Abstract
The Caenorhabditis elegans AWA, AWB, and AWC olfactory neurons are each required for the recognition of a specific subset of volatile odorants. lim-4 mutants express an AWC reporter gene inappropriately in the AWB olfactory neurons and fail to express an AWB reporter gene. The AWB cells are morphologically transformed toward an AWC fate in lim-4 mutants, adopting cilia and axon morphologies characteristic of AWC. AWB function is also transformed in these mutants: Rather than mediating the repulsive behavioral responses appropriate for AWB, the AWB neurons mediate attractive responses, like AWC. LIM-4 is a predicted LIM homeobox gene that is expressed in AWB and a few other head neurons. Ectopic expression of LIM-4 in the AWC neuron pair is sufficient to force those cells to adopt an AWB fate. The AWA nuclear hormone receptor ODR-7 described previously also represses AWC genes, as well as inducing AWA genes. We propose that the LIM-4 and ODR-7 transcription factors function to diversify C. elegans olfactory neuron identities, driving them from an AWC-like state into alternative fates.
Keywords: Olfactory neuron, cell fate specification, LIM homeobox gene, C. elegans
Olfactory neurons in worms, flies, and mice fall into a few structural classes, defined by characteristic cilia and axon morphologies. At a finer level, however, the diversity of signaling molecules, and especially of olfactory receptors, divides the olfactory neurons into many functionally distinct subtypes (for review, see Buck 1996). In the nematode Caenorhabditis elegans, the olfactory neurons reside in the amphids, a bilateral pair of sensory organs in the head of the animal that contain twelve sensory neurons each (White et al. 1986). The amphid sensory neurons extend dendrites to the tip of the nose, where specialized ciliated endings interact with the animal’s environment and send their axons into the nerve ring, a major neuropil in which synaptic connections are made. Each bilateral pair of amphid neurons has a particular function in the sensation of pheromones, temperature, mechanical stimulation, soluble chemicals, or volatile chemicals. Three pairs of neurons—AWA, AWB, and AWC—are required for olfaction, the detection of volatile odorants. The AWA and AWC neurons are required for chemotaxis toward distinct subsets of attractive odorants (Bargmann et al. 1993), whereas the AWB neurons are required for a repulsive behavioral response to at least one odorant (Troemel et al. 1997). Despite their shared olfactory function, the AWA, AWB, and AWC neurons are not closely related by cell lineage.
The AWA, AWB, and AWC olfactory neurons express overlapping but distinct sets of signal-transduction molecules that contribute to each cell’s unique identity. Chemotaxis to AWA-sensed odorants requires the G protein α subunit ODR-3 (Roayaie et al. 1998) and the predicted cation channel subunit OSM-9 (Colbert et al. 1997), which is related to TRP channels and the VR1 capsaicin receptor. Responses to AWC- and AWB-sensed odorants require ODR-3 and the TAX-2/TAX-4 cGMP-gated ion channel (Coburn and Bargmann 1996; Komatsu et al. 1996). C. elegans has a large number of seven transmembrane domain proteins expressed in sensory neurons that may function as chemoreceptors (Troemel et al. 1995). One of these genes, odr-10, has been shown to function in AWA as a receptor for the volatile odorant diacetyl (Sengupta et al. 1996; Zhang et al. 1997). Two additional odr-10-like genes have upstream promoter sequences that direct expression of GFP to other olfactory neurons, suggesting that they also encode olfactory receptors. The str-1 gene is expressed in the AWB neuron pair, whereas the str-2 gene is expressed asymmetrically in one of the two AWC olfactory neurons (Troemel et al. 1997; E.R. Troemel and C.I. Bargmann, in prep.). These putative receptors are useful both for investigating cell function and as markers of cell fate.
Little is known in any organism about how the specific identities of olfactory neurons are determined. Only two genes that participate in this process, odr-7 in C. elegans and acj6 in Drosophila, have been identified. The odr-7 nuclear hormone receptor has been implicated in the specification of AWA olfactory neuron identity in C. elegans (Sengupta et al. 1994). ODR-7 is expressed exclusively in the AWA neurons, is required for their function, and is required for full expression of the AWA-specific ODR-10 receptor (Sengupta et al. 1996). It therefore acts to promote the unique differentiated features of the AWA neuron. A similar function in Drosophila olfactory neurons may be provided by the acj6 POU homeobox gene (Clyne et al. 1999a,b). In acj6 mutants, some behavioral responses to odorants are lost, the electrophysiological responses of certain olfactory cells are altered, and the expression of several putative olfactory receptors is abolished.
Sensory neuron specification in C. elegans has been studied best in the mechanosensory system. A battery of transcription factors acts combinatorially to ensure that six neurons develop as touch receptor neurons (Mitani et al. 1993). At the core of this mechanosensory transcriptional program is the mec-3 gene, which is required for the neurons to exhibit the final differentiated features of touch receptors. mec-3 belongs to the LIM homeobox class of transcription factors (Way and Chalfie 1988; Freyd et al. 1990). These genes contain two metal-binding domains used for protein–protein interactions (LIM domains) and a DNA-binding homeodomain. Seven genes of this class have been found in the C. elegans genome and four of these now correspond to genetically defined mutants (Way and Chalfie 1988; Freyd et al. 1990; Hobert et al. 1997, 1999). All LIM homeobox genes are first expressed in post-mitotic neurons and are required for late aspects of neuron cell fate, such as axon pathfinding and neurotransmitter expression. Cells with mutant LIM homeobox genes therefore usually exhibit phenotypes that mimic loss of certain cell types. For example, the ttx-3 and lin-11 genes are required for the function of interneurons that act in different parts of the thermosensory circuit (Hobert et al. 1997, 1998b). Mutations in these genes result in phenotypes that resemble ablation of those interneurons (Mori and Ohshima 1995). Studies of LIM homeobox gene function in vertebrates and Drosophila have also implicated these genes in controlling various late aspects of cell fates. For instance, two vertebrate LIM homeobox genes, lhx3 and lhx4, as well as their Drosophila homolog, lim3, are required to maintain proper axon trajectories in a subset of motor neurons and to repress improper axon trajectories characteristic of different motor nuron classes (Sharma et al. 1998; Thor et al. 1999).
