Abstract
Isolation and amplification of primary lymphoma cells in vitro setting is technically and biologically challenging task. To optimize culture environment and mimic in vivo conditions, lymphoma cell lines were used as a test case and were grown in 3-dimension (3D) using a novel 3D tissue culture polystyrene scaffold with neonatal stromal cells to represent a lymphoma microenvironment. In this model, the cell proliferation was enhanced more than 200-fold or 20,000% neoplastic surplus in 7 days when less than 1% lymphoma cells were cocultured with 100-fold excess of neonatal stroma cells, representing 3.2-fold higher proliferative rate than 2D coculture model. The lymphoma cells grew and aggregated to form clusters during 3D coculture and did not maintained the parental phenotype to grow in single-cell suspension. The cluster size was over 5-fold bigger in the 3D coculture by day 4 than 2D coculture system and contained less than 0.00001% of neonatal fibroblast trace. This preliminary data indicate that novel 3D scaffold geometry and coculturing environment can be customized to amplify primary cancer cells from blood or tissues related to hematological cancer and subsequently used for personalized drug screening procedures.
1. Introduction
Emerging technologies in tissue engineering can be used to create 3-dimensional (3D) diseased tissue or organ models for screening therapeutic drugs and studying disease biology. 3D model is becoming more attractive to researchers as they realized the shortcomings of traditional two-dimensional (2D) in vitro tissue culture models. 2D culture does not closely mimic in vivo environment and often overlooks important variables such as dimensionality and microenvironment signaling [1, 2], which has an effect on cancer phenotype, aggressiveness, and drug resistance [3–10]. The use of 3D scaffolds to engineer 3D solid tumor models has been successful [11–15]. These in vitro 3D tumor models showed their potential values in oncology drug screening and tumor biology studies.
Previous works have demonstrated a direct link between 3D tumor microenvironment and cancer behavior. However, a direct application of polymer-based tissue engineering approach to recreate microenvironment for enrichment of primary blood cancer cells has not been explored. From the perspective of engineered cancer microenvironments, the stroma is an ubiquitous and necessary component that has been implicated during in vivo cancer progression [16–19]. The stromal compartment can be found throughout the body as a form of tissue support [20], covering internal conduits of secreting glands [21, 22], and increasing surface contact during paracrine-mediated maturation of cell populations in the bone marrow and lymphatic tissues [23, 24]. Therefore, in vitro models that include 3D stroma architecture offer the most native representation of complex cancer signatures during cancer progression.
A part of personalized cancer treatment for hematological malignancies requires culturing of primary cancer cells from the patient and use the cells to identify drugs that are most effective in cell killing. However, patient specimens that are derived from core biopsies, postoperative resection, and peripheral blood typically generate an insufficient number of primary cancer cells for the purpose of screening drugs. Consequently, identifying the “personalized” drugs for the patient will not be practical and has been an extremely difficult task with conventional methods for cell culture. The survival and the amplification of primary cancer cells are mainly due to suboptimum environment in vitro and inefficient 2-dimensional cell culture conditions. We have investigated multiple 3-dimensional cell culture systems to optimize the growth of cancer cells by using mantle cell lymphoma cell lines.
Hematological cancers are more complex than solid cancers due to its ability to efficiently proliferate in suspension and can proliferate or differentiate in stromal compartments such as the bone marrow, lymph nodes, spleen, and thymus. Lymphoma is a blood cancer type that involves both tissue and lymph system and can progressively become worse when cancer cells adapt to proliferate in the blood compartment. Mantle cell lymphoma (MCL) is an aggressive B-cell type lymphoma that represents up to 7% of all Non-Hodgkin's lymphomas in the USA and occurs more in older male patients with a median age of 60 years [25]. MCL arises from peripheral CD5-positive B-cells of the inner mantle zone of secondary follicles and is diagnosed typically in advanced stage (III/IV) to exhibit an aggressive B-cell lymphoma characteristic that has a broad morphologic spectrum [26]. Cytogenetic and immunohistochemical studies show MCL to carry the hallmark chromosomal translocation, t(11;14)(q13;q32) which causes overexpression of cyclinD1 and consequently implicates on disordered progression of cell cycle [27]. To this date, MCL does not have standard therapy for curative treatments but a combination of Hyper-CVAD with Rituximab has shown promising clinical outcomes as the front line therapy [28].
To treat MCL more effectively, an amplified primary MCL cells derived from tissue or blood can be screened with a short list of clinically available drugs, and the most effective drug or a combination of drugs can be considered for the patient. In this report, MCL cell lines were used to study conditions of 3-dimensional tumor microenvironments mediated by stromal components in 3D polymer scaffolds. The 3D coculture model presented here suggests that MCL cells that still retain tumor microenvironment phenotype may need to interact with stroma to unleash their maximal growth potential. Thus, these findings may help recreate true blood cancer phenotypes, minimizing the burden to amplify primary MCL tumor cells from tissue biopsies and surgical resections for high throughput drug screening effort.
2. Materials and Methods
2.1. Fabrication of 3D Polystyrene Polymer Scaffold
The 3D 12-well polystyrene (PS) scaffolds were constructed using 3D Precision Microfabrication system (3DPM) developed by 3D Biotek (Figure 1). 3DPM generates porous 3D scaffolds from polymers thermoplastic polymers with well-controlled pore size and porosity. Each molten polystyrene fiber is laid down with designated spacing on a flat surface and the subsequent layers are added at a 90° angle from the previous reference layer. This 3D scaffolds have interconnected porous structure with uniform pore sizes and polymer fibers. The 3D surface area created by layering fibers that are orthogonal to each other increases in surface area, but every spatial point is still close to each other. As opposed to the flat surface with the same surface area, the 3D scaffold keeps any two points in close proximity. For example, the total surface area of the 3D insert with dimensions (diameter 2.2-cm and thickness 0.06-cm) is 21.08 cm2, which is comparable to a 2D disk with a 5.2-cm diameter. Thus, every two points in the 2D disk will have a distribution of distances with a maximum distance at 5.2 cm, while in 3D for the same surface area the distribution of distances will have a maximum distance of 2.2 cm. For the cell culture experiment, the scaffold with a fiber size of 300-um in diameter and a pore size of 400-um diameter was used. To make these 3D scaffolds suitable for suspension cell culture, polystyrene legs were introduced to the 3D polystyrene scaffolds around the circumference of the bottom of the scaffolds. These legs will raise the scaffolds 0.5 mm from the bottom surface of the cell culture wells to minimize the contact with the underneath cell culture well surface. After finishing the scaffold fabrication, these scaffolds were plasma-treated using a Harrick Plasma Cleaner (Model PDC-001, Ithaca, NY) and gamma-ray-radiated before use.
Figure 1.
