Abstract
Switching of Saccharomyces mating type by replacement of sequences at the MAT locus involves a choice between two donors, HML and HMR. MATα cells inhibit recombination along the entire left arm of chromosome III, including HML, whereas MATa cells activate this same region. MATa-dependent activation of HML depends on a small, cis-acting DNA sequence designated the recombination enhancer (RE), located 17 kb centromere-proximal to HML. A comparison of RE sequences interchangeable between Saccharomyces cerevisiae and Saccharomyces carlsbergensis defines a minimum RE of 244 bp. RE activity is repressed in MATα cells by binding of the Matα2–Mcm1 corepressor to a site within the RE. Mutation of the two Matα2 binding sites removes most, but not all, of this repression, and RE chromatin structure in MATα cells becomes indistinguishable from that seen in MATa. Surprisingly, a 2-bp mutation in the Mcm1 binding site completely abolishes RE activity in MATa cells; moreover, RE chromatin structure in the MATa mutant becomes very similar to that seen in MATα cells with a normal RE, displaying highly ordered nucleosomes despite the absence of Matα2. Further, a mutation that alters the ability of Mcm1 to act with Matα2 in repressing a-specific genes also alters donor preference in either mating type. Thus, Mcm1 is critically responsible for the activation as well as the Matα2-Mcm1-mediated repression of RE activity.
Keywords: S. cerevisiae, donor preference, recombination enhancer, Mcm1, mating-type switching
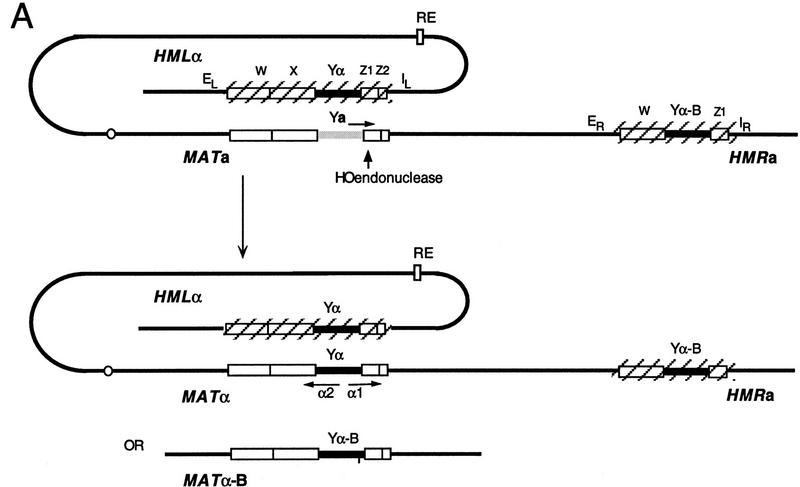
Saccharomyces cerevisiae is able to change its mating type as often as every cell division by a highly choreographed recombination mechanism. The induction of the HO endonuclease creates a double-strand break (DSB) at the MAT locus. This initiates the excision of ∼700 bp of the Ya or Yα-specific sequences that define MATa or MATα, respectively, and to initiate their replacement by sequences of the opposite mating type, provided by one of two donor loci, HML or HMR (Fig. 1). These two donors, located near the ends of chromosome III, are unexpressed and maintained in a chromatin structure that prevents their cleavage by HO (for review, see Strathern 1989; Haber 1992; Laurenson and Rine 1994). Normally, HML carries Yα, whereas HMR carries Ya, but each donor may harbor either mating-type allele.
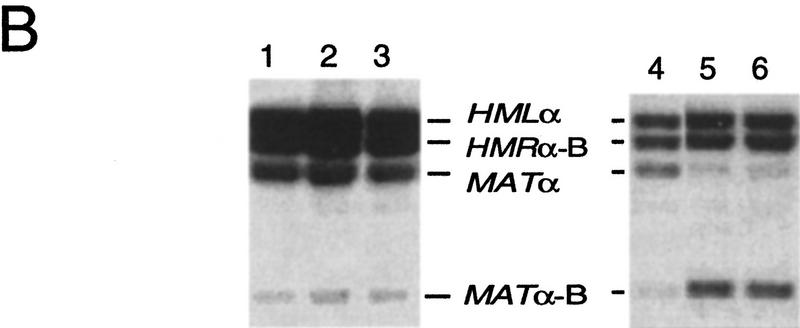
Figure 1.
(A) MATa switching to MATα, with HMLα as a donor. Expression of the HO endonuclease creates a DSB in the MAT locus, promoting repair by gene conversion with the donor HMLα. The two donor loci are maintained in a transcriptionally inactive chromatin structure whose maintenance depends on adjacent E and I silencer sequences. The preferential use of HML in MATa cells depends on the presence of a cis-acting RE, located 17 kb centromere-proximal to HML and which stimulates the use of a donor anywhere on the left arm of chromosome III. In MATα cells, the use of HML is actively repressed by an unknown mechanism, leaving HMR as the only available donor. Donor preference does not depend on the sequences present at HML or HMR. The alternative product of switching, with HMRα-B as a donor, would yield MATα-B, carrying a BamHI site. (B) Donor preference assayed in populations of cells induced to switch from MATa to either MATα or MATα-B. Normal donor preference in S. cerevisiae is shown in lane 1 (XW641) and in lane 4 (WX652). (Lane 2) 2.5-kb deletion of RE replaced by 753-bp REcer (CWU150). (Lane 3) 2.5-kb deletion re-placed by 813-bp REcarl (CWU61). (Lane 5) Identical to lane 2 except with a 2-bp GG > CC mutation of the Mcm1 binding site of REcer (CWU56). (Lane 6) Identical to lane 5 except the 753-bp fragment carrying the Mcm1 binding site mutation is in the inverted orientation.
One of the most remarkable features of this system is that MATa cells select the HML donor ∼90% of the time, whereas 90% of MATα cells select HMR, independent of whether the donor carries Ya or Yα (Klar et al. 1982; Weiler and Broach 1992; Wu and Haber 1995; Wu et al. 1996). Donor preference is not influenced by sequences immediately surrounding HML and HMR, as either locus can be replaced by a cloned copy of the other without altering the outcome (Weiler and Broach 1992). Donor choice is accomplished by two different mechanisms. In MATα cells, HML—in fact the entire 112-kb left arm of chromosome III—is rendered inaccessible, so that cells use HMR by default (Wu et al. 1996). However, in MATa cells, this unusually cold state is reversed and the entire left arm of the chromosome becomes activated, so that HML outcompetes HMR (Wu and Haber 1996; Wu et al. 1997). The activation of HML in MATa also occurs during spontaneous recombination between two alleles of leu2 when one of the leu2 alleles is located in place of HML (Wu and Haber 1996). There is little or no mating type-dependent difference in the use of HMR (Wu and Haber 1996; Wu et al. 1997).
Activation of the left arm requires a small, cis-acting DNA sequence that we have termed the recombination enhancer (RE) (Wu and Haber 1996). The RE was initially defined as a 700-bp orientation independent sequence, although a 361-bp segment retained much of the activity (Wu and Haber 1996). The RE region contains several distinctive features. First, there are no apparent ORFs in the RE or in the surrounding 2-kb region. Second, the DNA sequence of the RE contains a highly conserved binding site for the Matα2–Mcm1 repressor complex as well as an unusual stretch of TTT(A/G) repeats (Wu and Haber 1996). In MATα cells, there is strong evidence that RE activity is directly and negatively regulated by the binding of the Matα2–Mcm1 repressor complex to the site within the RE (Tanaka et al. 1984; Szeto and Broach 1997; Szeto et al. 1997; Weiss and Simpson 1997). In MATa cells (i.e., in the absence of Matα2 protein), the RE is distinguished by an unusual micrococcal nuclease-hypersensitive region flanked by two protein-binding footprints. In MATα cells, these features are lost, and the region is covered by highly phased nucleosomes extending from the site in which Matα2–Mcm1 is bound (Weiss and Simpson 1997). When the sites in which the two Matα2 proteins bind are mutated, this repression is partially lost (Szeto et al. 1997; see below). This result suggests that the activation of recombination does not absolutely depend on a-specific genes, as these genes should still be repressed in a MATα cell. There are two sterile (noncoding) transcripts in the activated RE region that are repressed in MATα. Szeto et al. (1997) further demonstrated that binding of Mcm1 protein to the Matα2–Mcm1 binding region of RE functions in reporter gene constructs as a transcriptional activator, as Mcm1 does at other sites (Elble and Tye 1991; Althoefer et al. 1995; McInerny et al. 1997). Thus, Mcm1 may be important, not only as a corepressor of RE activiy in MATα cells, but as a key activator in MATa.
When the RE is deleted, donor preference does not become 50:50 between HML and HMR; instead, MATa cells use the wrong donor, HMR, 90% of the time (Wu and Haber 1996; Wu et al. 1997). Thus, there is a constitutively cold state on the left arm of chromosome III, in MATa as well as in MATα, against which RE works to activate HML in MATa cells. At present, nothing is known about how the left arm is rendered inaccessible for recombination in MATα cells, or in MATa when the RE is deleted. The unavailability of HML in MATα is seen even when this donor is not silenced (Wu et al. 1995), so its lack of accessibility is unlikely to involve a change in the local chromatin structure around HML. Moreover, although there is a profound effect on recombination involving a leu2 gene situated in place of HML, there is no detectable mating-type dependent difference in its transcription (Wu and Haber 1996), nor are there any evident changes in the chromatin structure of the HML donor itself (K. Weiss and R.T. Simpson, unpubl.). We have suggested that the left arm of chromosome III in MATα may be folded into a higher-order chromatin structure or attached to the nuclear envelope in a way that makes it difficult to contact MAT (Wu and Haber 1996). An alternative model, based on the spreading of a change in chromosome structure, has been advanced by Szeto et al. (1997).
