Abstract
Histone modification has been implicated in the regulation of mammalian spermatogenesis. However, the association of differently modified histone H3 with a specific stage of germ cells during spermatogenesis is not fully understood. In this study, we examined the localization of variously modified histone H3 in paraffin-embedded sections of adult mouse testis immunohistochemically, focusing on acetylation at lysine 9 (H3K9ac), lysine 18 (H3K18ac), and lysine 23 (H3K23ac); tri-methylation at lysine 4 (H3K4me3) and lysine 27 (H3K27me3); and phosphorylation at serine 10 (H3S10phos). As a result, we found that there was a significant fluctuation in the modifications; in spermatogonia, the stainings for H3K9ac, H3K18ac, and H3K23ac were strong while that for H3K4me3 was weak. In spermatocytes, the stainings for H3K9ac, H3K18ac, H3K23ac, and H3K4me3 were reduced in the preleptotene to pachytene stage, but in diplotene stage the stainings for H3K18ac, H3K23ac, and H3K4me3 seemed to become intense again. The staining for H3K27me3 was nearly constant throughout these stages. In the ensuing spermiogenesis, a dramatic acetylation and methylation of histone H3 was found in the early elongated spermatids and then almost all signals disappeared in the late elongated spermatids, in parallel with the replacement from histones to protamines. In addition, we confirmed that the staining of histone H3S10phos was exclusively associated with mitotic and meiotic cell division. Based upon the above results, we indicated that the modification pattern of histone H3 is subject to dynamic change and specific to a certain stage of germ cell differentiation during mouse spermatogenesis.
Keywords: immunohistochemistry, histone H3 modification, epigenetics, spermatogenesis, mouse
I. Introduction
Histone modification is one of the major epigenetic parameters. Histone H3, a member of core histones (H2A, H2B, H3 and H4), is subject to various covalent modifications, including acetylation, methylation and phosphorylation [5]. All of these modifications have a substantial influence on gene expression as well as chromatin structure, the roles of which seem to be different depending on the type of modification and the location of the modified amino acid. Particularly, acetylation of histone H3 at lysine 18 has been known to be correlated with active transcription and was actually found at the 5' ends of transcribed sequences in human and mouse cells [2]. Moreover, knocking-out histone H3 lysine 9 specific histone methyltransferases (Suv39h HMTases) brought about chromosomal instabilities which are often associated with an increased tumor risk, and severely impaired viability of mouse embryo [23]. More importantly, tri-methylation of histone H3 at lysine 4 and lysine 27 were known to switch-on or -off the transcription of specific genes, respectively. Furthermore, as predicted by “histone code” hypothesis, a pattern of histone modification, which might be evolutionarily conserved, was involved in the expression of a distinct set of genes structurally [29]. For instance, methylation of histones H3 at lysine 9 [12] and phosphorylation of histone H3 at serine 10 [8] were required for faithful chromosome segregation in mitosis, presumably by preserving the more compacted chromatin structure of pericentromeric heterochromatin.
Mammalian spermatogenesis is a unique process with successive differentiation, consisting of spermatogonial mitosis, spermatocytic meiosis and spermiogenesis [24]. Recently, several investigators have reported the critical roles of histone modification in spermatogenesis [13, 15, 26]. The knock-out of histone H3 lysine 9 demethyltransferase (Jmjd1a) caused extensive germ cell apoptosis and disturbed the formation of elongated spermatids, resulting in severely shrunken testes and infertility in mice [19]. The XY body, which is condensed from X and Y chromosomes during meiosis, also revealed characteristic changes in histone modifications including deacetylation of histone H3 and methylation of H3K9 during pachytene stage [13]. Although the pattern of histone H3 modification may be important to better understand the involvement of epigenetic regulation in spermatogenesis, our knowledge of it has been limited.
In the present study, we examined the modification pattern of histone H3 in various stages of spermatogenic cells in mice immunohistochemically, using specific antibodies against H3K9ac, H3K18ac, H3K23ac, H3K4me3, H3K27me3, and H3S10phos. Consequently, we found differentiation stage-specific patterns of the modifications in spermatogenesis. In addition, we showed here that phosphorylation of histone H3S10 was closely associated with not only mitosis, but also meiosis.
