Abstract
Ribonuclease P (RNase P) is a ribonucleoprotein enzyme that cleaves precursor tRNA transcripts to give mature 5′ ends. RNase P in eubacteria has a large, catalytic RNA subunit and a small protein subunit that are required for precursor tRNA cleavage in vivo. Although the eukaryotic holoenzymes have similar, large RNA subunits, previous work in a number of systems has suggested that the eukaryotic enzymes require a greater protein content. We have purified the Saccharomyces cerevisiae nuclear RNase P to apparent homogeneity, allowing the first comprehensive analysis of an unexpectedly complex subunit composition. Peptide sequencing by ion trap mass spectrometry identifies nine proteins that copurify with the nuclear RNase P RNA subunit, totaling 20-fold more protein than in the bacterial enzyme. All of these proteins are encoded by genes essential for RNase P activity and for cell viability. Previous genetic studies suggested that four proteins might be subunits of both RNase P and RNase MRP, the related rRNA processing enzyme. We demonstrate that all four of these proteins, Pop1p, Pop3p, Pop4p, and Rpp1p, are integral subunits of RNase P. In addition, four of the five newly identified protein subunits, Pop5p, Pop6p, Pop7p, and Pop8p, also appear to be shared between RNase P and RNase MRP. Only one polypeptide, Rpr2p, is unique to the RNase P holoenzyme by genetic depletion and immunoprecipitation studies. The large increase in the number of protein subunits over eubacterial RNase P is consistent with an increase in functional complexity in eukaryotes. The degree of structural similarity between nuclear RNase P and RNase MRP suggests that some aspects of their functions in pre-tRNA and pre-rRNA processing pathways might overlap or be coordinated.
Keywords: Ribonuclease P, ribonuclease MRP, RNP, tRNA
Precursor tRNAs (pre-tRNAs) are processed at their 5′ and 3′ ends to yield mature tRNAs. Ribonuclease P (RNase P) is the enzyme responsible for the 5′ endonucleolytic cleavage in eubacteria, archaea, and eukaryotes (Altman et al. 1995; Pace and Brown 1995; Chamberlain et al. 1996b). In most instances the enzyme is composed of both RNA and protein, but it is the RNA subunit of the eubacterial RNase P that is capable of both recognizing and cleaving substrates efficiently and accurately (Guerrier-Takada et al. 1983). The protein component serves as a cofactor and is absolutely required in vivo (Reich et al. 1988). Although the eubacterial RNAs are divergent in primary sequence, characterization of the RNA structure has resulted in proposed tertiary models of the RNase P RNA subunit (Westhof and Altman 1994; Harris et al. 1997). Experimental conditions that would allow catalysis by the archaeal and eukaryotic RNase P RNAs have not been defined, although the archaeal and eukaryotic RNAs appear to fold in a secondary structure resembling the eubacterial RNA subunit (Forster and Altman 1990; Tranguch et al. 1994; Chen and Pace 1997). The eubacterial RNA subunit (350–410 nucleotides) is similar in size to the eukaryotic nuclear RNA (280–380 nucleotides), but the protein content of the eukaryotic enzyme is much greater, estimated to be 50%–70% of the eukaryotic holoenzyme as compared to 10% of the eubacterial enzyme (Lawrence et al. 1987; Doria et al. 1991; Jayanthi and Van Tuyle 1992). The increase in protein in the eukaryotic enzyme suggests that the eukaryotic RNase P may represent a transition from an early RNA enzyme, still maintained in bacteria, to an enzyme dependent on protein for activity. The large increase in the protein complexity might also be necessary for function in the increased organizational complexity of the eukaryotic cell.
RNase P activities are found in the nuclei (Bartkiewicz et al. 1989; Lee and Engelke 1989; Doria et al. 1991), mitochondria (Doersen et al. 1985; Hollingsworth and Martin 1986; Y. Lee et al. 1996), and chloroplasts (Thomas et al. 1995) of eukaryotes. Purifications of Saccharomyces cerevisiae mitochondrial and Schizosaccharomyces pombe nuclear RNase P identified single proteins of ∼100 kD that cofractionated with RNase P activity (Morales et al. 1992; Zimmerly et al. 1993). The nuclear gene encoding the S. cerevisiae mitochondrial protein subunit RPM2 has been found essential and necessary for mitochondrial pre-tRNA processing (Dang and Martin 1993). Other mitochondrial enzymes might have higher protein content, however. The mitochondrial enzyme from Aspergillus nidulans appears to have several proteins that cofractionate with activity (Y.C. Lee et al. 1996).
Several polypeptides have been implicated in S. cerevisiae nuclear RNase P function by direct and indirect experiments. Three of these proteins were first identified by genetic methods that were intended to identify proteins associated with the activity of a related ribonucleoprotein enzyme, RNase mitochondrial RNA processing (RNase MRP). RNase MRP has an essential RNA subunit that is evolutionarily related to RNase P (Gold et al. 1989; Morrissey and Tollervey 1995) and of similar size. This enzyme, however, cleaves rRNA precursors found in nucleoli (Shuai and Warner 1991; Schmitt and Clayton 1993; Chu et al. 1994; Lygerou et al. 1996) and has also been implicated in mitochondrial replication (Stohl and Clayton 1992). The three genes originally identified as affecting RNase MRP function, POP1, POP3, and POP4 (Lygerou et al. 1994; Chu et al. 1997; Dichtl and Tollervey 1997) encode proteins that are also required for RNase P activity in vivo. Immunoprecipitation of these Pop proteins shows that they are physically associated with both RNase P and RNase MRP RNAs. Another candidate protein subunit, Rpp1p, recently has been identified in yeast by its homology with a human protein, Rpp30, that cofractionates with HeLa cell RNase P along with a number of other polypeptides (Eder et al. 1997). Like the Pop proteins, depletion and immunoprecipitation studies suggest that Rpp1p is a subunit of both RNase P and RNase MRP (Stolc and Altman 1997). None of these subunits has obvious homology with either the 14-kD bacterial RNase P subunit C5 (Hansen et al. 1985) or the 105-kD mitochondrial RNase P subunit Rpm2p (Dang and Martin 1993). Only one protein, Snm1p, has been identified that is physically associated with only RNase MRP (Schmitt and Clayton 1994).
Confirmation of the Pop proteins as integral components of the RNase P holoenzyme has been difficult. Both enzymes might be part of a large complex in vivo such that mutations in subunits of either enzyme could indirectly affect the activity of the other. If the two enzymes are associated, it would be expected that they colocalize in cells, but in situ hybridization with probes specific for RNase P or RNase MRP probes has given conflicting results. The established cellular location of RNase MRP is primarily nucleolar, where it functions in pre-rRNA processing (Jacobson et al. 1995; Matera et al. 1995). There is disagreement in reports as to whether mammalian RNase P RNA is primarily nucleoplasmic after transient nucleolar association (Jacobson et al. 1997) or is mostly found in focused loci at the edges of nucleoli (Matera et al. 1995; B. Lee et al. 1996). Nuclear RNase P and RNase MRP have been shown to have similar chromatographic properties but can be well separated by high-resolution anion exchange chromatography (Chamberlain et al. 1996a; Lygerou et al. 1996). It has not been previously clear from these biochemical separations which subunits are integral to the RNase P holoenzyme.
Given the complexity of candidates for RNase P and RNase MRP subunits and limited information regarding the structure and function of the nucleolar processing enzymes, we have undertaken the purification of nuclear RNase P from S. cerevisiae. Partial purification previously identified the RNA subunit of nuclear RNase P, separated RNase P from RNase MRP, and suggested a link between RNase P and pre-rRNA processing (Lee and Engelke 1989; Lee et al. 1991; Chamberlain et al. 1996a). In this report we present the purification of RNase P to apparent homogeneity. We identify nine proteins that copurify with activity, confirm their functional association with RNase P, and determine which newly identified RNase P proteins are associated with RNase MRP activity.
Results
Ten subunits copurify with nuclear RNase P activity
The early steps in the purification of nuclear RNase P followed essentially those described previously (Chamberlain et al. 1996a). Cellular extract was prepared from 100 liters of yeast culture and high-salt extraction was performed on the particulate fractions that included the nuclei. Ion exchange chromatography was initiated by step elution from SP–Sepharose and DEAE–cellulose before attempting high-resolution chromatography by FPLC. Linear salt gradient elution was used with Mono Q that resulted in a peak of RNase P activity between 0.4 and 0.5 m KCl. The activity from Mono Q was subjected to high-resolution Mono S chromatography. RNase P enzymatic activity eluted between 0.2 and 0.3 m KCl in a linear salt gradient. The last step in the purification was velocity sedimentation in 15%–25% glycerol. RNase P migrated as a large complex and was well separated from contaminants that formed the majority of the input material of the glycerol gradients.
Table 1 summarizes the steps for purification of RNase P activity. The specific activity was increased ∼4000-fold over the starting yeast extract by completion of the Mono S chromatography step. Beyond this stage protein concentrations were too dilute for reliable spectrophotometric measure, and protein concentration in the glycerol gradient peak fractions had to be estimated based on quantitation of the RNA subunit. Known quantities of RNase P RNA synthesized in vitro were loaded in parallel with RNA from glycerol gradient peak fractions on a denaturing polyacrylamide gel and were analyzed by Northern blot using an antisense RNase P RNA probe. Although we cannot be sure on the basis of these data that each protein subunit is present in a molar ratio of 1, if we assume a 1:1 molar ratio of RNA to protein subunits, the final total protein content is ∼1 μg. The 100-fold purification resulting from velocity sedimentation is an estimate but is consistent with protein abundance by both silver and by Coomassie brilliant blue staining (see below).
