Abstract
Genetic recombination can lead to the formation of intermediates in which DNA molecules are linked by Holliday junctions. Movement of a junction along DNA, by a process known as branch migration, leads to heteroduplex formation, whereas resolution of a junction completes the recombination process. Holliday junctions can be resolved in either of two ways, yielding products in which there has, or has not, been an exchange of flanking markers. The ratio of these products is thought to be determined by the frequency with which the two isomeric forms (conformers) of the Holliday junction are cleaved. Recent studies with enzymes that process Holliday junctions in Escherichia coli, the RuvABC proteins, however, indicate that protein binding causes the junction to adopt an open square-planar configuration. Within such a structure, DNA isomerization can have little role in determining the orientation of resolution. To determine the role that junction-specific protein assembly has in determining resolution bias, a defined in vitro system was developed in which we were able to direct the assembly of the RuvABC resolvasome. We found that the bias toward resolution in one orientation or the other was determined simply by the way in which the Ruv proteins were positioned on the junction. Additionally, we provide evidence that supports current models on RuvABC action in which Holliday junction resolution occurs as the resolvasome promotes branch migration.
Keywords: Recombination, DNA repair, branch migration, crossover, nuclease
Homologous recombination is an essential cellular process that is required for the provision of genetic diversity, the maintenance of genomic stability after DNA-damage, and the restart of blocked or incomplete replication forks. Recombination can occur by a number of different mechanisms, all of which lead to the formation of heteroduplex DNA. Strand invasion mechanisms of recombination involve an intermediate structure, the Holliday junction, in which two DNA molecules are linked by a reciprocal exchange of strands (Holliday 1964; Dressler and Potter 1982). Movement of the Holliday junction, by a process known as branch migration, extends the length of heteroduplex DNA; resolution of the junction completes the recombination process and allows the separation of recombinant products.
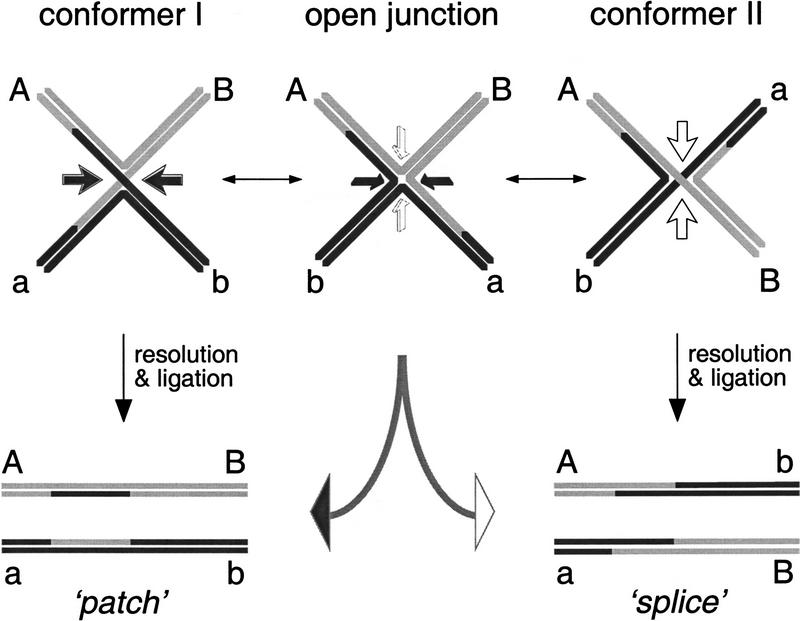
Genetic studies of the products of homologous recombination indicate that Holliday junction resolution occurs in either of two orientations, resulting in noncrossover products (‘patch’ recombinants) or crossover products (‘splice’ recombinants) in which flanking markers have been exchanged (Szostak et al. 1983). Because Holliday junctions can isomerize between two equivalent forms (differing only in the pair of strands that exchange at the crossover point; see Fig. 1), the classical view has been that the two types of recombinants are produced by cleavage of the two isomers (Holliday 1964; Meselson and Radding 1975; Szostak et al. 1983). As these species correspond to the two alternative choices of stacking partners, they are more properly termed ‘alternative conformers’ (Altona 1996). In vitro studies with synthetic four-way junctions have defined the physical properties of Holliday junctions (for review, see Lilley and Clegg 1993) and have shown that the isomeric form of a junction can be biased by localized sequence effects at the crossover point (Murchie et al. 1991; Chen and Chazin 1994; Miick et al. 1997). Such a ‘crossover preference’ could influence the outcome of a recombination event at any particular locus by biasing the ratio of crossover to noncrossover product formation. There is evidence that, in some circumstances at least, Holliday junctions may be resolved preferentially in one orientation. For example, whereas the frequent outcome of meiotic recombination is the formation of crossovers, mitotic recombination events often favor noncrossover product formation (McGill et al. 1989).
Figure 1.
Classical view of the formation of patch (i.e., noncrossover) and splice (crossover) recombination products by resolution of pairs of crossed strands present within the two isomeric forms (conformers) of the Holliday junction. Recent studies have shown that resolvases can bind Holliday junctions and cause them to adopt an open square-planar structure (middle). The studies described in this report indicate that the type of recombinant formed is dependent on the way that the resolvase assembles on the four fold symmetric open junction. Nucleolytic cleavage to give rise to patch or splice recombinants is indicated by the solid and open arrows, respectively.
Our understanding of the mechanism of Holliday junction resolution has been advanced by the isolation and characterization of junction-specific endonucleases from bacteriophage (T4 endonuclease VII, T7 endonuclease I), Escherichia coli (RuvC, RusA), yeast (Cce1), and humans (for reviews, see West 1993, 1996; Kemper 1998). These enzymes bind specifically to Holliday junctions and promote their cleavage by introducing two symmetrically related nicks at, or close to, the site of the crossover. Resolution occurs in either of the two possible orientations resulting in the formation of nicked duplex products that can be repaired by DNA ligase.
Using purified enzymes from E. coli, it is now possible to reconstitute the late stages of the recombination process. RecA protein promotes homologous pairing and strand exchange reactions leading to the formation of Holliday junctions, which are subsequently acted on by the RuvABC proteins (Eggleston et al. 1997). The RuvA and RuvB proteins have a dual role, first by promoting branch migration of the junction (Shiba et al. 1991; Iwasaki et al. 1992; Parsons et al. 1992; Tsaneva et al. 1992b), and second, by facilitating RuvC-mediated Holliday junction resolution (Mandal et al. 1993; van Gool et al. 1998; Zerbib et al. 1998).
Tetrameric RuvA protein binds specifically to Holliday junctions (Iwasaki et al. 1992; Parsons et al. 1992; Tsaneva et al. 1992a) and causes them to adopt a fourfold symmetric open structure in which the four arms are extended toward the corners of a square (Parsons et al. 1995; Rafferty et al. 1996; Hargreaves et al. 1998; Roe et al. 1998). RuvA also targets RuvB to the junction (Parsons and West 1993). RuvB is a hexameric ring helicase that acts as the motor of branch migration (Tsaneva et al. 1993; Stasiak et al. 1994). Electron microscopic visualization of the RuvAB–Holliday junction complex revealed that the junction lies sandwiched between two RuvA tetramers and that branch migration is driven by two oppositely oriented RuvB rings that bind to opposing arms (Parsons et al. 1995; Yu et al. 1997). During branch migration, the DNA passes through RuvA and exits the complex through the cavity in each RuvB ring (Hiom et al. 1996).
The RuvC protein promotes Holliday junction resolution in E. coli (Connolly et al. 1991; Dunderdale et al. 1991; Iwasaki et al. 1991). In vitro, dimeric RuvC binds specifically to Holliday junctions and catalyzes their resolution (Bennett et al. 1993; Takahagi et al. 1994). Incision occurs at the 3′-side of thymidine, preferentially at the consensus 5′-A/TTT↓C/G -3′ where ↓ indicates the site of cleavage (Shah et al. 1994; Shida et al. 1996; Shah et al. 1997; Hagan et al. 1998). In accord with genetic data indicating that RuvA, RuvB, and RuvC function in concert (Benson et al. 1991; Mandal et al. 1993), RuvC-mediated Holliday junction resolution is stimulated in vitro by the presence of RuvAB (van Gool et al. 1998; Zerbib et al. 1998). RuvC may therefore be regarded as the resolution subunit of a RuvABC–Holliday junction complex that is capable of both branch migration and Holliday junction resolution (Rafferty et al. 1996; Whitby et al. 1996; Davies and West 1998; van Gool et al. 1998; West 1998; Zerbib et al. 1998). In this report, we provide evidence for coupled branch migration and resolution, and show that the assembly of the RuvABC resolvasome on the open square Holliday junction directs the orientation of Holliday junction resolution.