Very little information is available presently to explain what proteins activate the RE in MATa cells. Deletion of the CHL1 gene, which causes a general increase in mitotic chromosome instability (Liras et al. 1976; Gerring et al. 1990), modestly reduces MATa donor preference but has no detectable effect on MATα switching (Weiler et al. 1995). chl1 does not alter HMLα or RE chromatin structures (K. Weiss and R.T. Simpson, unpubl.). However, no genes have been identified yet that are essential either for the activation of RE in MATa or for the inactivation of chromosome III’s left arm in MATα. In this report, we present evidence that the Mcm1 binding site is essential for MATa donor preference.
Results
The RE from S. carlsbergensis functions in S. cerevisiae
The DNA sequence from part of the left arm of chromosome III of S. carlsbergensis was determined as described in Materials and Methods. Two ORFs flanking the RE region, YCL055 (KAR4) and YCL052c, share ∼85% sequence identity, whereas the sequences of the 700-bp RE regions were only 60% identical between the two species (Fig. 2). However, four parts of this region, designated A, B, C, and D, were much more highly conserved. Included in the C region is a 29/31 match in the Matα2–Mcm1 consensus binding site (Smith and Johnson 1994). The D region contains many TTT(A/G) repeats, in which there were several A↔G transitions. The conserved regions were limited to a 244-bp region.
Figure 2.

Comparison of REcer and REcarl. Identical residues in the alignment of the DNA sequences of REcarl (top) and REcer (bottom) are indicated by vertical lines. The four most conserved subregions (A,B,C,D) are underlined. Region C contains the Matα2–Mcm1 consensus binding site (double underlined). Several of the TTT(A/G) repeats in region D differ in the purine position. The more highly conserved 3′ end of the YCL055 (KAR4) ORF is also shown. The beginning and end of the 753- and 270-bp REcer and the beginnings and ends of the 813- and 244-bp REcarl are also indicated.
An 813-bp RE-homologous region of S. carlsbergensis (REcarl) was inserted into S. cerevisiae at the site of a 2.5-kb deletion that completely removes the endogenous RE (REcer). To assess donor preference, MATa cells carrying a galactose-inducible HO gene were grown in galactose medium for 1.5 hr to induce gene conversion of MATa by use of sequences from wild-type HMLα or from HMRα–B. The latter carries a single base pair mutation that creates a BamHI site (Wu and Haber 1996). REcarl has comparable activity, in either orientation, to REcer in MATa cells (Table 1A). As shown in Figure 1B, HMLα is used 75% of the time. In strains lacking the RE, HML is used only 5% of the time. Moreover, in MATα, REcarl was nearly as completely repressed as REcer. When the C in the center of the Matα2–Mcm1-binding site of REcarl was changed to the T found in the S. cerevisiae sequence, the repression was indistinguishable (Table 1A).
Table 1.
Effect of RE sequence on donor preference
| Strain
|
Allele
|
Percent HML usage
|
|
|---|---|---|---|
|
MATa
|
MATα
|
||
| A. | |||
| XW641/XW643 | wild type | 85 | 15 |
| XW663/XW683a | 2.5-kb Δ | 5 | 5 |
| CWU150/CWU151 | 753-bp REcer | 75 | 15 |
| CWU61/CWU67 | 813-bp REcarl | 75 | 24 |
| CWU109/CWU111b | 813-bp REcarl w/1-bp change | 75 | 16 |
| B. | |||
| CWU123/CWU122c | 1.8-kb Δ | 5 | 5 |
| CWU81/CWU101 | 270-bp REcer | 45 | 8 |
| CWU141/CWU147 | 244-bp REcarl | 50 | 20 |
| C. | |||
| CWU70 | 270-bp REcer ABCDd | 45 | |
| CWU81/CWU101 | 270-bp REcer ABCDe | 45 | 8 |
| CWU71/CWU115 | 270-bp REcer AB
|
45 | 12 |
| CWU83/CWU103 | 270-bp REcer ACD | 45 | 12 |
| CWU118/CWU119 | 270-bp REcer BCDe | 20 | 2 |
| CWU120/CWU121 | 270-bp REcer ABDe | 10 | 10 |
| CWU82/CWU102 | 270-bp REcer ABCe | 12 | 2 |
| CWU72 | 270-bp REcer ABC | 10 | |
Donor preference was analyzed as described in Materials and Methods and as illustrated in Fig. 1. The percentage usage of HML is based on an average of at least three independent measurements.
As described in Wu and Haber (1996) the 2.5-kb deletion of the RE region in strains XW641 and XW643 is lethal; hence, these strains carry either the complementing TRP1-marked plasmid XW261 with a large segment of the left arm of chromosome III or URA3-marked plasmid pJR1684 (Loo et al. 1995) that complements the essential gene YCL054 that lies at the right end of the deletion. All sequences were inserted in the wild-type orientation.
A 1-bp C→T change in the middle of the Matα2-Mcm1 binding site of REcarl converts this sequence to that found in REcer.
Strains carrying a 1.8-kb deletion (see Materials and Methods) are viable and carry no additional plasmids containing DNA sequences in the RE region.
The four highly conserved domains shared by REcer and REcarl are designated A, B, C, and D (Fig. 2). Deletions removing one or more of these regions and the inversion of two regions relative to each other are described in Materials and Methods.
ABCD are in opposite orientation relative to wild type.
In a similar fashion, smaller DNA segments containing primarily the conserved sequences were introduced in place of the normal RE: a 270-bp region from REcer or a 244-bp segment from REcarl. We conclude that these small RE segments, from either species, retained most of the donor preference activity, because strains carrying these inserts used HML ∼50% of the time, whereas strains with only the 1.8-kb deletion of RE used HML 5% of the time (Table 1B). Thus, the 270-bp region of REcer and the 244 bp REcarl define a minimum enhancer region that is amenable to further mutational analysis (see below).
Deletion analysis of the 270-bp REcer
To investigate the importance of each of the four highly conserved regions of the RE, we deleted each of the regions, as described in Materials and Methods. As shown in Table 1C, region B is dispensable for MATa’s use of HML, whereas deletion of either region A, C, or D resulted in a complete loss of RE activity. In addition, we found that regions C and D could be inverted relative to A and B without loss of RE activity.
Chromatin structure of REcer and REcarl
We compared the chromatin structure of the 813-bp REcarl with that of the 753-bp REcer, in both MATa and MATα cells (Fig. 3). The chromatin structure of the 753-bp REcer is identical to the organization of the chromosomal RE in wild-type cells (Fig. 3B) (Weiss and Simpson 1997). In MATa cells, the uniquely nuclease hypersensitive region (HS), with a cleavage pattern distinct from protein-free DNA digests, is flanked by areas less susceptible to nuclease cutting, FP1 and FP2. Two regions, HS and FP1, containing multiple repeats of the TTT(G/A) sequence, have distinctive structures. In contrast, in MATα cells, these regions are protected from nuclease cleavage. Regions of strong protection extending ∼150 bp alternate with nuclease hypersensitive sites. This pattern suggests arrays of precisely positioned nucleosomes flanking the Matα2 binding site on both sides. The highly organized chromatin structure in MATα cells may exclude factors that can access the RE in MATa.
Figure 3.