II. Materials and Methods
Chemicals and biochemicals
Paraformaldehyde (PFA) was purchased from Merck (Darmstadt, Germany). 3,3'-Diaminobenzidine-4HCl (DAB) was from Dojin Chemical Co. (Kumamoto, Japan). OCT compound was from Sakura Finetek Japan Co., Ltd. (Tokyo, Japan). 3-Aminopropyltriethoxysilane, bovine serum albumin (BSA, minimum 98% pure), and Brij 35 were purchased from Sigma Chemical Co. (St. Louis, MO, USA). All other reagents used in this study were from Wako Pure Chemicals (Osaka, Japan) and were of analytical grade.
Antibodies
Rabbit polyclonal anti-H3 (1:100) specifically detects endogenous levels of total histone H3 protein and it is irrespective of any modification in histone H3. Acetylation, tri-methylation and phosphorylation of histone H3 at certain residues of histone H3 were detected by rabbit polyclonal anti-H3K9ac (1:50), anti-H3K18ac (1:100), anti-H3K23ac (1:25), anti-H3K4me3 (1:100), anti-H3K27me3 (1:100) and anti-H3S10phos (1:100), respectively. All of above antibodies were obtained from Cell Signaling Technology (Beverly, MA, USA). Their specificities have been determined by the company and they do not cross-react with other histone modifications. HRP-conjugated goat anti-rabbit IgG (1:200) were from Chemicon International Inc. (Temecula, CA, USA). Normal rabbit IgG and normal goat IgG were purchased from Sigma Chemical Co. (St. Louis, MO).
Animals and tissue preparation
Male ICR mice weighing 26–30 g (7 to 8-week-old) were used in the present study. All experiments were conducted according to the principles and procedures outlined in the Guidelines for Animal Experimentation of Nagasaki University with the approval of the Institutional Animal Care and Use Committee (#1004010843). For tissue sampling, mice were killed by cervical dislocation. Testes were fixed with 4% PFA in phosphate buffered saline (PBS), pH 7.4, and then embedded in paraffin according to a routine protocol. Sections (5-µm-thick) were prepared and then mounted on 3-aminopropyltriethoxysilane-coated glass slides and used for immunohistochemistry [10, 30, 31].
Immunohistochemistry
Immunohistochemistry was performed with the indirect enzyme-labelled antibody method, as described previously [1, 17, 27]. For the detection of histone H3, H3K4me3, H3K9ac, H3K18, H3K23ac, H3K27me3, and H3S10phos, paraffin sections of mouse testis were deparaffinized with toluene and dehydrated in serially graded ethanol solutions. The sections were autoclaved in a 10 mM citrate buffer (pH 6.0) for 15 min at 120°C. Then, the sections were preincubated with 500 µg/ml normal goat IgG dissolved in 1% BSA in PBS, pH 7.4, for 1 hr. Unless otherwise specified, all reactions were conducted at room temperature. Then the sections were reacted 16 hr with first antibodies in 1% BSA in PBS. After incubation, the sections were washed with 0.075% Brij 35 in PBS three times for 15 min each. The sections were then incubated with HRP-conjugated goat anti-rabbit IgG in 1% BSA in PBS for 1 hr. After washing with 0.075% Brij 35 in PBS three times for 15 min each, the sites of HRP were visualized with DAB and H2O2 in the presence of nickel and cobalt ions [4]. As a negative control, some sections were reacted with normal rabbit IgG instead of the specific antibodies.
III. Results
Immunohistochemical detection of histone H3 in mouse testis
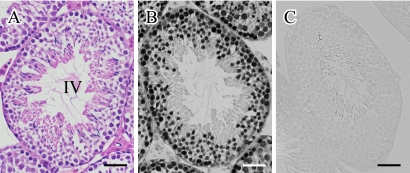
To assess the localization pattern of histone H3 in spermatogenesis, we conducted immunohistochemistry using anti-H3 antibody. As shown in Figure 1, the nuclei of most germ cells were strongly stained and there was no remarkable difference in the intensity of H3 positive germ cells. The staining of histone H3 could not be detected in late elongated spermatids and mature sperms. Sections that were reacted with rabbit IgG as a negative control gave no positive staining (Fig. 1C).