Results
Formation of RuvAB- and RuvABC–Holliday junction complexes in vitro
The RuvA, RuvB, and RuvC proteins are thought to form two types of Holliday junction processing complexes (West 1998): (1) RuvAB form a tripartite complex that is capable of efficient branch migration (Fig. 2A), and (2) RuvABC have been proposed to form a ‘resolvasome’ that promotes both branch migration and Holliday junction resolution (Fig. 2B).
Figure 2.
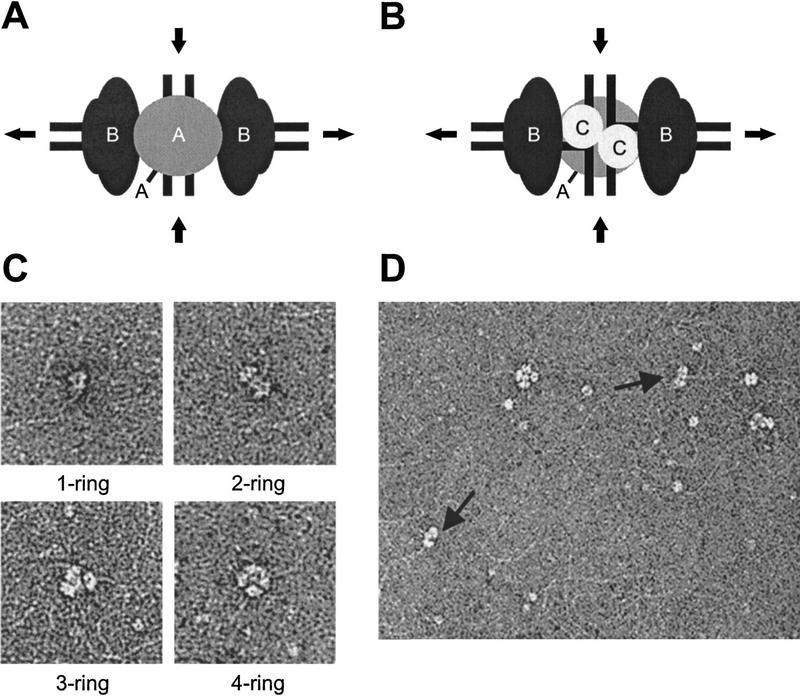
Visualization of RuvAB- and RuvABC–Holliday junction complexes. (A) Schematic diagram illustrating a model of the RuvAB–junction complex that promotes branch migration. In this complex, the Holliday junction DNA lies sandwiched between two RuvA tetramers. Hexameric rings of RuvB lie oppositely oriented on two arms of the junction, where they encircle the DNA. In association with RuvA, the RuvB rings promote ATP-dependent branch migration, as indicated by the arrows. (B) Model of a RuvABC complex capable of branch migration and resolution. The Holliday junction lies sandwiched between one RuvA tetramer and one RuvC dimer, each binding to a distinct face of the junction. The two RuvB hexamers are positioned as in the RuvAB complex and promote branch migration, thereby allowing RuvC to scan the DNA for preferred cleavage sites as they pass through the complex. (C) Electron microscopic visualization of RuvAB–junction complexes containing one, two, three, or four RuvB rings. χ-form DNA (1 nm) was incubated with RuvA (60 nm) and RuvB (25–200 nm) as described in Material and Methods and complexes were analyzed by EM after negative staining with uranyl acetate. (D) EM image showing a variety of protein–DNA complexes formed by incubation of χ-DNA (1 nm) with RuvA (60 nm), RuvB (200 nm), and RuvC (120 nm). Several complexes can be identified here, which include a four-ring complex, a three-ring complex, 2 two-ring complexes (arrows), and a doublet comprising two RuvB rings.
In an attempt to visualize these complexes by electron microscopy (EM) we used χ-form DNA containing Holliday junctions. As described previously (Parsons et al. 1995), incubation of RuvA (60 nm) and RuvB (25–300 nm) with χ-form DNA (1 nm) resulted in the formation of complexes in which RuvA was flanked by one, two, three, or four RuvB rings (Fig. 2C). The transition from one-, to two-, to three-, to four-ringed structures was dependent on the concentration of RuvB protein (Table 1). At the highest RuvB concentrations (200–300 nm), four-ringed complexes were the dominant species. Two-ringed complexes (shown schematically in Fig. 2A), which are thought to be the functional form of the RuvAB branch migration motor (Hiom and West 1995), were the predominant species at intermediate protein concentrations (50 nm RuvB; Table 1).
Table 1.
Effect of RuvC protein on the formation of multiring RuvAB–Holliday junction complexes
| RuvA (nM) | 60 | 60 | 60 | 60 | 60 | 60 |
|---|---|---|---|---|---|---|
| RuvB (nM) | 25 | 50 | 100 | 200 | 300 | 200 |
| RuvC (nM) | — | — | — | — | — | 120 |
| 1-ring | 53 | 0 | 0 | 0 | 0 | 0 |
| 2-ring | 46 | 96 | 29 | 2 | 5 | 32 |
| 3-ring | 0 | 4 | 62 | 36 | 19 | 43 |
| 4-ring | 0 | 0 | 10 | 62 | 77 | 25 |
Reactions contained χ-form DNA (1 nm) and the indicated concentrations of RuvA, RuvB and RuvC; the resultant complexes were spread directly onto carbon grids and analyzed by electron microscopy. Complexes containing one, two, three, or four RuvB rings (as shown in Fig. 1C) were quantified and expressed as a percentage of the total of protein–junction complexes encountered by random analysis of the grid.
To determine whether RuvC affected the formation of multi-ring RuvAB complexes, possibly by favoring the formation of a twofold symmetric complex containing RuvA, RuvB, and RuvC (as shown in Fig. 2B), EM studies were carried out in the presence of all three enzymes. We found that addition of 120 nm RuvC protein, to reactions containing 60 nm RuvA and 200 nm RuvB, increased the number of two-ring complexes from 2% to 32% (Fig. 2D, arrows; and Table 1). Unfortunately, our EM studies did not reveal any obvious differences between RuvAB complexes (624 kD) and the presumptive RuvABC complexes (572 kD). When the complexes were immunolabeled with gold-conjugated anti-RuvC antisera, however, we found that 35% of the complexes in the RuvABC sample became labeled compared with only 2% of those present in the RuvAB sample (data not shown). Because stable RuvBC-junction complexes were not detected by electron microscopy (data not shown), these results indicate that a significant portion of the two-ring complexes represent RuvABC complexes.
Assembly of the RuvABC complex directs the orientation of Holliday junction resolution
The classical view that the orientation of Holliday junction resolution is a consequence of the cleavage of one, or the other, isomeric form (conformer) of the junction is difficult to correlate with data indicating that many Holliday junction resolving proteins unfold junctions (Bennett and West 1995; Duckett et al. 1995; Pöhler et al. 1996; White and Lilley 1997, 1998). We therefore tested an alternative hypothesis—that the asymmetric binding of the resolvase (in this case the RuvABC resolvasome) to the open junction could direct the orientation of resolution. To do this, it was necessary to construct DNA substrates, which would limit the assembly of the RuvABC resolvasome to a single orientation. We therefore prepared two synthetic Holliday junctions (defined as junctions X1 and X2), each with two long (45 bp) and two short (21 bp) arms. The substrates were designed such that the long and the short arms were the opposite in each case (Fig. 3A). Because hexameric rings of RuvB require 24–27 bp of DNA to allow stable binding (Hiom and West 1995), X1 permits binding to the opposite pair of arms compared with X2. The RuvABC complex will therefore be positioned in two alternative configurations (see schematic in Fig. 3A). We reasoned that these alternative configurations might favor resolution in one orientation or the other.