Table 1.
Isolation and enrichment of RNase P
| Fraction
|
Volume (ml)
|
Total units (103)a
|
Total protein (mg)
|
Sp. Act. (U/mg)
|
Enrichment (fold)
|
|---|---|---|---|---|---|
| Yeast extract | 498 | 41.1 | 23,700 | 1.74 | 1 |
| SP—Sepharose | 775 | 53.9 | 852 | 63.3 | 36 |
| DE52 | 94 | 16.4 | 167 | 97.9 | 56 |
| Resource Q | 10.5 | 8.1 | 11.7 | 693 | 400 |
| Resource S | 2.9 | 2.3 | 0.3 | 6760 | 3900 |
| Glycerol gradient peak | 16 | 0.65 | [0.001]b | [650,000]b | [370,000]b |
1 Unit = relative RNase P activity required to convert 10% of 1.0 pmole of pre-tRNAASP in 15 min under standard assay conditions.
Quantity of total protein in glycerol gradient peak estimated on the basis of total RNA (see Materials and Methods).
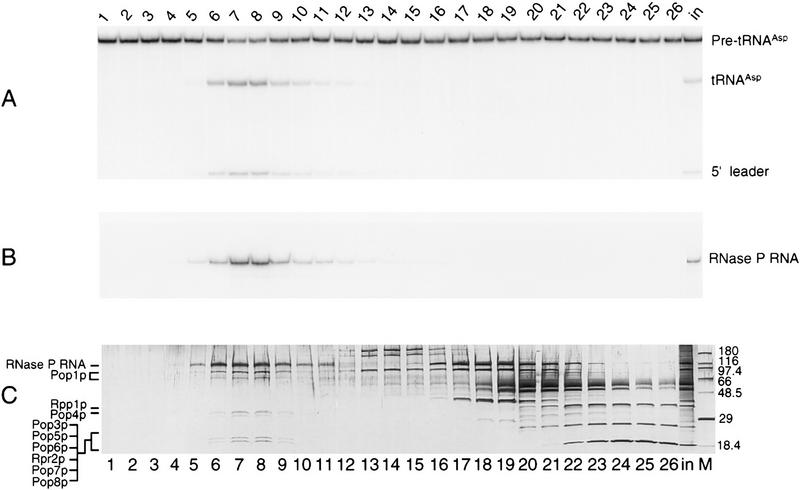
The peak of RNase P activity from the final glycerol gradient fractions (Fig. 1A) coincided exactly with the peak of RNase P RNA (RPR1 RNA) detected by Northern blots (Fig. 1B). A single form of the RPR1 RNA was observed (Fig. 1B), corresponding to the mature 369-nucleotide form generated by 5′ and 3′ processing (Lee and Engelke 1989). The pre-RPR1 form, removed by the earlier Mono Q separation (Chamberlain et al. 1996a), appeared to be assembled into a ribonucleoprotein complex, as judged by its copurification with the mature form through most chromatographic steps. As expected in this final glycerol gradient, we were unable to detect pre-RPR1, the RNase MRP RNA subunit, or the mitochondrial RNase P RNA subunit.
Figure 1.
RNase P activity and RPR1 RNA comigrate with a defined set of proteins in glycerol gradient velocity sedimentation. Contents of individual fractions from the final purification step, glycerol gradient sedimentation, were analyzed to locate RNase P activity and the RNA subunit, and to determine the profile of proteins that coincide with the enzyme. (A) RNase P activity was assayed by cleaving 32P-labeled pre-tRNAAsp to give tRNAAsp and the 5′ leader sequence. RNA products were separated electrophoretically through 12% denaturing polyacrylamide and analyzed quantitatively by PhosphorImager. Fraction numbers are provided at the top; the input to the gradient is marked in. Sedimentation was from right (higher-numbered fractions) to left (lower-numbered fractions). (B) RNA extracted from fractions was separated on 8% denaturing polyacrylamide gels, blotted to nylon membranes, and probed with 32P-labeled RPR1 antisense RNA to detect the RNase P RNA subunit. Amounts of RNA were estimated by quantitative comparison of the signal strength to known quantities of RPR1 sense-strand RNA prepared by in vitro transcription and blotted in parallel (not shown). (C) Protein and nucleic acid contents of fractions were visualized by denaturing PAGE in the presence of SDS and staining with silver. (Lane M) contains protein size markers with molecular mass (in kD) at right. The largest band comigrates with RPR1 RNA and does not stain with Coomassie brilliant blue in preparative gels, suggesting that it contains only the RNA subunit. The identities of proteins in lower regions of the gel were determined by excision of Coomassie-staining bands from a preparative-scale gel and exhaustive peptide analysis by ion trap mass spectrometry as described in Materials and Methods. Two bands migrating at ∼100 and 80 kD contained only Pop1p protein. The region of the two lowest visible bands contained six distinct RNase P subunits. The positions of the confirmed protein subunits relative to the polypeptide pattern are indicated at left; nomenclature is explained in the text and in Table 1.
Analysis of the same glycerol gradient fractions by SDS-PAGE (Fig. 1C) showed one RNA (upper band) and 6 bands of protein coincident with the peak of RNase P activity and with RPR1-containing fractions (lanes 5–10). The intact, large RNA appeared as an orange–brown color predicted for silver staining of nucleic acid. This band did not appear with Coomassie brilliant blue staining of subsequent preparative polyacrylamide gels, and its identity as the RNase P RNA also was confirmed by blotting and hybridization of an antisense RPR1 RNA probe (data not shown). In addition to the RNA, multiple protein bands appeared as 100, 80, 32, 30, 20, and 18 kD relative to protein standards. It appeared likely that all of these polypeptides are associated with a single large complex because of their comigration at this position of high apparent molecular mass in the gradient. No additional, prominent protein bands overlap the peak of the RNase P complex, although many proteins are clearly visible in higher numbered fractions (Fig 1C, lanes 12–26) corresponding to lighter gradient fractions.
Protein sequence determination by ion trap mass spectrometry
Because the protein sample was limiting (4 pmoles), amino acid sequence determination of protein subunits excised from preparative SDS-PAGE gels was accomplished by electrospray ionization ion trap mass spectrometry (see Materials and Methods). Exhaustive analysis of peptide fragments in gel slices identified a total of 9 proteins in the sample that were confirmed to be RNase P subunits by genetic depletion studies. All of the polypeptides were identified within the S. cerevisiae genome database (SGD) (Cherry et al. 1996; Goffeau et al. 1996) using Sequest and FASTA algorithms (Lipman and Pearson 1985; Pearson and Lipman 1988; Eng et al. 1994). Previously unidentified subunits were named according to whether they were shown by depletion and immunoprecipitation (see below) to be specific to RNase P (Rpr2p) or also necessary for function of RNase MRP (Pop5p, Pop6p, Pop7p, and Pop8p). The SDS-PAGE migrations of the RNase P subunits are labeled in Figure 1C, and a summary of all subunits is shown in Table 2.
Table 2.
Composition of the purified RNase P holoenzyme and summary of functional analysis of each protein subunit
| Gene
|
Subunit type
|
Estimated size (kd)
|
Processing defect with subunit depletion
|
Reduction of RNA with subunit depletion
|
Immunoprecipitated RNA
|
Referencea
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pre-tRNA
|
pre-rRNA
|
pre-P
|
P
|
MRP
|
pre-P
|
P
|
MRP
|
||||
| RPR1 | RNA | 120 | — | — | — | — | — | — | — | — | Lee et al. (1991) |
| POP1 | protein | 100.5 | Y | Y | Yb | Y | Y | Y | Y | Y | Lygerou et al. (1994) |
| POP4 | protein | 32.9 | Y | Y | N | Y | Y | Y | Y | Y | Chu et al. (1997) |
| RPP1 | protein | 32.2 | Y | Y | N | Y | Y | Y | Y | Y | Stolc and Altman (1997) |
| POP3 | protein | 22.6 | Y | Y | N | N | N | Y | Y | Y | Dicht and Tollervey (1997) |
| POP5 | protein | 19.6 | Y | Y | N | Y | Y | Y | Y | Y | this report |
| POP6 | protein | 18.2 | Y | Y | N | Y | Y | N | N | N | this report |
| RPR2 | protein | 16.4 | Y | N | N | Y | N | Y | Y | N | this report |
| POP7 | protein | 15.8 | Y | Y | N | Y | Y | Y | N | Y | this report |
| POP8 | protein | 15.5 | Y | N | N | Y | Y | Y | Y | Y | this report |
(Y) Yes; (N) no; (pre) precursor.
Information for Pop1p, Pop3p, Pop4p, and Rpp1p are derived from the references cited. The information reported for the remaining protein subunits is shown in Figs. 3 and 4.
A pop1 temperature-sensitive strain was examined at a nonpermissive temperature for RNA reduction. Relative to the mature RNase P RNA the precursor RNase P RNA levels declined less rapidly, and it is unclear whether this depletion is a result of the general decline of functional RNA levels under this condition.