Fig. 1.
Hematoxylin and eosin (H-E) staining (A) and immunohistochemical detection of histone H3 (B) in mirror sections of mouse testis. A stage IV seminiferous tubule is shown. (C) As a negative control, an adjacent section was reacted with normal rabbit IgG instead of histone H3 antibody. Bar=50 µm.
Acetylation of histone H3 at lysine 9, 18 and 23 in mouse testis
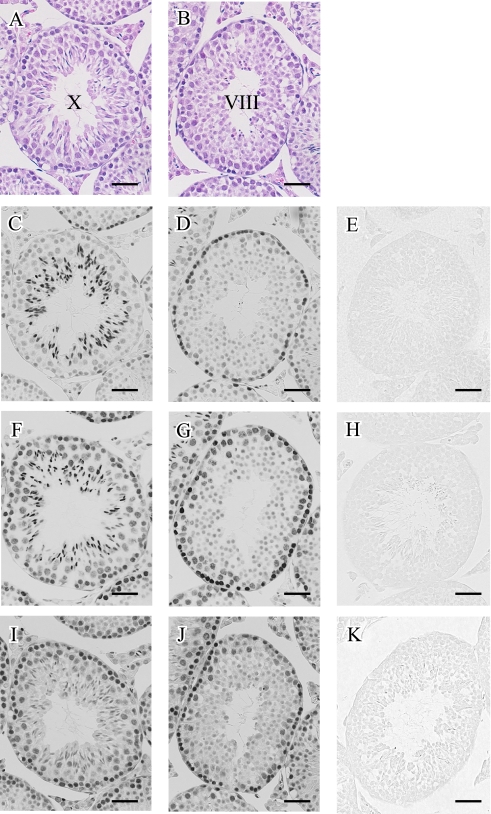
To examine the localization pattern of acetylated histone H3 at lysine 9, 18, and 23, we conducted immunohistochemistry on serial sections of mouse testes (Fig. 2). Although histone H3 in itself was consistently abundant in germ cells, we observed that the localization pattern of acetylated histone H3 was much more heterogenous. As summarized in Table 1, the stainings for H3K9ac, H3K18ac, and H3K23ac were variable among the differentiation stages of germ cells (Table 1). In spermatogonia, H3K9ac, H3K18ac, and H3K23ac were strongly expressed in any of the stages. Then the staining was reduced in pachytene spermatocytes, while the staining for H3K18ac and H3K23ac seemed to be restored in diplotene stages. During the spermiogenesis, the staining intensity for the three differently modified histone H3 reached a peak around step 9 spermatids. None of them were detected in elongated spermatids after step 13. No staining was found in sections that were reacted with rabbit IgG as a negative control (Fig. 2E, H, K).
Fig. 2.
Immunohistochemical detection of acetylated histone H3 at lysine 9, 18 and 23 in paraffin-embedded mouse testis. (A, C, F, I) Serial sections were used for H-E staining (A), immunohistochemical staining for H3K9ac (C), H3K18ac (F), and H3K23ac (I). A stage X seminiferous tubule was shown. (B, D, G, J) Another set of serial sections with a stage VIII seminiferous tubule is shown. (E, H, K) As a negative control, sections adjacent to B were reacted with normal rabbit IgG instead of anti-H3K9ac, anti-H3K18ac, and anti-H3K23ac, respectively. Bar=50 µm.
Table 1.