Figure 3.
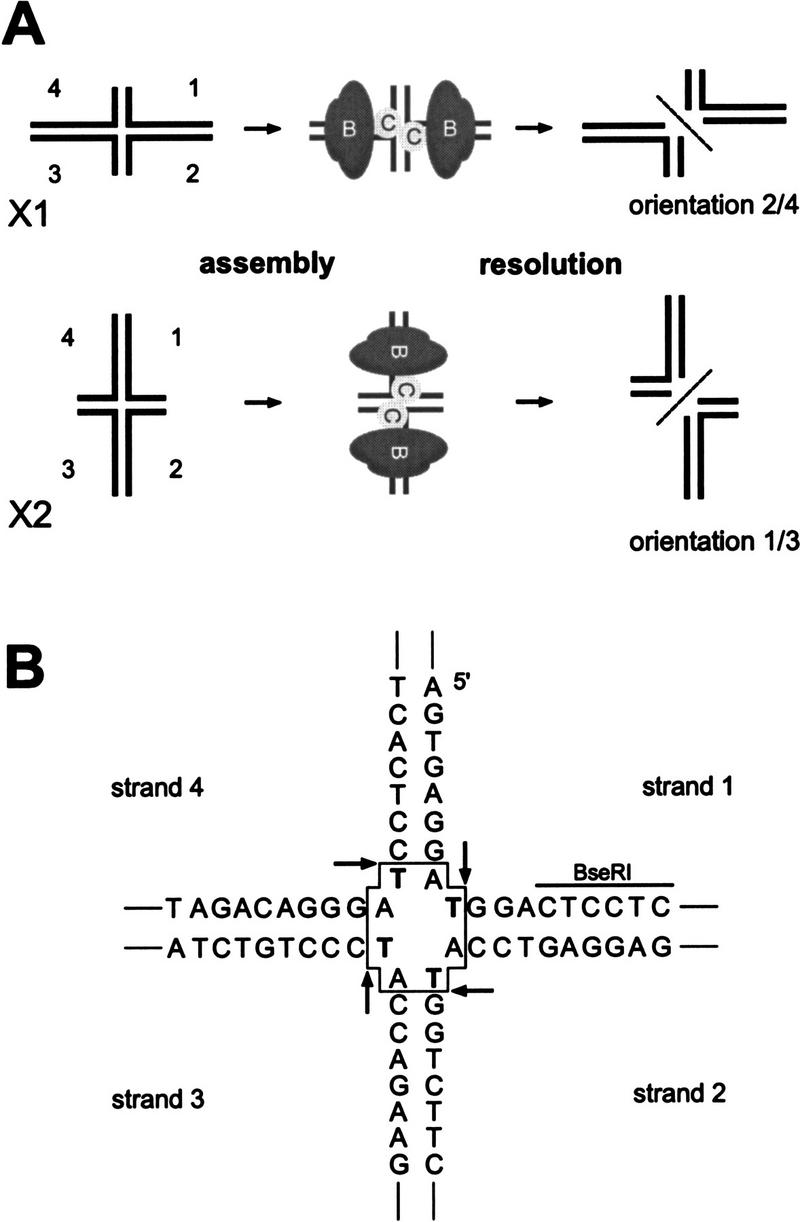
Design of the experimental system. (A) Junctions X1 and X2 are related by an identical core region, but differ in the pair of long arms, as indicated. Because RuvB has a minimal length requirement for DNA binding, it can only bind to the long arms. The experimental rationale was to determine whether the binding of RuvB to these arms would direct the loading of RuvC resolvase, thereby influencing the orientation of Holliday junction resolution (either 2/4 or 1/3). For simplicity, the presence of RuvA is not indicated on this diagram. (B) DNA sequence at the central core of junctions X1 and X2. All strands contain a thymidine residue at crossover point, at which RuvC can cleave at the 3′-side (as indicated by arrows). The presence of the BseRI restriction site in strand 1 enabled us to show that both conformers of each junction were present in the various DNA preparations.
To avoid any sequence or contextual complications, the central cores of junctions X1 and X2 were identical (Fig. 3B) and each of the four strands contained a thymidine residue at which RuvC would be expected to cleave (albeit with a reduced efficiency attributable to the lack of a true consensus sequence). In addition, by incorporating a BseRI restriction site in one of the junction arms (Fig. 3B), we confirmed that each preparation of junction contained equal amounts of the two conformers (as described in Murchie et al. 1991; data not shown).
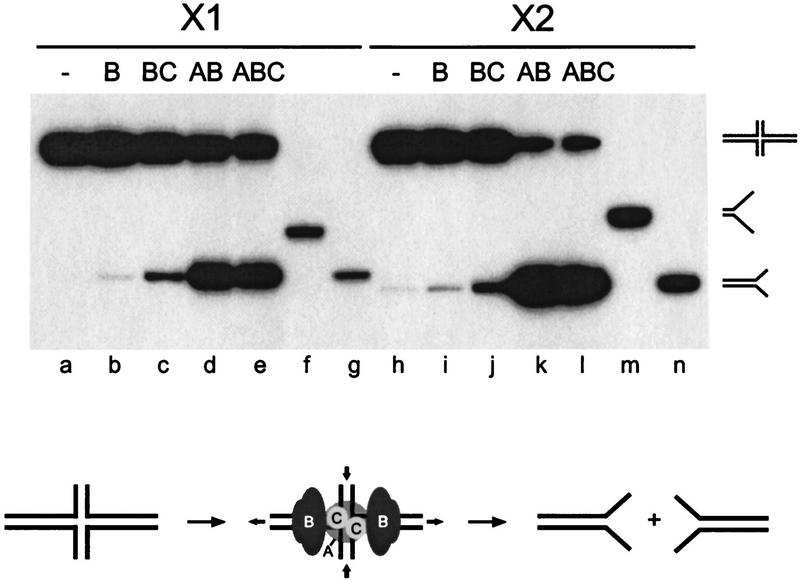
When X1 was incubated with RuvC alone and the cleavage patterns were analyzed by denaturing PAGE (Fig. 4A, lanes b–e), we observed that resolution occurred in both orientations (i.e., by cleavage of strands 1/3 and strands 2/4). The efficiency of resolution was low and a twofold preference for cleavage in orientation 2/4 was observed (Table 2A). The presence of RuvB resulted in a fourfold increase in resolution in strands 2/4, whereas cleavage of strands 1/3 remained unchanged (Fig. 4A, lanes g–j; Table 2A). As these reactions were carried out in the presence of a nonhydrolyzable ATP analog, ATPγS, to inhibit branch migration, it appears that this bias in resolution is attributable to the assembly of the RuvB rings on the long arms of the junction. When similar reactions were carried out using RuvA, RuvB, and RuvC, further stimulation of cleavage in strands 2/4 was observed and in this case resolution in the opposite orientation was barely detectable (Fig. 4A, lanes l–o; Table 2A). Novel cleavage sites were also observed in strands 2 and 4 (Fig. 4A, lanes m, o) and their nature will be discussed later. In summary, using junction X1, the presence of RuvAB resulted in a 16-fold bias in the orientation of Holliday junction resolution by RuvC.
Figure 4.