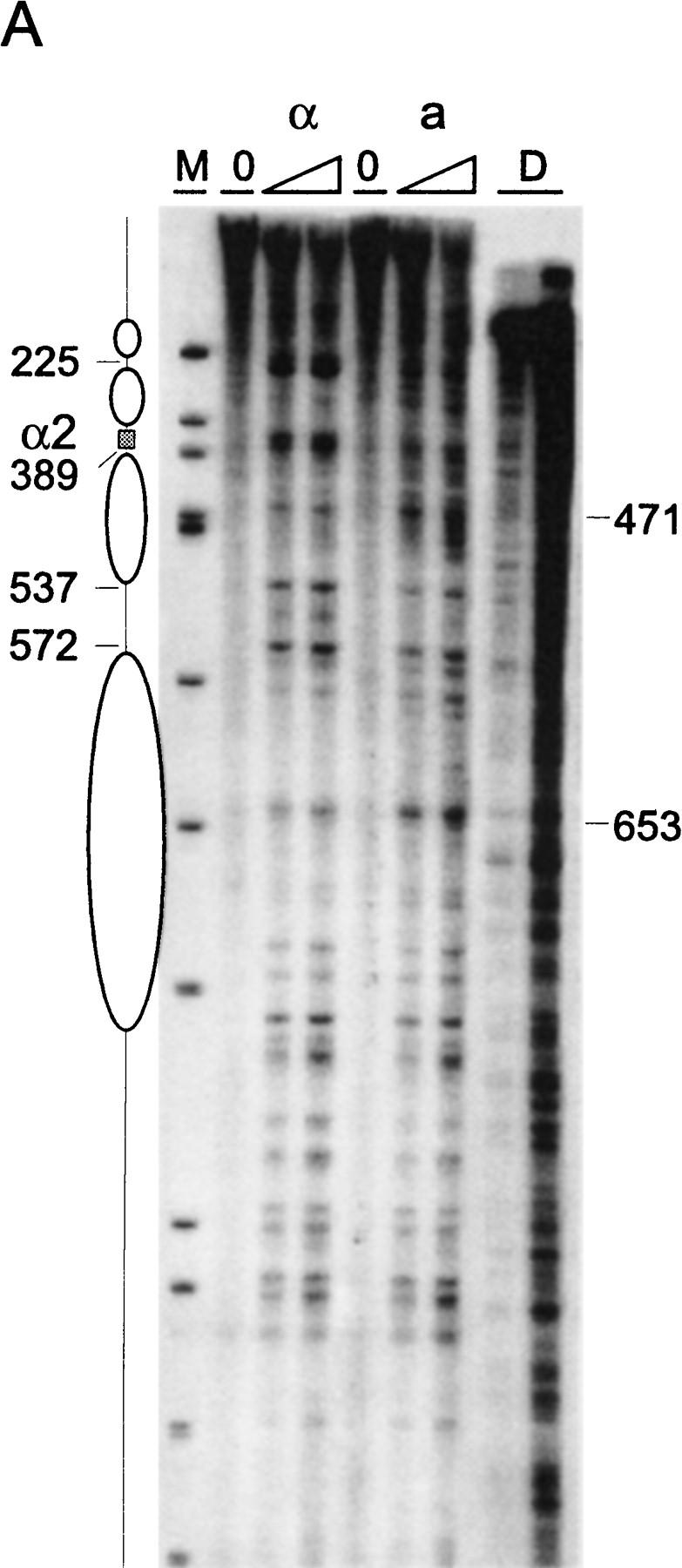
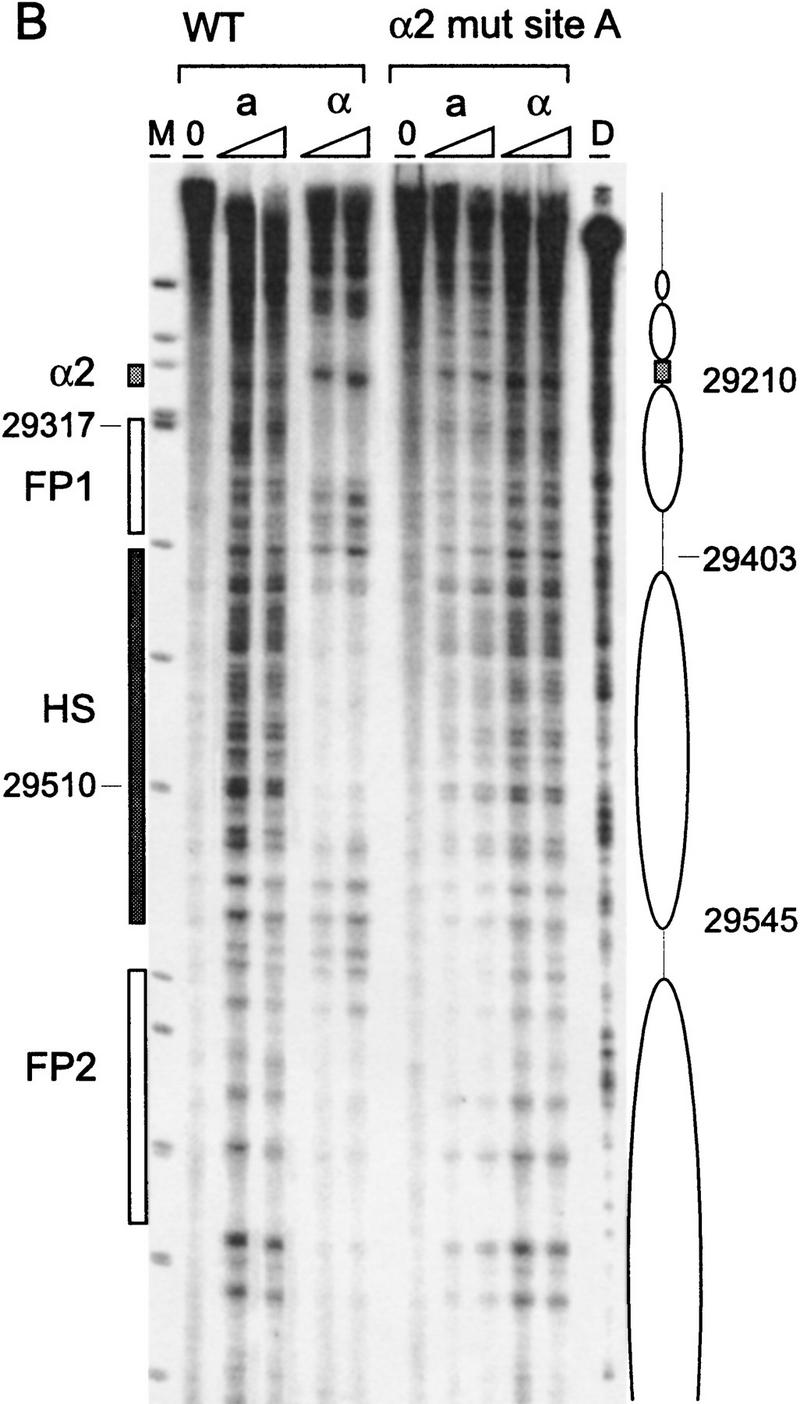
Chromatin structure of REcer and REcarl. (A) Chromatin structure of the S. carlsbergensis 813-bp RE fragment inserted in place of the S. cerevisiae RE at location 28.8 kb of chromosome III. Chromatin structure was mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer I11. MATa and MATα cells are as indicated (CWU61, CWU67). Coordinates correspond to the inserted fragment of REcarl with increasing values for centromere-proximal sequences. Extensions of undigested chromatin (0) and chromatin digested by use of two levels of micrococcal nuclease are presented. (D) Protein-free DNA digests to control for micrococcal nuclease sequence specificity. (M) φX174 DNA digested with HinfII kinased fragments as size standards. The Matα2–Mcm1 binding site (α2) is represented by a shaded box. Inferred positions for nucleosomes in MATα cells are identified by ellipses. (B) Chromatin structure of the wild-type S. cerevisiae 753-bp RE fragment (WT) and the same region containing a 2-bp mutation (GT > G at positions 29198–29199) in Matα2 binding site A. Chromatin structure was mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer b297. MATa and MATα cells (CWU150 and CWU151 for wild type and CWU51 and CWU54 for the Matα2 binding site A mutant) are as indicated. Coordinates correspond to the published sequence of chromosome III (Oliver et al. 1992). Structural elements of the wild-type MATa RE are shown, including two footprints (FP1 and FP2) and a hypersensitive region (HS), both corresponding to DNA regions having numerous repeats of the TTT(A/G) sequence (Weiss and Simpson 1997), are depicted by open boxes. Inferred positions for nucleosomes in wild-type MATα cells are indicated by ellipses.
Chromatin organization of the 813-bp REcarl is also notably different in the two cell types (Fig. 3A). Some features are similar to the structural elements characterizing the RE in S. cerevisiae. In MATα cells, two positioned nucleosomes abut the Matα2–Mcm1 binding site, although the precision of their location is reproducibly not as high as in REcer. One of them, protecting the area between 378 bp and 537 bp of the S. carlsbergensis sequences, protects the highly conserved TTT(G/A) sequence repeat from nuclease cleavage. In contrast to the S. cerevisiae chromatin in MATa cells, the region around 471 bp of the S. carlsbergensis RE is readily accessible to nuclease cleavage. In fact, higher resolution analysis (data not shown) reveals a cut at every T/A step. However, the region flanking this repeat is rather protected and could, in combination with the hypersensitivity of site 653, reflect protein binding. The structure of REcarl in MATa cells has a less extensive hypersensitive region than REcer; the region between 572 and 653 encompasses most of the nuclease sensitive sites.
Mutations in the Matα2 binding sites relieve RE repression
On the basis of the analysis of the Matα2–Mcm1 transcriptional repression site in the promoter of the STE6 gene (Smith and Johnson 1994), we created site-directed mutations of the Matα2 binding sites in the 753-bp segment of REcer (Table 2A). Two-base pair alterations of either Matα2 site A or Matα2 site B resulted in a similar phenotype: There was no effect on MATa donor preference, but there was a distinct weakening of repression in MATα (Table 2B), so that HML was used ∼35% of the time instead of 15%. A double mutation of both sites A and B increased HML usage to 55%. This is still less than the 75% usage of HML in a MATa strain and leaves open the possibility that there is a contribution to MATa donor preference by one or more a-specific genes that are still repressed in MATα cells.
Table 2.
