Summary
The troponin complex plays an essential role in the thin filament regulation of striated muscle contraction. Of the three subunits of troponin, troponin I (TnI) is the actomyosin ATPase inhibitory subunit and its effect is released upon Ca2+-binding to troponin C. The exon 8-encoded COOH-terminal end segment represented by the last 24 amino acids of cardiac TnI is highly conserved and critical to the inhibitory function of troponin. Here we investigated the function and calcium regulation of the COOH-terminal end segment of TnI. A TnI model molecule was labeled with Alexa fluor 532 at a Cys engineered at the COOH-terminal end and used to reconstitute tertiary troponin complex. A Ca2+-regulated conformational change in the COOH terminus of TnI was shown by a sigmoid-shape fluorescence intensity titration curve similar to that of the circular dichroism calcium titration curve of troponin C. Such corresponding Ca2+-responses are consistent with the function of troponin as a coordinated molecular switch. Reconstituted troponin complex containing a mini-troponin T lacking its two tropomyosin-binding sites showed a saturable binding to tropomyosin at pCa 9 but not at pCa 4. This Ca2+-regulated binding was diminished when the COOH-terminal 19 amino acids of cardiac TnI were removed. These results provided novel evidence for suggesting that the COOH-terminal end segment of TnI participates in the Ca2+-regulation of muscle thin filament through an interaction with tropomyosin.
Keywords: Troponin I, COOH-terminus, Tropomyosin, Calcium
INTRODUCTION
The contraction of striated (skeletal and cardiac) muscles is activated by calcium through troponin and tropomyosin associated with the sarcomeric actin thin filament [1]. Troponin is a protein complex composed of three subunits [2,3]: troponin C (TnC, the calcium receptor subunit), troponin T (TnT, the tropomyosin-anchoring subunit) and troponin I (TnI, the actomyosin ATPase inhibitory subunit) [4,5].
Three TnI genes (cardiac, fast skeletal muscle and slow skeletal muscle) are present in vertebrates to encode three muscle type-specific TnI isoforms [6,7]. Based on structural and functional studies, the TnI polypeptide chain had been dissected into six regions [8]: (i) The Cardiac TnI-specific NH2-terminal extension (cardiac TnI1–30); (ii) the NH2-terminal region (cardiac TnI34–71 and fast TnI1–40); (iii) the TnT-binding-region (cardiac TnI80–136 and fast TnI50–106); (iv) the inhibitory peptide (cardiac TnI128–147 and fast TnI96–115); (v) the switch or triggering region (cardiac TnI147–163 and fast TnI115–131) that binds the N-domain of TnC; and (vi) the remaining COOH-terminal region (cardiac TnI164–210 and fast TnI132–180). X-ray crystallography has determined high-resolution 3D structure for the majority of the TnI molecule in human cardiac troponin [9] and in chicken fast skeletal muscle troponin [10]. The troponin crystal structure of neither calcium-activated nor calcium-free state included the cardiac TnI-specific NH2-terminal extension (cardiac TnI1–39) or the COOH-terminal end segment of TnI (cardiac TnI192–210 and fast TnI162–181).
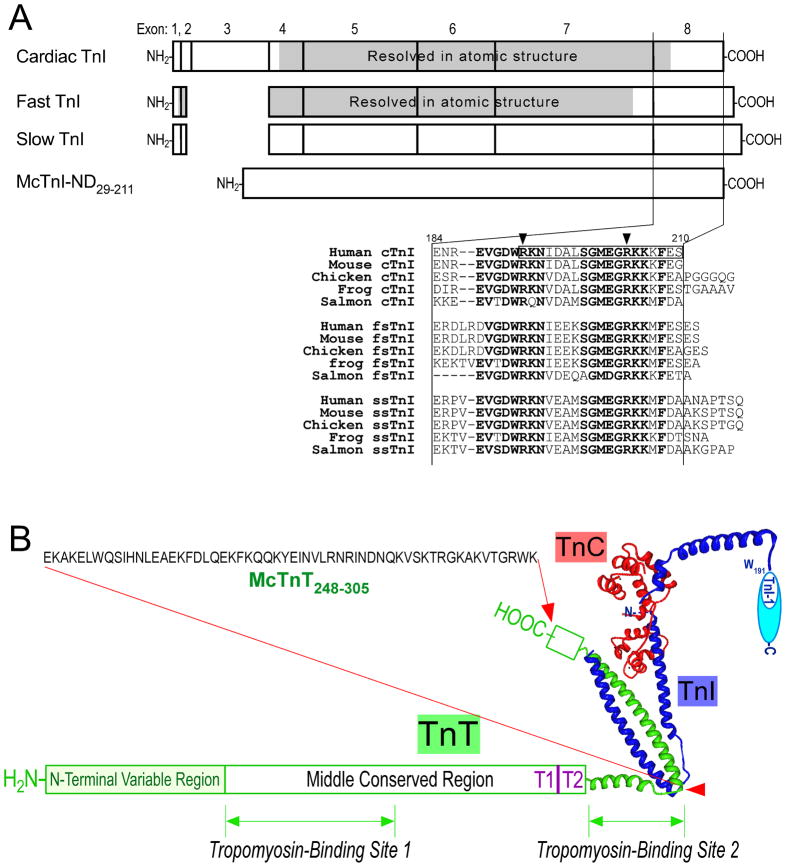
The COOH-terminal end segment of TnI is highly conserved in the amino acid sequences among the three TnI isoforms and across species (Fig. 1A). Exon 8 that encodes this segment (i.e., amino acids 184–210 in human cardiac TnI) is the most conserved exon among the TnI isoform genes [7,11]. The high degree of structural conservation implies a conserved essential function for COOH-terminal end segment of TnI.
Figure 1. TnI and TnT constructs.
A. The exon organizations of cardiac, fast and slow skeletal muscle TnI genes show that the NH2-terminal extension of cardiac TnI is a unique additional structure. The COOH-terminal end segment of TnI encoded by exon 8 has the most conserved primary structure among isoforms and across species [11], indicating a conserved function. In the sequence alignment, the most conserved residues are in bold letters, the two residues, of which mutations in human cardiac TnI caused restrictive cardiomyopathy, are indicated by arrows, and the COOH-terminal end segment, of which truncation resulted in severe cardiac muscle dysfunction, is boxed. Cardiac TnI has the shortest COOH-terminal end segment. The shaded areas indicate regions of known crystallographic structure [9,10]. McTnI29–211 is mouse cardiac TnI with the NH2-terminal extension removed and McTnI29–192 has an additional deletion of the COOH-terminal 19 amino acids. McTnI29-Cys212 is a site-specific triple modification of McTnI29–211 with two internal Cys substituted (Cys81Ile and Cys98Ser) and a Cys added to the COOH terminus (Cys212). B. This illustration outlines the position and amino acid sequence of the McTnT248–305 fragment in the troponin complex (its beginning and ending points are indicated with red arrows). The two known tropomyosin-binding sites of TnT [27] were removed in order to selectively study the interaction between tropomyosin and TnI in a troponin complex. The high resolution structure of partial troponin complex as determined with crystallography was redrawn from published data [9]. The conserved epitope in the COOH-terminal end segment of TnI recognized by mAb TnI-1 [11] is also indicated.