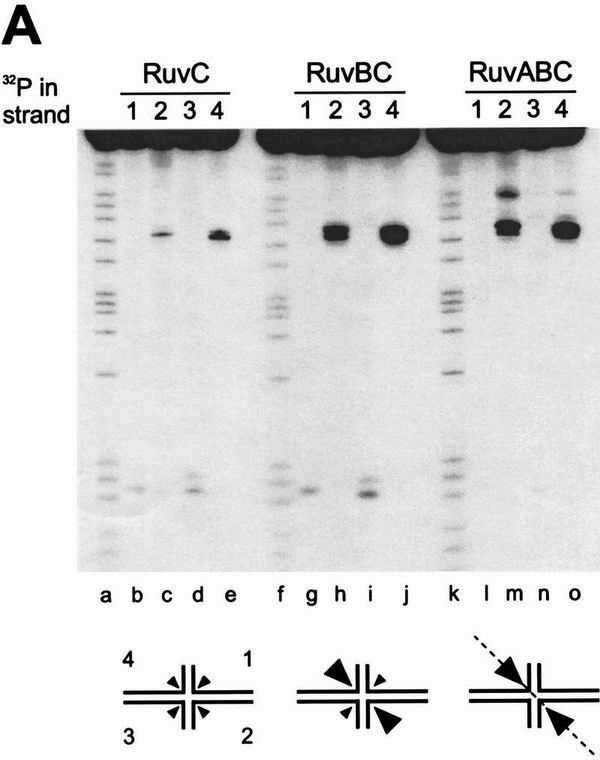
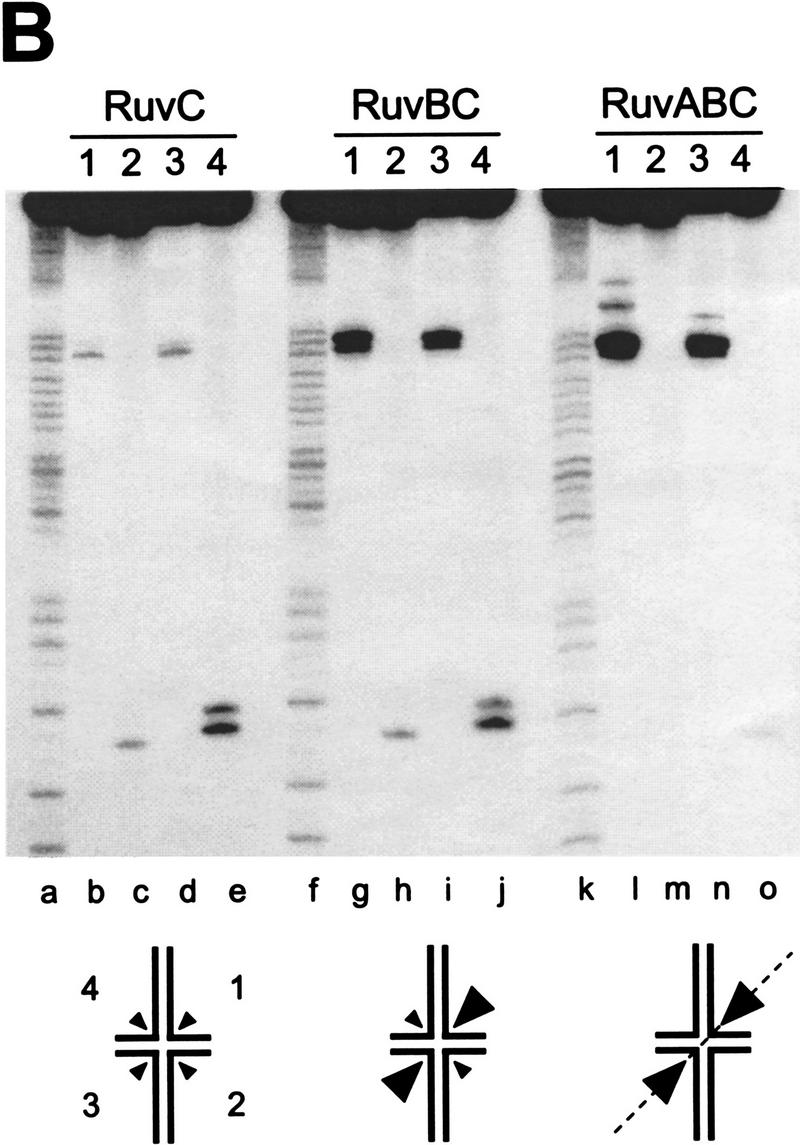
RuvAB impose an orientation bias on RuvC-mediated Holliday junction resolution. Synthetic junctions X1 (A) or X2 (B) were 5′-32P-end labeled in strands 1, 2, 3, or 4, as indicated, and incubated with RuvC (lanes b–e), with RuvB and RuvC (lanes g–j), or with RuvA, RuvB, and RuvC (lanes l–o) for 30 min at 37°C in cleavage buffer, as described in Materials and Methods. RuvA, RuvB, and RuvC were present at 20 nm, 600 nm, and 10 nm, respectively. The DNA concentration was 1 nm. Cleavage products were analyzed by denaturing PAGE, alongside A + G sequence ladders of strand 1 of each junction (lanes a,f,k). The schematic diagram indicates the relative levels of resolution in each orientation.
Table 2.
RuvAB directs the orientation of RuvC-mediated Holliday junction resolution
|
A X1 orientation
|
Resolution (%)
|
Fold stimulation
|
||||
|---|---|---|---|---|---|---|
|
C
|
BC
|
ABC
|
C
|
BC
|
ABC
|
|
| 2/4 | 2.6 | 10.1 | 12.8 | 1 | 3.9 | 4.9 |
| 1/3
|
1.1
|
1.2
|
0.3
|
1
|
1.1
|
0.3
|
| B X2 orientation | ||||||
| 2/4 | 1.9 | 2.3 | 0.4 | 1 | 1.2 | 0.2 |
| 1/3 | 1.3 | 6.9 | 12.9 | 1 | 5.3 | 9.9 |
The data shown in Fig. 3, produced by the cleavage of junctions X1 or X2 with RuvC, RuvBC, or RuvABC, was quantified by PhosphorImaging. Resolution in orientations 2/4 or 1/3 is expressed as a percentage of total resolution and relative to cleavage by RuvC alone.
To determine whether the orientation bias was imposed by the positioning of the RuvB rings onto one pair of junction arms, similar experiments were carried out with junction X2 (Fig. 4B). RuvC was found to cleave X2 in both orientations (Fig. 4B, lanes b-e; Table 2B) and the presence of RuvB resulted in a fivefold stimulation of resolution in orientation 1/3, but not in orientation 2/4 (Fig. 4B, lanes g–j; Table 2B). The presence of RuvA exaggerated this effect further and a near total bias for cleavage in orientation 1/3 was observed (Fig. 4B, lanes l–o; Table 2B). Overall, RuvAB imposed a 50-fold bias in the orientation of resolution compared with RuvC alone. This orientation bias was the opposite of that observed with junction X1.
Previously, it was shown that the pair of arms not bound by RuvB are pumped into the RuvAB complex and out through the RuvB rings (Hiom and West 1995). The same experimental rationale was used to confirm that the bias in orientation resolution was attributable to the way that the RuvABC resolvasome assembled on the Holliday junction. To do this, the reaction conditions were adjusted to inhibit resolution (by replacing Mg2+ with Ca2+) and the products of branch migration were analyzed. In comparison with defined DNA markers generated by annealing pairs of oligonucleotides, the products of branch migration were found to be representative of those expected from a reaction in which RuvB was targeted exclusively to the long arms of junctions X1 and X2 (Fig. 5). Taken together, the results presented in Figures 4 and 5 show that the orientation of assembly of the RuvABC resolvasome at the Holliday junction determines the direction of resolution.
Figure 5.
Specific targeting of RuvB to the long arms of junctions X1 and X2. Junctions X1 and X2 (5′-32P-end labeled in strand 1) were incubated with combinations of RuvA, RuvB, and RuvC for 30 min at 37°C, as indicated, in branch migration buffer containing Ca2+ (to inhibit resolution) and ATP (1 mm). Branch migration products were deproteinized and analyzed by neutral PAGE, alongside branch migration markers generated by annealing X1 strands 1 + 4 (lane f); X1 strands 1 + 2 (lane g); X2 strands 1 + 2 (lane m); or X2 strands 1 + 4 (lane n). The cartoon below indicates those branch migration products formed when the RuvB rings bind to the longer junction arms and promote branch migration to release splayed duplex products.
Branch migration by RuvAB, RuvBC, and RuvABC complexes
The experiment in Figure 5 also permitted a comparison of the relative branch migration activities of the various Ruv complexes. As observed previously (van Gool et al. 1998), RuvC enhanced the branch migration activity of RuvB (Fig. 5A, cf. lane b with c, and i with j), presumably by helping to stabilize the interaction of RuvB with the DNA. As expected, however, RuvA protein stimulated RuvB-mediated branch migration more effectively than RuvC (Fig. 5; lanes d, k). The branch migration activities of RuvABC were comparable with RuvAB under these experimental conditions (Fig. 5; lanes e, l). At low RuvB concentrations (<30 nm), we observed that the RuvABC proteins were less effective at branch migration than RuvAB (data not shown). The reason for this difference is unknown, but might involve competition between RuvA and RuvC for RuvB protein.