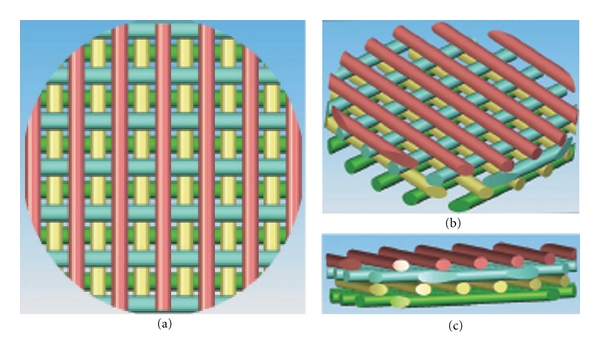
CAD models of 3D scaffolds. The scaffold is made of four layers (different color represent different layers from bottom to top) 90° and offset with respect to each other. This geometry generates the characteristic 3D porous structure of the PS scaffold (a)–(c).
2.2. Cell Lines
Two mantle cell lymphoma (MCL) cell lines, HBL2 (derived from lymph node, CyclinD1+, CD5−, CD23+) and Z138 (derived from bone marrow, CyclinD1+, CD5−, CD23−), were cocultured with human dermal fibroblasts (hDFb) in this 3D coculture study. These cell lines were chosen in this study because they are derived from nonperipheral blood, proliferate efficiently and remain as single cells suspensions in regular tissue culture conditions. To prepare fibroblast layer for coculture, hDFb were isolated from human neonatal foreskins as described previously [29]. The fibroblast preparation at passage 4 was grown in Gibco DMEM/F12 + GlutaMAX-I (Invitrogen, Carlsbad, CA) media supplemented with 10% fetal calf serum (PAA, Etobicoke Ontario, Canada) and 1% Penicillin/Streptomycin (Cellgro, Herndon, VA). Stroma cells had a surface area of 1.5 × 10−5 cm2 and a doubling time of 2.5 days.
2.2.1. HBL2 and Z138 MCL Culture
For lymphoma cell lines, HBL2 and Z138 mantle cell lymphoma cell lines were grown in RPMI-1640 (with L-glutamine and NaHCO3, Sigma St. Louis, MO) and supplemented with 10% fetal bovine serum (PAA, Etobicoke Ontario, Canada) and 1% Penicillin/Streptomycin. All cells were grown in water-jacketed incubator (Forma Scientific, Marietta, OH) with 5% CO2 and at 37°C.
2.2.2. 3D and 2D Coculture
To determine the effect of dimensionality and presence of stroma (3D coculture condition), a mixture of MCL and hDFb cells at the ratios of 1,000 : 100,000 (0.9%), 10,000 : 100,000 (9.0%), or 50,000 : 100,000 (33%) was initially seeded at 200-μL in volume. Based on the surface area of stroma and the available surface area of growth on 12-well 2D plates and 3D PS scaffold, initial seeding was 37% in 2D and 7% in 3D PS scaffolds. The surrounding empty wells of the 12-well plate were filled with DPBS to maintain high humidity and avoid volume loss of the initial mixture volume. After 3 hours seeding, the plates were placed on an Adams Nutator (Model 1105, 50–60 Hz, Becton Dickson, Parsippany, NJ) and rocked for 12 hours inside the incubator. The cells were subsequently supplemented with 1.5-mL of conditioned medium that was produced by coculturing hDFb and MCL cells at 50/50 ratio. For 2D-coculture, 200-uL of cell suspension at the same MCL : hDFb ratios as 3D-coculture were added 1.5-mL of conditioned media and placed on the shaker simultaneously with the 3D coculture plate. For routine feeding, 66% media from each 12-well was removed and changed for fresh coculture media (supplemented RPMI 1640 and DMEM/F12 + GlutaMAX-I) every 48 hours for 6 days. MCL cell aggregates that settle at the bottom of the plate were collected at day 7 for quantification and analysis.
2.3. Cell Proliferation and Viability
To harvest, cells were collected from each well (12-well plates (2D) and 12-well nontreated plates containing 3D PS scaffolds. 10-μL of cell suspension was mixed with 10-μL of 0.4% Tryphan Blue dye (Sigma St. Louis, MO). Cells were counted using a hemacytometer for each sample collected. Cells stained blue were counted as nonviable. Based on initial lymphoma seeding and current cell count after 7 days, the neoplastic percent surplus was calculated following the equation: (Final Cell Number/Initial Cell Number)∗100. This allowed us to compare the proliferative capacity of the two MCL cell lines under conditions of dimensionality and initial lymphoma seeding percent. The data was graphed and processed using Excel (Microsoft Excel Version 12.1.0) and Prism software (GraphPad Software, Inc, Version 4.0c).
2.4. Lymphoma Cluster Analysis
During the course of the experiment, pictures of the wells were taken at days 2 and 4 for 3D and 2D coculture of lymphoma and stroma conditions. Clusters images were taken with an Insight 2 Mp Monochrome Model 18.0 Camera attached to a Nikon, Eclipse TS 100 inverted light microscope (Micron-Optics, Cedar Knolls, NJ). Pictures were analyzed using Spot Software (Diagnostic Instruments, Detroit, MI) and Image J 1.43u (N.I.H. USA) for cluster size and graphed using Prism software (GraphPad Software, Inc, Version 4.0c).
2.5. Fibroblastic Contamination after Lymphoma Harvesting
Collected lymphoma cells from 2D and 3D coculture conditions were plated on treated T-75 flaks and allowed to incubate for 24 hours under regular tissue culture conditions. The next day, microscopic observation was made and pictures were taken on the harvested lymphoma cells, particularly looking for the presence of adherent dermal fibroblast. We took several pictures using an Insight 2 Mp Monochrome Model 18.0 Camera attached to a Nikon, Eclipse TS 100 inverted light microscope (Micron-Optics, Cedar Knolls, NJ). Pictures were analyzed using Spot Software (Diagnostic Instruments, Detroit, MI) and Image J to determine the pixel density of adherent dermal fibroblast after harvesting from 2D and 3D coculture. The data was tabulated and graphed using Prism software.
3. Results
3.1. 3DPM-Based 3D Polystyrene Polymer Scaffold Provides Excellent Structure Integrity for Cancer Tissue Engineering
Three-dimensional polystyrene scaffolds with a 4-layer porous structure were fabricated using 3DPM technology (Figure 1). As shown in the CAD drawing of Figure 1, each layer of struts is represented by a different color and the struts of each layer are layered in a 90-degree angle to its immediate adjacent layer. Each fiber has a diameter of 300-um and 400-um fiber-to-fiber spacing. The strut-to-strut and layer-to-layer spacing is optimized to provide both increased 3D surface area and 3D space for cell attachment and growth. For example, a four-layered scaffold provides with 16.4 cm2 of the total surface area, which is 4-fold larger than the growth surface area of a 12-well with a diameter of 2.1 cm.