Supporting the predicted functional importance, abnormalities in this region of TnI have been found to cause severe diseases. A nonsense mutation in the cardiac TnI gene truncating entire exon 8 and a part of exon 7-encoded segments was found in restrictive cardiomyopathy [12]. A proteolytic truncation of the COOH-terminal 19 amino acids of cardiac TnI was found in rat hearts after ischemia-reperfusion injury [13]. Over-expression of this COOH-terminal truncated cardiac TnI in transgenic mouse hearts produced severely weakened ventricular function [14]. A single amino acid substitution, R192H, in cardiac TnI was highly potent in causing restrictive cardiomyopathy [15,16]. A nonsense mutation in cardiac TnI gene deleting the COOH-terminal 8 amino acids was associated with familial hypertrophic cardiomyopathy [17], further supporting the critical role of the COOH-terminal end segment of TnI in maintaining normal muscle function,.
The fact that the COOH-terminal end segment of TnI was not resolved with high-resolution structure in crystallographic studies implies its mobility or flexibility in the troponin complex [9,10]. To test the hypothesis that the COOH-terminal end segment of TnI is a dynamic structure related to the physiological function of troponin, we previously used a site-specific monoclonal antibody (mAb) probe to demonstrate calcium-induced conformational change in a COOH-terminal epitope on cardiac TnI. The result suggested that the COOH-terminal end segment of TnI is a functional domain of troponin, which responds to calcium regulation with spatial and/or conformational changes [11].
To further understand the role of the COOH-terminal end segment of TnI in regulating muscle contraction, it is necessary to demonstrate the biochemical basis for its function and Ca2+-responsiveness. An earlier study observed a weak binding between TnI in the troponin complex and tropomyosin in the absence of Ca2+ [18]. This information led us to test a hypothesis that the COOH-terminal end segment of TnI may function in the thin filament regulation by Ca2+-controlled interactions with tropomyosin. Supporting this hypothesis, a recent study using electron microscopy and 3D image reconstruction showed that a COOH-terminal truncated human cardiac TnI (amino acids 1–192) in reconstituted troponin complex altered tropomyosin position in the thin filament toward an enhanced Ca2+-activated closed state (C-state) that exposes more of the myosin-binding site on actin than that of reconstituted thin filament containing troponin with full-length TnI [19]. In addition to TnI’s established inhibitory role of promoting the blocked-state (B-state) [20], this study suggested that TnI participates in positioning tropomyosin in the C-state via the COOH-terminal end segment.
In the present study, we analyzed reconstituted tertiary troponin complex containing TnI that was fluorescently labeled at the COOH terminus. The experiments detected Ca2+-regulated conformational changes with a responding curve similar to that of the circular dichroism calcium titration curve of TnC. Protein binding studies using reconstituted troponin complex containing a mini-TnT in which the two tropomyosin-binding sites were deleted demonstrated a saturable binding of TnI to tropomyosin at pCa 9 but not at pCa 4. This Ca2+-regulated binding was abolished when the COOH-terminal 19 residues were removed. The data provided strong evidence for the physiological function of the COOH-terminal end segment of TnI, involving a Ca2+-regulated interaction with tropomyosin.
RESULTS
Engineered TnI and TnT proteins
The COOH-terminal end of TnI varies in length by several amino acids among the three muscle type isoforms (Fig. 1A). Cardiac TnI has the shortest COOH terminus and therefore is chosen in our study to identify the basic structure required for the conserved function. However, cardiac TnI has an NH2-terminal extension that is not present in fast and slow skeletal muscle TnI [4–6]. To have a representative model molecule for functional studies of the COOH-terminal end segment of TnI, we used a modified cardiac TnI in which the NH2-terminal extension is removed (McTnI29–211, in which M indicates mouse, c indicates cardiac, the amino acid residue numbers indicate the beginning and ending position of the protein fragment) [21,22] (Fig. 1). Similar nomenclature is used for other TnI and TnT proteins described below.
The NH2-terminal extension of cardiac TnI can be naturally removed in vivo by restrictive proteolysis with up-regulations in myocardial adaptation to simulated microgravity [21] or β-adrenergic deficiency [23]. Previous studies have demonstrated that the NH2-terminal truncation of cardiac TnI does not destroy the interactions with TnC and TnT [24]. Over-expression of McTnI29–211 in transgenic mouse hearts did not cause destructive phenotype [23]. Transgenic expression of McTnI29–211 completely rescued the postnatal lethality of mice that had the endogenous cardiac TnI gene deleted [25], verifying its preserved functionality.
In order to place a unique fluorescence probe at the COOH-terminus of TnI, we mutated the two endogenous Cys residues of mouse cardiac TnI and added a Cys at the COOH terminus (McTnI29-Cys212). Previous studies have demonstrated that cardiac TnI engineered with the same C81I and C98S mutations had preserved primary function in regulating muscle thin filaments [26].
To examine the potential interaction of the COOH-terminal end segment of TnI and tropomyosin, we constructed a COOH-terminal truncated cardiac TnI (McTnI29–192), in which the last 19 amino acids are deleted.
In order to investigate the biochemical interaction between tropomyosin and TnI in a Ca2+-regulated troponin complex, the dominant high affinity binding between TnT and tropomyosin [4] needs to be minimized. Based on the known crystal structure of the troponin complex and the recent mapping data for the two tropomyosin-binding sites of TnT [27], we engineered a TnT fragment, McTnT248–305, which lacks the two tropomyosin-binding sites but retains the TnI and TnC binding regions (Fig. 1B). This engineered mini-TnT allowed us to reconstitute troponin complex without the dominant tropomyosin-binding sites of TnT, so we could detect the binding between the COOH-terminal end segment of TnI and tropomyosin in a Ca2+-regulated experimental system.
Recombinant McTnI29–211, McTnI29-Cys212, McTnI29–192, and McTnT248–305 proteins were successfully expressed in E. coli and purified for use in the experimental studies (Fig. 2).
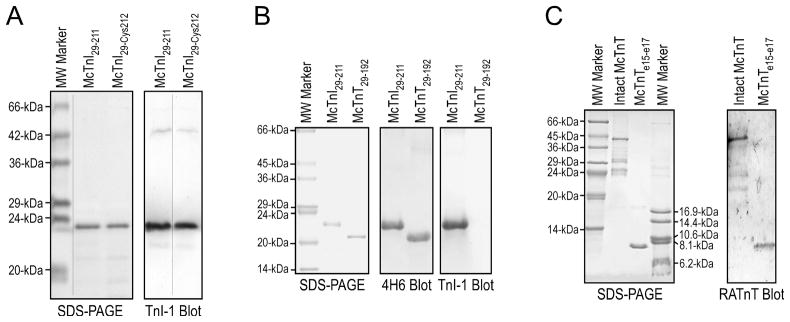
Figure 2. McTnI29–211, McTnI29-Cys212, McTnI29–192 and McTnT248–305 proteins.
