transformants harboring the rescued plasmid were selected on ampicillin plates.
Regeneration of W67 mutant from the wild-type strain DH1 was performed by homologous recombination using pRW67 redigested with BglII. The recombinants were selected by Southern analysis from the URA+ transformants.
Molecular genetics
The Ddmyb2–null strains were generated by gene targeting. The targeting construct pMKO1 contains a genomic fragment that encompasses the DdMYB2 coding region in which an E. coli vector pBluescript SK(−) (Stratagene) and a BSR cassette (Sutoh 1993) are inserted at the SacI site. pMKO1 is linearized with NheI and BglII and introduced into wild-type Ax3 cells. Transformants were selected for blasticidin resistance. Single colonies were isolated on bacterial plates and analyzed by Southern blots. Clone MD255 was identified as the Ddmyb2–null strain and used for the experiments in this study.
CAR1 or ACA were expressed in Ddmyb2–null cells by transformation of MD255 with pJK3 and pCP43, respectively (gifts from P. Devreotes, Johns Hopkins University, Baltimore, MD); both plasmids replicate extrachromosomally, contain a neo selection marker, and express the genes of interest under the control of an actin-15 promoter.
Southern and Northern blotting
Genomic DNA from AX3 and W67 was purified by CsCl centrifugation after nuclei isolation, phenol/chloroform extraction, and ethanol precipitation (Nellen et al. 1987). To screen for the recombinants generated by gene targeting, genomic DNA from individual clones was prepared similarly but without CsCl purification. Approximately 0.1 μg of genomic DNA was used for Southern analysis; 0.4 μg of genomic DNA from Ax3 was used for wild-type control. DNA was digested with BglII, separated on a 0.6% agarose gel, and transferred on Nytran filter (Schleicher & Schuell).
Total RNA from Ax3 and Ddmyb2–null cells was prepared as described by Mann and Firtel (1987); 20 μg was used for Northern analysis. Samples were separated by electrophoresis on a 1% agarose gel containing formaldehyde and transferred on Nytran filter.
Prehybridization of Southern and Northern blots was carried out at 65°C in the prehybidization solution (5× SSC, 4× Denhardt’s, 0.04 mg/ml sperm DNA, 1% SDS). Hybridization was carried out at 65°C in hybridization buffer (7% SDS, 0.5 m phosphate buffer at pH 7.0) containing 32P-labeled probe made by the random primer method (High Prime: Boehringer Mannheim). After hybridization, filters were washed with 1% SDS, 0.5 m phosphate buffer (pH 7.0) at 65°C. The filters were imaged and quantified using a PhosphorImager (Molecular Dynamics).
Isolation of the myb cDNA
cDNA clone representing the 3′ end of DdMYB2 was isolated from a λgt11 cDNA library made from 5-hr starved cells (gift from P. Devereotes), using the 1.3-kb SacI–BglII fragment of pRW67. The obtained clone starts at nucleotide 1675 (Fig. 2A). The 5′ part of the cDNA (nucleotide -9 to 1283) was amplified from the same library by PCR using primers 1 and primer 2 (primer 1: 5′-AATAATCATATGACTGCTATATTCC-3′, primer 2: 5′-TTTGGTGGAGATCTTATACC-3′).
Biochemical assays
cAMP-binding to cell-surface receptors was determined by the ammonium sulfate stabilization assay (Van Haastert and Kien 1983). The incubation mixture (1 ml) contained 1 nm [3H]cAMP, 1 mm dithiothreitol (DTT), 85% saturated ammonium sulfate, 5 × 106 cells and 50 μg BSA. After 5 min of incubation at 0°C, the cells were pelleted and radioactivity in the pellet was determined.
For determination of the cGMP and cAMP response, cells were starved for the appropriate period and resuspended in phosphate buffer at 108 cells/ml. Cells were stimulated with 0.1 μm cAMP for inducing a cGMP response, and with 5 μm 2′-deoxy-cAMP and 5 mm DTT for the cAMP response. cAMP oscillations were measured without an exogenous stimulus. The reactions were terminated by the addition of an equal volume of 3.5% perchloric acid. The levels of cGMP or cAMP were determined in the cell lysates by isotope dilution assays (Snaar-Jagalska and Van Haastert 1994).