The scaffold was designed to raise the structure above the bottom of the plate and this feature was intended to provide a better flow of medium and gas throughout the structure and reduce medium volume losses during the first 15 hours of seeding the cell mixture. To construct this structural feature, a polystyrene loop was generated at the edge of the scaffold and polystyrene struts are laid orthogonal to form a mesh-like structure (Figure 2). Figures 2(c) and 2(d) show the second engineering feature that is represented by one side of the scaffold projecting polystyrene loop (black arrow). This loop allows the scaffold to be raised approximately 0.5 mm from the bottom of any surface, thus forming a rim around the circumference of the scaffold which reduces contact with the underneath surface. The scaffolds were plasma-treated using a Harrick Plasma Machine (Model PDC-001, Ithaca, NY) and gamma-radiated for sterility.
Figure 2.
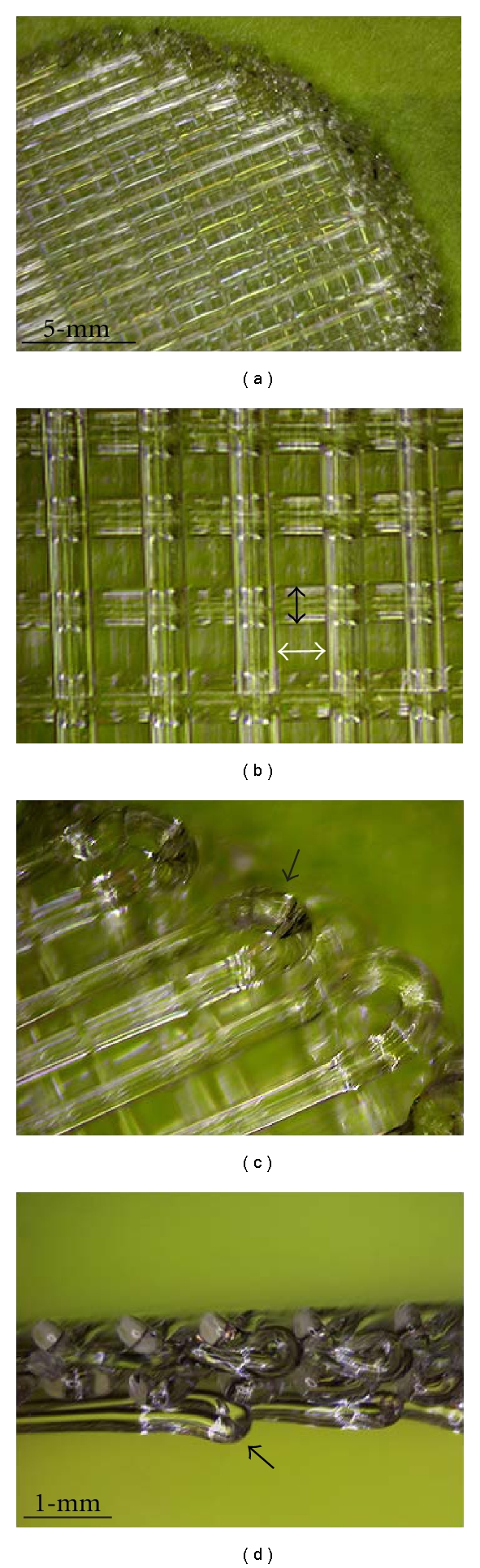
3D Polystyrene scaffold for 3D coculture model. Polystyrene fibers are laid down at 90° angles with respect and offset from each other, creating its characteristic 3D structure. There is spacing between each fiber (white arrow) to create the interconnected porous structure of the scaffold. The scaffold is made with fibers 300-um in diameter and 400-um spacing between the fibers (a, b). The scaffold also has a loop around the rim of the scaffold. The loop is about 0.5-mm height (black arrows) allowing the scaffold to have a raised conformation with reference to the bottom of the well (c, d).
3.2. Coculturing of HBL-2 MCL and Stromal Cells in 3D Environment Significantly Enhances Growth of Lymphoma Cells
In the absence of stroma cells, lymph-node-derived HBL-2 cells grow mostly in single cell suspension by loosely attaching to the polystyrene surface in both conventional 2D and 3D environment (Figure 3(a)). These cells expanded in unidirection as they proliferate without stroma cells, and the proliferation was independent of geometry because 3D culture did not show significant advantages over conventional 2D environment. Also, when seeded without the stroma component, HBL-2 cells initially floated in suspension within the interstitial space of the scaffold of the 3D environment (white arrows, Figure 3(a)), but most cells gradually settled to the bottom of the well over time. In the presence of stroma cells, the growth phenotypes of MCL cells in both 2D and 3D environments changed remarkably after coculturing with primary stromal cells that was generated from human neonatal skin (Figures 3(c) and 3(d)). These stroma cells (mostly fibroblast) were found to attach equally and firmly to both the 2D and 3D surfaces; the coculture caused HBL-2 cells to adopt a stratified organization and formed loosely aggregated cellular masses in both 2D and 3D cases. However, HBL-2 cells formed more clusters in 3D coculture model. Continued coculture produced large masses of cell clusters that are predominantly HBL-2 cells and the cluster formations were significantly more active on 3D when compared with 2D environment (Figure 4). In the 3D coculture environment, HBL-2 lymphoma cells initially adhered to stroma cells and start to form small aggregates. These small aggregates continued to grow into larger clusters within the porous structure of the 3D scaffolds. As the size of these clusters reached a critical mass, all clusters became separated from the stroma support and dropped eventually the bottom of the well. When HBL-2 aggregates were harvested from the 2D and 3D coculture models and placed in suspension in the absence of stroma, they do not go back to the morphology of single-cell suspension, suggesting that interacting with primary stroma cells may have changed the cellular programming of signaling pathways. In this setting, the expression of previously reported 3D markers (Actin, B2M, FMOD, TLR4, VTN, Collagen and Laminin and Integrins (ITGA1, ITGA4, ITGAL, ITGAM, ITGB1, ITGB2)) from various 3D spheroid cultures were tested by qRT-PCR but no significant differences were detected between HBL-2 cells grown in 2D and 3D (see Supplementary Material available online at doi: 10.1155/2011/362326).
Figure 3.