Effect on donor preference of Matα2 and Mcm1 binding to the RE
| A. Sequence of the Matα2–Mcm1 operator in REcer and mutations | |||||||||||||||||||||||||||||||
| C | A | T | G | T | A | A | T | T | A | C | T | T | A | A | A | T | A | G | G | A | A | G | T | T | T | A | C | A | C | G | |
| _ | _ | _ | |||||||||||||||||||||||||||||
| Mutation | T | G | C | C | T | A | |||||||||||||||||||||||||
| Matα2 site A | Mcm1 site | Matα2 site B | |||||||||||||||||||||||||||||
| Strain
|
Allele
|
Percent HML usage
|
|
|---|---|---|---|
|
MATa
|
MATα
|
||
| B. Mutations in Matα2 binding sitesa | |||
| CWU150/CWU151 | wild type | 75 | 15 |
| CWU51/CWU54 | GT → TG Matα2 site A | 75 | 35 |
| CWU79/CWU95 | AC → TA Matα2 site B | 75 | 35 |
| CWU128/CWU134 | GT → TG, AC → TA Matα2 sites A and B | 75 | 55 |
| C. Mutations in Mcm1 binding site in RE subfragments | |||
| CWU56/CWU93 | GG → CC 753-bp REcer | 15 | 12 |
| CWU137/CWU143 | GG → CC 270-bp REcer | 3 | 5 |
| CWU139/CWU145 | GG → CC 244-bp REcarl | 4 | 5 |
| D. Mutation of Mcm1 binding site in the complete REb | |||
| XW652 | wild type | 85 | |
| CWU157 | GG → CC | 20 | |
Donor preference was analyzed as described in Table 1.
Mutations were introduced into the 753-bp REcer.
An Mcm1 binding site mutation was introduced into chromosome III containing no other modifications of the chromosome.
As expected, mutation of one of the Matα2p binding half-sites abolished the phased nucleosome array in α cells (Fig. 3B). In fact, the nuclease digestion pattern of this mutated RE in both cell types is very similar to the pattern mapped in wild-type MATa cells. The hypersensitive region between 29403 and 29510 flanked by regions protected from nuclease cleavage is present. The absence of the nucleosome array, even when there is still some repression exerted by an intact second half-site, may mean that the RE fluctuates between an on and off state under these conditions, which cannot be detected by the analysis of chromatin structure in the average population. The wild-type RE map of micrococcal nuclease digests of MATa and MATα cell nuclei mixed 1:1 exhibits a largely MATa-like pattern (data not shown).
Mutation of the Mcm1 binding site abolishes MATa donor preference
In contrast to altering the Matα2 binding sites, mutation of the Mcm1 binding site in the 753-bp REcer had a profound effect on MATa switching. A GG to CC mutation of the Mcm1 binding site (Smith and Johnston 1994) had no effect in MATα cells (Table 2C); however, this mutation severely impaired MATa’s use of HML (15%). This holds true when this 2-bp mutation was introduced into the 753-bp and the 270-bp REcer, in either orientation, as well as into the 244-bp REcarl region (Table 2C).
The same 2-bp mutation in the Mcm1 binding site was also introduced into an otherwise unmodified chromosome III (Table 2D). In the context of the entire RE, this small change was sufficient to reduce MATa’s use of HML from 85% to 20%. MATα cells used HML as a donor ∼15% of the time. Thus, although MATα cells might lose Matα2–Mcm1 repression, there is no activation in either mating type in this mutant. The effect of the 2-bp mutation in the Mcm1 binding site thus resembles that of deleting the entire RE.
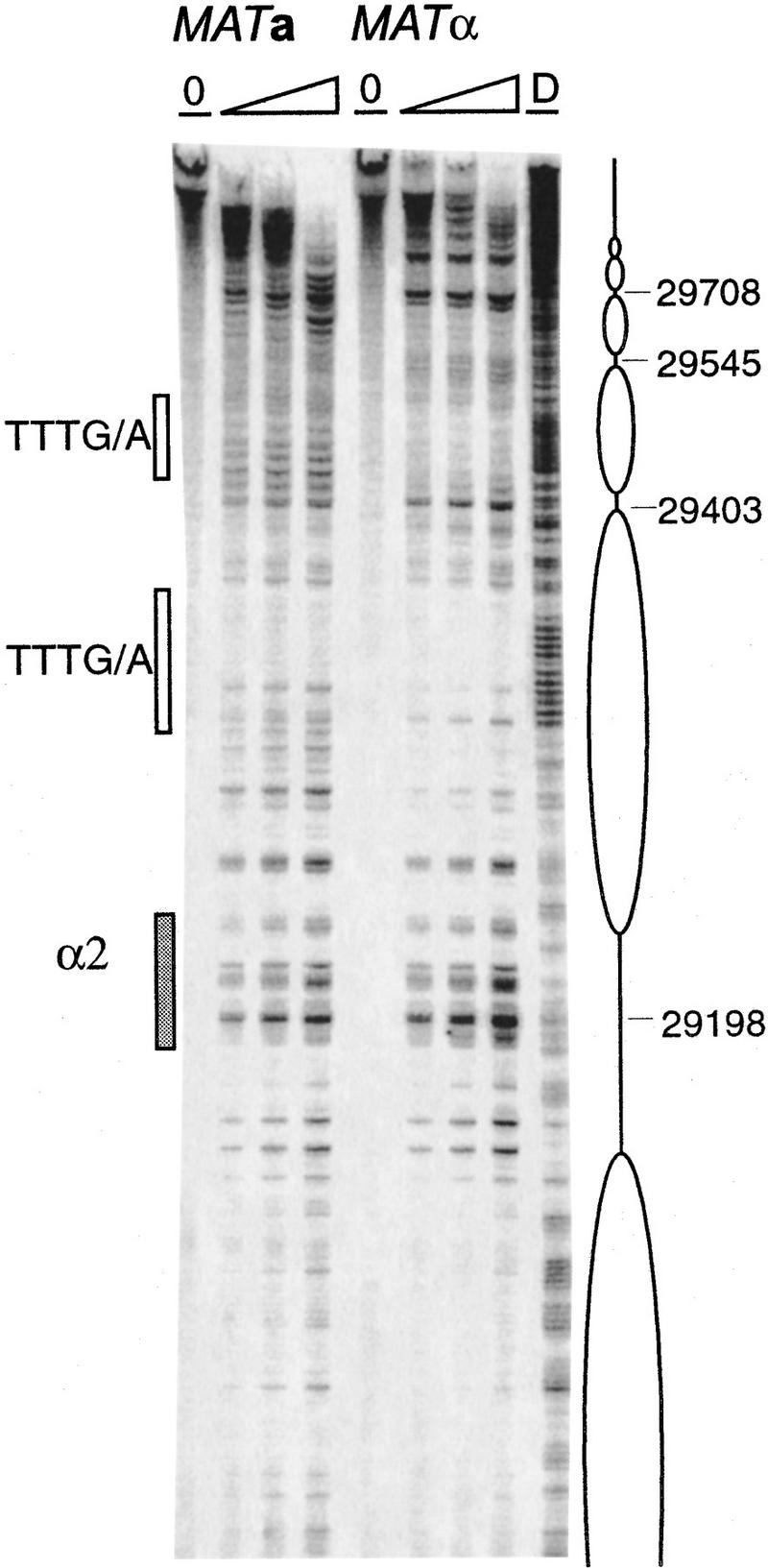
In keeping with these results, the chromatin structure of the 753-bp REcer carrying the 2-bp mutation in the Mcm1 binding site has lost all of its distinctive MATa features. In fact, the region is identical to the pattern seen in MATα cells. The RE near the mutated Matα2–Mcm1 operator exhibits a pattern of nucleosome-length areas of protection alternating with nuclease hypersensitive sites (Fig. 4A). A nucleosome array covers this inactive RE. Positioning appears less precise than in the wild-type MATα chromatin as some cutting can be seen inside areas protected by nucleosomes (central two nucleosomes, panel B). However, the pattern of DNaseI-digested chromatin between positions 29545 and 29425 (panel A) is reminiscent of the 10-bp repeat observed previously with bands decreasing in intensity from the edge of the region toward the center.
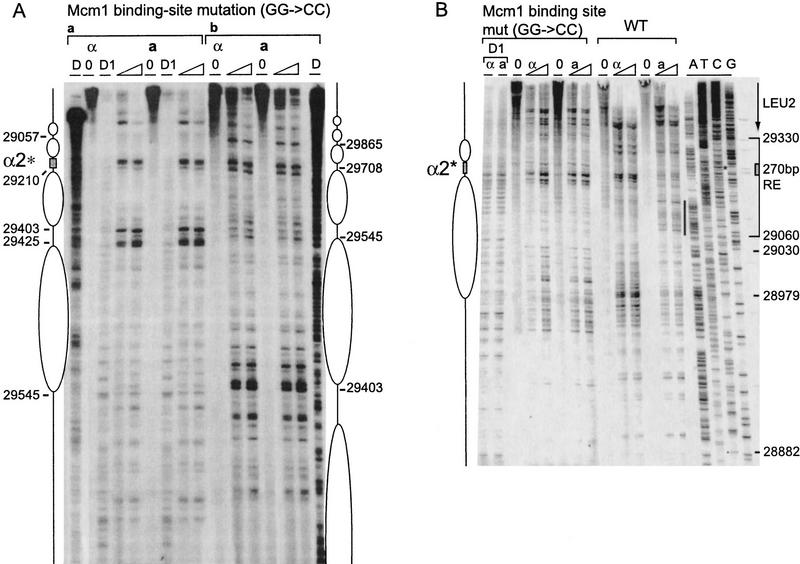
Figure 4.