The largest protein, estimated by SDS-PAGE to be 100 kD, was Pop1p. This protein previously was shown to be associated with both RNase P and RNase MRP from a functional genetic screen (Lygerou et al. 1994). The 80-kD protein band also contained only Pop1p, and the recovered tryptic peptides were consistent with this protein missing the carboxy-terminal 20 kD found in full-length Pop1p. It is not possible from current samples to determine whether this 80-kD form is an artifact produced by proteolysis during purification or an alternative product of the POP1 gene incorporated as a subunit. This question will be addressed once antibodies to Pop1p can be obtained and used to probe the subunit prior to purification. The protein with an apparent migration at a molecular mass corresponding to 32 kD was identified as the RNase P protein Rpp1p and has a predicted mass of 32.2 kD (Stolc and Altman 1997). This protein has been characterized as an RNase P and RNase MRP subunit from its similarity with a human protein that comigrates with RNase P activity, Rpp30 (Eder et al. 1997), and from functional studies in yeast that reveal pre-tRNA and pre-rRNA processing defects in cells depleted of Rpp1p. The protein migrating slightly more slowly than Rpp1p was yet another protein found to be functionally and physically associated with both RNase P and RNase MRP, Pop4p (Chu et al. 1997).
The two bands smaller than 20 kD actually contained six proteins (Fig. 1C; Table 2). Pop3p was suggested previously to be a subunit of both RNase P and RNase MRP (Dichtl and Tollervey 1997), but the remaining five proteins were predicted products of hypothetical open reading frames (ORFs) of unknown functions. Only the protein of an estimated 19.6 kD, a product of the FUN53 gene on chromosome I, had been shown to be essential (Harris et al. 1992). All but one of these five polypeptides were found in further analyses (Table 1; Figs. 3 and 4, see below) to affect both RNase P and RNase MRP activities and given Pop designations (Pop5p–Pop8p). A single protein was specific for RNase P and the gene is named RPR2 (RNase Pribonucleoprotein-2) according to our original nomenclature for yeast nuclear RNase P subunits.
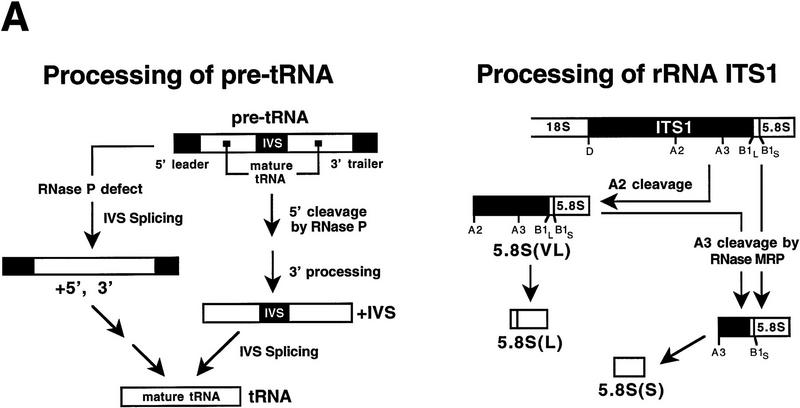
Figure 3.
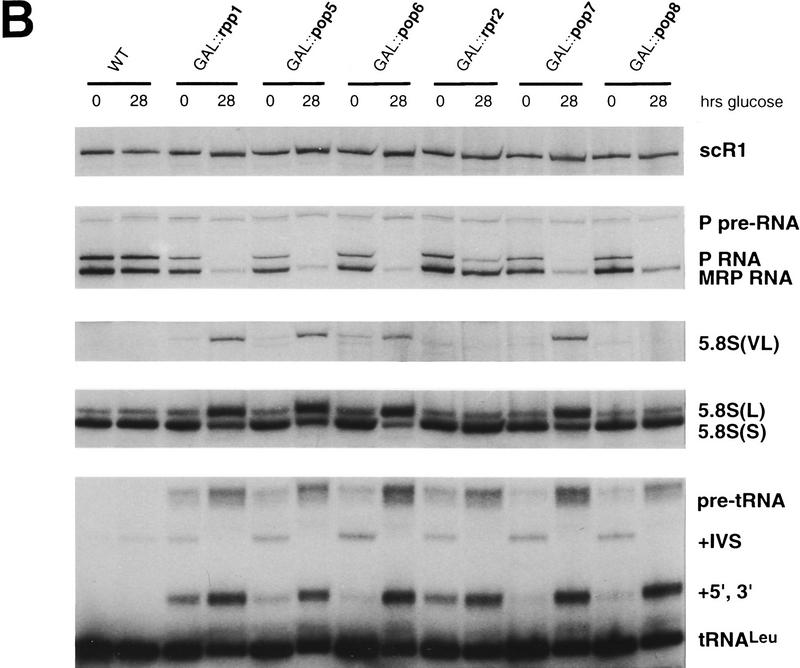
Depletion of the newly identified protein subunits shows that all are essential for RNase P activity, and several are essential for RNase MRP activity. The constructs described in Fig. 2 were used to deplete Pop5p, Pop6p, Rpr2p, Pop7p, and Pop8p by shifting to growth in glucose, which impairs growth by 6 hr and reaches maximal effect on RNA processing in these strains by 12 hr. Growth of the parent haploid strain and depletion of the previously identified Rpp1p subunit were used as controls. Steady-state levels of several different RNA types were determined at 0 and 28 hr after the shift to glucose. Strain names (GAL::gene name) indicate the subunit that is under Gal control in the strain. (A) The expected products for processing of pre-tRNALeu3 are shown for a strain with normal or defective nuclear RNase P. Nucleolytic processing normally proceeds at the 5′ end, at the 3′ end, and at intron removal. RNase P defects lead to an increase in intron removal prior to terminal processing (+5′, +3′ intermediate). The expected products for processing of 5.8S rRNA are shown for a strain with normal participation by RNase MRP or a deficiency in RNase MRP. Mutations cause accumulation of very long (VL) and long (L) forms of 5.8S at the expense of the short (S) form. (B) Whole cell RNA samples were prepared at 0 or 28 hr after shifting to glucose to deplete the indicated subunits. RNAs were separated by electrophoresis though denaturing polyacrylamide gels, electroblotted to nylon membranes, and probed to detect the indicated RNAs. Panels are taken from a single blot exposed to the following 32P-labeled probes (from the top): scR1 detects signal recognition particle RNA that should not be affected by either RNase P or RNase MRP defects; RNase P RNA detects both mature and precursor forms; RNase MRP RNA that detects a single form of the RNA; 5.8S rRNA detects the S, L, and VL forms; and tRNALeu3 detects the mature and all precursr forms. Depletion of all putative subunits strongly affects pre-tRNA processing. Effects on levels of RNase P RNA, RNase MRP RNA, and 5.8S rRNA-containing intermediates are variable with different subunit depletions and are discussed in the text.
Figure 4.
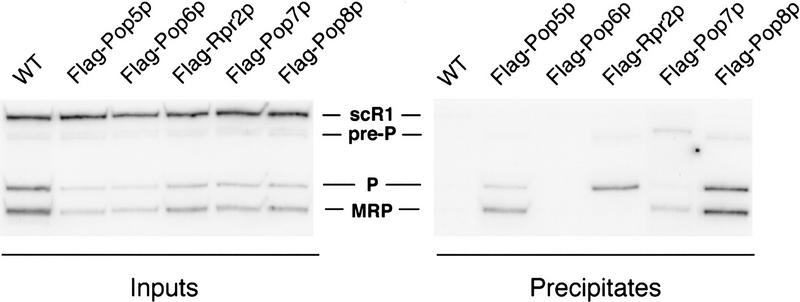
Immunoprecipitations of protein subunits selectively coprecipitate RNase P and RNase MRP RNA subunits. As shown schematically in Fig. 2, Rpp1p, Pop5p, Pop6p, Rpr2p, Pop7p, and Pop8p were each expressed from plasmid gene copies in a strain where the chromosomal copy of that gene had been deleted. Recombinant proteins contained Flag epitope at the carboxyl termini. Soluble extracts were made from actively growing cultures of each strain and the parental wild type (WT) strain. Immunoprecipitation with highly specific monoclonal antibodies against the Flag epitope was followed by Northern blot analysis for the abundance of precursor and mature RNase P RNA (pre-P RNA and P RNA) and RNase MRP RNA (MRP RNA). Signal recognition particle RNA (scR1) was used as a control for an RNA that was not expected to precipitate with any of the Flag-tagged proteins. (Left) The RNA content of the input fractions; (right) RNAs found in the precipitates. RNase P and RNase MRP RNAs were not precipitated in the wild-type (WT) strain or with the Flag tag on Pop6p. All three RNAs (pre-P, P, and MRP) were precipitated with the Flag tag on Pop5p or Pop8p. Rpr2p and Pop7p gave differential coprecipitation of the RNAs. Rpr2p coprecipitated only RNase P RNAs (pre-P and P). Pop7p coprecipitated with MRP and pre-P preferentially, bringing down relatively low levels of mature RNase P RNA.