Holliday junction resolution during RuvABC-mediated branch migration
Recent models for RuvABC action suppose that Holliday junction resolution occurs as the RuvC protein scans DNA sequences during RuvAB-mediated branch migration, within a RuvABC resolvasome complex (Shah et al. 1994; Rafferty et al. 1996; Whitby et al. 1996; Eggleston et al. 1997; van Gool et al. 1998; Zerbib et al. 1998). Direct evidence for resolution during branch migration, however, is presently lacking.
To determine whether branch migration and resolution are coupled, RuvABC-mediated cleavage reactions were carried out under conditions in which limited branch migration was permitted by inclusion of a mixture of ATP and the slowly-hydrolyzable ATP analog ATPγS. We observed that RuvAB stimulated junction cleavage by RuvC and that nicks were introduced at the 3′-side of the central thymidine and its adjacent nucleotide, and also at a site located six nucleotides away from the junction point (Fig. 6, lane d). The latter site mapped to the 3′-side of a TT dinucleotide, consistent with cleavage at a preferred site for RuvC-mediated cleavage (Shah et al. 1994) and was located in the short arms away from the initial position of the crossover. Cleavage at this site was not observed when reactions were carried out in the presence of AMP–PNP which inhibits branch migration (Fig. 6, lane c). In the absence of a nucleoside triphosphate cofactor, neither branch migration nor resolution were observed (Fig. 6, lane e), as observed previously (Zerbib et al. 1998). Similar results were obtained with junction X2 (data not shown).
Figure 6.
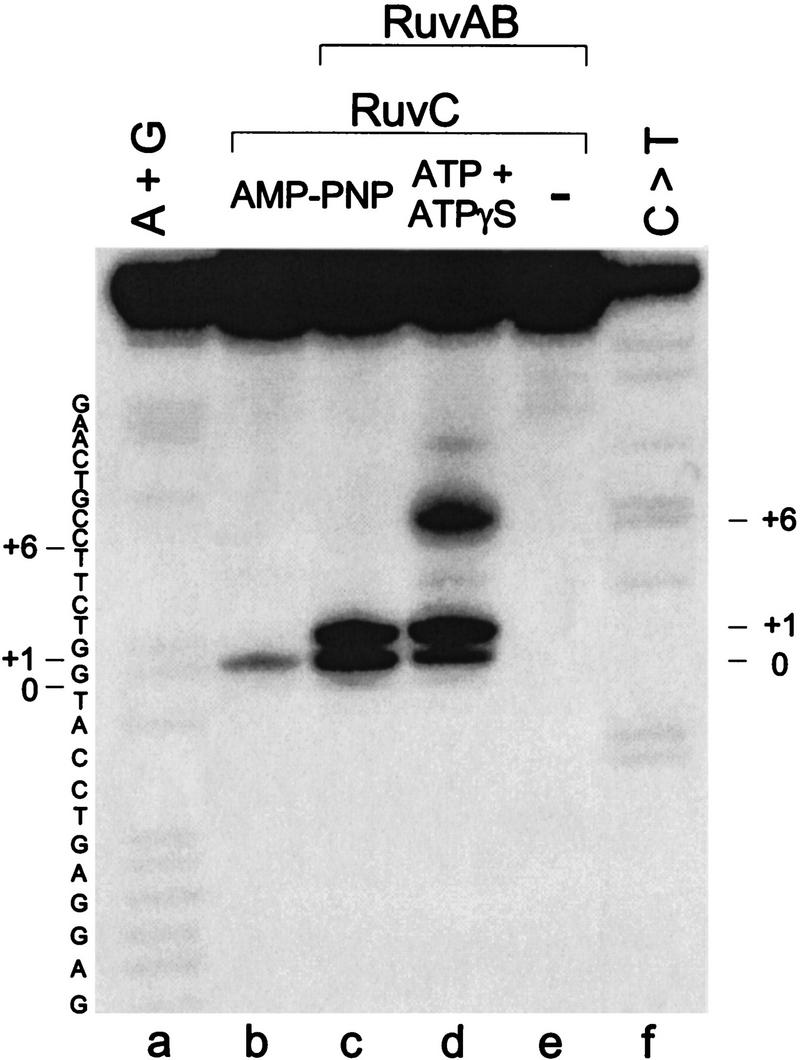
Holliday junction resolution can occur during branch migration. Junction X1 (5′-32P-end labeled in strand 2) was incubated with RuvC (10 nm), or with RuvA (20 nm), RuvB (600 nm) and RuvC (10 nm) for 30 min at 37°C, as indicated. In this experiment, resolution was permitted by inclusion of Mg2+, whereas branch migration was either blocked using AMP–PNP (lanes b and c) or slowed by the presence of a mixture of ATPγS and ATP (lane d). (Lane e) Reaction carried out in the absence of nucleoside triphosphates. 32P-Labeled products were analyzed by denaturing PAGE, alongside A + G and C > T sequence ladders, obtained by chemical cleavage of X1 strand 2. The thymine at the base of the junction is indicated by 0, and +1 and +6 indicate cleavage sites in the short arm away from the crossover.
Because the unique cleavage site was introduced in the short arms, away from the junction point, these results show that cleavage can take place as the DNA is pumped into and through the RuvABC–junction complex. The data are therefore indicative of a coupled branch migration/resolution reaction.
Discussion
Using purified RuvABC proteins and defined Holliday junction DNA substrates, we have shown that: (1) the presence of RuvC affects the assembly of RuvAB-junction complexes, favoring the formation of two-ring complexes that are thought to contain all three proteins assembled as a resolvasome, (2) the presence of RuvAB, presumably within the context of a resolvasome, directs the orientation of RuvC-mediated Holliday junction resolution; and (3) junction resolution by the resolvasome can occur at preferred DNA sequences located away from the initial junction point, most likely by means of a coupled branch migration/resolution reaction. The formation of a resolvasome complex, as indicated by the studies described here, is consistent with previous proposals that efficient Holliday junction resolution occurs as the resolution subunit (RuvC) scans the DNA for preferred sites of cleavage during RuvAB-mediated branch migration (Shah et al. 1994; Whitby et al. 1996; van Gool et al. 1998; Zerbib et al. 1998).
Recent structural studies have strengthened the argument that RuvAB and RuvABC complexes are both pres-ent in the cell (for review, see West 1998). Because the ruvA and ruvB genes are part of the E. coli SOS system, the induced synthesis of RuvA and RuvB (but not RuvC) in response to DNA damaging agents (Shurvinton and Lloyd 1982; Shinagawa et al. 1988) will affect the ratio of these two complexes. One consequence of RuvAB induction will be the formation of more branch migration motors that promote the formation of heteroduplex DNA. Because RuvAB can drive branch migration through insertions and deletions that normally block RecA-mediated strand exchange (Morel et al. 1994; Adams and West 1996), branch migration through these regions could enhance the error-prone survival of the cell. Additionally, elevated levels of RuvA and RuvB may serve to activate the RuvC resolvase by favoring its assembly into RuvABC resolvasomes. The presence of an activated resolvasome would facilitate the completion of damage-induced recombinational repair in times of stress, consistent with the repair-defective phenotype exhibited by ruv mutants (Sharples et al. 1990).
Models for Holliday junction resolution that assume that patch and splice recombinants arise by cleavage of the two alternative junction conformers have been questioned by structural studies showing that resolvases (Bennett and West 1995; Duckett et al. 1995; Pöhler et al. 1996; White and Lilley 1997, 1998; Chan et al. 1998; Giraud-Panis and Lilley 1998), and junction-binding proteins in general (Parsons et al. 1995; Rafferty et al. 1996; Hargreaves et al. 1998; Pöhler et al. 1998; Roe et al. 1998), alter junction structure, usually causing them to adopt an open-square structure (Fig. 1). In the case of RuvA protein, in particular, the junction within the protein–DNA complex lies in a fourfold symmetric configuration in which the four DNA arms are extended toward the corners of a square (Parsons et al. 1995; Hargreaves et al. 1998; Roe et al. 1998). Given such a structure, what factors determine resolution orientation?