To investigate how the unique features of olfactory neurons are determined, we performed a screen for altered expression of str-2::GFP, a reporter gene normally expressed in one of the two AWC olfactory neurons. Mutations in a LIM homebox gene, lim-4, were found to cause ectopic expression of str-2::GFP in the AWB neurons. Molecular, morphological, and functional analyses indicate that in this mutant, AWB neurons are strongly transformed into AWC neurons. Moreover, expression of LIM-4 in AWC is sufficient to force AWC neurons to adopt an AWB fate. LIM-4 thus acts as a molecular switch that distinguishes between alternative sensory neuron fates. We suggest that the LIM-4 and ODR-7 transcription factors function to drive neurons in the C. elegans amphid from an AWC-like fate into alternative olfactory neuron fates.
Results
The LIM homeobox gene lim-4 affects olfactory receptor expression, movement, and foraging behavior
A 4-kb region upstream of the putative seven transmembrane domain olfactory receptor STR-2 directs expression of GFP to a single AWC olfactory neuron (E.R. Troemel and C.I. Bargmann, in prep.). To study the mechanisms by which AWC cell fate is confined to a single neuron, we isolated mutant animals with altered expression of the str-2::GFP transgene (see Materials and Methods). Three mutants recovered from the screen expressed GFP in both AWB olfactory neurons as well as the normal AWC neuron (Fig. 1A,B). Conversely, expression of the AWB-specific marker gene str-1::GFP was severely reduced in these mutants (Fig. 1D,E). GFP markers for six other amphid sensory neuron cell types (ADLL/R, ADFL/R, ASER, ASEL, ASIL/R, and AWAL/R) were expressed normally, indicating that the mutants have a selective defect in sensory gene expression.
Figure 1.
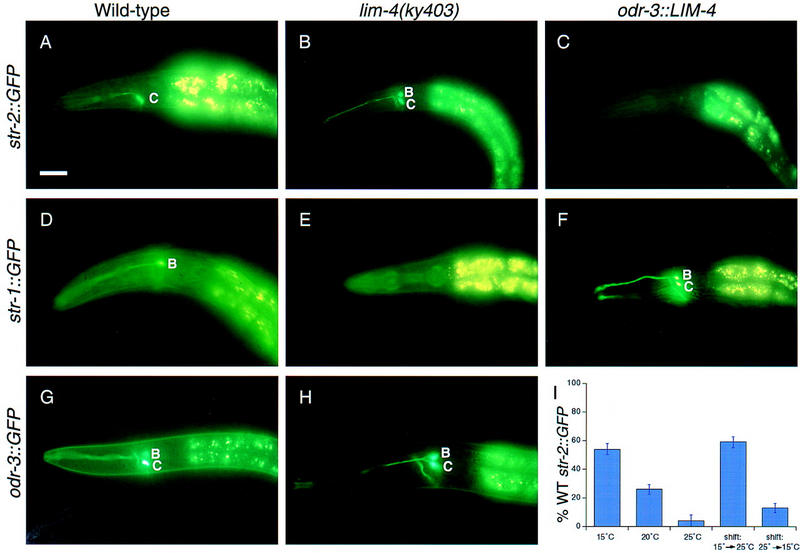
LIM-4 controls expression of putative receptor and signaling genes and is required early to repress str-2::GFP expression. (A) str-2::GFP is expressed in a single AWC neuron in wild-type animals. (B) lim-4 mutants express str-2::GFP in AWC and AWB. (C) Transgenic animals carrying the odr-3::LIM-4 transgene do not express str-2::GFP in either cell. (D) str-1::GFP is expressed in the AWB neurons in wild-type animals. (E) In lim-4 mutants, str-1::GFP expression is severely reduced. (F) Transgenic animals expressing odr-3::LIM-4 express str-1::GFP in both AWC and AWB. (G) In wild-type animals, odr-3::GFP fluoresces more brightly in AWC than in AWB. (H) In lim-4 mutants, the odr-3::GFP signal is equally bright in AWB and AWC. Quantitative analysis of these phenotypes is presented in Tables 1 and 2, except for str-2::GFP in an odr-3::LIM-4 strain (no GFP expression in 84% of animals, n = 80). Anterior is left and dorsal is up in all panels. Scale bar, 40 μm and applies to A–H. (I) LIM-4 is required early in development to repress str-2::GFP in AWB. The y axis indicates the percentage of animals that express str-2::GFP in its wild-type pattern, in a single AWC neuron. Animals grown at different temperatures have different penetrances for the str-2::GFP misexpression phenotype (first three columns). Animals were shifted from one temperature to another at the L1 larval stage and assayed as adults. Temperature-shifted animals exhibited the phenotype characteristic of the temperature they experienced during the embryonic and L1 stages (last two columns; shifts are indicated by arrows). Error bars, standard error of the proportion.