The RNase P subunit gene products are essential for spore viability
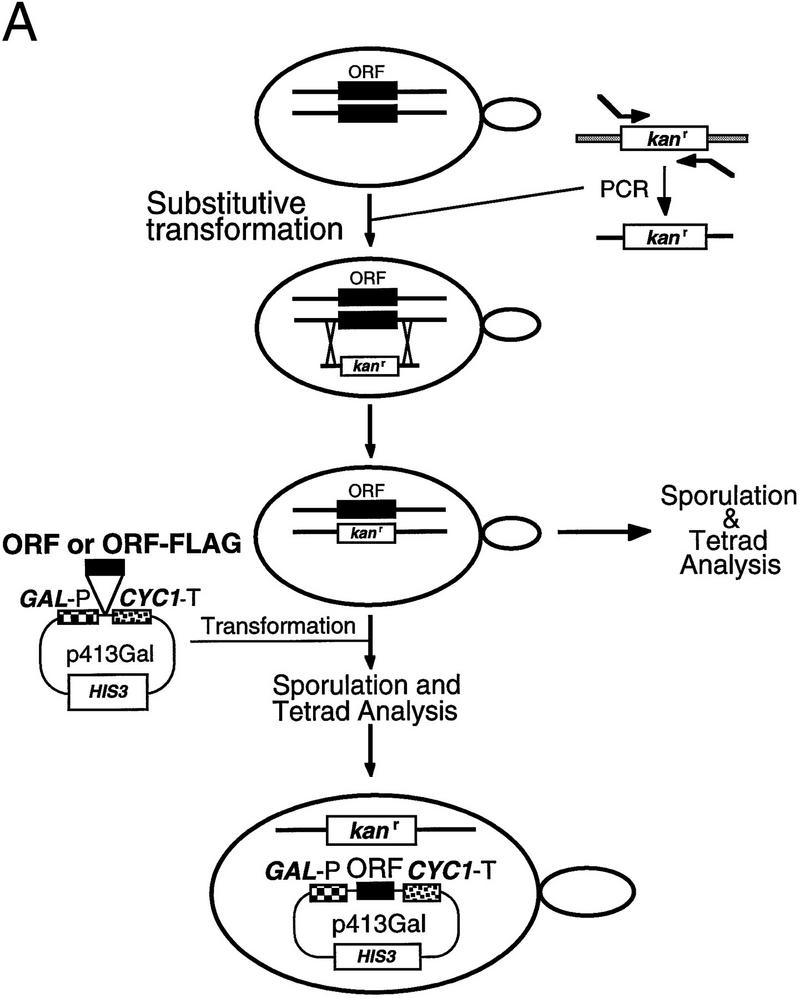
To determine whether the five previously uncharacterized polypeptides were essential cellular components, the genes encoding the candidate RNase P proteins were disrupted with a kanr gene in diploid S. cerevisiae (Fig. 2A). The kanr gene was generated as a PCR fragment with 50 bp of flanking homologous sequences to the gene candidate for disruption. Transformation of diploids and subsequent homologous recombination resulted in a diploid strain with only one functional copy of the gene of interest. Sporulation and tetrad analysis of resultant haploid spores revealed a 2:2 segregation of spore viability with kanamycin resistance in all cases, indicating that all protein products were essential (Fig. 2B).
Figure 2.
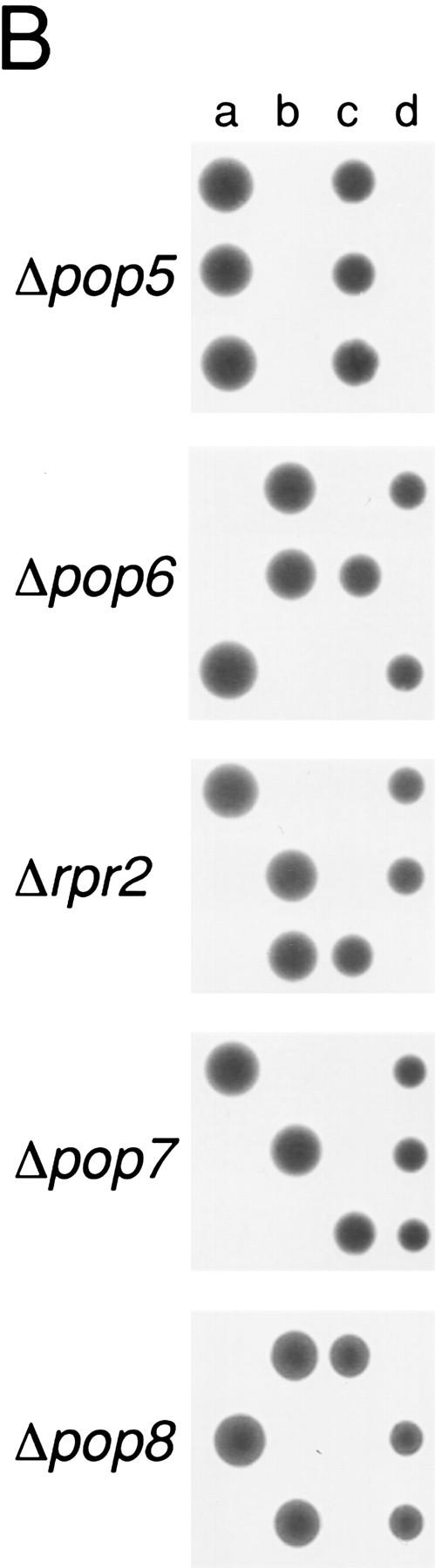
All of the newly identified protein subunit candidates are encoded by essential genes. Four of the proteins that copurify with RNase P activity (Pop1p, Rpp1p, Pop4p, and Pop3p) had been identified previously as candidate RNase P subunits, and their genes had been shown to be essential. The genes for the remaining five subunits (Pop5p, Pop6p, Rpr2p, Pop7p, and Pop8p) were tested to determine whether they were essential for viability and if depletion of their gene products affected RNase P or RNase MRP function. (A) The experimental design for deleting the coding regions and substituting a gene under the regulation of a GAL1 promoter is shown schematically. Homologous recombination was used to replace one copy of each coding region in diploids with a gene for kanamycin resistance. The resulting heterodiploids were first sporulated and tested for spore viability. B shows that all tetrads segregated as two viable and two nonviable spores, the result expected for essential genes. The diagram also shows a schematic for creation of a haploid strain in which the genomic copy of the gene of interest is disrupted, and production of the gene product from a plasmid is placed under Gal control for the depletion studies shown in Fig. 3. These recombinant gene products were also all made in two versions, both of which conferred viability. One version, used for the RNA blot analyses in Fig. 3, contains only the coding region for the gene. A second version, used in the immunoprecipitation experiments in Fig. 4, also contains the 8-amino-acid Flag epitope at the carboxyl terminus.
Depletion of RNase P candidate proteins indicates they are necessary for RNase P activity in vivo
To test for function of the candidate RNase P subunits as components of RNase P and RNase MRP activities in the cell, their expression was placed under the control of the GAL1 promoter (Fig. 2A). The ORFs to be expressed were inserted in a plasmid with the GAL1 promoter upstream and the CYC1 terminator placed downstream. Transformation of the disrupted diploid, followed by sporulation and tetrad analysis of the resultant spores, yielded a haploid yeast strain containing the sole copy of the gene of interest under GAL promoter control of expression. Shift of cultures to glucose medium depleted the protein gene products, with maximum effect on growth and RNA levels reached by 12 hr.
RNA biogenesis and processing defects resulting from depletion of these proteins were assessed by Northern blot analysis of RNA recovered from each strain at 0 and 28 hr after the shift to glucose-containing media (Fig. 3). The signal recognition particle RNA (scR1) was probed as a normalization control. The nucleolytic processing intermediates expected for the pre-tRNALeu and 5.8S pre-rRNA region in either wild-type or RNase P-deficient cells are represented in the schematic shown in Figure 3A. The wild-type strain does not accumulate significant levels of pre-tRNALeu either before or after the shift to glucose. In contrast, the control placing the previously characterized Rpp1p subunit in the Gal-driven expression vectors does show a low-level pre-tRNA accumulation even when grown in galactose, with a significant increase in pre-tRNAs after depletion of Rpp1p in glucose. This result was typical of pre-tRNA accumulation for all of the newly identified subunits as well. Despite minor accumulation of tRNA precursors in strains where subunits were expressed from the GAL promoter at 0 time points, depletion of the candidate RNase P subunits for glucose resulted in increased accumulation of pre-tRNALeu typical of an RNase P mutant (Lee et al. 1991). A pre-tRNALeu probe showed accumulation of the primary transcript (“pre-tRNA”) and spliced but not end-processed pre-tRNALeu (+5′, +3′). As noted previously, end processing normally precedes splicing in the pre-tRNA maturation pathway, but accumulation of the +5′, +3′ intervening sequence (IVS) intermediate in RNase P mutants suggests that the processing order is not obligatory.
RNase P protein subunit depletions differentially affect the endogenous levels of RNase P and RNase MRP RNAs
The Northern blot described above was also probed for RNase P and RNase MRP RNA subunits (Fig. 3B). As observed previously upon depletion of Pop1p, Pop4p, and Rpp1p proteins (Lygerou et al. 1994; Chu et al. 1997; Stolc and Altman 1997), a decrease in the levels of both mature RNase P and RNase MRP RNAs was observed for four of the five newly identified RNase P proteins: Pop5p, Pop6p, Pop7p, and Pop8p. Depletion of a single subunit, Rpr2p, resulted in almost complete loss of the RNase P RNA, but a relatively small reduction of RNase MRP RNA (twofold) with no detectable effect on RNase MRP function. Depletion of Pop8p protein also led to an RNase MRP RNA reduction, but only 4-fold, compared with a 7- to 13-fold loss of RNase MRP RNA observed with the other Pop protein and Rpp1p depletions. Persistence of the RNase P RNA precursor band in all the depletions could reflect continuing synthesis of the precursor but failure of the holoenzyme to assemble and mature properly. The chromatographic behavior of the ribonucleoprotein containing the RPR1 RNA precursor previously caused us to speculate that it was assembled into an RNP similar to the mature enzyme before terminal processing. Lack of accumulation of mature RNAs is consistent with the inability of the precursor to assemble into an RNP complex in the depleted strains.