Distribution pattern of histone H3 and its modified molecules during mouse spermatogenesis
| Spermatogonia | Spermatocytes | Spermatids | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | In+B | L+Z | P I–III | P IV–VI | P VII–X | D+M | Step 1–8 | Step 9–10 | Step 11–12 | Step 13 | Step 14–16 | |||
| H3 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | – | – | ||
| H3K9ac | +++ | +++ | ++ | + | + | + | + | + | +++ | +++ | – | – | ||
| H3K18ac | +++ | +++ | +++ | ++ | ++ | ++ | +++ | + | +++ | +++ | – | – | ||
| H3K23ac | +++ | +++ | ++ | ++ | ++ | ++ | +++ | + | ++ | + | – | – | ||
| H3K4me3 | + | ++ | +++ | + | + | + | +++ | ++ | ++ | + | – | – | ||
| H3K27me3 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ | ++ | – | – | ||
| H3S10phos | +++* | +++* | – | – | – | – | +++ | – | – | – | – | – | ||
A; type A spermatogonia, In+B; intermediate and type B spermatogonia, L+Z; leptotene and zygotene spermatocytes, P; pachytene spermatocytes, D+M; diplotene, meiosis I and II spermatocytes, Sp; spermatids.
Staining intensity: –; negative, +; weak, ++; moderate, and +++; intense.
*; only the cells undergoing mitotic cell division were positive.
Methylation of histone H3 at lysine 4 and 27 in mouse testis
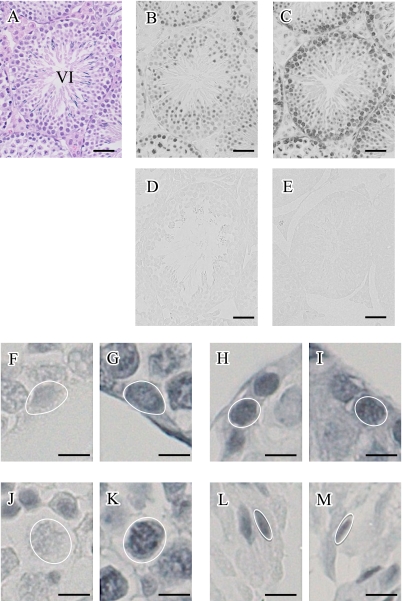
We examined the tri-methylation of histone H3 at lysine 4 and 27 in serial sections of mouse testes. As shown in Figure 3A–C, the staining patterns for H3K4me3 and H3K27me3 were clearly different. Contrary to the constitutive manner of H3K27me3 staining, H3K4me3 revealed much more dynamic staining pattern in mouse testes (Fig. 3B and Table 1). To gain further insight into the stainings for these two methylation modifications, we carefully examined them in mirror sections (Fig. 3F–M). The staining for H3K4me3 was weak in type A spermatogonia (Fig. 3F) and pachytene spermatocytes (Fig. 3J), while the staining was strong in leptotene spermatocytes (Fig. 3H) and spermatids of step 10 (Fig. 3L). On the other hand, the staining for H3K27me3 was almost constant in these cells (Fig. 3G, I, K, M). No staining was found in sections that were reacted with rabbit IgG as a negative control (Fig. 3D and E).
Fig. 3.
Immunohistochemical detection of tri-methylated of histone H3 at lysine 4 and 27 in paraffin-embedded mouse testis. (A–C) Serial sections were used for H-E staining (A), immunohistochemical staining for H3K4me3 (B) and H3K27me3 (C). A stage VI seminiferous tubule is shown. (D and E) As a negative control, adjacent sections were reacted with normal rabbit IgG instead of anti-H3K4me3 and anti-H3K27me3, respectively. (F–M) A set of mirror sections was used for the detection of H3K4me3 and H3K27me3. Identical spermatogonia (F and G), leptotene spermatocytes (H and I), pachytene spermatocytes (J and K) and elongating spermatids (L and M) are shown. Bar=50 µm (A–E) or 10 µm (F–M).