The three mutants failed to complement each other and formed an allelic series of increasingly defective str-2::GFP expression (Table 1). In the weaker mutant (ky395), a substantial fraction of animals expressed str-2::GFP in only one AWB neuron, whereas in the more severe mutants, most animals expressed GFP in both AWB neurons. When only one AWB neuron expressed str-2::GFP, the GFP-expressing AWB neuron was sometimes on the same side as the GFP-expressing AWC neuron and sometimes on the opposite side. All three alleles were slightly semidominant for their str-2::GFP misexpression phenotype and were more severe in hermaphrodites than males (data not shown). The mutants also moved in a coily manner (Fig. 3E–F, below), and exhibited defects in foraging behavior, as manifested by aberrant head movements (McIntire et al. 1993).
Table 1.
str-2::GFP expression in lim-4 mutant alleles
| Strain
|
Percent str-2::GFP expression
|
|||
|---|---|---|---|---|
| AWC
|
AWC + 1 AWB
|
AWC + 2 AWB
|
No.a
|
|
| Wild type, 20°C | 100 | 0 | 0 | >1000 |
| lim-4(ky403), 20°C | 0 | 10 | 90 | 223 |
| lim-4(ky402), 20°C | 4 | 15 | 81 | 209 |
| lim-4(ky395), 25°C | 4 | 35 | 60 | 116 |
| lim-4(ky395), 20°C | 30 | 43 | 27 | 241 |
| lim-4(ky395), 15°C | 54 | 32 | 14 | 162 |
No. of animals scored.
Figure 3.
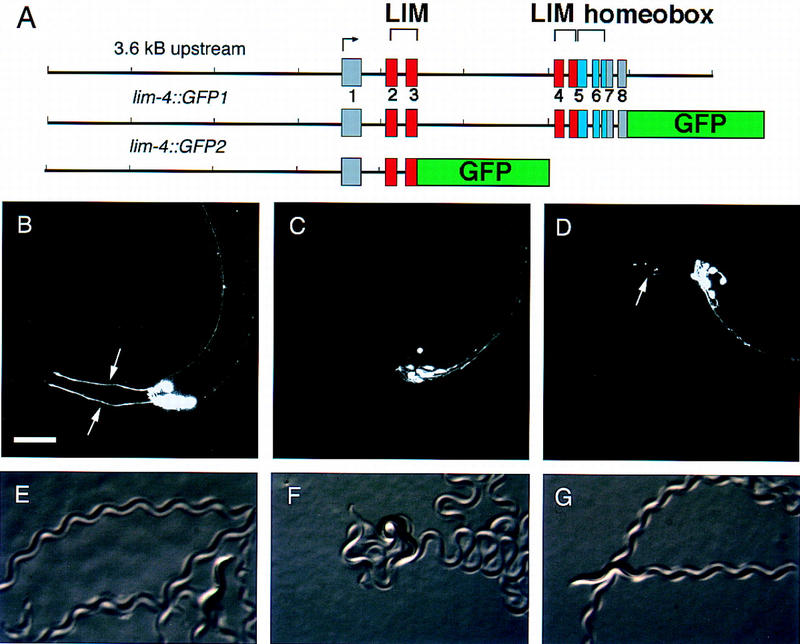
lim-4 is expressed in anterior neurons. (A) Two GFP fusion genes used to analyze LIM-4 expression. lim-4::GFP1 is a fusion of GFP to the end of the last amino acid in LIM-4, lim-4::GFP2 truncates the gene after the third exon. lim-4::GFP2 was integrated into the genome to minimize mosaicism. (B) lim-4::GFP2 expression in wild-type animals is confined to the anterior ganglia. lim-4::GFP2 is expressed in the two AWB neurons, which send dendrites to the tip of the nose (arrows), two of six RMD neurons (RMDL and RMDR), the RIV neuron, four SAA neurons, and four SIA neurons. lim-4::GFP1 is expressed in all the same cells as well as RMEV and RID (not shown). (C,D) In lim-4 mutants, AWB expression is eliminated, as is most evident by the absence of sensory dendrites. Sometimes ectopic sprouting of anterior processes from the SAA axons is visible in lim-4 mutants (D, arrow), these processes are normally unbranched. Anterior is left and dorsal is up in B–D. Scale bar, 20 μm, applies to B–D. (E–G) The full-length lim-4::GFP1 construct is capable of rescuing the lim-4 movement defect. Wild-type animals move in a characteristic manner, leaving regular, shallow sinusoidal tracks (E). lim-4 animals move in a coily manner, leaving exaggerated high amplitude tracks (F). This movement defect is rescued with a full-length lim-4::GFP1 transgene (G). The adult worms shown are ∼1 mm long.
The most severe allele in this complementation group (ky403) was mapped to a small region on the left arm of the X chromosome. Rescue of the str-2::GFP expression phenotype was obtained with a cosmid containing the LIM homeobox gene lim-4 (Fig. 2), and the coily movement phenotype was rescued with a LIM-4 transgene tagged at its 3′ end with GFP (Fig. 3E–G). To confirm the identification of lim-4, the coding region was sequenced in the lim-4 mutants and mutations were found in all three alleles (Fig. 2). The weakest allele, lim-4(ky395), was associated with a missense mutation in a well-conserved homeodomain residue. lim-4(ky402) had a mutation in the splice acceptor site before the fifth exon, which contains portions of the second LIM domain and the beginning of the homeodomain. The most severe lim-4 allele (ky403) had a stop codon within the DNA recognition helix of the homeodomain that would be predicted to disrupt the DNA-binding activity of the LIM-4 protein. All experiments were performed with the ky403 allele unless otherwise noted.
Figure 2.