In these studies, we have shown that the orientation of loading of the RuvABC complex onto the junction determines the direction of Holliday junction resolution. Using DNA substrates containing two long and two short arms, we found that the presence of RuvA and RuvB resulted in a strong (i.e., up to 50-fold) bias for resolution in one orientation compared with the other. This bias was shown to be attributable to the binding of RuvB to the long arms and the consequential unidirectional binding of the RuvC dimer. It is of interest to note that resolution in one orientation can only occur when the resolvase binds a unique face of the junction, as binding in a similar orientation to the other face would result in cleavage of the other pair of DNA strands. The co-crystal structure of the RuvA–junction complex (Hargreaves et al. 1998) indicates that RuvA binds exclusively to the major groove face of the junction, and it is tempting to suggest that, in the presence of RuvA, RuvC binds to the minor groove of the junction. The notion that RuvA and RuvC bind opposing faces is consistent with previous data on the formation of a RuvAC–Holliday junction complex (Whitby et al. 1996), and is supported by our finding that RuvA enhances cleavage in one orientation, whereas it inhibits cleavage in the other. Our observation that RuvAB stimulates the activity of RuvC is also consistent with the idea that the resolvasome scans DNA sequences for suitable resolution sites. New cleavage sites located several bases away from the original junction point were observed when limited branch migration was permitted.
Using E. coli proteins, we have shown that assembly of the resolvasome can influence the outcome of a recombination event. Although eukaryotic Holliday junction resolvases have not yet been identified, related activities have been observed during the fractionation of mammalian extracts (Elborough and West 1990; Hyde et al. 1994), indicating that similar junction-processing events are likely to occur in eukaryotes. Our data indicate that factors that affect the assembly of the resolvase at the site of the junction may affect the outcome of a recombination event. For example, DNA supercoiling, chromatin structure, or the presence of a nearby Holliday junction, could all affect resolvasome assembly and bias resolution in one orientation or the other. Previous in vitro studies with RuvC protein showed that catenated supercoiled figure-eight DNA molecules were resolved more efficiently than relaxed figure-eights and in a different orientation (Zerbib et al. 1997). Other studies showed that the orientation of Holliday junction resolution by RuvC was affected by the presence of RecA (Dunderdale et al. 1991) and that this bias was removed by RuvAB (Eggleston et al. 1997). Presumably, topological constraints imposed on the DNA, either by catenation or by the presence of the RecA filament, are capable of holding the junction in a configuration that favors resolution in one orientation. In E. coli, recombination proteins such as RecF, RecO, RecR, and SSB may influence the resolution reaction (Umezu et al. 1993; Webb et al. 1997). In eukaryotes, where we are only just beginning to understand the roles that the Rad51, Rad52, Rad54, Rad55, Rad57, and Dmc1 proteins have in recombination (Shinohara and Ogawa 1995; Baumann and West 1998; Kanaar et al. 1998), the situation is likely to be more complex because of the presence of chromatin and the interplay between DNA replication and recombination (Sonoda et al. 1998; Holmes and Haber 1999). To understand these processes in more detail, it will be necessary to identify enzymes involved in the processing of Holliday junction in eukaryotic cells and determine whether they act in a manner analogous to their bacterial counterparts.
Materials and methods
Proteins and DNA
RuvA (Tsaneva et al. 1992a), RuvB (Tsaneva et al. 1992a), and RuvC (Dunderdale et al. 1994) were purified as described. Protein concentrations are expressed in moles of protein monomers.
χ-Form DNA was prepared by restriction digestion of figure-eight DNA (McCulloch et al. 1994) with ScaI and StyI, essentially as described (Zerbib et al. 1997). Concentrations of χ-form DNA are expressed in moles of junctions.
Synthetic Holliday junctions X1 and X2 were generated by annealing four 66-mer oligonucleotides and purified as described (Parsons and West 1990). Before annealing, one oligonucleotide was 5′-32P-end labeled using T4 polynucleotide kinase. X1 and X2 contain one pair of long (45 bp) and one pair of short (21 bp) arms. In each, the core sequence (42 bp) is identical. The four oligonucleotides used to prepare junction X1 were: strand 1 (5′-GCCGAATTCTACCAGTGAGGATGGACTCCTCACCTGCAGGTTCACCGTTGTATGCCCACGTTGACC); strand 2 (5′-GGTCAACGTGGGCATACAACGGTGAACCTGCAGGTGAGGAGTCCATGGTCTTCCGTCAAGCTCGAT); strand 3 (5′-ATCGAGCTTGACGGAAGACCATCCCTGTCTAGAGGATCCGACTATCTTGACTAGACGGATTCGATA); and strand 4 (5′-TATCGAATCCGTCTAGTCAAGATAGTCGGATCCTCTAGACAGGGATCCTCACTGGTAGAATT-CGGC). Similarly, junction X2 was made from the following four strands: strand 1 (5′-TATCGAATCCGTCTAGTCAACGCTGCCGAATTCTACCAGTGAGGATGGACTCCTCACCTGCAGGTT); strand 2 (5′-AACCTGCAGGTGAGGAGTCCATGGTCTTCCGTCAAGCTCGATGCCGGTTGTATGCCCACGTTGACC); strand 3 (5′-GGTCAACGTGGGCATACAACCGGCATCGAGCTTGACGGAAGACCATCCCTGTCTAGAGGATCCGAC); and strand 4 (5′-GTCGGATCCTCTAGACAGGGATCCTCACTGGTAGAATTCGGCAGCGTTGACTAGACGGATTCGATA). Concentrations of junction DNA are expressed in moles of junctions. Branch migration markers were prepared by annealing strands 1 + 4 or 1 + 2, of which strand 1 was 5′-32P-end labeled.
Electron microscopy
Binding reactions (20 μl) contained 0.3 nm (50 ng) χ-form DNA in 40 mm triethanolamine-HCl (pH 7.5), 10 mm Mg(OAc)2, 1 mm DTT, 1 mm ATPγS, 60 nm RuvA, 25–300 nm of RuvB and 0–120 nm RuvC. Reactions were set up at room temperature as follows: DNA + RuvA (3 min), followed by addition of RuvC (3 min), and addition of RuvB (15 min). When RuvC was not added, reactions were supplemented with an equal volume of RuvC storage buffer. Samples were spread using the magnesium acetate droplet technique followed by adsorption onto glow-discharged carbon-coated grids. The specimens were negatively stained with 2% aqueous uranyl acetate (Sogo et al. 1987) and analyzed using a Philips CM100 electron microscope.
Resolution assays
5′-32P-Labeled synthetic Holliday junctions (1 nm) were incubated with RuvC in cleavage buffer (50 mm Tris-acetate, pH 8.0, 15 mm Mg(OAc)2, 1 mm DTT, 1 mm ATPγS and 50 μg/ml BSA; total volume 20 μl) for 30 min at 37°C. RuvC (10 nm final concentration) was added alone or premixed with RuvB (600 nm) or with RuvA (20 nm) and RuvB (600 nm). Labeled DNA products were analyzed by 12% denaturing PAGE after addition of 1 volume of sample buffer (80% formamide, 0.1% bromophenol blue, 0.1% xylene cyanol) and heating at 80°C for 3 min. Sites of cleavage were determined by comparison with sequence ladders produced by chemical cleavage of the appropriate 5′-32P-labeled strand (Maxam and Gilbert 1980). Because of the chemistry of chemical cleavage, markers ran ∼1 nucleotide faster than fragments of equivalent length produced by enzymatic cleavage. Gels were dried onto paper and exposed to Kodak Biomax films or quantified using a PhosphorImager (Molecular Dynamics).
To study coupled branch migration/resolution, RuvC (10 nm), or a mixture of RuvA (20 nm), RuvB (600 nm) and RuvC (10 nm), was incubated with 5′-32P-labeled junction (1 nm) in cleavage buffer supplemented with 0.25 mm ATP. In control reactions, the 1 mm ATPγS in the cleavage buffer was either omitted, or was replaced with 1 mm AMP–PNP. DNA products were analyzed by denaturing PAGE as described above.