Effect of Mcm1 binding site mutation on RE chromatin structure. (A) Chromatin structure of the S. cerevisiae 753-bp RE fragment containing a 2-bp mutation (GG to CC at positions 29212–29213) in the Mcm1 binding site of the α2 operator mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer b297 (a) and a292 (b). Mata and Matα cells are as indicated (CWU57, CWU94). Extensions of undigested chromatin (0) and chromatin digested with two levels of micrococcal nuclease are presented. (D) Protein-free DNA digests. The mutated Mcm1–Matα2 binding site consensus sequence (α2*) is represented by a shaded box. (B) Chromatin structure of the S. cerevisiae 270-bp minimal RE fragment containing the wild-type α2 consensus sequence (29194–29225) or a point mutation (GG-CC at positions 29212–29213) in the Mcm1 binding site mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer a288. MATa and MATα cells are as indicated (CWU81 and CWU101 for wild-type 270 bp, CWU137 and CWU143 for Mcm1 binding site mutation, respectively). Extensions of undigested chromatin (0) and two levels of micrococcal nuclease-digested chromatin are presented. (D1) DNase I-digested chromatin. (*) The position of the mutation in the Mcm1 binding sequence. The limits of the 270-bp fragment are indicated by a bracket. The ellipse corresponds to the inferred position of a nucleosome. The vertical black stripe emphasizes the area of increased nuclease cleavage in wild-type MATa vs. MATα cells. The 3′ end of the LEU2 transcript is indicated by an arrow. The positions of the sequences were determined by four dideoxy sequencing reactions with the same primer (lanes A, T, G, and C).
Surprisingly, the Matα2–Mcm1 binding site is still nuclease sensitive and appears to lie in the linker region between two nucleosomes. The general pattern of nucleosome positions in MATa is similar to the one found in MATα for this mutant and—more surprisingly—very similar to the nucleosome array mapped in wild-type MATα cells. However, the nucleosome adjacent to the mutated Matα2–Mcm1 site (panel A) adopts a different position than when the original operator is in MATα: The protected region is between 29210 and 29403, whereas in the presence of a functional operator, a nucleosome is located between 29210 and 29365. This shift in nucleosome placement likely reflects inherent nucleosome positioning ability of underlying sequences that is overruled by binding of the Matα2–Mcm1 complex to the operator, which locks the nucleosome in a position closer to the Matα2 site. Such a shift actively caused by Matα2p binding has been observed previously (Simpson 1990). This suggests that the second centromere-proximal nucleosome from the α2 site has the same translational and rotational position in wild type and Mcm1-operator mutant cells.
We also examined the chromatin structure of the 270-bp minimum REcer (Fig. 4B, WT). The 270-bp wild type REcer shows similar features to the 753-bp REcer (Fig. 3B). In MATα, the Matα2 binding site, which is nuclease sensitive, is flanked by a region protected from MNase cleavage and delimited by a site of strong sensitivity to cleavage (28979). We interpret this as being the footprint of a positioned nucleosome in MATα cells. In MATa, MNase cleavage sites of varying frequency can be seen throughout the region, yet no hypersensitive sites are observed. Hence, the nucleosome is disrupted when the RE is active. Because the 2-bp mutation of the Mcm1 binding site abolishes RE activity, the chromatin becomes identical in MATa and MATα: It resembles the inactive state with large areas of protection abutting the mutated Matα2–Mcm1 binding site (Fig 4B, Mcm1 binding site mut). The pattern is very different from the wild-type chromatin in MATa cells and is very similar to the one mapped in wild-type MATα cells. Nevertheless, the hypersensitivity of site 28979 is lost and adjacent sites are more readily cleaved. If a nucleosome is present in MATα cells, the mutation of the operator causes its position to be less precise. This observation is consistent with the nucleosome shift in the 753-bp Mcm1 binding site mutant analyzed above.
A mutant Mcm1 protein is unable to activate MATa donor preference
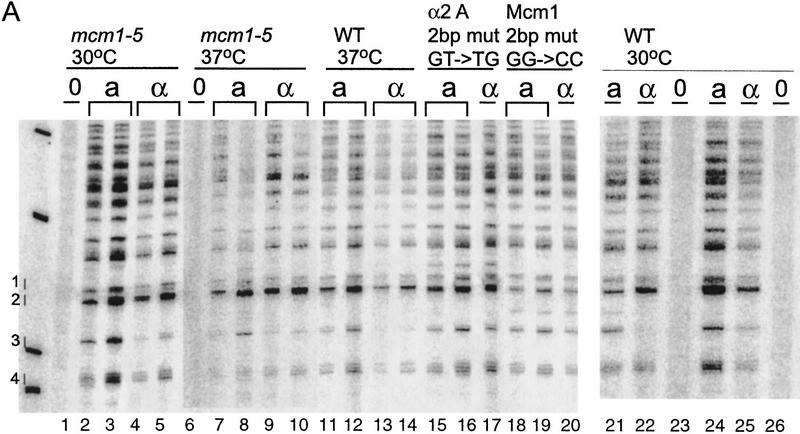
The site-directed mcm1-5 mutation, R87A, was shown to be defective in its ability repress an a-specific reporter gene in MATα cells (M.A. Harris and B.-K. Tye, unpubl.). Residues near this site have been implicated in other Mcm1 protein interactions (Mueller and Nordheim 1991; Primig et al. 1991) and the recent crystal structure of Matα2/Mcm1/DNA ternary complex shows that this residue is in close proximity to Matα2 (Tan and Richmond 1998). mcm1-5 MATα cells are sterile at 37°C, apparently because a-specific genes are expressed and/or because α-specific gene expression is reduced (Elble and Tye 1991; Bruhn et al. 1994). The 2-bp mcm1-5 mutation was introduced into strains CWU116 (HMLα MATa HMRα-B GAL::HO) and CWU117 (HMRa–BglII(X) MATα-B HMRa GAL::HO) and assayed for its effect on MAT switching at 37°C, as described previously (Wu and Haber 1996). HO endonuclease induction was normal and >75% of cells switched. The mcm1-5 MATa cells used HML only 50% of the time, whereas mcm1-5 MATα cells used HML 30% of the time. Wild-type cells showed no change at 37°C, using HML >85% of the time in MATa and <15% in MATα. Thus, the mcm1-5 mutant reduces donor selectivity in both MATa and MATα cells. Qualitatively, the results are similar to what we observed for MATa cells lacking the Mcm1 DNA binding site and for MATα cells with a reduced ability to bind the Matα–Mcm1 repressor. These results strongly suggest that it is the Mcm1 protein itself and not some other protein that competes for an overlapping site that activates RE in MATa cells.
The chromatin of the RE was analyzed in the mcm1-5 mutant strains grown at 37°C (Fig. 5). In MATα, nucleosomes are positioned as in wild-type MATα cells. However, a slight increase in cutting within nucleosome-protected regions can be noticed. A more fragile chromatin structure could be the result of a defect in the binding ability of the mutated Mcm1 protein or to the effect of higher temperature on nucleosome stability. The structure in mcm1-5 MATa cells is intermediate between wild-type MATa and MATα chromatin. This differs from the chromatin structure of the mutated Mcm1 binding site in the RE, which is much more similar to the wild-type MATα RE structure (see above), or to the mutation of Matα2 binding site A, which resembles wild-type MATa structure. At 37°C, the mcm1-5 mutation causes the hypersensitive region to be less pronounced. In addition, certain sites are identical in MATa and MATα and correlate with nucleosome linker regions. Possibly, nucleosomes are slightly disrupted in MATa, but not sufficiently disturbed to allow much RE activation.
Figure 5.
Chromatin structure of the S. cerevisiae 753-bp RE fragment in the mcm1-5 mutant strain grown at 37°C mapped by primer extension analysis of micrococcal nuclease cleavage sites using primer a290. MATa and MATα cells (CWU116 and CWU117) are as indicated. Extensions of undigested chromatin (0) and chromatin digested using two or three levels of micrococcal nuclease are presented. (D) Protein-free DNA digests. Coordinates refer to the published sequence of chromosome III. The Mcm1–Matα2 binding site (α2) is represented by a shaded box. DNA regions containing TTT(A/G) repeats are depicted in open boxes.
Finally, we assayed protein binding to the RE by UV photofootprinting in vivo. Protein binding to Matα2–Mcm1 operator was determined by comparing the formation of thymidine dimers in mutants with the pattern seen in wild-type cells (Fig. 6). By the same means, we examined the effect of the mcm1-5 mutant protein in protein binding to wild-type RE sequences. As shown previously (Murphy et al. 1993), three sites inside the Matα2–Mcm1 operator are subject to thymidine dimer formation. In wild-type cells, the three thymidine repeat (site 3) in Matα2 binding site B is more strongly protected in MATα than in MATa (lane 21 vs. lane 22) This site serves as an indicator of Matα2 protein binding. As expected, in the Matα2 binding site A mutant, site 3 becomes equally modified in MATa and MATα cells, indicating that Matα2 protein binding is impaired (lane 16 vs. lane 17). There is a small effect of the mcm1-5 mutation at this site, at both 30°C and 37°C.
Figure 6.