A. SDS-gels and anti-TnI mAb Western blots of purified McTnI29–211 and McTnI29-Cys212. B. SDS-gel and Western blots of purified McTnI29–211 and McTnI29–192. The deletion of the COOH-terminal 19 amino acids abolished the binding of the anti-COOH-terminal mAb TnI-1 but not that of mAb 4H6 recognizing a middle region epitope. C. SDS-gel and Western blot using a polyclonal anti-TnT antiserum RATnT [42] identified the McTnT248–305 protein purified from E. coli culture.
McTnI29-Cys212 retained normal COOH-terminal conformation and binding affinities for TnC and TnT
The McTnI29-Cys212 protein with substitutions of two internal Cys residues and addition of a Cys at the COOH-terminus (Fig. 1) was examined by epitope analysis using enzyme-linked immunosorbent assay (ELISA) for effect of the modifications on COOH-terminal conformation. The results in Fig. 3A demonstrated that McTnI29-Cys212 gave an affinity titration curve for the anti-COOH-terminal segment mAb TnI-1 comparable to that of the McTnI29–211 control, indicating retained wild type molecular conformation in the COOH-terminal end segment.
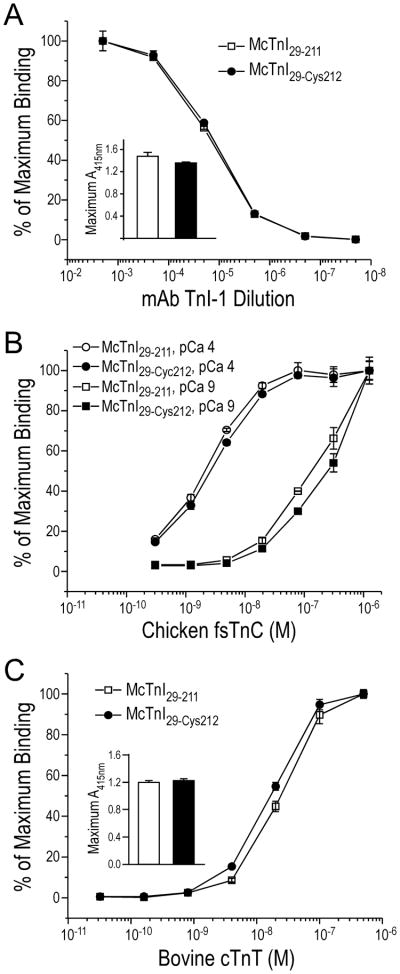
Figure 3. Preserved conformation and function of McTnI29-Cys212.
A. Affinity titration using mAb TnI-1 recognizing a COOH-terminal epitope [11] showed a binding curve identical to the McTnI29–211 control, indicating that the modifications in McTnI29-Cys212 (Fig. 1A) did not alter the conformation of the COOH-terminal end segment. B. The TnC-binding curves showed that McTnI29-Cys212 had a Ca2+-enhanced binding to TnC, similar to that of McTnI29–211. C. The TnT-binding curves showed that McTnI29-Cys212 bound TnT similarly to that of McTnI29–211.
The functionality of McTnI29-Cys212 was examined for its binding to TnC and TnT. Protein binding results in Fig. 3B showed that McTnI29-Cys212 bound chicken fast TnC in a Ca2+-enhanced manner as anticipated [4] with same affinities as that of the McTnI29–211 control. McTnI29-Cys212 also bound bovine cardiac TnT same as that of McTnI29–211 control (Fig. 3C). These data verified that the McTnI29-Cys212 model molecule has retained normal biochemical activity, suitable for functional studies.
Calcium-regulated conformational change in the COOH-terminal end segment of TnI
Fluorescence image of SDS-gel (data not shown) verified that McTnI29-Cys212 was effectively labeled with Alexa Fluor 532 dye at the COOH-terminal end Cys212 through maleimide-thiol group reactions.
The fluorescently labeled McTnI29-Cys212 was able to reconstitute tertiary troponin complex in vitro with chicken fast TnC and bovine cardiac TnT. Fluorescence image of SDS-gel (data not shown) further verified fluorescently labeled TnI in the troponin complex purified by gel filtration chromatography, which was used for fluorescence study of Ca2+-induced conformational changes in the COOH-terminal end segment of TnI.
As specified in the manufacturer’s datasheet, the emission spectral scans of Alexa Fluor 532 free dye detected a peak at 552 nm. The fluorescently labeled troponin complex showed a small red-shift of the emission peak to 558 nm (data not shown). The fluorescence emission peak did not change further during calcium titrations, indicating the 552 nm to 558 nm red-shift was due to the dye-protein conjugation.
Fluorescence intensity of the free dye exhibited mild decreases during the titration, reflecting a quenching effect. In contrast, TnI COOH terminus-labeled troponin complex showed increases in fluorescence intensity when [Ca2+] was increased during the titration (data not shown). The results indicated conformational changes in the COOH terminus of TnI when calcium binds to TnC in the troponin complex.
Ca2+ induced conformational responses in the COOH-terminal end segment of TnI
Plotted from the fluorescence spectral data, the TnI-COOH-terminus-labeled troponin complex demonstrated a sigmoid shape calcium titration curve (Fig. 4). EGTA back-titration from pCa 4 to pCa 8 showed that the effects of calcium were reversible.
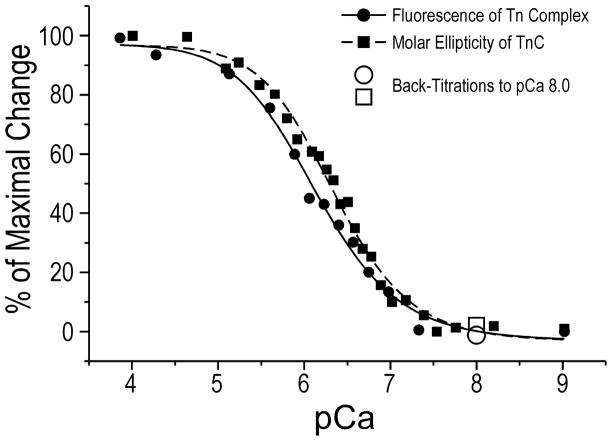
Figure 4. Calcium regulated conformational change in the COOH-terminal end segment of TnI.
Calcium titration curve of TnI-COOH-terminus-labeled troponin complex was plotted from the fluorescent intensity data after correction for the dye quenching effect. Calcium titration of secondary structure of free TnC was performed using far-UV CD spectral analysis. The molar ellipticity trough values at 222 nm indicating increases in α-helical contents in TnC when calcium is bound were collected to plot the calcium titration curve. Both curves were corrected for dilution factors, normalized to the maximal change, and fitted to the Hill equation (see Experimental Procedures). The open symbols represent EGTA back-titration from pCa 4 to 8, demonstrating the reversibility of the calcium-induced conformational changes in TnI COOH-terminal end segment and TnC. The results demonstrated very similar shapes of the calcium responding curves for the molecular conformations of TnI COOH-terminal end segment and TnC with pCa50 values of 6.10±1.02 and 6.32±1.19, respectively, with no statistical difference.