Branch migration assays
Synthetic junctions (1 nm), 5′-32P-labeled in strand 1, were incubated with combinations of RuvA (20 nm), RuvB (600 nm), and RuvC (10 nm) in 50 mm Tris-acetate at pH 8.0, 15 mm CaCl2, 1 mm DTT, 1 mm ATP, and 50 μg/ml BSA for 30 min at 37°C (total volume 20 μl). The DNA products were deproteinized and analyzed by 8% neutral PAGE, as described previously (van Gool et al. 1998).
Acknowledgments
We thank our colleagues in the ICRF genetic recombination laboratory for discussions and comments on the manuscript and Jacques Dubochet for his interest in the work. These studies were supported by the ICRF, the Human Frontiers Science Program, the Swiss National Foundation, and the Swiss-British Council Joint Research Program. A.v.G was supported in part by an European Molecular Biology Organization fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL s.west@icrf.icnet.uk; FAX (44)-(0)-171-269-3811.
References
- Adams DE, West SC. Bypass of DNA heterologies during RuvAB-mediated three- and four-strand branch migration. J Mol Biol. 1996;263:582–596. doi: 10.1006/jmbi.1996.0600. [DOI] [PubMed] [Google Scholar]
- Altona C. Classification of nucleic acid junctions. J Mol Biol. 1996;263:568–581. doi: 10.1006/jmbi.1996.0599. [DOI] [PubMed] [Google Scholar]
- Baumann P, West SC. Role of the human Rad51 protein in homologous recombination and double-stranded break repair. Trends Biol Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Dunderdale HJ, West SC. Resolution of Holliday junctions by RuvC resolvase: Cleavage specificity and DNA distortion. Cell. 1993;74:1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, West SC. Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J Mol Biol. 1995;252:213–226. doi: 10.1006/jmbi.1995.0489. [DOI] [PubMed] [Google Scholar]
- Benson FE, Collier S, Lloyd RG. Evidence of abortive recombination in ruv mutants of Escherichia coli K-12. Mol Gen Genet. 1991;225:266–272. doi: 10.1007/BF00269858. [DOI] [PubMed] [Google Scholar]
- Chan SN, Vincent SD, Lloyd RG. Recognition and manipulation of branched DNA by the RusA Holliday junction resolvase of Escherichia coli. Nucleic Acids Res. 1998;26:1560–1566. doi: 10.1093/nar/26.7.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SM, Chazin WJ. 2-dimensional H1-NMR studies of immobile Holliday junctions: Nonlabile proton assignments and identification of crossover isomers. Biochemistry. 1994;33:11453–11459. doi: 10.1021/bi00204a007. [DOI] [PubMed] [Google Scholar]
- Connolly B, Parsons CA, Benson FE, Dunderdale HJ, Sharples GJ, Lloyd RG, West SC. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc Natl Acad Sci. 1991;88:6063–6067. doi: 10.1073/pnas.88.14.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, West SC. Formation of RuvABC-Holliday junction complexes in vitro. Curr Biol. 1998;8:725–727. doi: 10.1016/s0960-9822(98)70282-9. [DOI] [PubMed] [Google Scholar]
- Dressler D, Potter H. Molecular mechanisms in genetic recombination. Ann Rev Biochem. 1982;51:727–762. doi: 10.1146/annurev.bi.51.070182.003455. [DOI] [PubMed] [Google Scholar]
- Duckett DR, Giraud-Panis MJE, Lilley DMJ. Binding of the junction-resolving enzyme T7 endonuclease I to DNA; Separation of binding and catalysis by mutation. J Mol Biol. 1995;246:95–107. doi: 10.1006/jmbi.1994.0069. [DOI] [PubMed] [Google Scholar]
- Dunderdale HJ, Benson FE, Parsons CA, Sharples GJ, Lloyd RG, West SC. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991;354:506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- Dunderdale HJ, Sharples GJ, Lloyd RG, West SC. Cloning, over-expression, purification and characterization of the Escherichia coli RuvC Holliday junction resolvase. J Biol Chem. 1994;269:5187–5194. [PubMed] [Google Scholar]
- Eggleston AK, Mitchell AH, West SC. In vitro reconstitution of the late steps of genetic recombination in E. coli. Cell. 1997;89:607–617. doi: 10.1016/s0092-8674(00)80242-1. [DOI] [PubMed] [Google Scholar]
- Elborough KM, West SC. Resolution of synthetic Holliday junctions in DNA by an endonuclease activity from calf thymus. EMBO J. 1990;9:2931–2936. doi: 10.1002/j.1460-2075.1990.tb07484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud-Panis MJE, Lilley DMJ. Structural recognition and distortion by the DNA junction-resolving enzyme RusA. J Mol Biol. 1998;278:117–133. doi: 10.1006/jmbi.1998.1681. [DOI] [PubMed] [Google Scholar]
- Hagan NFP, Vincent SD, Ingleston SM, Sharples GJ, Bennett RJ, West SC, Lloyd RG. Sequence-specificity of Holliday junction resolution: Identification of RuvC mutants defective in metal binding and target site recognition. J Mol Biol. 1998;281:17–29. doi: 10.1006/jmbi.1998.1934. [DOI] [PubMed] [Google Scholar]
- Hargreaves D, Rice DW, Sedelnikova SE, Artymiuk PJ, Lloyd RG, Rafferty JB. Crystal structure of E. coli RuvA with bound DNA Holliday junction at 6 Å resolution. Nat Struct Biol. 1998;5:441–446. doi: 10.1038/nsb0698-441. [DOI] [PubMed] [Google Scholar]
- Hiom K, Tsaneva IR, West SC. The directionality of RuvAB-mediated branch migration: In vitro studies with three-armed junctions. Genes Cells. 1996;1:443–451. doi: 10.1046/j.1365-2443.1996.d01-253.x. [DOI] [PubMed] [Google Scholar]
- Hiom K, West SC. The mechanism of branch migration in homologous recombination: Assembly of a RuvAB-Holliday junction complex in vitro. Cell. 1995;80:787–793. doi: 10.1016/0092-8674(95)90357-7. [DOI] [PubMed] [Google Scholar]
- Holliday R. A mechanism for gene conversion in fungi. Genet Res Camb. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- Holmes AM, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Hyde H, Davies AA, Benson FE, West SC. Resolution of recombination intermediates by a mammalian endonuclease activity functionally analogous to Escherichia coli RuvC resolvase. J Biol Chem. 1994;269:5202–5209. [PubMed] [Google Scholar]
- Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Takahagi M, Nakata A, Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes & Dev. 1992;6:2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JHJ, van Gent DC. Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- Kemper B. Branched DNA resolving enzymes (X-solvases) In: Nicoloff JA, Hoekstra MF, editors. DNA damage and repair. Totowa, NJ: Humana Press Inc; 1998. pp. 179–204. [Google Scholar]
- Lilley DMJ, Clegg RM. The structure of the four-way junction in DNA. Annu Rev Biophys Biomol Struct. 1993;22:299–328. doi: 10.1146/annurev.bb.22.060193.001503. [DOI] [PubMed] [Google Scholar]
- Mandal TN, Mahdi AA, Sharples GJ, Lloyd RG. Resolution of Holliday intermediates in recombination and DNA repair: Indirect suppression of ruvA, ruvB, and ruvC mutations. J Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCulloch R, Coggins LW, Colloms SD, Sherratt DJ. Xer-mediated site-specific recombination at cer generates Holliday junctions in vivo. EMBO J. 1994;13:1844–1855. doi: 10.1002/j.1460-2075.1994.tb06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill C, Shafer B, Strathern J. Coconversion of flanking sequences with homothallic switching. Cell. 1989;57:459–467. doi: 10.1016/0092-8674(89)90921-5. [DOI] [PubMed] [Google Scholar]
- Meselson MM, Radding CM. A general model for genetic recombination. Proc Natl Acad Sci. 1975;72:358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miick SM, Fee RS, Millar DP, Chazin WJ. Crossover isomer bias is the primary sequence-dependent property of immobilized Holliday junctions. Proc Natl Acad Sci. 1997;94:9080–9084. doi: 10.1073/pnas.94.17.9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel P, Stasiak A, Ehrlich SD, Cassuto E. Effect of length and location of heterologous sequences on RecA-mediated strand exchange. J Biol Chem. 1994;269:19830–19835. [PubMed] [Google Scholar]