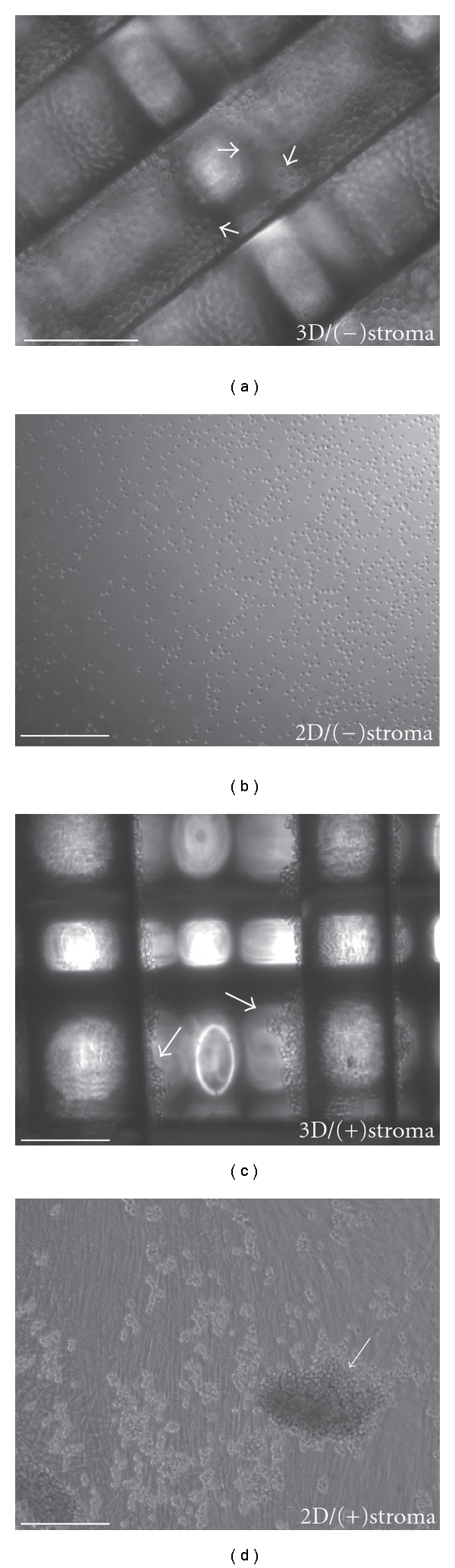
In vitro 3D and 2D MCL coculture models. HBL-2 lymphoma cells remain as single suspension cells (white arrows) in the absence of stroma for 2D and 3D models (a, b). Mixtures of stroma and HBL-2 cells seeded on 3D PS scaffolds and 2D PS plate causes aggregation of HBL-2. Over time, HBL-2 cells grow as weakly bound clusters on the surface of plates and from the internal fibrous structure of the scaffold (c, d). Scale-bar 250-μm.
Figure 4.
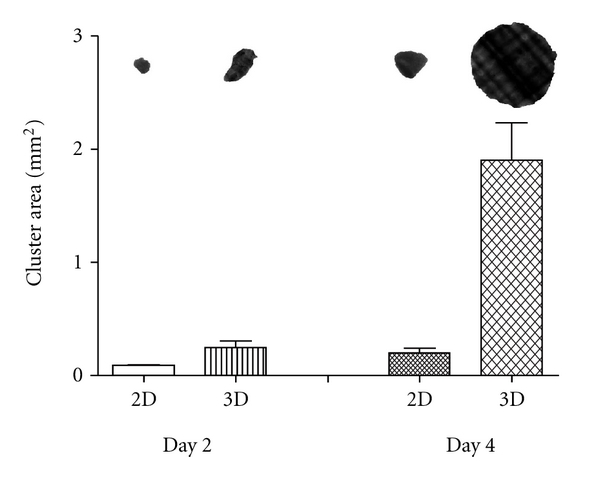
The effect of 3D stroma on cluster size. After 2 days in coculture, HBL-2 clusters from 3D stromal coculture were 0.25 ± 0.1 (STDEV) mm2 and HBL-2 clusters from 2D stromal coculture 0.1 ± 0.02 (STDEV) mm2. The difference in size became more apparent at day 4, 3D stroma model yielded HBL-2 clusters with an average size of 1.9 ± 0.7 (STDEV) mm2, approximately 10-fold bigger than HBL-2 clusters from 2D stroma coculture at day 4 (±SEM in the graph) (n = 5).
3.3. HBL-2 Cells Proliferate with Greater Efficiency in 3D Than 2D Coculture Environment
HBL-2 cells grown in 3D with stroma cells had a significantly higher efficiency in generating cell clusters when compared with cells grown in 2D (Figure 4). At this seeding condition, HBL-2 clusters were more than 2-fold larger in 3D than 2D within 2 days and more than 10-fold larger by day 4. Three-dimensional coculturing enhanced the proliferation of cancer cells and a higher rate of clustering, as a result of faster dislodging of clusters from the stroma support. Thus, a combination of the geometry, greater surface area, and 3D spatial environment provides significantly better growth condition for MCL cells derived from lymph node. The cluster size measurements in this setting exclude continuously growing HBL-2 clusters on the scaffold (Figure 4).
To further investigate the optimum growth condition, the seeding density of HBL-2 cells was tested at 0.9%, 9.0%, and 33% with respect to the number of cocultured stroma cells (Table 1). After 7 days of coculture, the proliferation rate of HBL-2 cells seeded in 0.9% category showed the fastest growth followed by 9% and 33% categories (unpaired t-test P = 0.001) in 3D than 2D (Figure 5). The cell viability was 100% by day 7 and the cell division is calculated to be 9.6 hours per division when the initial HBL-2 seeding ration was 0.9%. At this condition, 3D coculture was able to amplify 1,000 HBL-2 to 177,722 cells in a week, accounting for a 17,722 ± 2,394% (STDEV) neoplastic surplus, which is about 3.2-fold greater than HBL-2 proliferation in 2D coculture (Table 1). This suggested that the cell cycle of HBL-2 cells was remarkably enhanced by the direct interaction with primary stroma in 3D compartments, mimicking the tumor microenvironment. HBL-2 cells viability for the 3 seeding conditions showed a decreasing trend with increasing HBL-2 seeding ratio. The cell viability in 9% MCL condition was 2-fold better than 33% MCL, but both cases failed to show proliferation advantage on both 3D and 2D culture conditions. We believe that the decreased proliferation and cell viability was likely caused by the inability to supply sufficient nutrients as a result of increased initial HBL-2 seeding density and its increased proliferation potential in the 3D coculture condition. When HBL-2 cells from 2D and 3D environments were analyzed for the cell proliferation markers, it was found that the expressions of NFκB members NFκB2, RELA, and RELB were overexpressed up to 2-fold in 3D setting (see Supplementary Material).
Table 1.
Proliferation of HBL-2 and Z138 on 3D and 2D stromal compartments.
| Mantle cell lymphoma | ||||
|---|---|---|---|---|
| 0.90% HBL-2: stroma | 0.90% Z138: stroma | |||
| Geometry | 3D | 2D | 3D | 2D |
| Day 0 | 1,000 | 1,000 | 1,000 | 1,000 |
| Day 7 | 177,722 ± 23,940 | 55,777 ± 8,071 | 4,640 ± 1,282 | 2,260 ± 533 |
| % surplus | 17,722 | 5,577 | 464 | 226 |
| % viability | 100% | 100% | 100% | 100% |
Figure 5.