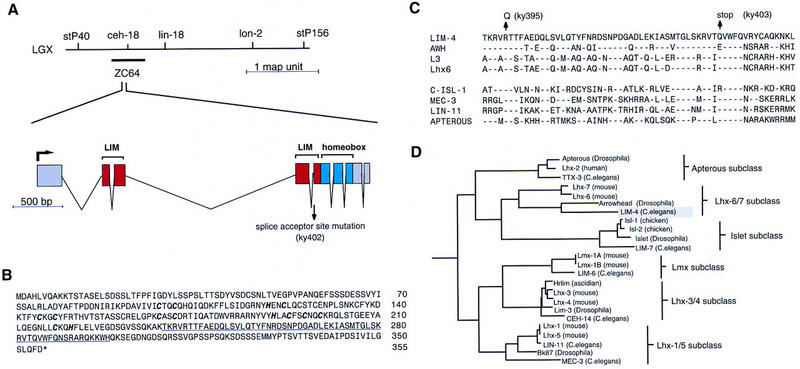
lim-4 gene structure. (A) Genetic map and genomic structure of the lim-4 locus. The splice acceptor site mutation (G to A conversion) in the ky402 allele is shown. Mutations in the other two lim-4 alleles, ky395 and ky403, are shown in C. The gene structure was confirmed by isolating a cDNA by PCR from a mixed-stage cDNA library with primers that match the amino and carboxyl terminus of the predicted ZC64.4 gene. The predicted ZC64.4 gene on cosmid ZC64 (GenBank accession no. U39740) contains a 17-amino-acid insertion at amino acid position 289. This insertion most likely results from a misprediction of the gene structure, as we could not detect it in cDNAs that we isolated. (B) Protein sequence of LIM-4. The Zn-coordinating residues of the two LIM domains are in bold and italics. The homeodomain is underlined. The GenBank accession no. for lim-4 is U72348. (C) The homeodomain of LIM-4 aligned with the homeodomains of seven other LIM homeobox genes. The mutations in the ky395 and ky403 alleles are shown. LIM-4 is 67% identical to Drosophila Arrowhead in its homeodomain and 50% identical to mouse L3. Broken lines indicate amino acid identities. (D) Phylogenetic relationship of LIM homeodomain proteins. The analysis was performed with individual homeodomains, with the pileup, distances, and grow-tree algorithms of the GCG software package. Only a limited number of vertebrate representatives from each subgroup are shown.
lim-4 is most similar to the Drosophila arrowhead gene (Curtiss and Heilig 1997) and the vertebrate L3/Lhx7 and Lhx6 genes (Fig. 2C,D) (Matsumoto et al. 1996; Grigoriou et al. 1998). Although these genes were presumably derived from a common ancestor, their degree of identity to each other is lower than that observed within LIM homeodomain protein subclasses, suggesting a greater divergence of function. Both arrowhead and L3 are expressed in subsets of neurons of the central nervous system (Matsumoto et al. 1996; Curtiss and Heilig 1997).
The weakest lim-4 allele (ky395) was temperature sensitive for its str-2::GFP misexpression phenotype (Table 1). The proportion of animals expressing str-2::GFP in its wild-type pattern in this allele ranged from 54% at 15°C to 4% at 25°C. However, the defect in str-1::GFP expression was not affected by temperature in the lim-4(ky395) allele (at 15°C, 66% of ky395 animals had defective str-1::GFP expression, n = 99; and at 25°C, 69% of animals had defective expression, n = 112). At high temperature, there must therefore be some lim-4(ky395) animals in which the cells that normally develop into AWB express neither the AWB nor the AWC marker gene. We made use of the lim-4(ky395) allele’s temperature sensitivity for the str-2::GFP expression phenotype to determine when during development LIM-4 performs this function. Animals carrying the str-2::GFP reporter were grown at 15°C or 25°C, shifted to the other temperature during the L1 larval stage, and scored for GFP expression as young adults, 2 or 3 days later. Temperature-shifted animals exhibited the phenotype characteristic of the temperature they experienced during the embryonic and L1 stages (Fig. 1I). LIM-4 was therefore required in the AWB neurons for a discrete interval of early development to repress the AWC fate, and was not needed later in life to perform this function.
LIM-4 is expressed in head neurons and regulates its own expression in AWB
The expression pattern of LIM-4 was determined with two GFP fusion genes, one that included 3.6 kb of upstream sequence, all exons and introns, and had GFP fused to the carboxyl terminus (lim-4::GFP1), and one fusion gene that truncated the protein after the third exon (lim-4::GFP2) (Fig. 3A). In larvae and adults, lim-4::GFP expression was confined to neurons in the worm’s anterior ganglia (Fig. 3B); embryonic expression was not examined in detail. Expression from both transgenes was observed in the AWB neurons but not in other sensory neurons. lim-4::GFP1 was also expressed in one RME motor neuron (RMEV), two RMD motor neurons (RMDL and RMDR), and the RID, RIV, SAA and SIA interneurons. lim-4::GFP2 was expressed in the same neurons, except for RID and RMEV. Expression in the GABAergic RME motor neurons, which control head movement, is consistent with the mutants’ foraging defect (McIntire et al. 1993). Because the functions of the interneurons and motor neurons expressing LIM-4 are poorly understood, lack of LIM-4 function in any of these cells could be responsible for the coily movement of lim-4 mutants.
To ask whether LIM-4 regulates its own expression, the short lim-4::GFP2 transgene was examined in the lim-4(ky403) mutant. In lim-4 mutants, lim-4::GFP2 expression was abolished in the AWB neurons (Fig. 3C,D), but unaffected in the other lim-4::GFP2-expressing cells. The lim-4::GFP2 fusion gene also revealed defects in neurite morphology in lim-4 mutants. For example, the SAA neurons often sent thick sprouting processes into the head in place of their normal unbranched processes (Fig. 3D). Ectopic axon sprouting and misrouted axon trajectories are common defects in LIM homeobox mutants (Way and Chalfie 1988; Lundgren et al. 1995; Hobert et al. 1997, 1998b, 1999; Sharma et al. 1998).