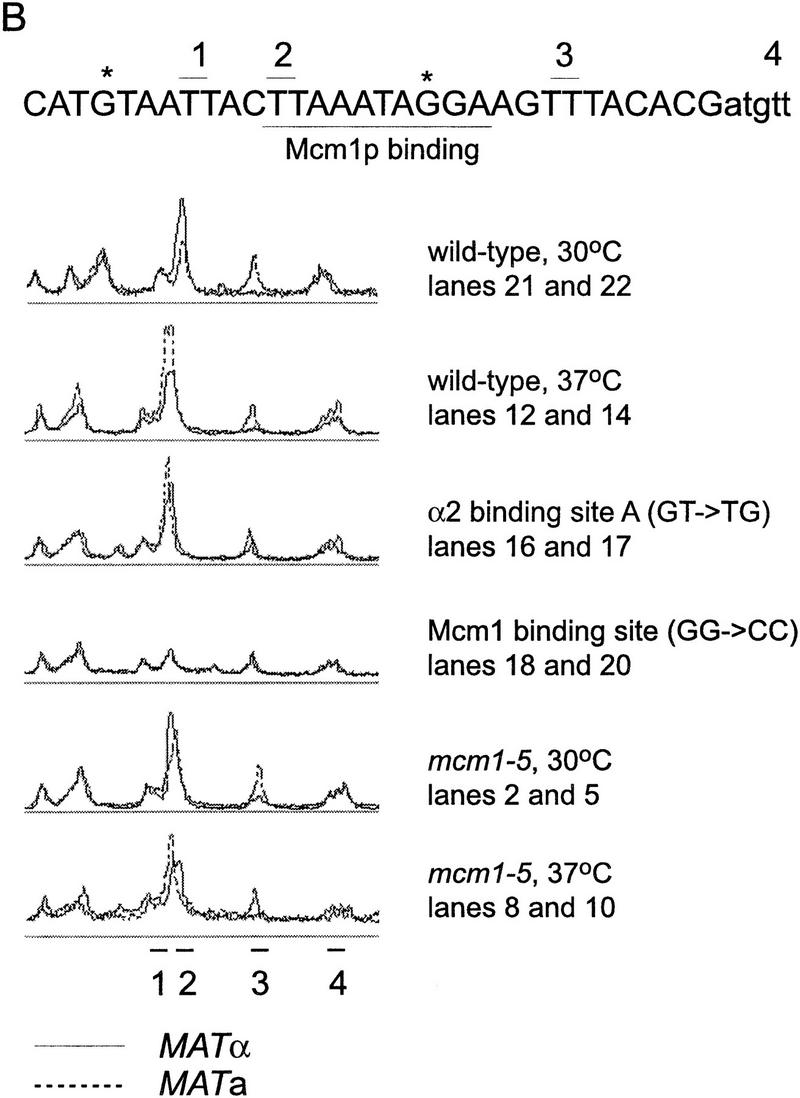
UV photofootprint of the wild-type and mutant Matα2–Mcm1 binding site. α2 operator (29194–29224) occupancy was assessed by primer extension analysis of UV-induced thymidine dimers using primer b294. (B) Scans of the operator region directly comparing levels of modifications in MATa and MATα cells in wild-type and selected mutants. The different strains and the temperature of culture are as indicated: mcm1-5 is a Mcm1 protein mutant with a more severe mating defect at 37°C (CWU116, CWU117); α2mut(GT-TG) carries a mutation in Matα2 binding site A indicated (*) next to the sequence (CWU51, CWU54); Mcm1 binding site mut (GG-CC) carries a mutation in the Mcm1 binding site indicated (*) next to the sequence (CWU57, CWU94). (O) Non-UV-treated chromatin to control for DNA degradation and primer extension artifacts. MATa and MATα cells are as indicated (a and α). (1,2,3) Modifiable thymidine pairs inside the operator sequence. Site 4 flanks the operator. The Mcm1 recognition sequence is underlined. Solid lines correspond to scans of MATα photofootprints; broken lines to MATa scans. Data are taken from two identical, but independent experiments.
The level of modification of site 2 is a good indicator of Mcm1 binding. Mcm1 is a MADS box protein that bends the DNA of its recognition sequence such that the thymidine dimer of site 2 is in an extended major groove away from the protein–DNA contact (Acton et al. 1997), a situation that favors pyrimidine dimer formation (Pehrson and Cohen 1992). In wild-type cells, this site is apparently occupied in both MATa and MATα. In the Matα2 binding site A mutant, occupancy of Mcm1 is essentially normal, supporting the idea that Mcm1 binds to this site in the absence of Matα2 (in MATa cells) or when Matα2 cannot bind. This is consistent with the activation of HML usage in MATα cells with the Matα2 binding site mutation. In mcm1-5 strains at 37°C, there is not much change relative to the wild type, suggesting that the mutant Mcm1-5 protein may be able to bind, but not interact properly with other proteins that interact with RE.
Discussion
The results we present here represent a major advance in understanding the structure and activity of the donor preference RE in Saccharomyces. First, we show that this sequence is functionally conserved in another Saccharomyces species, thus defining a region of only 244 bp that harbors the essential sequences for RE activity. Three of four highly conserved subregions are essential for RE activity. Surprisingly, even within this small region, the sequence orientation of some elements can be inverted with no deleterious effect.
Second, we confirm and extend previous observations (Szeto et al. 1997) that the absence of Matα2 binding allows HML usage in a MATα strain by removing the ordered nucleosomal positioning that appears to repress RE activity. Although the alteration of one of the two Matα2 binding sites is sufficient to abolish the ordered chromatin structure of this region and create a MATa-like pattern, it only partially relieves Matα2 repression. This suggests that, with a single Matα2 binding site, the RE may exist in a mixed population of both MATa and MATα chromatin structures. When both Matα2 binding sites are mutated, usage of HML is 55% compared with 75% for MATa cells. The remaining preference for MATa versus MATα could suggest the action of an a-specific gene, which would still be repressed in MATα cells. The difference could also be caused by a change in the amount of free Mcm1 that can associate with its activation site when no Matα2 protein is present.
Third, and most significant, we show that a 2-bp mutation abolishes RE activity on an otherwise unaltered chromosome III. This strongly suggests that Mcm1 is itself the essential activator of MATa donor preference, though one could not entirely rule out that some other protein occupies this site in MATa cells. However, the fact that a single amino acid substitution in the Mcm1 protein itself causes a reduction in HML usage in MATa cells leads us to conclude that Mcm1 is an essential activator of the RE. Previous studies have found that Mcm1 protein is a transcriptional activator of both a- and α-specific genes as well as of other, essential genes (Elble and Tye 1991; Althoefer et al. 1995; McInerny et al. 1997). In MATα cells, the Matα1 protein acts as a coactivator with Mcm1 protein. Whether Mcm1 has a coactivator for a-specific genes has not yet been established. We note that the sequences necessary for Mcm1’s activation of a-specific genes are apparently only 14 bp (Elble and Tye 1991) compared with the 244 bp of the RE operator needed to activate HML in a MATa cell.
Previously, Szeto et al. (1997) showed that the RE could act as a transcriptional activator when placed in front of a reporter gene and that this activity depended on the Matα2–Mcm1 binding site. They suggested that RE’s activation of HML might depend on two apparently sterile (noncoding) transcripts of 500 and 800 bp that extend rightward from subregion D. Although the Mcm1 binding and the act of transcription may be a key feature of RE regulation, it seems unlikely that the nucleotide sequence of the transcript is significant. First, the transcribed region is nearly 50% diverged in REcer and REcarl. Second, both of the two transcripts extend much further than the border of the 270-bp minimum REcer that we have defined. In fact, the smaller transcript begins outside of the 270 (or 244) minimum RE we have defined and the larger would begin within the D region, which can be inverted relative to regions A and B. Nevertheless, binding of transcription factors and initiation of transcription might be an important aspect of RE activation. Mcm1 protein could remodel or disrupt the repressed, inactive RE, thus allowing other proteins access to binding sites in other regions of the RE.
Nucleosome positioning in the RE
The fourth, very surprising result is that a very ordered nucleosome organization is found when the Mcm1 binding site is ablated, even in the absence of Matα2. However, we note that nucleosomes are more precisely positioned in a wild-type MATα cell than in MATa or MATα cells with the 2-bp Mcm1 binding site mutation. In addition, in the wild-type RE, the nucleosome flanking the operator centromere proximal is 155-bp long, whereas the protected region in the Mcm1 binding site mutant extends over 190 bp. The second nucleosome adopts an identical position in both wild-type and mutant RE. This argues that other sequences of the RE may have the capacity to impart nucleosome positioning. Other, unidentified proteins may bind to the RE (and to the mutated Matα2–Mcm1 operator) and contribute to nucleosome phasing. It is possible that some of the important organizing sequences lie within the Matα2–Mcm1 operator region itself, because a 31-bp deletion of the entire operator (Szeto et al. 1997; C. Wu et al., unpubl.) has a much less profound effect on donor preference than does the 2-bp mutation of the Mcm1 binding site. We suggest that the Matα2–Mcm1 repressor complex plays a role in locking in the highly ordered nucleosome structure that is found in the wild-type RE in MATα cells. Inactivation would then be further stabilized by binding of the corepressor protein Tup1 (Szeto and Broach 1997).