Defects in 5.8S rRNA precursor accumulation are detectable upon depletion of Pop5p, Pop6p, and Pop7p
The characterized precursors of the 5.8S rRNA mature forms containing internal transcribed spacer (ITS1) sequences are depicted in Figure 3A. In wild-type strains a ratio of ∼10:1 exists between the 5.8S(S) (short) and 5.8S(L) (long) mature rRNAs. RNase MRP mutations typically result in a shift in the ratios of the short and long forms to approximately 1:1. In addition, a 5′-extended form of the 5.8S rRNA with additional ITS1 sequences, 5.8S(VL) (very long), accumulates in RNase MRP mutants (Lygerou et al. 1994; Chu et al. 1997; Dichtl and Tollervey 1997). Depletion of three of the five uncharacterized candidate RNase P subunits, Pop5p, Pop6p, and Pop7p, in addition to the previously characterized Rpp1p, shows a clear shift in the ratio of the 5.8S(S) to 5.8S(L) rRNAs and accumulation of the very long form of the 5.8S rRNA intermediate, 5.8S(VL), relative to the wild-type strain (Fig. 3B). Neither GAL::rpr2 nor GAL::pop8 strains show a 5.8(S):5.8(L) shift, suggesting the depleted proteins might be RNase P-specific subunits. However, further immunoprecipitation experiments showed that Pop8p, but not Rpr2p, was physically associated with both RNase P and RNase MRP RNA subunits. From these data it appears that the moderate lowering of the RNase MRP RNA level seen with Pop8p depletion is not sufficient to cause RNase MRP cleavage to become rate-limiting and lead to the accumulation of alternately processed 5.8S forms.
Epitope-tagged Rpr2p immunoprecipitated in association with only RNase P RNA
To characterize further the association of the large number of protein subunits shared between RNase P and RNase MRP, each of the carboxyl termini of the newly identified subunits was tagged with a Flag epitope. Soluble cell extracts were prepared from each of the haploid yeast strains harboring an epitope-tagged RNase P protein as the sole source of that subunit. The immunoprecipitates obtained from the extracts using anti-Flag antibody were analyzed by Northern blot using probes to RNase P and RNase MRP RNAs (Fig. 4). Both Pop5p and Pop8p were precipitated in association with RNase P and RNase MRP RNAs, indicating that these proteins are subunits of both complexes. Even though Pop8p depletion had little observable effect on RNase MRP function, it has been tentatively designated a Pop protein on the basis of physical association with both RNase P and RNase MRP RNAs, in addition to the partial RNase MRP RNA reduction seen with Pop8p depletion. Pop7p was found in a complex with the precursor to RNase P RNA (pre-P RNA) and RNase MRP RNA, but little or no mature RNase P RNA coprecipitated. Although this might indicate an association with only the pre-RNase P RNA, the purification of Pop7p with mature RNase P holoenzyme supports the likelihood that the Flag epitope is not exposed in the mature RNase P complex. Rpr2p immunoprecipitated with only RNase P RNAs, mature and precursor forms, consistent with the observation that its depletion affects RNase P activity in pre-tRNA processing, but not RNase MRP activity in pre-rRNA processing. Of the newly identified yeast subunits, only the Pop6p Flag tag could not be used to precipitate either RNase P or RNase MRP RNAs. These data, and the purification of these subunits with RNase P alone, are not consistent with a higher-order complex between RNase P and RNase MRP in cell extracts. In particular, immunoprecipitation results with tagged Rpr2p and Pop7p suggest that the two enzymes can be immunoprecipitated independently.
Amino acid sequence of Pop5p–Pop8p and Rpr2p does not show significant similarity to any known proteins or RNA-binding motifs
All of the newly identified RNase P subunits were analyzed for homology by pairwise alignment to the eubacterial RNase P proteins, their signature sequence (Bairoch et al. 1995), and the previously identified nuclear and mitochondrial RNase P protein subunits. In addition, pairwise alignment was attempted between a variety of known tRNA processing enzymes and nucleolar antigens, including the genetically identified Snm1p subunit of RNase MRP. None of the BLAST searches (Altschul et al. 1990) of the SGD, the SWISS–PROT database (Rodriguez-Tome et al. 1996), and PIR databases (Sidman et al. 1988) yielded significant matches, and only one pairwise alignment between Pop7p and nucleolar antigen Rrp7p (Baudin-Baillieu et al. 1997) showed regions of significant similarity. Even in this case, however, no motif in the alignment could be identified (not shown).
A search for known motifs and sequence patterns has yielded limited information. No known RNA-binding motifs could be identified (Burd and Dreyfuss 1994), but most subunits possess regions rich in basic amino acids. The lysines and arginines in the newly identified RNase P subunit sequences are underlined in the sequences shown in Figure 5. Of the RNase P proteins only Pop8p is acidic overall with a pI of 4.8; the remaining basic subunits have an average pI of 9.5. Every RNase P protein subunit, except Rpp1p, contains a pattern of two contiguous lysine residues preceded or followed by an additional lysine at a distance of 3–8 intervening residues. The lysines that conform to this pattern are indicated by bold lettering in Figure 5. Pop6p, Rpr2p, and Pop7p contain three of these regions. This lysine pattern can be found in a number of ribosomal proteins with some in highly conserved regions of S8, S7, S21, and L25 (Burton et al. 1983; Rutgers et al. 1991; Salazar et al. 1993; Engemann et al. 1995). These residues might be involved in protein–RNA association, as proposed for positively charged residues conserved in the Escherichia coli C5 protein subunit (Gopalan et al. 1997). Alternatively, the charged regions could be sites of protein–protein interactions or nuclear localization signals (Garcia-Bustos et al. 1991; Dingwall and Laskey 1992). The proposed lysine motif can also be found in Pop1p at amino acids 616–623 (Lygerou et al. 1994), Pop3p at positions 10–20 and 183–192 (Dichtl and Tollervey 1997), and Pop4p as two overlapping occurrences at positions 97–111 (Chu et al. 1997).
Figure 5.

Sequences of the protein subunits have little similarity to bacterial or mitochondrial RNase P proteins. The amino acid sequences of the five newly identified subunits are shown, as derived from the SGD (http://genome-www.stanford.edu/Saccharomyces/). Of the five ORFs, only POP8 contains a predicted intron, composed of 75 bp between the first and second base pair of the sixteenth codon of the ORF. PCR analyses using primers flanking the intron with genomic DNA and cDNA library confirm that the POP8 cDNA gives a smaller PCR product by the expected amount. As discussed in the text, these sequences do not display strong similarities to other gene sequences in current databases and do not display notable similarities when aligned pairwise with each other or with other identified RNase P or RNase MRP proteins. Sequence features that are discussed in the text are indicated by underlining (lysines and arginines) or boldface type (lysine residues that form a short pattern in all RNase P subunits except Rpp1p).
Discussion
We have isolated eukaryotic nuclear RNase P to resolve the question of what components are associated with and necessary for function of the isolated RNase P enzyme. The candidate polypeptides from previous work included Pop1p, Pop3p, Pop4p, and Rpp1p (Chu et al. 1997; Dichtl and Tollervey 1997; Stolc and Altman 1997). All four of these proteins were found in the purified fractions. Pop1p, Pop3p, and Pop4p had been identified first in genetic screens, and Rpp1p had been identified by similarity to a human RNase P-associated protein, Rpp30. It has been reported that Pop1p did not appear to be a member of the RNase P complex from a purification of the human enzyme (Eder et al. 1997; Stolc and Altman 1997). The data presented in this report demonstrate that Pop1p is part of the yeast nuclear RNase P holoenzyme.
In addition to Pop1p, Pop3p, Pop4p, and Rpp1p, five new protein subunits were identified with peptide sequences obtained by ion trap spectrometric analysis. The protein sequences of the corresponding ORFs were used to search databases but revealed no significant homology to any known proteins from other organisms. In an attempt to reveal potential functional domains, pairwise alignments were performed with proteins associated with other RNase P enzymes and RNase MRP, including Snm1p (yeast RNase MRP-specific protein), Rpm2p (yeast mitochondrial protein), and the C5 protein (E. coli RNase P protein subunit), as well as the sequence consensus for the bacterial RNase P proteins (Gopalan et al. 1997). None of these pairwise alignments revealed regions of obvious similarity to the RNase P protein subunits.
Because one or more of the subunits must interact with the RNA subunit and could interact with the RNA substrate, a concerted attempt was made to identify RNA binding domains in the RNase P proteins. None of the reported RNA-binding domains were present. However, most of the proteins are very basic, and it was noted that eight of the nine RNase P protein components contain a pattern of a lysine doublet with a third lysine spaced 3–8 residues away. This region has charged residues in addition to lysine in a number of these proteins. A pattern search (Chervitz et al. 1997) revealed the presence of clustered lysine residues in conserved regions of several ribosomal proteins. The eubacterial RNase P protein does not contain the lysine doublet pattern but does contain a conserved sequence between the single proteins of the different enzymes. The eubacterial consensus sequence is composed of positively charged residues thought to be involved in protein–RNA contact (Gopalan et al. 1997). Only one nucleolar yeast protein, Rrp7p (Baudin-Baillieu et al. 1997) was found to possess regions of notable homology to one of the eukaryotic subunits, Pop7p. Rrp7p is involved in production of the 18S rRNA and is thought to be required for correct assembly of the ribosomal protein S27 into the preribosomal particle. In a pairwise alignment amino acids 10–34 of Pop7p showed 33% identity with positions 10–36 in Rrp7p, with a gap of 2 amino acids at positions 17–18 in the Pop7p sequence. Another region, amino acids 106–121 of Pop7p, was 44% identical to the Rrp7p sequence spanning amino acids 195–210. These regions of identity do not contain the lysine doublet motif or other notable functional motifs.