As a control, Ca2+-induced conformational changes in free TnC was titrated using far-UV CD spectral analysis and compared with the fluorescence titration of TnI-COOH-terminus-labeled troponin complex using the same set of pCa buffers. Consistent with published data [28,29], the far-UV CD spectra of TnC showed that Ca2+-binding increased the α-helix content of TnC as shown by the decreases in molar ellipticity. The sigmoid shape calcium titration curve of TnC derived from far-UV CD measurements (Fig. 4) was similar to that previously reported [28,29].
Superimposing the calcium titration curves for the fluorescence intensity of TnI-COOH-terminus-labeled troponin and the CD spectra of free TnC demonstrated similar patterns of calcium responsiveness (Fig. 4).
The binding with TnI was previously shown to increase the Ca2+ affinity of TnC at the high affinity sites III and IV (30). However, our assay buffer contained 3 mM Mg2+ to minimize this effect. Therefore, although the Ca2+ titration curves of free TnC and TnI COOH-terminal fluorescence represented individual conformational changes, their similar shapes suggested that the COOH-terminal end segment of TnI underwent conformational changes corresponding to the binding of Ca2+ to TnC in a troponin complex, supporting a physiological significance.
The COOH-terminal end segment of TnI binds tropomyosin in a Ca2+-regulated manner
The protein binding results in Fig. 5 showed that McTnT248–305 had very much reduced binding to tropomyosin in comparison to that of intact mouse cardiac TnT. The result verified the removal of the two known tropomyosin-binding sites in this mini-TnT, justifying its use in the reconstitution of tertiary troponin complex for the study of Ca2+-regulated interaction between TnI and tropomyosin.
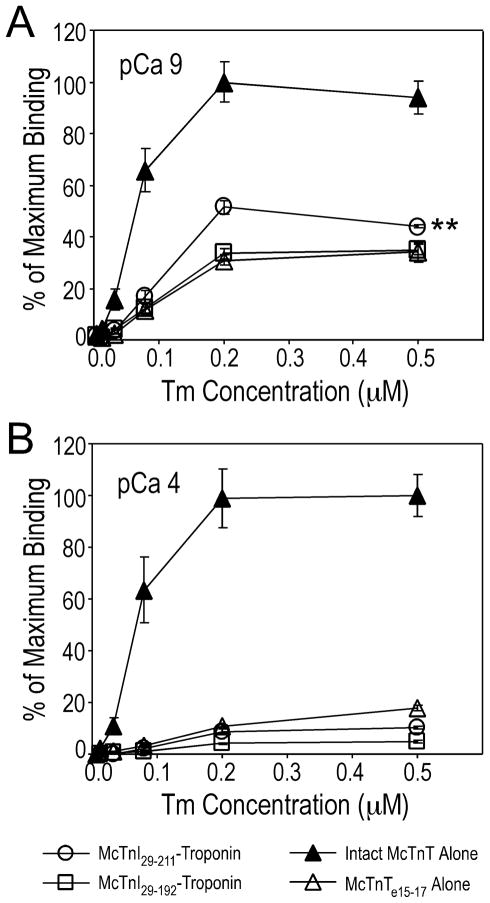
Figure 5. Ca2+-regulated binding between the COOH-terminal end segment of TnI and tropomyosin.
Protein binding experiments using reconstituted troponin complex containing McTnI29–211 or McTnI29–192 and McTnT248–305 in which the known tropomyosin-binding sites of TnT were deleted showed a saturable binding to tropomyosin at pCa 9 depending on the COOH-terminal 19 amino acids of TnI (A). This binding was abolished at pCa 4 (B). Intact mouse cardiac TnT alone and McTnT248–305 alone were used as controls to show the strong structural binding between intact TnT and tropomyosin and the much diminished binding between McTnT248–305 and tropomyosin.
Significantly above this background level, McTnT248–305-troponin containing McTnI29–211 exhibited a saturable binding to tropomyosin at pCa 9. This binding disappeared at pCa 4. In contrast, McTnT248–305-troponincontaining McTnI29–192 showed only a background level of binding to tropomyosin at pCa 9 similar to that of McTnT248–305 alone. In the solid phase protein-binding assay, the plateau level of the binding curve reflected the coupling strength (Kd) of the two proteins studied. The higher plateau of the McTnI29–211 curve reflecting the binding of the COOH-terminal end segment of TnI to tropomyosin at pCa 9 indicated stronger binding versus that at pCa 4. The results demonstrated that TnI directly interacts with tropomyosin under Ca2+-regulation and the COOH-terminal 19 amino acids of TnI plays a critical role in this interaction.
It was observed that the background binding of McTnI29–192-troponin to tropomyosin as well as the binding of free McTnT248–305 to tropomyosin in the absence of TnC was also diminished at pCa 4 (Fig. 5). It is possible that the weak binding of cTnT248–305 (free or in troponin complex) at pCa 9 but not at pCa 4 may be due to a direct conformational effect of calcium. The underlying mechanism remains to be investigated.
DISCUSSION
Although the importance of TnI in muscle diastolic function through its inhibitory effect on actomyosin ATPase was well established [4,5], the structural and functional significance of the highly conserved COOH-terminal end segment of TnI remains to be fully understood. Previous studies had suggested that the COOH-terminal half of the TnI polypeptide is involved in binding to the actin-tropomyosin thin filament. In the absence of calcium, TnI weakly bound TnC and tropomyosin. In the presence of calcium, the binding to TnC increased but the binding to tropomyosin diminished [18]. Removal of the COOH-terminal 17 amino acids decreased fast skeletal muscle TnI’s affinity for the thin filament [31]. These studies suggested that this region of TnI undergoes a Ca2+-dependent equilibrium between interactions with TnC and tropomyosin-actin thin filament [31,32]. These observation lead to a proposed model that the COOH-terminal terminal half of TnI interacts with actin-tropomyosin in the absence of Ca2+ and switches to TnC in the presence of Ca2+ [32].
Showing that the COOH-terminal end segment of TnI is a structure responding to Ca2+-regulation, our previous study using a site specific mAb as structural probe found that a COOH-terminal epitope involving the last 19 amino acids of TnI switched conformations between the presence and absence of calcium [11]. Recent studies by others further implied that the C-terminal domain of TnI is involved in the regulation of thin filament function [19,37]. Our present study is the first quantitative investigation of the calcium-regulated conformational change of the very COOH-terminal end of TnI and its binding to tropomyosin. Fluorescence intensity titration specifically demonstrated in reconstituted troponin complex the effect of calcium-binding to TnC on the conformation of the COOH-terminal end segment of TnI. This conformational modulation is corresponding to the Ca2+-responses of TnC, consistent with the function of troponin as a coordinated molecular switch.