The repulsive olfactory neuron AWB is transformed into the attractive olfactory neuron AWC in lim-4 mutants
To determine the extent to which the AWB neurons were transformed towards the AWC cell fate, we characterized several aspects of AWB fate in the lim-4(ky403) mutant. As noted above, expression of the AWB-specific marker str-1::GFP was severely reduced or absent in the lim-4(ky403) mutant (Fig. 1D,E; Table 2). AWC and AWB neurons share most of their signal transduction components, but the ODR-3 Gα protein is expressed at notably different levels in AWC and AWB. In wild-type animals, an odr-3::GFP reporter is expressed most intensely in AWC, more weakly in AWB, and very faintly in the AWA, ADF, and ASH neurons (Roayaie et al. 1998). In lim-4 animals, however, odr-3::GFP expression was usually equally bright in AWB and AWC (Fig. 1G,H; Table 2).
Table 2.
Cell fate transformations caused by mutations in lim-4 and ectopic expression of LIM-4
| Phenotype (%)
|
||||
|---|---|---|---|---|
| Strain
|
str-1::GFPb
|
|||
| no GFP
|
AWB
|
AWB + AWC
|
No.a
|
|
| Wild type | 0 | 100 | 0 | >1000 |
| lim-4(ky403) | 100 | 0 | 0 | 206 |
| odr-3::LIM-4c | 0 | 5 | 95d | 93 |
|
|
odr-3::GFPe
|
||
|---|---|---|---|
| AWB ≥ AWC
|
AWC > AWB
|
|
|
| Wild type | 14 | 86 | 73 |
|
lim-4(ky403)
|
97
|
3
|
79
|
|
|
cilia morphologyf
|
||
 |
 |
||
| Wild type AWB | 0 | 100 | 106 |
| Wild type AWC | 98 | 2 | 96 |
| lim-4(ky403) AWB | 85 | 15 | 68 |
| odr-3::LIM-4 AWC | 14 | 86 | 78 |
|
|
axon morphologyg
|
|||
|---|---|---|---|---|
| both S-shape
|
AWB = U-shape AWC = S-shape
|
both U-shape
|
|
|
| Wild type | 0 | 100 | 0 | 34 |
| lim-4(ky403) | 30 | 70 | 0 | 33 |
|
odr-3::LIM-4
|
6
|
60
|
34
|
32
|
| dye fillingh | ||||
| no cells | AWB | AWB + AWC | ||
| Wild type | 6 | 94 | 0 | 81 |
| lim-4(ky403) | 97 | 3 | 0 | 98 |
| odr-3::LIM-4 | 0 | 6 | 94 | 79 |
No. of animals scored for str-1::GFP and odr-3::GFP expression, and number of neurons scored for cilia, axons, and dye-filling.
GFP was scored as present if fluorescence was visible in the cell bodies and processes, dim GFP only in the cell body was considered “off.”
lim-4 str-1::GFP with extrachromosomal odr-3::LIM-4 rol-6.
23% expressed GFP in one ectopic cell; 77% in two ectopic cells.
For each animal, the relative intensities of GFP expression in the AWB and AWC neurons was compared. (AWB ≥ AWC) GFP expression in AWB was as intense or more intense than in AWC.
(Wild type) str-1::GFP and str-2::GFP were scored for AWB and AWC, respectively. (lim-4) lim-4 str-2::GFP was scored using animals in which only one neuron expressed str-2::GFP. (odr-3::LIM-4) lim-4 lin-15 with extrachromosomal odr3::LIM-4 str-1::GFP lin-15.
Determined in mosaics with GFP expressed in two cells on one side. (Wild type) str-2::GFP with extrachromosomal str-1::GFP rol-6 (lim-4) lim-4 odr-10 lin-15 with extrachromosomal str-2::GFP str-2::odr-10 lin15 (The odr-10 strain was used in this experiment simply because it was available and should not affect the results.) (odr-3::LIM-4) lim-4 lin-15 with extrachromosomal odr-3::LIM-4 str-1::GFP lin-15.
Determined by filling with DiD and comparing to GFP expression. (Wild type) str-1::GFP str-2::GFP. (lim-4) lim-4 str-2::GFP. (odr-3::LIM-4) lim-4 lin-15 with extrachromosomal odr-3::LIM-4 str-1::GFP lin-15.