- Murchie AIH, Portugal J, Lilley DMJ. Cleavage of a 4-way DNA junction by a restriction enzyme spanning the point of strand exchange. EMBO J. 1991;10:713–718. doi: 10.1002/j.1460-2075.1991.tb08001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons CA, West SC. Specificity of binding to four-way junctions in DNA by bacteriophage T7 endonuclease I. Nucleic Acids Res. 1990;18:4377–4384. doi: 10.1093/nar/18.15.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Formation of a RuvAB-Holliday junction complex in vitro. J Mol Biol. 1993;232:397–405. doi: 10.1006/jmbi.1993.1399. [DOI] [PubMed] [Google Scholar]
- Parsons CA, Tsaneva I, Lloyd RG, West SC. Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc Natl Acad Sci. 1992;89:5452–5456. doi: 10.1073/pnas.89.12.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons CA, Stasiak A, Bennett RJ, West SC. Structure of a multisubunit complex that promotes DNA branch migration. Nature. 1995;374:375–378. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- Pöhler JRG, Giraud-Panis MJE, Lilley DMJ. T4 endonuclease VII selects and alters the structure of the four-way junction; binding of a resolution-defective mutant enzyme. J Mol Biol. 1996;260:687–696. doi: 10.1006/jmbi.1996.0430. [DOI] [PubMed] [Google Scholar]
- Pöhler JRG, Norman DG, Bramham J, Bianchi ME, Lilley DMJ. HMG box proteins bind to 4-way DNA junctions in their open conformation. EMBO J. 1998;17:817–826. doi: 10.1093/emboj/17.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafferty JB, Sedelnikova SE, Hargreaves D, Artymiuk PJ, Baker PJ, Sharples GJ, Mahdi AA, Lloyd RG, Rice DW. Crystal structure of the DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science. 1996;274:415–421. doi: 10.1126/science.274.5286.415. [DOI] [PubMed] [Google Scholar]
- Roe SM, Barlow T, Brown T, Oram M, Keeley A, Tsaneva IR, Pearl LH. Crystal structure of an octameric RuvA-Holliday junction complex. Mol Cell. 1998;2:361–372. doi: 10.1016/s1097-2765(00)80280-4. [DOI] [PubMed] [Google Scholar]
- Shah R, Bennett RJ, West SC. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell. 1994;79:853–864. doi: 10.1016/0092-8674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Shah R, Cosstick R, West SC. The RuvC dimer resolves Holliday junctions by a dual incision mechanism that involves base-specific contacts. EMBO J. 1997;16:1464–1472. doi: 10.1093/emboj/16.6.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples GJ, Benson FE, Illing GT, Lloyd RG. Molecular and functional analysis of the ruv region of Escherichia coli K-12 reveals three genes involved in DNA repair and recombination. Mol Gen Genet. 1990;221:219–226. doi: 10.1007/BF00261724. [DOI] [PubMed] [Google Scholar]
- Shiba T, Iwasaki H, Nakata A, Shinagawa H. SOS-inducible DNA repair proteins, RuvA and RuvB, of Escherichia coli: Functional interactions between RuvA and RuvB for ATP hydrolysis and renaturation of the cruciform structure in supercoiled DNA. Proc Natl Acad Sci. 1991;88:8445–8449. doi: 10.1073/pnas.88.19.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Iwasaki H, Saito A, Kyogoku Y, Shinagawa H. Analysis of substrate specificity of the RuvC Holliday junction resolvase with synthetic Holliday junctions. J Biol Chem. 1996;271:26105–26109. doi: 10.1074/jbc.271.42.26105. [DOI] [PubMed] [Google Scholar]
- Shinagawa H, Makino K, Amemura M, Kimura S, Iwasaki H, Nakata A. Structure and regulation of the Escherichia coli ruv operon involved in DNA repair and recombination. J Bacteriol. 1988;170:4322–4329. doi: 10.1128/jb.170.9.4322-4329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Homologous recombination and the roles of double-strand breaks. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- Shurvinton CE, Lloyd RG. Damage to DNA induces expression of the ruv gene of Escherichia coli. Mol Gen Genet. 1982;185:352–355. doi: 10.1007/BF00330811. [DOI] [PubMed] [Google Scholar]
- Sogo J, Stasiak A, De Bernadin W, Losa R, Koller T. Binding of protein to nucleic acids. In: Sommerville J, Scheer U, editors. Electron microscopy in molecular biology. Oxford, UK: IRL Press; 1987. pp. 61–79. [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51 deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak A, Tsaneva IR, West SC, Benson CJB, Yu X, Egelman EH. The Escherichia coli RuvB branch migration protein forms double hexameric rings around DNA. Proc Natl Acad Sci. 1994;91:7618–7622. doi: 10.1073/pnas.91.16.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Takahagi M, Iwasaki H, Shinagawa H. Structural requirements of substrate DNA for binding to and cleavage by RuvC, a Holliday junction resolvase. J Biol Chem. 1994;269:15132–15139. [PubMed] [Google Scholar]
- Tsaneva IR, Illing GT, Lloyd RG, West SC. Purification and properties of the RuvA and RuvB proteins of Escherichia coli. Mol Gen Genet. 1992a;235:1–10. doi: 10.1007/BF00286175. [DOI] [PubMed] [Google Scholar]
- Tsaneva IR, Müller B, West SC. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992b;69:1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- Tsaneva IR, Müller B, West SC. The RuvA and RuvB proteins of Escherichia coli exhibit DNA helicase activity in vitro. Proc Natl Acad Sci. 1993;90:1315–1319. doi: 10.1073/pnas.90.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K, Chi NW, Kolodner RD. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded-DNA binding-protein. Proc Natl Acad Sci. 1993;90:3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool AJ, Shah R, Mézard C, West SC. Functional interactions between the Holliday junction resolvase and the branch migration motor of Escherichia coli. EMBO J. 1998;17:1838–1845. doi: 10.1093/emboj/17.6.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb BL, Cox MM, Inman RB. Recombinational DNA repair: The RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell. 1997;91:347–356. doi: 10.1016/s0092-8674(00)80418-3. [DOI] [PubMed] [Google Scholar]
- West SC. The nucleases of genetic recombination. In: Linn SM, Lloyd RS, Roberts RJ, editors. Nucleases. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 145–169. [Google Scholar]
- ————— The RuvABC Proteins and Holliday junction processing in Escherichia coli. J Bacteriol. 1996;178:1237–1241. doi: 10.1128/jb.178.5.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— RuvA gets X-rayed on Holliday. Cell. 1998;94:699–701. doi: 10.1016/s0092-8674(00)81729-8. [DOI] [PubMed] [Google Scholar]
- Whitby MC, Bolt EL, Chan SN, Lloyd RG. Interactions between RuvA and RuvC at Holliday junctions: Inhibition of junction cleavage and formation of a RuvA-RuvC-DNA complex. J Mol Biol. 1996;264:878–890. doi: 10.1006/jmbi.1996.0684. [DOI] [PubMed] [Google Scholar]
- White MF, Lilley DMJ. The resolving enzyme Cce1 of yeast opens the structure of the 4-way DNA junction. J Mol Biol. 1997;266:122–134. doi: 10.1006/jmbi.1996.0795. [DOI] [PubMed] [Google Scholar]
- White MF, Lilley DMJ. Interaction of the resolving enzyme YDC2 with the four-way DNA junction. Nucleic Acids Res. 1998;26:5609–5616. doi: 10.1093/nar/26.24.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, West SC, Egelman EH. Structure and subunit composition of the RuvAB-Holliday junction complex. J Mol Biol. 1997;266:217–222. doi: 10.1006/jmbi.1996.0799. [DOI] [PubMed] [Google Scholar]
- Zerbib D, Colloms SD, Sherratt DJ, West SC. Effect of DNA topology on Holliday junction resolution by Escherichia coli RuvC and bacteriophage T7 endonuclease I. J Mol Biol. 1997;270:663–673. doi: 10.1006/jmbi.1997.1157. [DOI] [PubMed] [Google Scholar]
- Zerbib D, Mézard C, George H, West SC. Co-ordinated actions of RuvABC in Holliday junction processing. J Mol Biol. 1998;281:621–630. doi: 10.1006/jmbi.1998.1959. [DOI] [PubMed] [Google Scholar]