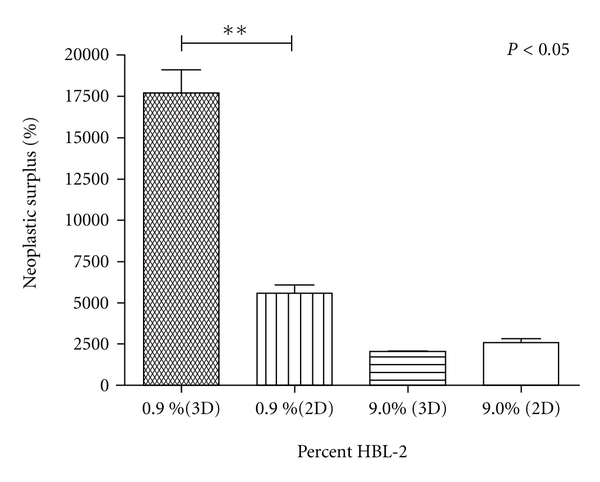
Neoplastic growth of HBL-2 on 3D and 2D stromal models. HBL-2 efficiently proliferated at lower initial seeding densities in 3D coculture with stroma. 0.9% HBL-2 mixture in neonatal dermal fibroblast showed statistically significant greater proliferation potential with a percent neoplastic surplus of 17,722 compared to 5,577 in 2D stroma. Increasing percent HBL-2 seeding density did not show the same trend and cell viability was greatly reduced (±SEM) (n = 4).
In order to further explore the effect of stroma on other suspension MCL types, we investigated the proliferative capacity of Z-138, a cell line derived from bone marrow, in identical conditions as HBL-2 cell line (Figure 5). At day 7, Z-138 cells showed lower proliferation with a percent surplus of 464 ± 128% for 3D and 226 ± 53% for 2D and with no clustering behavior, indicating that these cells may require a specific bone marrow stroma-signaling signature. Despite the lower proliferation rates, in comparison to HBL-2, Z-138 cells seeded at 0.9% in stroma had a higher percent surplus in 3D than 2D at day 7. However, Z-138 in absence of stroma was nonviable for same initial cell seeding and time in culture (Figure 5). These findings elucidate the synergy between dimensionality and stroma microenvironment, particularly its effect on liquid cancer neoplastic growth.
3.4. Three-Dimensional Coculture Efficiently Separates HBL-2 from MCL-Stroma Mixtures
Besides enhancement of HBL-2 proliferation on 3D stromal compartments, we determined that harvesting of HBL-2 cells could be achieved with minimal stroma contamination in the 3D coculture condition. Figure 6 compares the effect of geometry, that is, 2D and 3D, and HBL-2 harvesting from stroma cell mixtures. After 24 hours in static culture, HBL-2 remains as weakly bound suspension clusters for both 2D and 3D culture conditions (Figures 6(a) and 6(b)). However, there was a marked difference in the amount of stroma cells adhering to the bottom of the harvesting culture plates (Figures 6(c) and 6(d)). In the 2D coculture condition, there was a large number of stroma clusters (<10 clusters) and single adherent stroma to the bottom of the culture dish. Quantification of the stromal contamination after harvesting shows that, at the scale of single stroma cluster to single stroma cell, the gray pixel value is 20-fold between the cell cluster to single cell (Figures 6(e) and 6(f)). Thus, HBL-2 harvesting from 3D coculture condition offers both amplification and efficient separation of MCL cells from heterogeneous cell mixtures at minimal stroma contamination.
Figure 6.
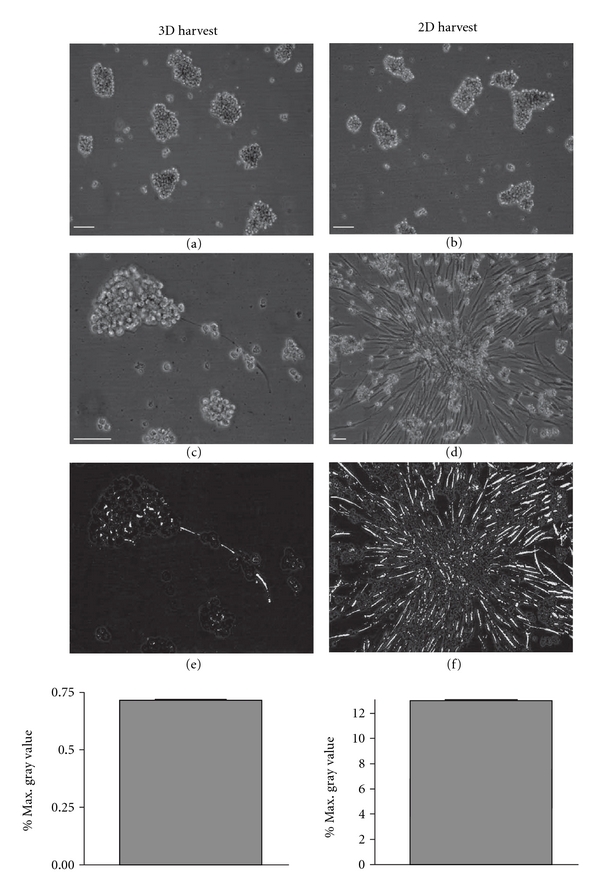
Stroma traces after HBL-2 harvesting from 2D and 3D coculture. After harvesting, HBL-2 remained as single aggregates in suspension and in the absence of stroma (a, b). There were traces of stroma from the 3D and 2D coculture systems adhering to the HBL-2 harvesting plates (c, d). Quantification of stroma traces showed that there is a characteristic singe stroma to stroma clusters in 3D and 2D coculture models, which is about 20-fold difference between single cell to single stroma cluster.