Using engineered troponin complex lacking the dominant tropomyosin-binding sites of TnT, we further demonstrated in the present study that the COOH-terminal end segment of TnI binds tropomyosin at pCa 9 in a saturable manner (Fig. 5). This binding is abolished at pCa 4, which may indicate a Ca2+-induced release of the restriction of tropomyosin movement by TnI. Providing a novel biochemical evidence for the function of TnI in the core mechanisms of striated muscle contraction, this finding supports the notion that the COOH-terminal end segment of TnI is a dynamic structure in the calcium regulation of muscle thin filament.
Supporting the model that the Ca2+-regulated binding between the COOH-terminal end segment of TnI and tropomyosin is a part of the physiological function of troponin, the Ca2+-induced conformational change in the COOH terminus of TnI exhibited a titration curve with similar shape to that of the titration curve of Ca2+-induced conformational change in TnC (Fig. 4). Although indicating two individual allosteric changes in separate experiments, the similarly shaped calcium titration curves of free TnC and TnI COOH terminus in troponin complex suggest correlated conformational responses transmitting the Ca2+ signal from the calcium to TnC to the COOH-terminal end segment of TnI and the tropomyosin-actin thin filament. The potentially corresponding conformational modulations of TnC and the COOH-terminal end segment of TnI are plausible for the troponin complex to function as a molecular switch in the Ca2+-regulation of striated muscle contraction.
The COOH-terminal end segment of TnI has the most conserved amino acid sequence (Fig. 1). Phylogenetic analysis of the amino acid sequences of the three muscle type-specific TnI isoforms from a wide range of vertebrate species showed that this segment has been conserved even better than the adjacent segment that contains the TnT and TnC binding sites [11]. This evolutionarily stringent conservation of structure supports the hypothesis that the COOH-terminal end segment of TnI has a conserved fundamental function and likely interacts with a partner of evolutionarily conserved structure, such as sarcomeric tropomyosins [33], in the thin filament regulatory system of striated muscle.
Our biochemical evidence for the interaction of the COOH-terminal end segment of TnI with tropomyosin supports the previous electron microscopic imaging reconstruction studies showing that a larger COOH-terminal fragment of human cardiac TnI (residues 131–210) drove tropomyosin movement to the blocking position to inhibit myosin-cross bridge association at low calcium [34,35]. An atomic structure model obtained from docking NMR spectroscopy structure into cryo-electron microscopy density map also indicated a mobile domain in the troponin complex corresponding to a similarly large COOH-terminal fragment of chicken fast TnI (residues 131–182), in which changes in calcium concentration generated shifts in the azimuthal position of tropomyosin on the actin filament [36]. These results support the hypothesis that the COOH-terminal end segment of TnI may restrict tropomyosin movement at the resting condition when calcium is low, contributing to the inhibition of actomyosin ATPase and muscle relaxation.
A more recent electron microscopy and 3D image reconstruction study [37] found that whereas the direction of the tropomyosin movement was retained in thin filament containing human cardiac TnI with a COOH-terminal truncation equivalent to that of McTnI29–192, the movement was greater in the absence of the COOH-terminal end segment of TnI [19]. Our data showing Ca2+-regulated binding between the COOH-terminal end segment of TnI and tropomyosin would support the hypothesis that the mobile COOH-terminal end of TnI plays a role in positioning the Ca2+-regulated switching of tropomyosin in the thin filament, which merits further investigation.
We previously observed that affinity chromatography using mAb TnI-1 against the COOH-terminal end segment of TnI readily isolated tertiary troponin complex from crude muscle homogenate [21]. Therefore, the COOH-terminal end segment of TnI appeared as an exposed structure in native troponin complex. The increases in fluorescence intensity of the TnI-COOH-terminus-labeled troponin when calcium concentration rose suggested that the COOH-terminal end segment of TnI became more exposed in the troponin complex when calcium bound to TnC. This observation is consistent with our previous finding that binding of calcium to troponin made the COOH-terminal end segment of TnI more accessible by mAb TnI-1 [11]. The increased freedom of TnI COOH-terminal end segment at high calcium may allow the movement of tropomyosin and the release of thin filament inhibition. These experimental results added new understanding of the function of TnI and the molecular mechanism of myofilament regulation. The calcium-regulated conformational change in the COOH-terminal end segment of TnI and its binding to tropomyosin laid a biochemical foundation for further studies on the function of TnI in the Ca2+-regulation of muscle contraction.
EXPERIMENTAL PROCEDURES
Expression and purification of McTnI29–211
cDNA encoding McTnI29–211 was cloned into pAED4 plasmid for protein expression in BL21(DE3)pLysS E. coli [22]. Recombinant McTnI29–211 was first fractionated on a CM52 cation-exchange column in 6 M urea, 0.1 mM EDTA, 6 mM β-mercaptoethanol and 10 mM imidazole-HCl, pH 7.0. Eluted with 0–500 mM linear KCl gradient, the protein peaks were examined by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting using anti-TnI mAb TnI-1 [11]. Fractions containing McTnI29–211 protein were dialyzed, concentrated by lyophilization, and further purified using Sepharose G75 gel filtration chromatography in 6 M urea, 500 mM KCl, 0.1 mM EDTA, 6 mM β-mercaptoethanol, 10 mM imidazole-HCl, pH 7.0. The fractions containing pure McTnI29–211 was dialyzed against 0.1% formic acid and lyophilized [24]. The purification procedures were performed at 4°C.
The cDNA encoding McTnI29–211 was modified to place a sole Cys at the COOH terminal end (McTnI29-Cys212) for fluorescence labeling. Site-specific modifications were carried out in two rounds of recombinant polymerase chain reaction (PCR) and cloning. The first round modification mutated endogenous Cys81 to Ile and added a Cys to the COOH terminus (Cys212). The second round PCR mutagenesis further replaced endogenous Cys98 with Ser. The modified cDNA clones were verified by sequencing. McTnI29-Cys212 protein was expressed and purified as described above for McTnI29–211.
COOH-terminal truncated TnI
The cDNA template encoding McTnI29–211 was modified by PCR to produce a COOH-terminal truncated TnI (McTnI29–192) [38]. The McTnI29–192 protein was expressed in E. coli, purified as above, and verified by Western blotting using mAb 4H6 that recognizes a middle region epitope [16].
Engineering a mini-TnT lacking the tropomyosin-binding sites
A cDNA corresponding to the exons 15–17-encoded COOH-terminal segment of mouse cardiac TnT (McTnT248–305) was constructed using PCR from the full length cDNA and inserted into pAED4 plasmid for non-fusion protein expression. As shown in the crystal structures of troponin [9,10], this fragment of TnT fully retains the binding sites for TnI and TnC (Fig. 1B) and, therefore, can be used to reconstitute tertiary troponin complex.