The protein products of the newly identified RNase P subunit ORFs, Pop5p–Pop8p and Rpr2p, were shown to be essential for cell survival (Fig. 2). Depletion of these proteins from the cell resulted in the same pre-tRNA processing defects seen with mutation or depletion of previously proposed components, Pop1p, Pop3p, Pop4p, and Rpp1p (Fig. 3). Depletion of Pop5p–Pop7p of the newly identified subunits is also concomitant with the defect in pre-rRNA processing at site A3 by RNase MRP. This observation is consistent with cleavage defects observed at A3 with the depletion of Pop1p, Pop3p, Pop4p, and Rpp1p (Lygerou et al. 1994; Chu et al. 1997; Dichtl and Tollervey 1997; Stolc and Altman 1997). Depletion of Pop8p did not give the characteristic ratio change of 5.8S(L) to 5.8S(S) as a result of a defect in processing at A3 but did cause a four-fold reduction of the RNase MRP RNA level upon extended depletion of Pop8p. The reduction in endogenous RNA levels from protein subunit depletion might not have reached a threshold level required for the 5.8S precursor processing defect or Pop8p might not be necessary for the 5.8S precursor processing function of RNase MRP. Because RNase P and RNase MRP RNAs also coimmunoprecipitate with epitope-tagged Pop8p, we have tentatively assigned Pop8p as a subunit of both enzymes.
Only one protein, Rpr2p, gave results entirely consistent with an RNase P subunit but not an RNase MRP subunit. Depletion of Rpr2p did not give defects in RNase MRP RNA levels or pre-rRNA processing but did deplete mature RNase P RNA, and caused expected pre-tRNA accumulations. Also, Rpr2p was found in association with precursor and mature RNase P RNA, but not RNase MRP RNA, by immunoprecipitation of epitope-tagged Rpr2p. Therefore, Rpr2p is the only protein specific to RNase P. Similarly, there has been a report of one yeast protein specific to RNase MRP, Snm1p (Schmitt and Clayton 1994), suggesting that the two enzymes might differ only in their divergent RNA subunit and one protein subunit with eight proteins held in common. Physical isolation of RNase MRP will be necessary to determine its composition.
A profile of the purified nuclear RNase P holoenzyme and subsequent characterization of the protein subunits resolves the question of what proteins are biologically relevant to the function of this enzyme. However, there is remaining uncertainty as to whether RNases P and MRP are separate enzymes with many identical protein subunits or whether the enzymes share some subunits in vivo as part of a larger complex. From the data presented in this report it does not appear that subunits are shared on the basis of a common complex that is required for RNase P activity against pre-rRNA. Similarly, it appears from the work of others (Lygerou et al. 1996) that active RNase MRP can be separated from the majority of RNase P. Immunoprecipitation results with Flag-tagged Rpr2p and Pop7p suggest that the two enzymes are distinct complexes and can be precipitated independently.
The precise cellular location of nuclear RNase P is not clear at present. RNase MRP is a nucleolar enzyme (Jacobson et al. 1995; Matera et al. 1995), but there are reports that most of the mammalian RNase P is either nucleoplasmic (Jacobson et al. 1997) or at the periphery of nucleoli (Matera et al. 1995; B. Lee et al. 1996). Several observations would be consistent with nucleolar RNase P localization. It was shown previously that a yeast RNase P RNA subunit mutation causes a pre-rRNA processing defect (Chamberlain et al. 1996a). Although this could be an indirect effect, the existence of eight proteins common to both RNase P and RNase MRP suggests that at least some of the two enzymes might colocalize by virtue of signals in the proteins. Nucleolar localization of RNase P would suggest that some aspects of pre-tRNA processing occur at the nucleolus.
Although the eukaryotic RNase P activity requires a greater contribution from protein than is provided by the single, small eubacterial protein subunit, it seems unlikely that nine protein subunits totaling 275 kD compared with the 14-kD subunit in the eubacterial enzyme are all necessary for this activity. The complex subunit composition suggests that some of the protein subunits are important for functions other than pre-tRNA cleavage and might be necessary to promote structural integrity or direct substrate interaction in compensation for loss of catalytic competence in the RNA subunit. In addition, the protein subunits could be responsible for broadening the RNA substrate specificity. The bacterial RNase P cleaves a number of substrates including other functional RNAs (Bothwell et al. 1976; Brown et al. 1990; Komine et al. 1994), a pre-mRNA (Alifano et al. 1994), and certain viral RNAs (Green et al. 1988; Kikuchi et al. 1990; Mans et al. 1990). This substrate range appears to be dependent on its protein subunit (Liu and Altman 1994). Although yeast nuclear RNase P cleaves at several positions in the ITS regions of pre-rRNA in vitro, it is not clear that the minor effects RNase P mutations can have on 5.8S rRNA maturation are directly related to those cleavage sites (Chamberlain et al. 1996a). We have not yet been able to demonstrate that RNase P mutations specifically affect maturation or turnover of any additional small RNA or pre-mRNA substrates.
The additional protein subunits in the nuclear RNase P might also have other functions including signaling, non-nucleolytic enzymatic activities, and subcellular localization. The identification of the protein subunits and the corresponding genes will facilitate a detailed investigation of the functions of the eukaryotic RNase P holoenzyme.
Materials and methods
RNase P purification
The protease-deficient strain PP1002 (MATα ade2 leu2-3,112 pep4-3 rna3 rna82) was used as a source of RNase P (Lee and Engelke 1989). Extracts were prepared from 100 liters of saturated yeast culture in batches of 6 or 12 liters as described previously (Evans and Engelke 1990). The ammonium sulfate precipitate from the last step of the extraction protocol was dissolved in 25 ml of HGMND (20 mm HEPES at pH 7.9, 10 mm MgCl2, 10% glycerol, 0.5% NP-40, 1 mm DTT), diluted to a conductivity equivalent to 0.15 m KCl, and applied to 8 ml of SP–Sepharose (Pharmacia) per original liter of cell culture. The resin was washed with HGMND containing 0.15 m KCl and eluted in a single step with HGMND containing 0.4 m KCl. The peak protein fractions were pooled, adjusted to 0.1 m KCl, and applied to a 300 ml DEAE cellulose (Whatman DE52) column and eluted with 0.45 m KCl–HGMND in a single step. RNase P activity was assayed as described below starting at this stage.
Fractions containing RNase P activity were pooled, diluted to 0.28 m KCl and half was applied to each of two 6 ml Mono Q columns (Pharmacia). Following extensive washing with 0.28 m KCl–HGMND, an 80-ml linear gradient of 0.28–0.85 m KCl was used for elution. Fractions of 1 ml were collected and analyzed for activity, protein, and RNA content. The RNase P activity eluted between 0.38 and 0.5 m KCl. Fractions containing the peak of RNase P activity from the two Mono Q columns were applied to a 1-ml Mono S column in HGMND containing 0.12 m KCl. After washing with 0.12 m KCl–HGMND, a 25-ml linear gradient of 0.12–0.9 m KCl resulted in elution of RNase P activity between 0.2 and 0.3 m KCl. Fractions of 0.5 ml were collected and analyzed. The Mono S fractions with RNase P activity were applied to four 34-ml glycerol gradients of 15%–25% in HGMND (DTT at 0.1 mm). The first half and second half of the Mono S activity peak fractions were layered on top of different glycerol gradients because protein gels showed that the two halves of the Mono S peak had different contaminants. The same protein bands were observed in the glycerol gradient peak activity fractions in both cases. Gradients were spun at 96,500g (27,000 rpm) for 30 hr at 4°C in a SW28 rotor (Beckman). Fractions of 1 ml were collected by puncture of the Ultraclear ultracentrifuge tubes near the bottom with a 12-gauge hypodermic needle.
RNase P activity assay
An internally 32P-labeled pre-tRNAAsp substrate (Hollingsworth and Martin 1987) was prepared, and RNase P activity assays were conducted as described (Chamberlain et al. 1996a). Specific activity determinations were obtained by electrophoretic separation of the substrate and products on 12% denaturing polyacrylamide gels (Sequegel) and analysis of products by PhosphorImager (Molecular Dynamics). One unit of activity was defined as the amount of RNase P activity necessary to cleave 10% of 1 pmole of the substrate under standard assay conditions.
Protein quantitation and analysis
Quantitation of yeast extract and column fractions was performed using the Micro BCA protein quantitation kit with a range of sensitivity of 0.5–20 μg (Pierce). Estimates for final total protein from the glycerol gradient were made based on silver stain showing approximately equimolar levels of putative protein subunits and quantitation of the RNase P RNA subunit by Northern blot (see RNA analysis). All fractions from two glycerol gradients were analyzed by SDS–PAGE to assess the level of purification and to approximate the size of candidate subunits. The gel shown in Figure 2C was a Ready Gel (Bio-Rad) with gradient acrylamide concentration of 10%–20%. A sample size of 20 μl or 2% of each fraction was applied to each well of multiple gels. No protein was detected in the first five fractions from the bottom of the gradient and their analyses are not shown in Figure 2. The proteins and RNA were visualized using the Silver Stain Plus kit (Bio-Rad). Silver-stained electrophoretic protein standards (6.5–180 kD) were run in parallel (Sigma).