4. Discussion
Lymphoma cells reside and proliferate in environments represented by tissue, bone marrow, and blood. In each environment, lymphoma cells have adopted and utilize unique survival signaling pathways that consequently complicate cancer therapy options [30]. Among non-Hodgkin's lymphoma, mantle cell lymphoma (MCL) represents a disease that is indolent yet aggressive in proliferative nature that ultimately leads to unfavorable clinical outcome [31]. MCL cells can also metastasize from blood to multiple tissues as they enter advance stages, and this systemic spread further reduces targeted and personalized treatment options [32]. As the disease advances to leukemic phase, drugs that may be effective for MCL cells in circulating blood may not be as effective when MCL cells are partly protected in the tumor microenvironment in tissues. The proliferative potential and molecular signatures of MCL cells are different when the cells in nutrient-rich blood environment are compared with tumor environments that are localized to bone marrow or lymph nodes with stroma, in turn requiring specific targeting strategies [33]. The neighboring stroma cells secrete various growth factors to the local environment and directly support and simultaneously protect MCL cells from anticancer drugs [34–36]. To develop therapeutic options that is patient specific and target MCL cells in tissues, primary MCL cells must be initially grown and amplified in vitro cell culture condition, so that the cells can be used for screening a set of drugs that are currently available in the clinic. However, this task is not practical due to low proliferative potential and viability of MCL cells in conventional 2D tissue culture conditions and the lack of appropriate 3D culture environment that mimics the tumor microenvironment in vivo. Selected factors such as soluble CD40 ligands or IL10 were exogenously added to amplify primary MCL cell in 2D culture but the treatment promoted a limited cell growth potential [37, 38]. We report that 3-dimensional tumor microenvironment for MCL was successfully constructed by using MCL cells with primary neonatal fibroblast by culturing the combination of cells in a polymer-derived 3D scaffold. MCL cell lines derived from lymph node were used as a model system to optimize 3D culture conditions that can be implicated to amplify primary MCL cells from biopsies or tissues that originate from surgical procedures in the clinic.
MCL cells that derived from lymph node (HBL-2) proliferated with remarkable efficiency in the presence of neighboring stroma cells when grown in 3DPM -based 3D polystyrene polymer scaffold. In our experimental model, we treated 2D and 3D equally in terms of volume of medium, feeding schedule, the ratio of cell numbers/area and the availability of surface area to grow for the total time in culture. Particularly, the limitation of nutrition does not represent a limiting factor that may differentiate the cell proliferation between the two cases at 0.9% MCL: stroma because this seeding density represent the optimum for the 12-well format and cell culture time of 7 days. The analyses of previously reported markers from 3D spheroids on the mixture of cells grown in 2D and 3D showed no differential expressions of 3D markers involved in extracellular matrix, cell adhesion, and cytoskeleton, further indicating that the geometric effect of 3D-stroma produced superior environment for the observed accelerated growth of lymphoma cells. The cell doubling time of HBL-2 exceeded 3-fold in the optimized 3D environment when compared to previous reported doubling time in conventional 2D culture without stroma [39]. Although HBL-2 cells grow in single cell suspension in conventional 2D, the cells intrinsically retained signaling circuitry that takes an advantage of the neighboring stroma cells to proliferate. The interaction of HBL-2 cells and the supporting stroma cells was evident because they actively grew together throughout the culturing phase and until the cluster of HBL-2 cells began to separate apart from the stroma. The undetermined factors that are secreted by the neonatal stroma component may turn on genetic switches in MCL cells to promote cell-to-cell interaction with the stroma component to enhance the growth. Stroma cells were previously used in cell culture for ex vivo expansion of cord blood hematopoietic stem/progenitor cells [40] and long-term culture and maintenance of leukemia or stem cells [41, 42], suggesting the neonatal stroma contains sufficient number of stem-like cells to supplement growth factors to MCL cells in vitro [43, 44]. Recently, dermal cells isolated from foreskin can differentiate into functional epidermal melanocytes, indicating the presence of stem cell properties in dermis-derived stroma components [45].
These reports and the data from our laboratory suggest that the inclusion of the neonatal foreskin stroma in our 3D model plays a key role in the amplification of MCL cells. In our experimental setting, cell proliferation markers from NFκB family were moderately increased in the mixture of cells grown in 3D environment (see Supplementary Material), suggesting that the proliferation potential of the lymphoma cells (represented by <1% of total cell number in the mixture) is superior in 3D scaffold condition than 2D. Conversely, MCL cells derived from bone marrow (Z-138) proliferated poorly albeit in the presence of neonatal stroma, indicating that alternative factors may be needed to optimize the growth. Nonetheless, the effect of 3D stroma coculture was able to maintain better growth and viability of Z-138 for low seeding density in the absence of dimensionality and stroma. The results from HBL-2 and Z-138 differentiated the requirements of optimal culturing conditions for MCL cells that have dissimilar origins of tumor environment. Thus, Z-138 many require a microenvironment that mimics bone marrow and other supplementary factors besides neonatal stroma. Furthermore, our preliminary data indicated that MCL cell lines derived from peripheral blood, typically from leukemic phases, may require yet another optimization of culturing condition (data not shown). Our data suggest that in order to better mimic different types of tumor micro-environment, such as blood, bone marrow, and lymphatic tumors, different types of stroma or supporting cells are needed in the 3D scaffolds.
Patient specimens from surgical procedures normally have a mixture of cancer cells with a great excess of stromal cells. Tissue specimens of MCL from biopsies or surgical resections (i.e., spleen) can be mixed and cultured in the optimized 3D culture environment to selectively amplify MCL cells away from the stroma. In this setting, an addition of neonatal stroma may offer necessary supplementary factors to enhance the growth of MCL cells with minimal stroma contamination. In our 3D model, HBL-2 cells were amplified over 100-fold within 7 days period with 0.00001% contamination of stroma cells, indicating that the stroma cells prefer to stay attached to the 3D scaffold while MCL tumor cells undergo exponential growth phase as in aggregates, in turn producing highly purified cancer cell population. HBL-2 cells from the cluster did not go back to the parental phenotype (single cell suspension) when placed back in the 2D environment (data not shown), indicating that the phenotype of single-cell suspension may be directly associated with the absence of active stroma cells. Several reports indicate that stromal cells in tumor microenvironment play pivotal roles in B-cell malignancies by activating multiple signaling pathways [46–48]. Together, the combination of 3D system with polymer scaffold with neonatal stroma provides an excellent growth environment for the purpose of amplifying MCL cells, and similar 3D environment can be implicated on other liquid and solid tumor models. The model described here can improve decision making on cancer treatment options by providing a 3D cell-based platform to screen drugs that are currently in clinical use and identifying best-fit drug(s) for the patient.
We have shown the versatility of polymer-based scaffolds for the recreation of dimensionality and coculture signatures. Particularly, both contributions have biological relevance, seen as a superior proliferation of HBL-2 and maintenance of Z138 under the prescribed conditions. Further work is necessary to explore the versatility of the model using primary cell mixtures. This would increase the attractiveness of 3D polymer scaffolds for the recreation of personalized cancer signatures and effective drug screening methodologies in cancer therapies.
Supplementary Material
To investigate differential regulations of cytoskeletal proteins (Supplementary Figure 1), known 3D-Spheroids markers (Supplementary Figure 2), Integrin members (Supplementary Figure 3) and NFkB members (Supplementary Figure 4) between 2D and 3D culture model, conventional qRT-PCR was performed by using validated primer sets and ABI-9700 under a fast cycle. Representative members of each signaling group and 3D spheroid markers were quantified and normalized by actin and the expression of each gene is shown in fold change over the parent cell line without the stroma component. The differential expression of all genes were minimal, except for the NFkB members, between 2D and 3D culture models.