Phosphorylation of histone H3 at serine 10 in mouse testis
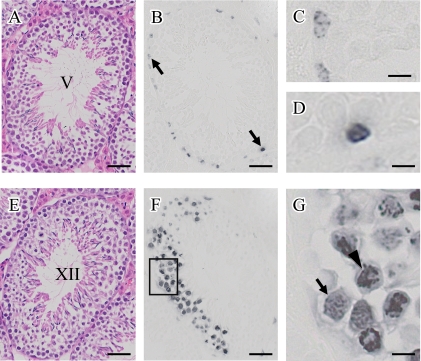
To investigate the phorsphorylation of H3S10 in spermatogenesis, we performed immunohistochemistry in mouse testes. The nuclei of mitotic spermatogonia were positive to H3S10phos (Fig. 4A–D). The phosphorylation of H3S10 began with a small number of discrete foci (Fig. 4C), and then it was expanded throughout the entire nucleus in the later stages (Fig. 4D). Furthermore, H3S10phos was also detected in meiotic spermatocytes (Fig. 4E–G). In stage XII seminiferous tubule, H3S10phos was specifically stained in spermatocytes which underwent meiotic division. H3S10phos was firstly found in diakinesis stage (Fig. 4G, arrow), and then dramatically spread over the chromatin in metaphase (Fig. 4G, arrowhead). Finally, H3S10phos could not be detected in newly formed step 1 spermatids just after second meiotic cell division.
Fig. 4.
H-E staining and immunohistochemical detection of phosphorylated histone H3 at serine 10 in mirror sections of mouse testis (A and B, E and F). (A and B) A stage V seminiferous tubule. (E and F) A stage XII seminiferous tubule. Arrows indicate H3S10phos positive spermatogonia in B. C and D are enlarged from B. (G) The inset of F is enlarged. Bar=50 µm (A, B, E, F) or 10 µm (C, D, G).
IV. Discussion
In the present study, we investigated the distribution of histone H3 and its modified molecules with acetylation (H3K9ac, H3K18ac, H3K23ac), methylation (H3K4me3, H3K27me3) and phosphorylation (H3S10phos) immunohistochemically in adult mouse testis, and we determined the differentiation stage-specific modification patterns of histone H3 in spermatogenic cells. We believe these basic findings are quite useful to better understand the roles of epigenetic factors in the regulation of mammalian spermatogenesis.
Spermatogonia undergo self-renewal to ensure a constant supply of germ cells to form spermatozoa [24]. During the process, many distinct genes have been identified with specific expression profiles and the expression was thought to be influenced by histone modifications [14]. In the case of somatic cells, acetylation of histone H3 is generally linked with active transcription [18, 32, 33], whereas methylated histone H3 at lysine 27 is associated with transcriptionally silent states of genes [20, 28]. Moreover, methylated histone H3 at lysine 4 is known to be closely linked with the transcriptionally active states of genes [25]. In the present study, we detected significant acetylation of histone H3 at lysine 9, 18, and 23 in spermatogonia, which is consistent with active chromatin patterns in somatic cells. However, the staining for methylated histone H3 at lysine 4 and 27 in spermatogonia seems to indicate transcriptionally silent states of genes, raising an apparent controversy. In this context, it is of interest to note that similar conflicting histone patterns were also identified in mouse embryonic stem cells that displayed both active and repressive histone modifications [3]. Considering that spermatogonia in adult mouse may retain the capacity to generate pluripotent cells which are able to differentiate into derivatives of three embryonic germ layers [7], the unique pattern of histone H3 modifications found in the present study may extend our understanding of pluripotency of spermatogonia.
In spermatocytic differentiation, the spermatocytes undergo a long prophase of meiosis, consisting of leptotene, zygotene, pachytene, and diplotene stage, which leads to meiotic cell division. During the differentiation, chromatin becomes continuously condensed and the homologous chromosomes are paired and then exchange DNA segments through a process of homologous recombination. In this stage, most of the genes are transcriptionally inactive [13, 16]. For this inactivation, a few studies reported potential requirements for specific histone modification during meiosis in mouse and Drosophila [11, 13]. In the present study, the signals for H3K9ac, H3K18ac, H3K23ac, and H3K4me3 were significantly reduced from preleptotene to pachytene stage, indicating that genes may be generally transcriptionally inactivated. In addition, the staining intensity for H3K18ac, H3K23ac and H3K4me3 was increased from the stage of diplotene spermatocytes. Although the roles of the modification change are still unknown, these changes should be involved in the progression of meiotic cell division.