RNA analysis
For detection of RNase P RNA in the glycerol gradient fractions an aliquot of each fraction was prepared and analyzed by Northern blot as described (Chamberlain et al. 1996a). Hybridization probes included 32P-labeled antisense RPR1 RNA (RNase P RNA) and NME1 RNA (RNase MRP RNA), and an oligodeoxynucleotide probe complementary to S. cerevisiae mitochondrial RNase P RNA. The absolute concentration of RNase P RNA in various fractions was determined by including, on the same blot, a titration of known concentrations of pure RPR1 RNA produced by T7 RNA polymerase in vitro using cRPR1–pGB plasmid. This plasmid construct contained an EcoRI–SmaI fragment of the RPR1 gene cloned into pGEM-3Z (Promega) (Lee and Engelke 1989). Oligodeoxynucleotide probes used for detection of 5.8 rRNA and scR1 RNA (Felici et al. 1989) were 5′-32P-CGCATTTCGCTGCGTTCTTCATCG-3′ and 5′-32P-GGCGTGCAATCCGTGTCT-3′, respectively.
Protein sequence and analysis
In preparation for SDS-PAGE separation, 10 ml of fractions 6–8 of the glycerol gradients were precipitated with a defined methanol/chloroform/water mixture (Wessel and Flügge 1984) and resuspended in 30 μl of 2% SDS. Following separation by SDS-PAGE (12%), proteins were stained with Coomassie blue and excised as individual band(s). After in gel reduction and S-carboxyamidomethylation, the band(s) were subjected to in gel tryptic digestion (Promega) and a single 10% or 20% aliquot from each was analyzed as follows. Sequence information was determined by microcapillary (75 μm × 10-cm column, packed in-house) reverse-phase chromatography coupled to the electrospray ionization (ESI) source of a quadrupole ion trap mass spectrometer (Finnigan LCQ, San Jose, CA) as in Nash et al. (1996). The instrument was programmed to acquire successive sets of three scan modes consisting of full scan mass spectrometry (MS) over the mass-to-charge ratio range 395–1200 atomic mass units; followed by two data-dependent scans on the most abundant ion in that full scan. These data-dependent scans allowed the automatic acquisition of a high-resolution (zoom) scan to determine charge state and exact mass and collisionally induced dissociation (CID) spectra for peptide sequence information. Interpretation of the resulting MS/MS spectra of the peptides was facilitated by searching the yeast protein database with the algorithm SEQUEST (Eng et al. 1994). Base peak zoom scan ion intensities corresponded to a load of 25–200 fmoles by comparison with the average ion abundance of a standard peptide mixture analyzed under identical conditions. Pairwise sequence alignments of proteins identified in the SGD (Cherry et al. 1996) were performed using the computer program Lasergene.
Gene disruption and epitope tagging of proteins
PCR-based gene disruptions were carried out as described previously in Wach et al. (1994). Gene sequence for candidate proteins from the SGD were disrupted by replacing their entire ORFs with the kanMx2 module. Previous identifications of the ORFs for the newly identified subunits in the SGD were YAL033W (Pop5p), YGR030C (Pop6p), YIR015W (Rpr2p), YBR167C (Pop7p), and YBL018C (Pop8p). The kanr gene was inserted at the indicated loci in diploid W303 (Lee et al. 1991) by homologous recombination using DNA fragment termini identical to the 50 bp immediately upstream and downstream of the ORF. Correct disruption of alleles was verified by yeast colony PCR using oligonucleotides within the kanr gene to oligonucleotides outside the deleted ORFs. Tetrads were dissected from sporulated heterozygous diploids to test viability.
The PCR fragments for indicated gene ORFs were subcloned into the XmaI and SalI site of yeast expression vector, p413GAL (gift of D. Thiele, University of Michigan, Ann Arbor) containing the GAL1 promoter and CYC1 terminator. RNA was isolated from these constructed strains (Rose et al. 1990). Two versions of each ORF were expressed, either precise reproduction of the endogenous sequence or with the Flag epitope tag (N-DYKDDDDK-C) added precisely to the carboxyl terminus. Both versions conferred viability for all ORFs tested. p413GAL was constructed from p413GPD (Mumberg et al. 1995) by replacing the SacI–BamHI GPD promoter fragment with EcoRI (end-filled)–BamHI GAL1 promoter fragment. The expression constructs were transformed into appropriate heterozygous disrupted diploids. Haploid spores carrying the disrupted genomic allele and the corresponding functional plasmid copy with the GAL1 promoter were identified by selecting spores resistant to G418 and unable to grow on selective media containing glucose. Whole-cell yeast protein extraction and immunoprecipitation of the Flag-tagged proteins were performed as described in Chu et al. (1997) and Schmitt and Clayton (1994), with minor modifications. A 10-ml culture was grown in SD–his to OD600 nm = 0.7 and washed with water and buffer A (50 mm Tris-HCl at pH 7.4, 150 mm NaCl, 5 mm EDTA, 0.1% Triton X-100, 10% glycerol, 1 mm DTT, 1 mm PMSF). Cells were opened and treated as described in Chu et al. (1997) and Schmitt and Clayton (1994). Anti-Flag M2 affinity gel (Kodak) (50 μl) was prepared according to the manufacturers recommendations. Whole-cell extract (40 μl) was applied and treated as described in Chu et al. (1997) and Schmitt and Clayton (1994) to obtain RNA from the bound complexes.
Acknowledgments
We are grateful to Ann Kendall and Paul Good for their technical assistance and R. Robinson, E. Spooner, K. Pierce, and D. Kirby for expert mass spectrometric sequence analysis. We thank Robert Fuller, Dennis Thiele, Ron Taussig, William Ziehler, and Felicia Houser-Scott for advice and critical evaluation of the manuscript. This work was supported by National Institutes of Health (NIH) Grant RO1 GM34869 (to D.R.E.). Oligonucleotide synthesis was subsidized by NIH grant P30CA46592 to the University of Michigan Cancer Center. J.R.C. was supported in part by NIH predoctoral training grant T32 GM07315 awarded to the Cellular and Molecular Biology Graduate Program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL engelke@umich.edu; FAX (313) 763-7799.
References
- Alifano P, Rivellini F, Piscitelli C, Arraiano CM, Bruni CB, Carlomagno MS. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes & Dev. 1994;8:3021–3031. doi: 10.1101/gad.8.24.3021. [DOI] [PubMed] [Google Scholar]
- Altman S, Kirsebom L, Talbot SJ. Recent studies of RNase P. In: Söll D, RajBhandary UL, editors. tRNA: Structure, biosynthesis, and function. Washington, D.C.: American Society of Microbiology; 1995. pp. 67–78. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1995. Nucleic Acids Res. 1995;24:189–196. doi: 10.1093/nar/24.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkiewicz M, Gold H, Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes & Dev. 1989;3:488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Tollervey D, Cullin C, Lacroute F. Functional analysis of Rrp7p, an essential yeast protein involved in pre-rRNA processing and ribosome assembly. Mol Cell Biol. 1997;17:5023–5032. doi: 10.1128/mcb.17.9.5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothwell AL, Garber RL, Altman S. Nucleotide sequence and in vitro processing of a precursor molecule to Escherichia coli 4.5 S RNA. J Biol Chem. 1976;251:7709–7716. [PubMed] [Google Scholar]
- Brown JW, Hunt DA, Pace NR. Nucleotide sequence of the 10Sa RNA gene of the β-purple eubacterium Alcaligenes eutrophus. Nucleic Acids Res. 1990;18:2820. doi: 10.1093/nar/18.9.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;256:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Burton ZF, Gross CA, Watanabe KK, Burgess RR. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983;32:335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- Chamberlain JR, Pagán-Ramos E, Kindelberger DW, Engelke DR. An RNase P RNA subunit mutation that affects ribosomal RNA processing. Nucleic Acids Res. 1996a;24:3158–3166. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Tranguch AJ, Pagán-Ramos E, Engelke DR. Eukaryotic nuclear RNase P: Structures and functions. In: Moldave K, Cohn W, editors. Progress in nucleic acids research and molecular biology. San Diego, CA: Academic Press; 1996b. pp. 87–119. [DOI] [PubMed] [Google Scholar]
- Chen JL, Pace NR. Identification of the universally conserved core of ribonuclease P RNA. RNA. 1997;3:557–560. [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Dwight S, Chervitz S, Jia Y, Juvik G, Weng S, Botstein D. Saccharomyces Genome Database. 1996. http://genome-www.stanford.edu/Saccharomyces/1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz SA, Cherry JM, Botstein D. Saccharomyces genome database on-line pattern matching program. 1997. http://genome-www.stanford.edu/Sacch3D/patmatch.html. [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Zengel JM, Lindahl L. A novel protein shared by RNase MRP and RNase P. RNA. 1997;3:382–391. [PMC free article] [PubMed] [Google Scholar]
- Dang YL, Martin NC. Yeast mitochondrial RNase P. Sequence of the RPM2 gene and demonstration that its product is a protein subunit of the enzyme. J Biol Chem. 1993;268:19791–19796. [PubMed] [Google Scholar]
- Dichtl B, Tollervey D. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 1997;16:417–429. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey R. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1992;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Doersen CJ, Guerrier Takada C, Altman S, Attardi G. Characterization of an RNase P activity from HeLa cell mitochondria. Comparison with the cytosol RNase P activity. J Biol Chem. 1985;260:5942–5949. [PubMed] [Google Scholar]