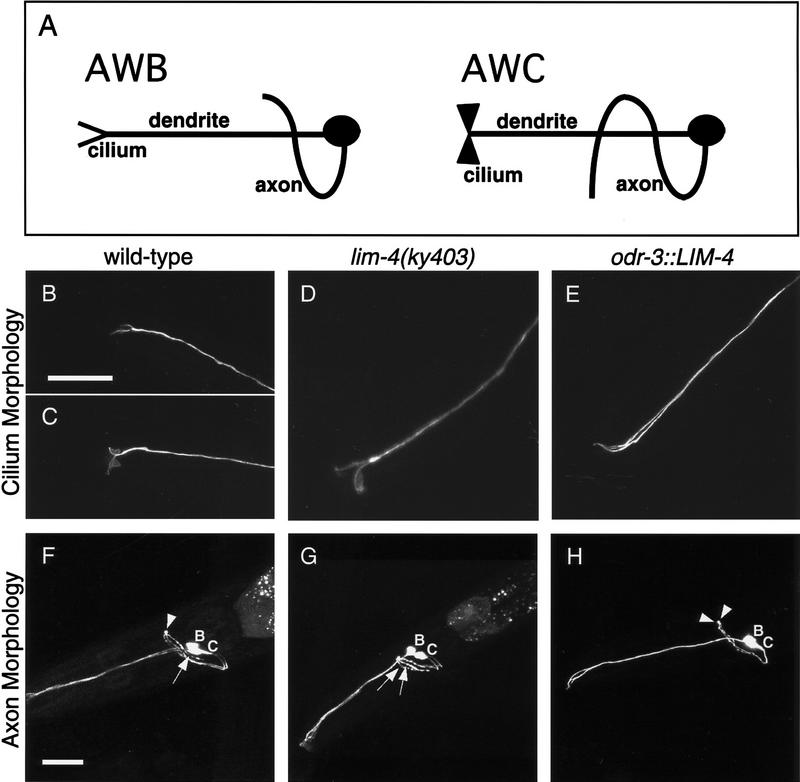
The cilia and axons of the AWB and AWC sensory neurons are strikingly different (Fig. 4A). Cilia are specialized structures at the tips of sensory dendrites that encounter environmental stimuli. AWB cells have a simple two-pronged cilium morphology reminiscent of a tuning fork, whereas AWC neurons have more elaborate, fan-like cilia. In lim-4 mutants, the AWB neurons often have a fan-like cilium morphology, like AWC (Fig. 4B–D; Table 2). The other notable difference in the morphology of AWB and AWC neurons is their axon trajectory. The AWB neurons extend a U-shaped axon that terminates at the dorsal midline, in which it makes contact with its homolog from the contralateral side. The AWC neurons have a similar initial trajectory, but then extend past the dorsal midline to the contralateral side of the nerve ring, resulting in an S-shaped axon. In lim-4 mutants, 30% of the AWB axons continue past the dorsal midline, forming an S shape like the AWC neurons (Fig. 4F,G; Table 2).
Figure 4.
LIM-4 controls the morphology of sensory cilia and axons. (A) Diagrams illustrating the wild-type morphologies of AWB and AWC neurons. Cilia are specialized sensory structures at the tips of dendrites. An AWB cilium is a simple two-pronged structure, resembling a tuning fork, whereas an AWC cilium is more membranous or fan-like. The U-shaped AWB axon extends into the nerve ring and stops at the dorsal midline. The S-shaped AWC axon continues past the dorsal midline to a ventral position on the contralateral side of the nerve ring. (B–E) Confocal Z-series projections of AWC and AWB cilia. Wild-type cilium morphologies of AWB (B) and AWC (C), visualized with str-1::GFP and str-2::GFP, respectively. (D) An AWB cilium in a lim-4(ky403) mutant, visualized with str-2::GFP, resembles the fan-like AWC cilia. (E) An odr-3::LIM-4-expressing animal visualized with str-1::GFP; both cilia exhibit the simple tuning fork morphology of AWB. (F–H) Confocal Z-series projections of GFP-expressing AWC and AWB neurons. Mosaic animals in which the neurons on only one side of the animal expressed GFP were chosen to simplify analysis of axon trajectories. (F) In animals with wild-type axon morphologies, the AWB axon stops at the ventral midline (arrowhead), whereas the AWC axon continues ventrally past the midline (arrow). This picture is of a lim-4 mutant expressing str-2::GFP displaying the wild-type axon morphologies for AWB and AWC. Wild-type animals expressing str-1::GFP and str-2::GFP exhibited an identical phenotype 100% of the time (Table 2). (G) A lim-4(ky403) mutant expressing str-2::GFP; both AWB and AWC axons exhibit the longer S-shaped AWC axon morphology (arrows). (H) An odr-3::LIM-4-expressing animals with str-1::GFP; both AWB and AWC axons stop at the midline (arrowheads). Quantitative analysis of these phenotypes is presented in Table 2. Anterior is left and dorsal is up in all panels. Scale bars, 20 μm. Bar in B applies to B–E; bar in F applies to F–H.
Another distinction between AWB and AWC cells is their abilities to take up certain fluorescent lipophilic dyes. It is not known what morphological or molecular aspects of the cells’ identity determine this property. In wild-type animals soaked in DiD or DiO, six pairs of amphid neurons, including AWB but not AWC, take up the dye (Fig. 5A–C). In lim-4 mutants, however, neither AWB nor AWC cells fill with dye, consistent with an AWB-to-AWC change in cell fate (Fig. 5D–F; Table 2).
Figure 5.