In spermiogenesis, a process of postmeiotic metamorphosis of male haploid germ cells, spermatids undergo the extreme condensation of chromatin into sperm head, in which histones are sequentially replaced by protamines [15]. In mouse, histones are firstly replaced with transition proteins, and subsequently with protamine 1 and 2. However, our knowledge about the mechanisms controlling the process of histone-protamine exchange is still rather limited. In the present study, we showed that histone H3 became significantly acetylated at lysine 9, 18, and 23 in spermatids. Since a few in vitro studies proposed that certain histone modifications could facilitate histone-protamine exchange [21, 22], the hyperacetylation of these residues, which is usually related to the relaxation of chromatin structure [6], might be a possible mechanism.
The phosphorylation of histone H3 at serine 10 was already well-known to be correlated with the M-phase in mitosis [9]. In this study, we also confirmed that phosphorylation of histone H3 at serine 10 was strictly associated with meiotic cell division of diplotene spermatocytes under a pattern similar to mitosis. Thus, we believe that the phosphorylation of histone H3 at serine 10 may be essential for the movement of condensed chromosomes during cell division to proceed.
In conclusion, we have shown here the unique pattern of acetylation, methylation, and phosphorylation of histone H3 in male germ cells, which was substantially different from that of somatic cells. Our results may extend our understanding of male germ cell specific epigenetic regulation in the mouse, which would therefore be an excellent source for the investigation of pluripotency, chromatin reorganization, and nucleosome disassembly machinery. However, a precise correlation of these modifications of histone H3 with the chromatin structure in spermatogenic cells remains to be clarified in the future.
V. Acknowledgments
This study was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (No. 18390060 to Koji, T.).
VI. References
- 1.An S., Hishikawa Y., Koji T. Induction of cell death in rat small intestine by ischemia reperfusion: differential roles of Fas/Fas ligand and Bcl-2/Bax systems depending upon cell types. Histochem. Cell Biol. 2005;123:249–261. doi: 10.1007/s00418-005-0765-6. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein B. E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D. K., Huebert D. J., McMahon S., Karlsson E. K., Kulbokas E. J., 3rd, Gingeras T. R., Schreiber S. L., Lander E. S. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein E., Duncan E. M., Masui O., Gil J., Heard E., Allis C. D. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damavandi E., Hishikawa Y., Izumi S., Shin M., Koji T. Involvement of Bax redistribution in the induction of germ cell apoptosis in neonatal mouse testes. Acta Histochem. Cytochem. 2002;35:449–459. [Google Scholar]
- 5.Goldberg A. D., Allis C. D., Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Govin J., Caron C., Lestrat C., Rousseaux S., Khochbin S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur. J. Biochem. 2004;271:3459–3469. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 7.Guan K., Nayernia K., Maier L. S., Wagner S., Dressel R., Lee J. H., Nolte J., Wolf F., Li M., Engel W., Hasenfuss G. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199–1203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- 8.Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., Brinkley B. R., Bazett-Jones D. P., Allis C. D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 9.Hirota T., Lipp J. J., Toh B. H., Peters J. M. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 10.Hishikawa Y., An S., Yamamoto-Fukuda T., Shibata Y., Koji T. Improvement of in situ PCR by optimization of PCR cycle number and proteinase k concentration: localization of x chromosome-linked phosphoglycerate kinase-1 gene in mouse reproductive organs. Acta Histochem. Cytochem. 2009;42:15–21. doi: 10.1267/ahc.09011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanovska I., Khandan T., Ito T., Orr-Weaver T. L. A histone code in meiosis: the histone kinase, NHK-1, is required for proper chromosomal architecture in Drosophila oocytes. Genes Dev. 2005;19:2571–2582. doi: 10.1101/gad.1348905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeppesen P., Mitchell A., Turner B., Perry P. Antibodies to defined histone epitopes reveal variations in chromatin conformation and underacetylation of centric heterochromatin in human metaphase chromosomes. Chromosoma. 1992;101:322–332. doi: 10.1007/BF00346011. [DOI] [PubMed] [Google Scholar]