A complete understanding of the action of RE to promote recombination on the left arm of chromosome III will depend on solving a number of riddles. What makes the region so cold in MATα cells or in MATa when the RE is deleted? How does the RE reverse this inactivation, which extends over 100 kb? Is it chromosome III specific? What trans-acting gene products are required both to activate and inactivate this region? And finally, what is the relation of this remarkably complex mechanism to other examples of regulation at a distance of gene expression, and even the initiation of DNA replication in other organisms (Kim et al. 1992; Capone et al. 1993; Bone et al. 1994; Brown and Willard 1994; Chuang et al. 1994; Aladjem et al. 1995).
Materials and methods
Strains
All strains were derived from DBY745 (HMLα MATα HMRa ade1 ura3-52 leu2-3,112). Strains used to monitor MATa donor preference carried HMRα-B, containing a single base pair substitution that creates a BamHI site (Wu and Haber 1995, 1996). In all the strains used in these experiments, a galactose-inducible HO endonuclease gene (Jensen and Herskowitz 1984) was inserted at the ADE3 locus (Sandell and Zakian 1993). Internal chromosomal deletions were accomplished by gene replacement of linearized DNA fragments (Rothstein 1983; Surosky and Tye 1985; Scheistl and Geitz 1989), which contained DNA homologous to two sites on chromosome III flanking a selectable marker such as URA3. These linearized fragments were constructed as described previously (Wu and Haber 1996).
Reintroduction of subfragments into a deletion of the recombination enhancer
Plasmids pXW279 and pCWU155, each of which contains two PCR fragments, each of ∼500 bp, that define the boundaries of either a 2.5-kb (nucleotides 28904–31409) or a 1.8-kb (nucleotides 29031–30878) deletion of the recombination enhancer, replaced by a LEU2 gene. A unique SalI site at the junction of the left end of the deletion and the LEU2 insert provides a site into which PCR-amplified subregions could be inserted by virtue of SalI or XhoI sites included in the PCR primers. The modified deletion was then liberated from pXW279 or pCWU155 derivative by digestion at BamHI and HindIII sites that border the entire construct, and the fragment was then transformed into a strain carrying a URA3-marked 2.5-kb or 1.8-kb deletion. Leu+ Ura− transformants were recovered.
Site-directed mutagenesis
Point mutations were introduced into the recombination enhancer by sequential PCR steps. Synthetic oligonucleotides were designed to incorporate a 2-bp change at one end of the two amplified fragments. Following first PCR, the two fragments encompassing the mutation were annealed with each other and extended by mutually primed synthesis. This fragment was then amplified by another PCR step. PCR products were subcloned into pXW279. The modified RE was then liberated from pXW279 and transformed into a strain of a 2.5-kb deletion. All point mutations were confirmed by sequencing. For the GG > CC introduced in chromosome III, the fragment was inserted into the SalI site of pJH18 (YIp30). The derivative plasmid (pCWU190) was then digested with BstXI to target integration to the RE locus and transformed into XW652. Transformants were counter-selected on media containing 5-FOA (Boeke et al. 1987) to identify cells retaining a single copy of RE. RE with the mutation was identified by sequencing.
The mcm1-5 mutation was constructed by site-directed mutagenesis with an oligonucleotide:5′-ACGCAGCAGG(A/C)AGGTGCAAACCTGATC-3′. Oligonucleotide-mediated site-specific mutagenesis was carried out by use of the Mut-a-Gene kit from Bio-Rad. The template plasmid pMH1 was constructed by adding the yeast selectable marker URA3 to KS–MCM1–NdeI (C. Christ and B.-K. Tye, unpubl.). A 1.5-kb BamHI–SpeI fragment bearing URA3 was isolated from YEp24 and ligated to KS–MCM1–NdeI digested with BamHI and XbaI. Escherichia coli strain CJ236 was transformed with pMH1 and single-stranded DNA containing uracil residues was isolated by infection with helper phage CVSM13. The presence of the GC mutation in codon 87 and the absence of a mutation at codon 85 was confirmed by double-strand dideoxy sequencing. The plasmid containing the mcm1-5 allele was digested with SphI to target integration to the MCM1 locus and transformed into XW643b(α). Single colony transformants were counterselected on medium containing 5-FOA to identify cells retaining a single copy of MCM1. Isolates retaining the mcm1-5 allele were identified by assaying mating at 37°C, a temperature restrictive for mating in mcm1-5 cells. The derivative strain CWU117 (MATα mcm1-5) was switched to MATa, to get isogenic CWU116 (MATa mcm1-5).
Analysis of S. carlsbergensis DNA
A library of BamHI fragments prepared from gel-purified S. carlsbergensis chromosome III was constructed in the vector pRS306. This library was screened by hybridization at low stringency with the 5.2-kb S. cerevisiae BamHI fragment (D10B, Newlon et al. 1991) that contains the RE and ARS304. The screen identified a plasmid, 8BH2, carrying a 6.7-kb S. carlsbergensis BamHI fragment. The ends of the fragment were sequenced and shown to have strong homology to portions of the YCL056 and YCL052 ORFs of S. cerevisiae. The RE of S. cerevisiae lies between YCL055 and ARS304. To determine the DNA sequence of the corresponding region in clone 8BH2, primers were designed beginning centromere distal to the EcoRI fragment that contains S. carlsbergensis ARS304 (C. Yang and C.S. Newlon, in prep.). Nine primers (sequences available upon request) were used to sequence both strands of a 1061-bp fragment shown in Figure 2A that extended from the edge of the EcoRI fragment into the YCL055 ORF.
Measurement of donor preference
MATa cells carrying HMLα and HMRα-B, or MATα cells carrying HMRα–BglII(X) and HMRa, or MATα-B cells carrying HMLα and HMRa, were induced to switch mating type by adding 2% galactose to cells grown in YEP–lactate medium, thus expressing a galactose-inducible HO gene (Wu and Haber 1995, 1996). After 1.5 hr, cells were washed and either plated or grown for a longer period of time in YEPD. When individual colonies were assayed, 40 MATα (or MATα-B), or 20 MATa [or MATα–BglII(X)] colonies resulting from switching were analyzed by Southern blot, as described previously (Wu and Haber 1995, 1996). The proportion of cells switching mating type was also analyzed by examining the entire population of cells. For MATa cells switching to MATα or MATα-B, DNA was digested with HindIII and BamHI, and probed with a Yα-specific probe. For MATα cells switching to MATa or MATα–BglII(X), DNA was digested with HindIII and BglII, and probed with a Ya-specific probe. For MATα-B cells switching to MATa or MATα, DNA was digested with BamHI and StyI, and probed with a MAT distal probe. The proportion of fragments corresponding to the switch products was then analyzed as described previously (Wu and Haber 1995, 1996).
Nuclei isolation, micrococcal nuclease digestion, DNA purification, and multiple cycle primer extension
Yeast were routinely grown in YEPD at 30°C to mid-log phase (OD600∼1). mcm1-5 mutant strains CWU116 and 117 were grown at 37°C to mid-log phase (OD600 = 0.8). Cells grown at 37°C were subsequently treated with 0.015% sodium azide for 10 min. Nuclei were isolated, digested with micrococcal nuclease or DNaseI (Worthington), and DNA was purified as described (Szent-Gyorgyi and Isenberg 1983; Roth and Simpson 1991) with modifications detailed in Weiss and Simpson (1997). Naked DNA controls were obtained by digesting a PCR product. About 3 kb of the sequences including the RE constructs were amplified by use of oligonucleotides a288 and b318 (see below) as primers. Approximately 100 ng of the PCR product was digested with 0.5 U/ml MNase or 0.05 U/ml DNaseI at 37°C for 3 min in the presence of 36 μg carrier DNA (calf thymus). After ethanol precipitation, DNA was resuspended in 50 μl of 0.1X TE.
MNase and DNaseI cleavage sites were located by primer extension assays with Taq polymerase as described (Shimizu et al. 1991) with minor modifications (Weiss and Simpson 1997). Oligonucleotides used as primers include [coordinates are base pair positions in the published sequence of S. cerevisiae chromosome III (Oliver et al. 1992)]: a288, 28766–28789; a290, 20031–29053; a292, 29242–29264; b294, 29376–29352; b297, 29710–29687; b318, 31829–31796. Primer I11 contains bases 905–804 of the S. carlsbergensis sequence shown in Figure 2.
UV photofootprinting
Yeast were grown in 10 ml YEPD at 30°C overnight, then reinoculated in 5 ml fresh medium and allowed to reach mid-log phase (OD600 = 1.5; 3 hr). Cells were pelleted and resuspended in 15 ml ice cold UV resuspension buffer (0.2 m NaCl, 2.7 mm KCl, 15.3 mm Na2HPO4, 1.5 mm KH2PO4, 0.7 mm CaCl2, and 0.5 mm MgCl2). Five milliliter samples were UV irradiated on ice in a Stratalinker for 0, 6, or 24 sec at 5 cm from the light source. Cells were pelleted, resuspended in 1 ml Sorbitol buffer (see nuclei preparation) and treated with 0.5 mg/ml zymolyase 100T (Seikagaku) for 90 min. Cells were pelleted, resuspended in 500 μl TE, vigorously mixed with a vortex and treated with 100 ng Proteinase K in 2% Sarkosyl, and 200 mm NaClO4 at 50°C for 1 hr. Two hundred microliters of 5 m potassium acetate was added, and precipitation was allowed to proceed on ice overnight. The precipitate was pelleted by centrifugation at 14,000 rpm for 5 min. Nucleic acids were precipitated by adding 700 μl isopropanol to the supernatant, followed by centrifugation. The pellet was washed with 70% ethanol and resuspended in 250 μl TE. DNA was purified by the following extractions (1/1 vol/vol): phenol/chloroform/isoamyl alcohol (25:24:1); chloroform/isoamyl alcohol (24:1); isobutanol; ether. After precipitation, nucleic acid was resuspended in 15 μl 0.1X TE for primer extension analysis (see above).