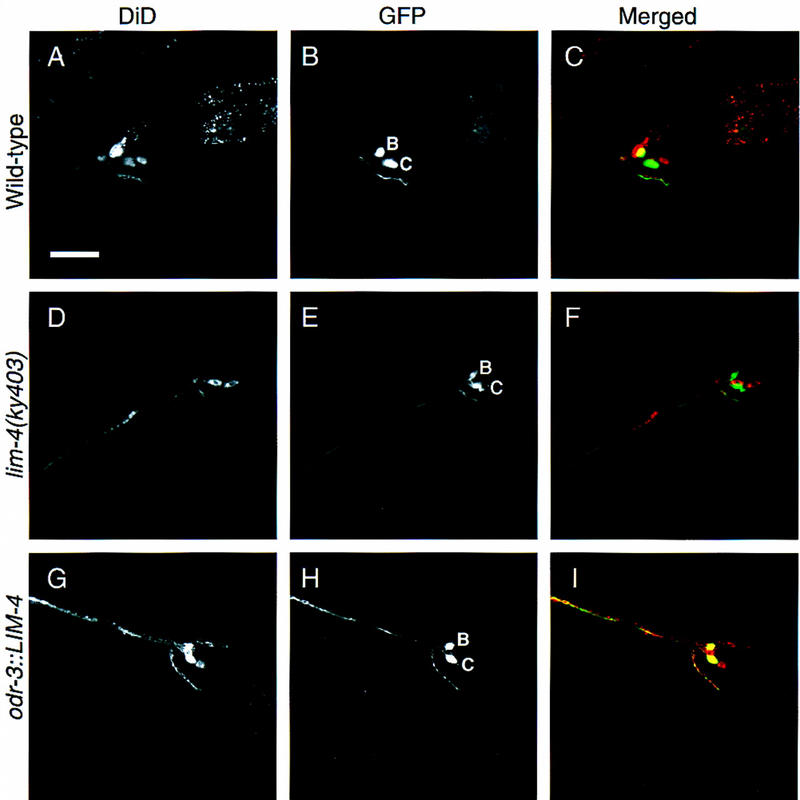
LIM-4 controls dye-filling properties of AWB and AWC. Confocal sections of worms expressing GFP in AWB and AWC neurons, soaked in the fluorescent lipophilic dye DiD. GFP is visualized with FITC wavelength filters (A,D,G), and DiD is visualized with TRITC wavelength filters (B,E,H). GFP was false-colored green, DiD was false-colored red and the images were merged (C,F,I); yellow indicates areas of overlap. In all cases, the more dorsal GFP-expressing cell is AWB and the more ventral cell is AWC. In wild-type dye-filled animals with str-1::GFP and str-2::GFP (A–C), only AWB fills with dye. In lim-4 mutant animals with str-2::GFP (D–F), neither cell fills with dye. In transgenic worms expressing odr-3::LIM-4 and str-1::GFP (G–I), both cells fill with dye. Quantitative analysis of these phenotypes is presented in Table 2. Anterior is left and dorsal is up in all panels. Scale bar, 20 μm in A–I.
The most profound physiological difference between the fates of AWB and AWC neurons is their function during behavior—the AWB neurons mediate repulsion from a subset of volatile odorants, whereas the AWC neurons mediate attraction to a different subset of odorants. These differences in neuron function may result from the distinct patterns of synaptic wiring characteristic of AWB and AWC (White et al. 1986). To ask whether the AWB neurons have changed their behavioral function in lim-4 mutants, we expressed the ODR-10 diacetyl receptor in these cells. In wild-type animals, ODR-10 expressed in AWB under the str-1 promoter mediates repulsion from diacetyl (Troemel et al. 1997; Fig. 6G). We used single animal behavioral assays to ask whether ODR-10 expression in AWB mediates attraction or repulsion in a lim-4 mutant.
Figure 6.
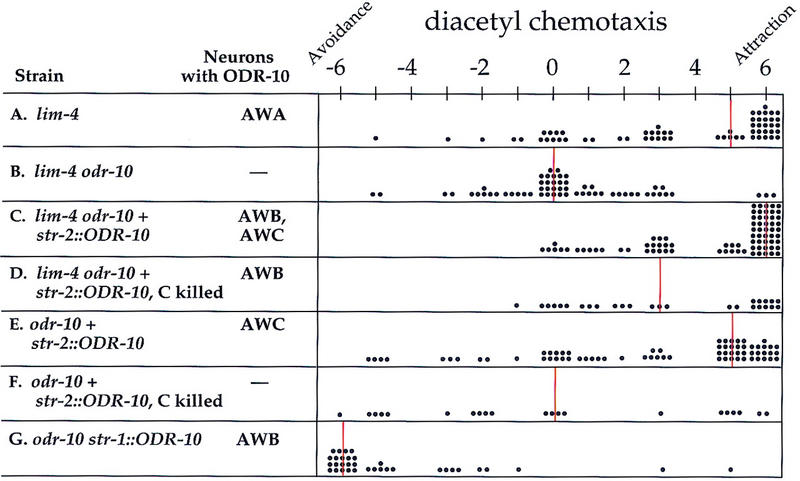
AWB mediates attractive chemosensory behaviors in lim-4. Graphic representation of single worm chemotaxis assays: Strains and neurons in those strains that express ODR-10 are at left, and assay results are at right. −6 indicates complete repulsion from diacetyl, 0 indicates indifference, and +6 indicates attraction. Each dot represents the chemotaxis score for a single worm, and vertical lines indicate the median score. All strains tested also carried a mutation in the lin-15 marker and were rescued with an extrachromosomal lin-15 gene. Assays were performed with (A) lim-4(ky403) mutants, (B) lim-4 odr-10(ky225) double mutants, (C) lim-4 odr-10 animals with extrachromosomal str-2::ODR-10 and str-2::GFP (only animals expressing GFP in AWC and AWB were assayed), (D) the same strain as in C following laser ablation of the GFP-expressing AWC neuron, (E) an odr-10 strain wild-type for LIM-4, expressing the str-2::ODR-10 and str-2::GFP transgenes, and F the same strain as in E following laser ablation of the GFP-expressing AWC neuron. (G) Data from a str-1::ODR-10 strain in a wild-type background, reproduced from Troemel et al. (1997) for comparison. Data were analyzed with a Mann-Whitney rank sum test, and significant differences (P ≤ 0.002) were observed between D and G, B and C, B and D, D and F, and C and E. D and E were not significantly different (P = 0.499).