- 13.Khalil A. M., Boyar F. Z., Driscoll D. J. Dynamic histone modifications mark sex chromosome inactivation and reactivation during mammalian spermatogenesis. Proc. Natl. Acad. Sci. U S A. 2004;101:16583–16587. doi: 10.1073/pnas.0406325101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalil A. M., Wahlestedt C. Epigenetic mechanisms of gene regulation during mammalian spermatogenesis. Epigenetics. 2008;3:21–28. doi: 10.4161/epi.3.1.5555. [DOI] [PubMed] [Google Scholar]
- 15.Kimmins S., Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–589. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- 16.Kniewel R., Keeney S. Histone methylation sets the stage for meiotic DNA breaks. EMBO J. 2009;28:81–83. doi: 10.1038/emboj.2008.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koji T., Kondo S., Hishikawa Y., An S., Sato Y. In situ detection of methylated DNA by histo endonuclease-linked detection of methylated DNA sites: a new principle of analysis of DNA methylation. Histochem. Cell Biol. 2008;130:917–925. doi: 10.1007/s00418-008-0487-7. [DOI] [PubMed] [Google Scholar]
- 18.Liang G., Lin J. C., Wei V., Yoo C., Cheng J. C., Nguyen C. T., Weisenberger D. J., Egger G., Takai D., Gonzales F. A., Jones P. A. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z., Zhou S., Liao L., Chen X., Meistrich M., Xu J. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J. Biol. Chem. 2010;285:2758–2770. doi: 10.1074/jbc.M109.066845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohn F., Weber M., Rebhan M., Roloff T. C., Richter J., Stadler M. B., Bibel M., Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Oliva R., Mezquita C. Marked differences in the ability of distinct protamines to disassemble nucleosomal core particles in vitro. Biochemistry. 1986;25:6508–6511. doi: 10.1021/bi00369a025. [DOI] [PubMed] [Google Scholar]
- 22.Oliva R., Bazett-Jones D., Mezquita C., Dixon G. H. Factors affecting nucleosome disassembly by protamines in vitro. Histone hyperacetylation and chromatin structure, time dependence, and the size of the sperm nuclear proteins. J. Biol. Chem. 1987;262:17016–17025. [PubMed] [Google Scholar]
- 23.Peters A. H., O’Carroll D., Scherthan H., Mechtler K., Sauer S., Schofer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 24.Roosen-Runge E. C. The process of spermatogenesis in mammals. Biol. Rev. Camb. Philos. Soc. 1962;37:343–377. doi: 10.1111/j.1469-185x.1962.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 25.Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 26.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 27.Shirendeb U., Hishikawa Y., Moriyama S., Win N., Thu M. M., Mar K. S., Khatanbaatar G., Masuzaki H., Koji T. Human papillomavirus infection and its possible correlation with p63 expression in cervical cancer in Japan, Mongolia, and Myanmar. Acta Histochem. Cytochem. 2009;42:181–190. doi: 10.1267/ahc.09030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sims R. J., 3rd, Nishioka K., Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Strahl B. D., Allis C. D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 30.Tajika Y., Takahashi M., Hino M., Murakami T., Yorifuji H. VAMP2 marks quiescent satellite cells and myotubes, but not activated myoblasts. Acta Histochem. Cytochem. 2010;43:107–114. doi: 10.1267/ahc.10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsukasaki S., Miyazaki M., Koji T., Abe K., Furusu A., Shin M., Suzuki D., Harada T., Ozono Y., Sakai H., Kohno S. Semi-quantitative non-radioactive in situ hybridization and its clinical application. Acta Histochem. Cytochem. 2000;33:39–47. [Google Scholar]
- 32.Turner B. M. Histone acetylation and an epigenetic code. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 33.Vettese-Dadey M., Grant P. A., Hebbes T. R., Crane-Robinson C., Allis C. D., Workman J. L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]