Acknowledgments
Research was supported by National Institutes of Health grants GM20056 to J.E.H., GM 52311 to R.T.S., GM35679 to C.S.N., and GM34190 to B.-K.T. Partial support for C.Y. was provided by a fellowship from UMDNJ–Graduate School of Biomedical Sciences. M.A.H. was supported by a predoctoral fellowship from the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL haber@hydra.rose.brandeis.edu; FAX (781) 736-2405.
References
- Acton TB, Zhong H, Vershon AK. DNA-binding specificity of Mcm1: Operator mutations that alter DNA-bending and transcriptional activities by a MADS box protein. Mol Cell Biol. 1997;17:188–1889. doi: 10.1128/mcb.17.4.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aladjem MI, Groudine M, Brody LL, Dieken ES, Fournier RE, Wahl GM, Epner EM. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- Althoefer H, Schleiffer A, Wassmann K, Nordheim A, Ammerer G. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5917–5928. doi: 10.1128/mcb.15.11.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Lacroute F, Fink GR. A postitive selection for mutants lacking orotidine 5′-phosphate decarboxylase activity in yeast. Mol & Gen Genet. 1987;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bone JR, Lavender J, Richman R, Palmer MJ, Turner BM, Kuroda MI. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes & Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Willard HF. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Capone M, Watrin F, Fernex C, Horvat B, Krippl B, Wu L, Scollay R, Ferrier P. TCR beta and TCR alpha gene enhancers confer tissue- and stage- specificity on V(D)J recombination events. EMBO J. 1993;12:4335–4346. doi: 10.1002/j.1460-2075.1993.tb06118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn L, Sprague GJ. MCM1 point mutants deficient in expression of alpha-specific genes: Residues important for interaction with alpha 1. Mol Cell Biol. 1994;14:2534–2544. doi: 10.1128/mcb.14.4.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone M, Watrin F, Fernex C, Horvat B, Krippl B, Wu L, Scollay R, Ferrier P. TCR beta and TCR alpha gene enhancers confer tissue- and stage- specificity on V(D)J recombination events. EMBO J. 1993;12:4335–4346. doi: 10.1002/j.1460-2075.1993.tb06118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang P-T, Albertson DG, Myer BJ. DPY-27: A chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–474. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- Elble R, Tye BK. Both activation and repression of a-mating-type-specific genes in yeast require transcription factor Mcm1. Proc Natl Acad Sci. 1991;88:10966–10970. doi: 10.1073/pnas.88.23.10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerring SL, Spencer F, Hieter P. The CHL1 (CTF1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990;9:4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Trends Genet. 1992;8:446–452. doi: 10.1016/0168-9525(92)90329-3. [DOI] [PubMed] [Google Scholar]
- Jensen R, Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harbor Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- Kim CG, Epner EM, Forrester WC, Groudine M. Inactivation of the human beta-globin gene by targeted insertion into the beta-globin locus control region. Genes & Dev. 1992;6:928–938. doi: 10.1101/gad.6.6.928. [DOI] [PubMed] [Google Scholar]
- Klar AJ, Hicks JB, Strathern JN. Directionality of yeast mating-type interconversion. Cell. 1982;28:551–561. doi: 10.1016/0092-8674(82)90210-0. [DOI] [PubMed] [Google Scholar]
- Laurenson P, Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liras P, McCusker JH, Mascioli S, Haber JE. Characterization of a mutation in yeast causing non-random chromosome loss during mitosis. Genetics. 1978;88:651–671. [PMC free article] [PubMed] [Google Scholar]
- Loo S, Laurenson P, Foss M, Dillin A, Rine J. Roles of ABF1, NPL3, and YCL54 in silencing in Saccharomyces cerevisiae. Genetics. 1995;141:889–902. doi: 10.1093/genetics/141.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerny CJ, Partridge JF, Mikesell GE, Creemer DP, Breeden LL. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes & Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- Mueller CG, Nordheim A. A protein domain conserved between yeast MCM1 and human SRF directs ternary complex formation. EMBO J. 1991;10:4219–4229. doi: 10.1002/j.1460-2075.1991.tb05000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MR, Shimizu M, Roth SY, Dranginis AM, Simpson RT. DNA-protein interactions at the S. cerevisiae alpha 2 operator in vivo. Nucleic Acids Res. 1993;21:3295–3300. doi: 10.1093/nar/21.14.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon CS, Lipchitz L R, Collins I, Deshpande A, Devenish RJ, Green RP, Klein HL, Palzkill TG, Ren RB, Synn S, et al. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: A physical map and identification and location of ARS elements. Genetics. 1991;129:343–357. doi: 10.1093/genetics/129.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SG, van der Aart QJ, Agostoni CM, Aigle M, Alberghina L, Alexandraki D, Antoine G, Anwar R, Ballesta JP, Benit P, et al. The complete DNA sequence of yeast chromosome III. Nature. 1992;257:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Pehrson JR, Cohen LH. Effects of DNA looping on pyrimidine dimer formation. Nucleic Acids Res. 1992;20:1321–1324. doi: 10.1093/nar/20.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M, Winkler H, Ammerer G. The DNA binding and oligomerization domain of MCM1 is sufficient for its interaction with other regulatory proteins. EMBO J. 1991;10:4209–4218. doi: 10.1002/j.1460-2075.1991.tb04999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SY, Simpson RT. Yeast minichromosomes. Methods Cell Biol. 1991;35:289–314. [PubMed] [Google Scholar]
- Rothstein, R.J. 1983. One-step gene disruption in yeast. Methods Enzymol. 202–211. [DOI] [PubMed]
- Sandell LL, Zakian VA. Loss of a yeast telomere: Arrest, recovery and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Roth SY, Szent GC, Simpson RT. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 1991;10:3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- Smith DL, Johnson AD. Operator-constitutive mutations in a DNA sequence recognized by a yeast homeodomain. EMBO J. 1994;13:2378–2387. doi: 10.1002/j.1460-2075.1994.tb06521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN. Control and execution of mating type switching in Saccharomyces cerevisiae. In: Kucherlapati R, Smith GR, editors. Genetic recombination. Washington, D.C.: American Society Microbiology; 1989. pp. 445–464. [Google Scholar]
- Surosky RT, Tye B-K. Site-directed chromosomal rearrangements in yeast. Methods Enzymol. 1987;153:243–253. doi: 10.1016/0076-6879(87)53057-9. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi c, Isenberg I. The organization of oligonucleosomes in yeast. Nucleic Acids Res. 1983;11:3717–3736. doi: 10.1093/nar/11.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto L, Broach JR. Role of alpha2 protein in donor locus selection during mating type interconversion. Mol Cell Biol. 1997;17:751–759. doi: 10.1128/mcb.17.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto L, Fafalios MK, Zhong H, Vershon AK, Broach JR. Alpha2p controls donor preference during mating type interconversion in yeast by inactivating a recombinational enhancer of chromosome III. Genes & Dev. 1997;11:1899–1911. doi: 10.1101/gad.11.15.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Richmond TJ. Crystal structure of the yeast MATα2/MCM1/DNA ternary complex. Nature. 1998;391:660–666. doi: 10.1038/35563. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Oshima T, Araki H, Harashima S, Oshima Y. Mating type control in Saccharomyces cerevisiae: A frameshift mutation at the common DNA sequence, X, of the HMLα locus. Mol Cell Biol. 1984;4:203–211. doi: 10.1128/mcb.4.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler KS, Broach JR. Donor locus selection during Saccharomyces cerevisiae mating type interconversion responds to distant regulatory signals. Genetics. 1992;132:929–942. doi: 10.1093/genetics/132.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler KS, Szeto L, Broach JR. Mutations affecting donor preference during mating-type switching in Saccharomyces cerevisiae. Genetics. 1995;139:1495–1510. doi: 10.1093/genetics/139.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Simpson RT. Cell type-specific chromatin organization of the region that governs directionality of yeast mating type switching. EMBO J. 1997;16:4352–4360. doi: 10.1093/emboj/16.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Haber JE. MATa donor preference in yeast mating-type switching: Activation of a large chromosomal region for recombination. Genes & Dev. 1995;9:1922–1932. doi: 10.1101/gad.9.15.1922. [DOI] [PubMed] [Google Scholar]
- ————— A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell. 1996;87:277–285. doi: 10.1016/s0092-8674(00)81345-8. [DOI] [PubMed] [Google Scholar]
- Wu X, Moore JK, Haber JE. Mechanism of MATα donor preference during mating-type switching of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:657–668. doi: 10.1128/mcb.16.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wu C, Haber JE. Rules of donor preference in Saccharomyces mating-type gene switching revealed by a competition assay involving two types of recombination. Genetics. 1997;147:399–407. doi: 10.1093/genetics/147.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]