References
- 1.Nyga A, Cheema U, Loizidou M. 3D tumour models: novel in vitro approaches to cancer studies. Journal of Cell Communication and Signaling. 1924;5(4):239–248. doi: 10.1007/s12079-011-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krause S, Maffini MV, Soto AM, Sonnenschein C. The microenvironment determines the breast cancer cells’ phenotype: organization of MCF7 cells in 3D cultures. BMC Cancer. 2010;10, article no. 263 doi: 10.1186/1471-2407-10-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olsen CJ, Moreira J, Lukanidin EM, Ambartsumian NS. Human mammary fibroblasts stimulate invasion of breast cancer cells in a three-dimensional culture and increase stroma development in mouse xenografts. BMC Cancer. 2010:p. 444. doi: 10.1186/1471-2407-10-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Starzec A, Briane D, Kraemer M, et al. Spatial organization of three-dimensional cocultures of adriamycin-sensitive and -resistant human breast cancer MCF-7 cells. Biology of the Cell. 2003;95(5):257–264. doi: 10.1016/s0248-4900(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 5.Leung M, Kievit FM, Florczyk SJ, et al. Chitosan-alginate scaffold culture system for hepatocellular carcinoma increases malignancy and drug resistance. Pharmaceutical Research. 2010;27(9):1939–1948. doi: 10.1007/s11095-010-0198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shekhar MPV. Drug resistance: challenges to effective therapy. Current Cancer Drug Targets. 2011;11(5):613–623. doi: 10.2174/156800911795655921. [DOI] [PubMed] [Google Scholar]
- 7.Fischbach C, Chen R, Matsumoto T, et al. Engineering tumors with 3D scaffolds. Nature Methods. 2007;4(10):855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 8.Mi K, Wang G, Liu Z, Feng Z, Huang B, Zhao X. Influence of a self-assembling peptide, RADA16, compared with collagen I and matrigel on the malignant phenotype of human breast-cancer cells in 3D cultures and in vivo. Macromolecular Bioscience. 2009;9(5):437–443. doi: 10.1002/mabi.200800262. [DOI] [PubMed] [Google Scholar]
- 9.Gurski LA, Jha AK, Zhang C, Jia X, Farach-Carson MC. Hyaluronic acid-based hydrogels as 3D matrices for in vitro evaluation of chemotherapeutic drugs using poorly adherent prostate cancer cells. Biomaterials. 2009;30(30):6076–6085. doi: 10.1016/j.biomaterials.2009.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benton G, Kleinman HK, George J, Arnaoutova I. Multiple uses of basement membrane-like matrix (BME/Matrigel) in vitro and in vivo with cancer cells. International Journal of Cancer. 2011;128(8):1751–1757. doi: 10.1002/ijc.25781. [DOI] [PubMed] [Google Scholar]
- 11.Burdett E, Kasper FK, Mikos AG, Ludwig JA. Engineering tumors: a tissue engineering perspective in cancer biology. Tissue Engineering Part B. 2010;16(3):351–359. doi: 10.1089/ten.TEB.2009.0676. [DOI] [PubMed] [Google Scholar]
- 12.Horning JL, Sahoo SK, Vijayaraghavalu S, et al. 3-D tumor model for in vitro evaluation of anticancer drugs. Molecular Pharmaceutics. 2008;5(5):849–862. doi: 10.1021/mp800047v. [DOI] [PubMed] [Google Scholar]
- 13.Dhiman HK, Ray AR, Panda AK. Three-dimensional chitosan scaffold-based MCF-7 cell culture for the determination of the cytotoxicity of tamoxifen. Biomaterials. 2005;26(9):979–986. doi: 10.1016/j.biomaterials.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Verbridge SS, Chandler EM, Fischbach C. Tissue-engineered three-dimensional tumor models to study tumor angiogenesis. Tissue Engineering Part A. 2010;16(7):2147–2152. doi: 10.1089/ten.tea.2009.0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergenstock MK, et al. Engineered polystirene scaffolds for in vitro three-dimensional diesease models. In: Proceedings of the Trans of the 34th Society for Biomaterials; 2010; Seatle, Wash, USA. [Google Scholar]
- 16.Tuxhorn JA, Ayala GE, Rowley DR. Reactive stroma in prostate cancer progression. Journal of Urology. 2001;166(6):2472–2483. [PubMed] [Google Scholar]
- 17.Franses JW, Baker AB, Chitalia VC, Edelman ER. Stromal endothelial cells directly influence cancer progression. Science Translational Medicine. 2011;3(66, article 66ra5) doi: 10.1126/scitranslmed.3001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiseman BS, Werb Z. Development: stromal effects on mammary gland development and breast cancer. Science. 2002;296(5570):1046–1049. doi: 10.1126/science.1067431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasanen K, Vaheri A. Activation of fibroblasts in cancer stroma. Experimental Cell Research. 2010;316(17):2713–2722. doi: 10.1016/j.yexcr.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 20.McCave EJ, Cass CAP, Burg KJL, Booth BW. The normal microenvironment directs mammary gland development. Journal of Mammary Gland Biology and Neoplasia. 2010;15(3):291–299. doi: 10.1007/s10911-010-9190-0. [DOI] [PubMed] [Google Scholar]
- 21.Kass L, Erler JT, Dembo M, Weaver VM. Mammary epithelial cell: influence of extracellular matrix composition and organization during development and tumorigenesis. International Journal of Biochemistry and Cell Biology. 2007;39(11):1987–1994. doi: 10.1016/j.biocel.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong YC, Wang XH, Ling MT. Prostate development and carcinogenesis. International Review of Cytology. 2003;227:65–130. doi: 10.1016/s0074-7696(03)01008-8. [DOI] [PubMed] [Google Scholar]
- 23.Di Maggio N, Piccinini E, Jaworski M, Trumpp A, Wendt DJ, Martin I. Toward modeling the bone marrow niche using scaffold-based 3D culture systems. Biomaterials. 2011;32(2):321–329. doi: 10.1016/j.biomaterials.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 24.Fletcher AL, Malhotra D, Turley SJ. Lymph node stroma broaden the peripheral tolerance paradigm. Trends in Immunology. 2011;32(1):12–18. doi: 10.1016/j.it.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin P, Coleman M, Leonard JP. Progress in mantle-cell lymphoma. Journal of Clinical Oncology. 2009;27(4):481–483. doi: 10.1200/JCO.2008.19.5032. [DOI] [PubMed] [Google Scholar]