- Doria M, Carrara G, Calandra P, Tocchini Valentini GP. An RNA molecule copurifies with RNase P activity from Xenopus laevis oocytes. Nucleic Acids Res. 1991;19:2315–2320. doi: 10.1093/nar/19.9.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder PS, Kekuda R, Stolc V, Altman S. Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc Natl Acad Sci. 1997;94:1101–1106. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormick AL, Yates JRI. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Engemann S, Herfurth E, Briesemeister U, Wittmann-Liebold B. Amino acid sequence of the ribosomal protein HS23 from the halophilic Haloarcula marismortui and homology studies to other ribosomal proteins. J Protein Chem. 1995;14:189–195. doi: 10.1007/BF01886759. [DOI] [PubMed] [Google Scholar]
- Evans CF, Engelke DR. Yeast extracts for transfer RNA gene transcription and processing. Methods Enzymol. 1990;181:439–450. doi: 10.1016/0076-6879(90)81142-h. [DOI] [PubMed] [Google Scholar]
- Felici F, Cesareni G, Hughes JM. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol Cell Biol. 1989;9:3260–3268. doi: 10.1128/mcb.9.8.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster AC, Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- Garcia-Bustos J, Heitman J, Hall MN. Nuclear protein localization. Biochim Biophys Acta. 1991;1071:83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldman H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gold HA, Topper JN, Clayton DA, Craft J. The RNA processing enzyme RNase MRP is identical to the Th RNP and related to RNase P. Science. 1989;245:1377–1380. doi: 10.1126/science.2476849. [DOI] [PubMed] [Google Scholar]
- Gopalan V, Baxevanis AD, Landsman D, Altman S. Analysis of the functional role of conserved residues in the protein subunit of ribonuclease P from Escherichia coli. J Mol Biol. 1997;267:818–829. doi: 10.1006/jmbi.1997.0906. [DOI] [PubMed] [Google Scholar]
- Green CJ, Vold BS, Morch MD, Joshi RL, Haenni AL. Ionic conditions for the cleavage of the tRNA-like structure of turnip yellow mosaic virus by the catalytic RNA of RNase P. J Biol Chem. 1988;263:11617–11620. [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hansen FG, Hansen EB, Atlung T. Physical mapping and nucleotide sequence of the rnpA gene that encodes the protein component of ribonuclease P in Escherichia coli. Gene. 1985;38:85–93. doi: 10.1016/0378-1119(85)90206-9. [DOI] [PubMed] [Google Scholar]
- Harris ME, Kazantsev AV, Chen JL, Pace NR. Analysis of the tertiary structure of the ribonuclease P ribozyme-substrate complex by site-specific photoaffinity crosslinking. RNA. 1997;3:561–576. [PMC free article] [PubMed] [Google Scholar]
- Harris SD, Cheng J, Pugh TA, Pringle JR. Molecular analysis of Saccharomyces cerevisiae chromosome I. On the number of genes and the identification of essential genes using temperature-sensitive-lethal mutations. J Mol Biol. 1992;225:53–65. doi: 10.1016/0022-2836(92)91025-k. [DOI] [PubMed] [Google Scholar]
- Hollingsworth MJ, Martin NC. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Mol Cell Biol. 1986;6:1058–1064. doi: 10.1128/mcb.6.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Alteration of a mitochondrial tRNA precursor 5′ leader abolishes its cleavage by yeast mitochondrial RNase P. Nucleic Acids Res. 1987;15:8845–8860. doi: 10.1093/nar/15.21.8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Wang YL, Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Taneja K, Singer RH, Wang YL, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Jayanthi GP, Van Tuyle GC. Characterization of ribonuclease P isolated from rat liver cytosol. Arch Biochem Biophys. 1992;296:264–270. doi: 10.1016/0003-9861(92)90571-d. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Sasaki N, Ando-Yamagami Y. Cleavage of tRNA within the mature tRNA sequence by the catalytic RNA of RNase P: Implication for the formation of the primer tRNA fragment for reverse transcription in copia retrovirus-like particles. Proc Natl Acad Sci. 1990;87:8105–8109. doi: 10.1073/pnas.87.20.8105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence N, Wesolowski D, Gold H, Bartkiewicz M, Guerrier-Takada C, McClain WH, Altman S. Characteristics of ribonuclease P from various organisms. Cold Spring Harbor Symp Quant Biol. 1987;52:233–238. doi: 10.1101/sqb.1987.052.01.028. [DOI] [PubMed] [Google Scholar]
- Lee B, Matera AG, Ward DC, Craft J. Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolus: A possible coordinate role in ribosome biogenesis. Proc Natl Acad Sci. 1996;93:11471–11476. doi: 10.1073/pnas.93.21.11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Engelke DR. Partial characterization of an RNA component that copurifies with Saccharomyces cerevisiae RNase P. Mol Cell Biol. 1989;9:2536–2543. doi: 10.1128/mcb.9.6.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Rohlman CE, Molony LA, Engelke DR. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol Cell Biol. 1991;11:721–730. doi: 10.1128/mcb.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Lee BJ, Hwang DS, Kang HS. Purification and characterization of mitochondrial ribonuclease P from Aspergillus nidulans. Eur J Biochem. 1996;235:289–296. doi: 10.1111/j.1432-1033.1996.00289.x. [DOI] [PubMed] [Google Scholar]
- Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Liu F, Altman S. Differential evolution of substrates for an RNA enzyme in the presence and absence of its protein cofactor. Cell. 1994;77:1093–1100. doi: 10.1016/0092-8674(94)90448-0. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Mitchell P, Petfalski E, Seraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes & Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Allmang C, Tollervey D, Séraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- Mans RM, Guerrier Takada C, Altman S, Pleij CW. Interaction of RNase P from Escherichia coli with pseudoknotted structures in viral RNAs. Nucleic Acids Res. 1990;18:3479–3487. doi: 10.1093/nar/18.12.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales MJ, Dang YL, Lou YC, Sulo P, Martin NC. A 105-kDa protein is required for yeast mitochondrial RNase P activity. Proc Natl Acad Sci. 1992;89:9875–9879. doi: 10.1073/pnas.89.20.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JP, Tollervey D. Birth of the snoRNPs: The evolution of RNase MRP and the eukaryotic pre-rRNA-processing system. Trends Biochem Sci. 1995;20:78–82. doi: 10.1016/s0968-0004(00)88962-8. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nash HM, Bruner SD, Scharer OD, Kawate T, Addona TA, Spooner E, Lane WS, Verdine GL. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- Pace NR, Brown JW. Evolutionary perspective on the structure and function of ribonuclease P, a ribozyme. J Bacteriol. 1995;177:1919–1928. doi: 10.1128/jb.177.8.1919-1928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich C, Olsen GJ, Pace B, Pace NR. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science. 1988;239:178–181. doi: 10.1126/science.3122322. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Tome P, Stoehr PJ, Cameron GN, Flores TP. The European Bioinformatics Institute (EBI) databases. Nucleic Acids Res. 1996;24:6–12. doi: 10.1093/nar/24.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rutgers CA, Rientjes JMJ, van’t Reit J, Raué HA. rRNA binding domain of yeast ribosomal protein L25. Identification of its borders and a key leucine residue. J Mol Biol. 1991;218:375–385. doi: 10.1016/0022-2836(91)90719-m. [DOI] [PubMed] [Google Scholar]
- Salazar CE, Mills-Hamm DM, Kumar V, Collins FH. Sequence of a cDNA from the mosquito Anopheles gambiae encoding a homologue of human ribosomal protein S7. Nucleic Acids Res. 1993;21:4147. doi: 10.1093/nar/21.17.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Characterization of a unique protein component of yeast RNase MRP: An RNA-binding protein with a zinc-cluster domain. Genes & Dev. 1994;8:2617–2628. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]
- Shuai K, Warner JR. A temperature sensitive mutant of Saccharomyces cerevisiae defective in pre-rRNA processing. Nucleic Acid Res. 1991;19:5059–5064. doi: 10.1093/nar/19.18.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman KE, George DG, Barker WC, Hunt LT. The protein identification resource (PIR) Nucleic Acids Res. 1988;16:1869–1871. doi: 10.1093/nar/16.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl LL, Clayton DA. Saccharomyces cerevisiae contains an RNase MRP that cleaves at a conserved mitochondrial RNA sequence implicated in replication priming. Mol Cell Biol. 1992;12:2561–2569. doi: 10.1128/mcb.12.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Altman S. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes & Dev. 1997;11:2414–2425. doi: 10.1101/gad.11.18.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BC, Gao L, Stomp D, Li X, Gegenheimer PA. Spinach chloroplast RNase P: A putative protein enzyme. Nucleic Acids Symp Ser. 1995;33:95–98. [PubMed] [Google Scholar]
- Tranguch AJ, Kindelberger DW, Rohlman CE, Lee JY, Engelke DR. Structure-sensitive RNA footprinting of yeast nuclear ribonuclease P. Biochemistry. 1994;33:1778–1787. doi: 10.1021/bi00173a022. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Westhof E, Altman S. Three-dimensional working model of M1 RNA, the catalytic RNA subunit of ribonuclease P from Escherichia coli. Proc Natl Acad Sci. 1994;91:5133–5137. doi: 10.1073/pnas.91.11.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerly S, Drainas D, Sylvers LA, Söll D. Identification of a 100-kDa protein associated with nuclear ribonuclease P activity in Schizosaccharomyces pombe. Eur J Biochem. 1993;217:501–507. doi: 10.1111/j.1432-1033.1993.tb18270.x. [DOI] [PubMed] [Google Scholar]