For assaying adenylyl cyclase activity, starved cells were harvested and resuspended in lysis buffer (3 mm MgCl2, 10 mm Tris-HCl at pH 8.0) at 7.5 × 107 cells/ml. Cell lysates were prepared by forced filtration of cells through a Nucleopore filter (pore size 3 μm). Twenty microliters of lysate was immediately mixed with 20 μl of lysis buffer containing 20 mm DTT, 1 mm ATP, and 10 mm MnSO4. The reaction was terminated after 0, 1, 2, and 4 min by the addition of 20 μl of 3.5% perchloric acid, and cAMP levels were determined.
Chemotaxis toward cAMP and folic acid was measured by the small population assay (Konijn 1970).
Acknowledgments
We thank P.N. Devereotes for providing plasmids pJB1, pJK3, and pCP43, the Gα2 probe, and the cDNA library; M.H.K. Linskens for helpful comments on the text; and J.G. Williams and P. Schaap for their contributions to the collection of aggregateless REMI mutants. W67 was isolated in the laboratory of J.G.W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL haastert@chem.rug.nl; FAX 31-50-3634165.
References
- Amaravadi L, King MW. Characterization and expression of the Xenopus c-Myb homolog. Oncogene. 1994;9:971–974. [PubMed] [Google Scholar]
- Clark KA, McKearin DM. The Drosophila stonewall gene encodes a putative transcription factor essential for germ cell development. Development. 1996;122:937–950. doi: 10.1242/dev.122.3.937. [DOI] [PubMed] [Google Scholar]
- Frampton J, Leutz A, Gibson TJ, Graf T. DNA-binding domain ancestry. Nature. 1989;342:134. doi: 10.1038/342134a0. [DOI] [PubMed] [Google Scholar]
- Garcia-Bustos J, Heitman J, Hall MN. Nuclear protein localisation. Biochim Biophys Acta. 1991;1071:83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Athma P, Peterson T. Alternatively spliced products of the maize P gene encode proteins with homology to the DNA-binding domain of myb-like transcription factors. Proc Natl Acad Sci. 1991;88:4587–4591. doi: 10.1073/pnas.88.11.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RL, Vaughan RA, Caterina MJ, Van Haastert PJM, Devreotes PN. Overexpression of the cAMP receptor 1 in growing Dictyostelium cells. Biochemistry. 1991;30:6982–6986. doi: 10.1021/bi00242a025. [DOI] [PubMed] [Google Scholar]
- Katzen AL, Kornberg TB, Bishop JM. Isolation of the proto-oncogene c-myb from D. melanogaster. Cell. 1985;41:449–456. doi: 10.1016/s0092-8674(85)80018-0. [DOI] [PubMed] [Google Scholar]
- Kimmel AR, Firtel RA. Sequence organization in Dictyostelium: Unique structure at the 5′-ends of protein coding genes. Nucleic Acids Res. 1983;11:541–552. doi: 10.1093/nar/11.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— cAMP signal transduction pathways regulating development of Dictyostelium discoideum. Curr Opin Genet Dev. 1991;1:383–390. doi: 10.1016/s0959-437x(05)80304-1. [DOI] [PubMed] [Google Scholar]
- Klein PS, Sun TJ, Saxe III CL, Kimmel AR, Johnson RL, Devreotes PN. A chemoattractant receptor controls development in Dictyostelium discoideum. Science. 1988;241:1467–1472. doi: 10.1126/science.3047871. [DOI] [PubMed] [Google Scholar]
- Knecht DA, Cohen SM, Loomis WF, Lodish HF. Developmental regulation of Dictyostelium discoideum actin gene fusions carried on low-copy and high-copy transformation vectors. Mol Cell Biol. 1986;6:3973–3983. doi: 10.1128/mcb.6.11.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konijn TM. Microbiological assay of cyclic 3′,5′-AMP. Experientia. 1970;26:367–369. doi: 10.1007/BF01896891. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Mann SKO, Pupillo M, Pitt G, Devreotes PN, Firtel RA. A molecular analysis of G proteins and control of early gene expression by the cell-surface cAMP receptor in Dictyostelium. Cold Spring Harbor Symp Quant Biol. 1988;53:675–685. doi: 10.1101/sqb.1988.053.01.077. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Pupillo M, Gundersen R, Miake-Lye R, Devreotes PN, Firtel RA. Regulation and function of G alpha protein subunits in Dictyostelium. Cell. 1989;57:265–275. doi: 10.1016/0092-8674(89)90964-1. [DOI] [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman DS. Eukaryotic transcription factors. Biochem J. 1990;270:281–289. doi: 10.1042/bj2700281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsick JS. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- Loomis WF. Genetic networks that regulate development in Dictyostelium cells. Microbiol Rev. 1996;60:135–150. doi: 10.1128/mr.60.1.135-150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis JM, Saxe III CL, Kimmel AR. Two transmembrane signaling mechanisms control expression of the cAMP receptor gene CAR1 during Dictyostelium development. Proc Natl Acad Sci. 1993;90:5969–5973. doi: 10.1073/pnas.90.13.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majello B, Kenyon LC, Dalla-Favera R. Human c-myb protooncogene: Nucleotide sequence of cDNA and organization of the genomic locus. Proc Natl Acad Sci. 1986;83:9636–9640. doi: 10.1073/pnas.83.24.9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann SKO, Firtel RA. Cyclic AMP regulation of early gene expression in Dictyostelium discoideum: Mediation via the cell surface cyclic AMP receptor. Mol Cell Biol. 1987;7:458–469. doi: 10.1128/mcb.7.1.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann SKO, Brown JM, Briscoe C, Parent C, Pitt G, Devreotes PN, Firtel RA. Role of cAMP-dependent protein kinase in controlling aggregation and postaggregative development in Dictyostelium. Dev Biol. 1997;183:208–221. doi: 10.1006/dbio.1996.8499. [DOI] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Nellen W, Datta S, Reymond C, Sivertsen A, Mann S, Crowley T, Firtel RA. Molecular biology in Dictyostelium: Tools and applications. Methods Cell Biol. 1987;28:67–100. doi: 10.1016/s0091-679x(08)61637-4. [DOI] [PubMed] [Google Scholar]
- Ness SA. The Myb oncoprotein: Regulating a regulator. Biochim Biophys Acta. 1996;1288:F123–F139. doi: 10.1016/s0304-419x(96)00027-3. [DOI] [PubMed] [Google Scholar]
- Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- Ohi R, McCollum D, Hirani B, Den Haese GJ, Zhang X, Burke JD, Turner K, Gould KL. The Schizosaccharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J. 1994;13:471–483. doi: 10.1002/j.1460-2075.1994.tb06282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent CA, Devreotes PN. Molecular genetics of signal transduction in Dictyostelium. Annu Rev Biochem. 1996;65:411–440. doi: 10.1146/annurev.bi.65.070196.002211. [DOI] [PubMed] [Google Scholar]
- Pitt GS, Milona N, Borleis J, Lin KC, Reed RR, Devreotes PN. Structurally distinct and stage-specific adenylyl cyclase genes play different roles in Dictyostelium development. Cell. 1992;69:305–315. doi: 10.1016/0092-8674(92)90411-5. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C, Schneider R. PHD-an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- Shen-Ong GLC. The myb oncogene. Biochim Biophys Acta. 1990;1032:39–52. doi: 10.1016/0304-419x(90)90011-o. [DOI] [PubMed] [Google Scholar]
- Snaar-Jagalska BE, Van Haastert PJM. G-protein assays in Dictyostelium. Methods Enzymol. 1994;237:387–408. doi: 10.1016/s0076-6879(94)37077-x. [DOI] [PubMed] [Google Scholar]
- Stober-Grässer U, Brydolf B, Bin X, Grässer F, Firtel RA, Lipsick JS. The Myb DNA-binding domain is highly conserved in Dictyostelium discoideum. Oncogene. 1992;7:589–596. [PubMed] [Google Scholar]
- Sun TJ, Devreotes PN. Gene targeting of the aggregation stage cAMP receptor cAR1 in Dictyostelium. Genes & Dev. 1991;5:572–582. doi: 10.1101/gad.5.4.572. [DOI] [PubMed] [Google Scholar]
- Sutoh K. A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid. 1993;30:150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- Theibert A, Devreotes PN. Surface receptor-mediated activation of adenylate cyclase in Dictyostelium. Regulation by guanine nucleotides in wild-type cells and aggregation deficient mutants. J Biol Chem. 1986;261:15121–15125. [PubMed] [Google Scholar]
- Van Haastert PJM, Kien E. Binding of cAMP and adenosine derivatives to Dictyostelium discoideum cells. Relationships of binding, chemotactic, and antagonistic activities. J Biol Chem. 1983;258:9643–9648. [PubMed] [Google Scholar]
- Wieser J, Adams TH. flbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes & Dev. 1995;9:491–502. doi: 10.1101/gad.9.4.491. [DOI] [PubMed] [Google Scholar]
- Witke W, Nellen W, Noegel A. Homologous recombination in the Dictyostelium alpha-actinin gene leads to an altered mRNA and lack of the protein. EMBO J. 1987;6:4143–4148. doi: 10.1002/j.1460-2075.1987.tb02760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]