- 26.Pileri SA, Falini B. Mantle cell lymphoma. Haematologica. 2009;94(11):1488–1492. doi: 10.3324/haematol.2009.013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alinari L, White VL, Earl CT, et al. Combination bortezomib and rituximab treatment affects multiple survival and death pathways to promote apoptosis in mantle cell lymphoma. MAbs. 2009;1(1):31–40. doi: 10.4161/mabs.1.1.7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suh KS, Goy A. Bortezomib in mantle cell lymphoma. Future Oncology. 2008;4(2):149–168. doi: 10.2217/14796694.4.2.149. [DOI] [PubMed] [Google Scholar]
- 29.Rittié L, Fisher GJ. Isolation and culture of skin fibroblasts. Methods in Molecular Medicine. 2005;117:83–98. doi: 10.1385/1-59259-940-0:083. [DOI] [PubMed] [Google Scholar]
- 30.Herreros B, Sanchez-Aguilera A, Piris MA. Lymphoma microenvironment: culprit or innocent? Leukemia. 2008;22(1):49–58. doi: 10.1038/sj.leu.2404970. [DOI] [PubMed] [Google Scholar]
- 31.Goy A, Feldman T. Expanding therapeutic options in mantle cell lymphoma. Clinical lymphoma & myeloma. 2007;7:S184–S191. doi: 10.3816/clm.2007.s.021. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez J, Gutierrez A, Obrador-Hevia A, Fernandez De Mattos S, Cabanillas F. Therapeutic concepts in mantle cell lymphoma. European Journal of Haematology. 2010;85(5):371–386. doi: 10.1111/j.1600-0609.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- 33.Kurtova AV, Tamayo AT, Ford RJ, Burger JA. Mantle cell lymphoma cells express high levels of CXCR4, CXCR5, and VLA-4 (CD49d): importance for interactions with the stromal microenvironment and specific targeting. Blood. 2009;113(19):4604–4613. doi: 10.1182/blood-2008-10-185827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Døsen-Dahl G, Munthe E, Nygren MK, Stubberud H, Hystad ME, Rian E. Bone marrow stroma cells regulate TIEG1 expression in acute lymphoblastic leukemia cells: role of TGFβ/BMP-6 and TIEG1 in chemotherapy escape. International Journal of Cancer. 2008;123(12):2759–2766. doi: 10.1002/ijc.23833. [DOI] [PubMed] [Google Scholar]
- 35.Lwin T, Hazlehurst LA, Li Z, et al. Bone marrow stromal cells prevent apoptosis of lymphoma cells by upregulation of anti-apoptotic proteins associated with activation of NF-κB (RelB/p52) in non-Hodgkin’s lymphoma cells. Leukemia. 2007;21(7):1521–1531. doi: 10.1038/sj.leu.2404723. [DOI] [PubMed] [Google Scholar]
- 36.Baglole CJ, Ray DM, Bernstein SH, et al. More than structural cells, fibroblasts create and orchestrate the tumor microenvironment. Immunological Investigations. 2006;35(3-4):297–325. doi: 10.1080/08820130600754960. [DOI] [PubMed] [Google Scholar]
- 37.Andersen NS, Larsen JK, Christiansen J, et al. Soluble CD40 ligand induces selective proliferation of lymphoma cells in primary mantle cell lymphoma cell cultures. Blood. 2000;96(6):2219–2225. [PubMed] [Google Scholar]
- 38.Visser HPJ, Tewis M, Willemze R, Kluin-Nelemans JC. Mantle cell lymphoma proliferates upon IL-10 in the CD40 system. Leukemia. 2000;14(8):1483–1489. doi: 10.1038/sj.leu.2401829. [DOI] [PubMed] [Google Scholar]
- 39.Tucker CA, Bebb G, Klasa RJ, et al. Four human t(11;14)(q13;q32)-containing cell lines having classic and variant features of Mantle Cell Lymphoma. Leukemia Research. 2006;30(4):449–457. doi: 10.1016/j.leukres.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 40.Da Silva CL, Gonçalves R, Crapnell KB, Cabral JMS, Zanjani ED, Almeida-Porada G. A human stromal-based serum-free culture system supports the ex vivo expansion/maintenance of bone marrow and cord blood hematopoietic stem/progenitor cells. Experimental Hematology. 2005;33(7):828–835. doi: 10.1016/j.exphem.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 41.Umiel T, Friedman S, Zaizov R. Long-term culture of infant leukemia cells: dependence upon stromal cells from the bone marrow and bilineage differentiation. Leukemia Research. 1986;10(8):1007–1013. doi: 10.1016/0145-2126(86)90253-5. [DOI] [PubMed] [Google Scholar]
- 42.Prowse ABJ, McQuade LR, Bryant KJ, Van Dyk DD, Tuch BE, Gray PP. A proteome analysis of conditioned media from human neonatal fibroblasts used in the maintenance of human embryonic stem cells. Proteomics. 2005;5(4):978–989. doi: 10.1002/pmic.200401087. [DOI] [PubMed] [Google Scholar]
- 43.Toma JG, McKenzie IA, Bagli D, Miller FD. Isolation and characterization of multipotent skin-derived precursors from human skin. Stem Cells. 2005;23(6):727–737. doi: 10.1634/stemcells.2004-0134. [DOI] [PubMed] [Google Scholar]
- 44.Michelini M, Franceschini V, Sihui Chen S, et al. Primate embryonic stem cells create their own niche while differentiating in three-dimensional culture systems. Cell Proliferation. 2006;39(3):217–229. doi: 10.1111/j.1365-2184.2006.00381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li N, Fukunaga-Kalabis M, Yu H, et al. Human dermal stem cells differentiate into functional epidermal melanocytes. Journal of Cell Science. 2010;123(6):853–860. doi: 10.1242/jcs.061598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dierks C, Grbic J, Zirlik K, et al. Essential role of stromally induced hedgehog signaling in B-cell malignancies. Nature Medicine. 2007;13(8):944–951. doi: 10.1038/nm1614. [DOI] [PubMed] [Google Scholar]
- 47.Gribben JG. Implications of the tumor microenvironment on survival and disease response in follicular lymphoma. Current Opinion in Oncology. 2010;22(5):424–430. doi: 10.1097/CCO.0b013e32833d5938. [DOI] [PubMed] [Google Scholar]
- 48.Fortney JE, Zhao W, Wenger SL, Gibson LF. Bone marrow stromal cells regulate caspase 3 activity in leukemic cells during chemotherapy. Leukemia Research. 2001;25(10):901–907. doi: 10.1016/s0145-2126(01)00051-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
To investigate differential regulations of cytoskeletal proteins (Supplementary Figure 1), known 3D-Spheroids markers (Supplementary Figure 2), Integrin members (Supplementary Figure 3) and NFkB members (Supplementary Figure 4) between 2D and 3D culture model, conventional qRT-PCR was performed by using validated primer sets and ABI-9700 under a fast cycle. Representative members of each signaling group and 3D spheroid markers were quantified and normalized by actin and the expression of each gene is shown in fold change over the parent cell line without the stroma component. The differential expression of all genes were minimal, except for the NFkB members, between 2D and 3D culture models.