After DNA sequencing to verify the expression plasmid, McTnT248–305 protein was expressed in E. coli culture. BL21(DE3)pLysS E. coli cells transformed with the recombinant plasmid were cultured, induced in the early log phase, and harvested. The cells were lysed in 5 mM EDTA, 15 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride, 50 mM Tris-HCl, pH 8.0, using a French press. The protein extract was fractionated with ammonium sulfate precipitation at 0°C. The fraction between 30–50% saturation was dialyzed against 0.1 mM EDTA, brought to 6 M urea, 15 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride, 20 mM imidazole, pH 7.0, clarified by centrifugation, and loaded on a CM52 column equilibrated with the same buffer. The column was eluted with 0–500 mM linear KCl gradient, and the protein peaks were analyzed with SDS-PAGE. The fractions containing McTnT248–305 were dialyzed against 0.1 mM EDTA and concentrated by lyophilization for further purification using Sephadex G75 gel filtration chromatography in 6 M urea, 0.5 M KCl, 0.1 mM EDTA, 6 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride, 10 mM imidazole, pH 7.0. Protein peaks were analyzed by SDS-PAGE and the fractions containing pure McTnT248–305 were dialyzed against de-ionized water and lyophilized.
Preparation of other proteins
Bovine cardiac TnT was purified from adult left ventricular muscle as previously described [39]. Chicken fast TnC was purified from E. coli expression as described previously [40]. Bovine cardiac α-tropomyosin was purified from adult ventricular muscle as described [41].
mAb epitope analysis
Based on the nature that antibody-antigen bindings are dependent on fits between 3D protein structures [42], mAb TnI-1 that recognizes a conserved epitope in the COOH-terminal end segment of TnI [11] was used as a 3D structural probe to examine the modified TnI molecules.
ELISA epitope analysis [42,43] was applied to detect whether the C81I and C98S substitutions and the addition of a Cys to the COOH terminus had any effect on the molecular conformation of the COOH-terminal end segment of TnI. McTnI29–211 and McTnI29-Cys212 were dissolved at 1 μg/mL in Buffer A (20 mM PIPES, 3 mM MgCl2, 100 mM KCl, pH 7.0) and coated separately in 96-well microtitering plates (100 μL/well) at 4°C overnight. After washing with Buffer T (Buffer A plus 0.05% Tween-20) to remove unbound protein, the immobilized TnI was incubated with serial dilutions of mAb TnI-1 in Buffer B (Buffer T plus 0.1% bovine serum albumin (BSA)) at room temperature for 2 hr. The plates were then processed with a standard ELISA procedure including Buffer T washes, horseradish peroxidase-conjugated anti-mouse immunoglobulin second antibody (Santa Cruz Biotechnology) incubation, and H2O2/2,2′-azinobis(3-ethylbenzothiazolinesulfonic acid) substrate reaction. Absorbance at 415 nm of each assay well was recorded at a series of time points during the substrate development using an automated microplate reader (Bio-Rad Benchmark). The A415nm values in the linear course of the color development were plotted to obtain mAb affinity curves that reflect the molecular conformation at the TnI-1 epitope in the COOH-terminal end segment of TnI. The experiments were performed in triplicate and repeated.
Protein binding assays
ELISA solid-phase protein binding experiments were performed to examine the interactions of McTnI29-Cys212 with TnT and TnC. Purified McTnI29–211 and McTnI29-Cys212 proteins were dissolved at 5 μg/mL in Buffer A containing 1 mM dithiothreitol (DTT) and non-covalently immobilized onto 96-well microtitering plates. After removing free protein and blocking the wells with 1% BSA in Buffer T, the immobilized TnI was incubated with serial dilutions of bovine cardiac TnT or chicken fast TnC in Buffer B containing 1 mM DTT. After Buffer T washes once with DTT and four times without DTT, the TnT or TnC bound to the immobilized TnI was quantified via an anti-TnT mAb CT3 [38] or an anti-TnC mAb 2C3 [40], respectively. The plates were then processed with ELISA procedure as described above to obtain binding affinity curves for TnT and TnC. The experiments were performed in triplicate and repeated.
Fluorescence labeling of McTnI29-Cys212
Purified McTnI29-Cys212 protein was dissolved in 100 mM KCl, 10 mM imidazole-HCl, pH 7.0, and mixed with Alexa Fluor 532 C5-maleimide dye in a protein:dye molar ratio of 1:10 for incubation with shaking at room temperature for 2 hr. The labeling mix was put through a Superose F12 column (GE Healthcare) to remove free dye. McTnI29-Cys212 labeled at the COOH-terminal Cys212 was examined by SDS-PAGE and fluorescence imaging using a Typhoon 9210 scanner (GE Healthcare) with excitation at 532 nm and recording the emission at 555 nm. Total protein in the gels was revealed with Coomassie Brilliant Blue R-250 staining.
Reconstitution of troponin complex
Tertiary troponin complex was reconstituted in vitro using a) McTnI29–211, McTnI29–192 or fluorescence-labeled McTnI29-Cys212, b) intact bovine cardiac TnT or McTnT248–305, and c) chicken fast TnC. The protein mixture was dissolved in 6 M urea, 1 M KCl, 1 mM DTT, 10 mM imidazole-HCl, pH 7.1, and dialyzed to gradually remove urea and high salt to reach the final assay condition of 100 mM KCl, 3 mM MgCl2, 10 mM imidazole-HCl, pH 7.1. To compensate for the predicted higher loss of McTnT248–305 and TnC during dialysis due to their smaller sizes, these two components were used in 2-fold and 1.4-fold molar excesses, respectively. The reconstituted troponin complex was purified using Superose F12 column in the assay buffer. The eluted troponin peak containing the three troponin subunits was identified by SDS-PAGE for use in structure-function studies.
Fluorescence measurements
Steady-state fluorescence measurements were carried out at room temperature using an ISS PC1 spectrofluorometer with the band pass of the excitation and emission monochromators both at 8 nm. Fluorescence intensity was recorded with the excitation at 528 nm for the TnI-COOH-terminus labeled troponin complex and Alexa Fluor 532 free dye control.
Far-UV CD measurements
Chicken fast TnC was dissolved at 0.5 mg/mL in 100 mM KCl, 3 mM MgCl2, and 10 mM sodium phosphate, pH 7.1. Far UV CD spectra of TnC were measured on a Jasco J715 spectropolarimeter at room temperature following a previously described method [28].
Calcium titration and fluorescence curve fitting
To avoid metal ion contaminations, all solutions were prepared with Milli-Q deionized water in HCl pre-treated plastic ware. Stocks of 10 and 20 mM CaCl2 were prepared from 1 M CaCl2 standard purchased from Fluka. Titrations were conducted by adding the CaCl2 stocks in serial increments. With 10 mM imidazole buffer in the fluorescence measurements of troponin complex and 10 mM sodium phosphate buffer in the far-UV CD measurements of TnC, no change in pH was detected before and after the calcium titrations. Free Ca2+ concentrations buffered by 1 mM EGTA were calculated using Maxchelator program (http://maxchelator.stanford.edu). pCa values were calculated as -log(free [Ca2+]).
For the TnI-COOH-terminus-labeled troponin complex, the fluorescence intensity at 558 nm was recorded and corrected by the dilution factor during the titration and the quenching rate of the free dye (Equation 1).
| Equation 1 |
In far-UV CD calcium titration of TnC, the molar ellipticity at 222 nm was recorded and corrected by the dilution factor (Equation 2).
| Equation 2 |
The corrected fluorescence intensity or molar ellipticity was normalized to the percentage of their maximal changes using Equation 3 and plotted against the pCa values.
| Equation 3 |
Curve fitting for the titration data was performed with nonlinear Hill equation (Equation 4) using the Origin software.
| Equation 4 |
in which Y represents the normalized fluorescence intensity or molar ellipticity, K is the apparent dissociation constant, and n is the Hill coefficient.
Detection of Ca2+-regulated binding between the COOH-terminal end segment and tropomyosin
Similar to the binary protein binding experiments described above, ELISA experiments were carried out to investigate the binding between TnI COOH-terminal end segment and tropomyosin. Reconstituted tertiary troponin complex containing McTnT248–305 and McTnI29–211 or McTnI29–192 were immobilized onto microtitering plates in Buffer A containing DTT at pCa 4 [40]. After washing and blocking the wells as above but at pCa 4, the immobilized troponin were incubated with serial dilutions of tropomyosin in Buffer B at pCa 4 or pCa 9. The plates were then processed as above with washing, incubation with anti-tropomyosin mAb CH1 (a gift from Dr. Jim Lin, University of Iowa), washing to remove excess CH1, incubation with horseradish peroxidase-second antibody, and final washes, in which the pCa remained to be controlled at 4 or 9 to examine the effect of Ca2+. After substrate color development, tropomyosin-binding curves were plotted with data from triplicate wells and repeated experiments. Immobilized intact mouse cardiac TnT alone and McTnT248–305 alone were examined as controls in the tropomyosin-binding experiments.
Data analysis
All values are presented as the mean ± S.D. Statistical analysis were performed using unpaired Student’s t test.
Acknowledgments
This study was supported by grants from the National Institutes of Health AR-048816 and HL-098945.
We thank Stephen Chong for participating in the construction of cTnI29–192, Annie Wang for technical assistance, Dr. Jim Lin for the CH1 mAb, and the Keck Biophysics Facility at Northwestern University for instrumental services.
Abbreviations
- TnC
troponin C
- BSA
bovine serum albumin
- CD
circular dichroism
- DTT
dithiothreitol
- ELISA
enzyme-linked immunosorbent assay
- mAb
monoclonal antibody
- PCR
polymerase chain reaction
- SDS-PAGE
SDS-polyacrylamide gel electrophoresis
- TnI
troponin I
- TnT
troponin T
References
- 1.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 2.Leavis PC, Gergly J. Thin filament proteins and thin filament-linked regulation of vertebrate muscle contraction. CRC Crit Rev Biochem. 1984;16:235–305. doi: 10.3109/10409238409108717. [DOI] [PubMed] [Google Scholar]
- 3.Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996;58:447–481. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- 4.Perry SV. Troponin I: inhibitor or facilitator. Mol Cell Biochem. 1999;190:9–32. [PubMed] [Google Scholar]
- 5.Solaro RJ, Rosevear P, Kobayashi T. The unique functions of cardiac troponin I in the control of cardiac muscle contraction and relaxation. Biochem Biophys Res Commun. 2008;369:82–87. doi: 10.1016/j.bbrc.2007.12.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hastings KE. Molecular evolution of the vertebrate troponin I gene family. Cell Struct Funct. 1997;22:205–211. doi: 10.1247/csf.22.205. [DOI] [PubMed] [Google Scholar]
- 7.Jin JP, Zhang Z, Bautista JA. Isoform diversity, regulation, and functional adaptation of troponin and calponin. Crit Rev Eukaryot Gene Expr. 2008;18:93–124. doi: 10.1615/critreveukargeneexpr.v18.i2.10. [DOI] [PubMed] [Google Scholar]
- 8.Li MX, Wang X, Sykes BD. Structural based insights into the role of troponin in cardiac muscle pathophysiology. J Muscle Res Cell Motil. 2004;25:559–579. doi: 10.1007/s10974-004-5879-2. [DOI] [PubMed] [Google Scholar]
- 9.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 10.Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ. Ca(2+)-regulated structural changes in troponin. Proc Natl Acad Sci U S A. 2005;102:5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin JP, Yang FW, Yu ZB, Ruse CI, Bond M, Chen A. The highly conserved COOH terminus of troponin I forms a Ca2+-modulated allosteric domain in the troponin complex. Biochemistry. 2001;40:2623–2631. doi: 10.1021/bi002423j. [DOI] [PubMed] [Google Scholar]
- 12.Kostareva A, Gudkova A, Sjöberg G, Mörner S, Semernin E, Krutikov A, Shlyakhto E, Sejersen T. Deletion in TNNI3 gene is associated with restrictive cardiomyopathy. Int J Cardio. 2009;131:410–412. doi: 10.1016/j.ijcard.2007.07.108. [DOI] [PubMed] [Google Scholar]
- 13.McDonough JL, Arrell DK, Van Eyk JE. Troponin I degradation and covalent complex formation accompanies myocardial ischemia/reperfusion injury. Circ Res. 1999;84:9–20. doi: 10.1161/01.res.84.1.9. [DOI] [PubMed] [Google Scholar]
- 14.Murphy AM, Kogler H, Georgakopoulos D, McDonough JL, Kass DA, Van Eyk JE, Marban E. Transgenic mouse model of stunned myocardium. Science. 2000;287:488–491. doi: 10.1126/science.287.5452.488. [DOI] [PubMed] [Google Scholar]
- 15.Mogensen J, Kubo T, Duque M, Uribe W, Shaw A, Murphy R, Gimeno JR, Elliott P, McKenna WJ. Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations. J Clin Invest. 2003;111:209–216. doi: 10.1172/JCI16336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du J, Liu J, Feng HZ, Hossain MM, Gobara N, Zhang C, Li Y, Jean-Charles PY, Jin JP, Huang XP. Impaired relaxation is the main manifestation in transgenic mice expressing a restrictive cardiomyopathy mutation, R193H, in cardiac TnI. Am J Physiol Heart Circ Physiol. 2008;294:H2604–2613. doi: 10.1152/ajpheart.91506.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morner S, Richard P, Kazzam E, Hainque B, Schwartz K, Waldenstrom A. Deletion in the cardiac troponin I gene in a family from northern Sweden with hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2000;32:521–525. doi: 10.1006/jmcc.1999.1099. [DOI] [PubMed] [Google Scholar]
- 18.Pearlstone JR, Smilli LB. Effects of troponin-I plus-C on the binding of troponin-T and its fragments to alpha-tropomyosin. Ca2+ sensitivity and cooperativity. J Biol Chem. 1983;258:2534–2542. [PubMed] [Google Scholar]
- 19.Galińska A, Hatch V, Craig R, Murphy AM, Van Eyk JE, Wang CL, Lehman W, Foster DB. The C terminus of cardiac troponin I stabilizes the Ca2+-activated state of tropomyosin on actin filaments. Circ Res. 2010;106:705–11. doi: 10.1161/CIRCRESAHA.109.210047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maytum R, Lehrer SS, Geeves MA. Cooperativity and switching within the three-state model of muscle regulation. Biochemistry. 1999;38:1102–1110. doi: 10.1021/bi981603e. [DOI] [PubMed] [Google Scholar]
- 21.Yu ZB, Zhang LF, Jin JP. A proteolytic NH2-terminal truncation of cardiac troponin I that is up-regulated in simulated microgravity. J Biol Chem. 2001;276:15753–15760. doi: 10.1074/jbc.M011048200. [DOI] [PubMed] [Google Scholar]
- 22.Barbato JC, Huang QQ, Hossain MM, Bond M, Jin JP. Proteolytic N-terminal truncation of cardiac troponin I enhances ventricular diastolic function. J Biol Chem. 2005;280:6602–6609. doi: 10.1074/jbc.M408525200. [DOI] [PubMed] [Google Scholar]
- 23.Feng HZ, Chen M, Weinstein LS, Jin JP. Removal of the N-terminal extension of cardiac troponin I as a functional compensation for impaired myocardial beta-adrenergic signaling. J Biol Chem. 2008;283:33384–33393. doi: 10.1074/jbc.M803302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biesiadecki BJ, Schneider KL, Yu ZB, Chong SM, Jin JP. An R111C polymorphism in wild turkey cardiac troponin I accompanying the dilated cardiomyopathy-related abnormal splicing variant of cardiac troponin T with potentially compensatory effects. J Biol Chem. 2004;279:13825–13832. doi: 10.1074/jbc.M314225200. [DOI] [PubMed] [Google Scholar]
- 25.Feng HZ, Hossain MM, Huang XP, Jin JP. Myofilament incorporation determines the stoichiometry of troponin I in transgenic expression and the rescue of a null mutation. Arch Biochem Biophys. 2009;487:36–41. doi: 10.1016/j.abb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong WJ, Xing J, Chandra M, Solaro J, Cheung HC. Structural mapping of single cysteine mutants of cardiac troponin I. Proteins. 2000;41:438–447. [PubMed] [Google Scholar]
- 27.Jin JP, Chong SM. Localization of the two tropomyosin-binding sites of troponin T. Arch Biochem Biophys. 2010;500:144–150. doi: 10.1016/j.abb.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golosinska K, Pearlstone JR, Borgford T, Oikawa K, Kay CM, Carpenter MR, Smillie LB. Determination of and corrections to sequences of turkey and chicken troponins-C. Effects of Thr-130 to Ile mutation on Ca2+ affinity. J Biol Chem. 1991;266:15797–15809. [PubMed] [Google Scholar]
- 29.Pearlstone JR, McCubbin WD, Kay CM, Sykes BD, Smillie LB. Spectroscopic analysis of a methionine-48 to tyrosine mutant of chicken troponin C. Biochemistry. 1992;31:9703–9708. doi: 10.1021/bi00155a025. [DOI] [PubMed] [Google Scholar]
- 30.Ngai SM, Pearlstone JR, Farah CS, Reinach FC, Smillie LB, Hodges RS. Structural and functional studies on Troponin I and Troponin C interactions. J Cell Biochem. 2001;83:33–46. doi: 10.1002/jcb.1204. Erratum in: J Cell Biochem (2002), 84, 847. [DOI] [PubMed] [Google Scholar]
- 31.Ramos CH. Mapping subdomains in the C-terminal region of troponin I involved in its binding to troponin C and to thin filament. J Biol Chem. 1999;274:18189–18195. doi: 10.1074/jbc.274.26.18189. [DOI] [PubMed] [Google Scholar]
- 32.Farah CS, Miyamoto CA, Ramos CH, da Silva ACR, Quaggio RB, Fujimori K, Smillie LB, Reinach FC. Structural and regulatory functions of the NH2- and COOH-terminal regions of skeletal muscle troponin I. J Biol Chem. 1994;269:5230–5240. [PubMed] [Google Scholar]
- 33.Perry SV. Vertebrate tropomyosin: distribution, properties and function. J Muscle Res Cell Motil. 2001;22:5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- 34.Galinska-Rakoczy A, Engel P, Xu C, Jung H, Craig R, Tobacman LS, Lehman W. Structural basis for the regulation of muscle contraction by troponin and tropomyosin. J Mol Biol. 2008;379:929–935. doi: 10.1016/j.jmb.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pirani A, Vinogradova MV, Curmi PM, King WA, Fletterick RJ, Craig R, Tobacman LS, Xu C, Hatch V, Lehman W. An atomic model of the thin filament in the relaxed and Ca2+-activated states. J Mol Biol. 2006;357:707–717. doi: 10.1016/j.jmb.2005.12.050. [DOI] [PubMed] [Google Scholar]
- 36.Murakami K, Yumoto F, Ohki SY, Yasunaga T, Tanokura M, Wakabayashi T. Structural basis for Ca2+-regulated muscle relaxation at interaction sites of troponin with actin and tropomyosin. J Mol Biol. 2005;352:178–201. doi: 10.1016/j.jmb.2005.06.067. [DOI] [PubMed] [Google Scholar]
- 37.Lehman W, Galińska-Rakoczy A, Hatch V, Tobacman LS, Craig R. Structural basis for the activation of muscle contraction by troponin and tropomyosin. J Mol Biol. 2009;388:673–681. doi: 10.1016/j.jmb.2009.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chong SM, Jin JP. To investigate protein evolution by detecting suppressed epitope structures. J Mol Evol. 2009;68:448–460. doi: 10.1007/s00239-009-9202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin JP, Lin JJ. Rapid purification of mammalian cardiac troponin T and its isoform switching in rat hearts during development. J Biol Chem. 1988;263:7309–7315. [PubMed] [Google Scholar]
- 40.Jin JP, Chong SM, Hossain MM. Microtiter plate monoclonal antibody epitope analysis of Ca2+- and Mg2+-induced conformational changes in troponin C. Arch Biochem Biophys. 2007;466:1–7. doi: 10.1016/j.abb.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smillie LB. Preparation and identification of alpha- and beta-tropomyosins. Methods Enzymol. 1982;85:234–241. doi: 10.1016/0076-6879(82)85023-4. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Jin JP. Conformational modulation of troponin T by configuration of the NH2-terminal variable region and functional effects. Biochemistry. 1998;37:14519–14528. doi: 10.1021/bi9812322. [DOI] [PubMed] [Google Scholar]
- 43.Jin JP, Chen A, Ogut O, Huang QQ. Conformational modulation of slow skeletal muscle troponin T by an NH(2)-terminal metal-binding extension. Am J Physiol Cell Physiol. 2000;279:C1067–1077. doi: 10.1152/ajpcell.2000.279.4.C1067. [DOI] [PubMed] [Google Scholar]