Despite their coily movement, lim-4 animals were able to chemotax towards diacetyl with the endogenous odr-10 gene expressed in AWA (Fig. 6A). The odr-10(ky225) null mutation abolished this attraction (Fig. 6B). Expression of ODR-10 in AWB in lim-4 odr-10 double mutants was accomplished by placing ODR-10 under the control of the str-2 promoter. In wild-type animals, this transgene expresses ODR-10 exclusively in a single AWC neuron, but in lim-4 mutants it is expressed in one AWC neuron and both AWB neurons. The str-2::ODR-10 transgene restored diacetyl chemotaxis to both odr-10 and lim-4 odr-10 animals (Fig. 6C,E). ODR-10 expression in AWC could therefore mediate chemotaxis towards diacetyl, and in lim-4 mutants this effect was enhanced by its simultaneous expression in AWB (P < 0.001).
To probe the specific function of the transformed AWB neuron in lim-4 mutants, the AWC neuron expressing ODR-10 was killed with a laser so that the only remaining ODR-10-expressing cells were the transformed AWB neurons. Although their attractive response was reduced, these animals were able to chemotax towards diacetyl (Fig. 6D). In contrast, killing the ODR-10-expressing AWC in wild-type str-2::ODR-10 animals abolished diacetyl chemotaxis (Fig. 6F). These results indicate that the AWB neurons in a lim-4 mutant can mediate attractive olfactory behaviors rather than repulsive ones.
LIM-4 is sufficient to repress the AWC cell fate and promote the AWB cell fate
LIM-4 is required to repress AWC fate and promote AWB fate in the AWB neurons. To ask whether it is also sufficient to perform these functions, LIM-4 was expressed ectopically in the AWC neurons. The lim-4 cDNA was placed under the control of the odr-3 promoter, which is expressed in AWB, AWC, and faintly in AWA, ADF, and ASH (Roayaie et al. 1998). Expression of odr-3::LIM-4 caused ectopic expression of the AWB-specific str-1::GFP marker and repression of str-2::GFP in the AWC neurons (Fig. 1C,F; Table 2). The odr-3::LIM-4 transgene was also able to rescue AWB defects in a lim-4(ky403) mutant, as assessed by its str-1::GFP and str-2::GFP expression phenotypes, but it did not rescue coily movement (Fig. 1C,F; data not shown). Thus, LIM-4 acts in sensory neurons, and probably acts cell autonomously within AWB or AWC to promote the AWB cell fate and repress the AWC cell fate. The AWA, ADF, and ASH neurons, which should also express a low level of LIM-4 from this transgene, did not express detectable str-1::GFP.
The morphology and dye-filling properties of the AWC neurons were also transformed in odr-3::LIM-4-expressing animals. Cilium morphology in AWC neurons changed from the typical fan-like AWC structure to the tuning fork appearance characteristic of AWB (Fig. 4E; Table 2). Similarly, the odr-3::LIM-4 transgene caused the longer S-shaped axon of AWC to adopt the U-shaped morphology appropriate for the AWB axon (Fig. 4H; Table 2). The lipophilic dye-filling properties of the AWC cell were also transformed by ectopic LIM-4 expression. In odr-3::LIM-4 animals exposed to DiD, both AWB and AWC neurons took up dye, like normal AWB neurons (Fig. 5G–I; Table 2). Transformation of AWC into AWB by the odr-3::LIM-4 transgene was therefore complete by several criteria, indicating that LIM-4 can repress an AWC fate and promote an AWB fate in the AWC neuron.
ODR-7 promotes the AWA olfactory neuron cell fate and represses the AWC fate, making AWA unresponsive to LIM-4
ODR-7 is a protein homologous to nuclear hormone receptors that is expressed exclusively in the AWA olfactory neurons. A null mutation in odr-7 causes defects in chemotaxis toward all AWA-sensed odorants and reduces expression of the AWA olfactory receptor ODR-10 (Sengupta et al. 1994, 1996). We observed that ODR-7 is also required to repress str-2::GFP expression in AWA, but does not affect str-1::GFP expression (Fig. 7A,B). In the odr-7(ky4) null mutant, 97% of the worms expressed str-2::GFP ectopically in at least one AWA neuron (n = 150). In the mild odr-7(ky55) missense mutation, however, str-2::GFP expression was restricted to AWC.
Figure 7.
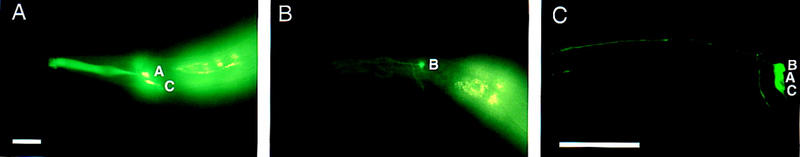
ODR-7 distinguishes between AWA and AWC-like cell fates. (A) An odr-7(ky4) mutant expresses str-2::GFP ectopically in AWA and in the appropriate AWC cell. (B) Expression of the AWB-specific str-1::GFP marker is not altered in an odr-7 mutant. (C) Ectopic expression of LIM-4 from the odr-3 promoter causes str-1::GFP to be expressed ectopically in both AWA and AWC, as well as the normal AWB cells in an odr-7 mutant. (C) A projection of a confocal Z-series. Anterior is left and dorsal is up in A–C. Scale bars, 40 μm. Bar in A applies to A–B; bar in C applies to C.
Because AWA was transformed toward an AWC fate in odr-7 mutants, it might have become sensitive to LIM-4’s ability to transform AWC neurons into AWB neurons. As noted above, odr-3::LIM-4 was not able to induce expression of the AWB marker str-1::GFP in AWA in a wild-type background, even though ODR-3 is expressed in AWA at a low level. However, odr-7(ky4) str-1::GFP animals carrying the odr-3::LIM-4 transgene expressed GFP in AWC, AWB, and AWA (Fig. 7C; 41% of the time three cells on one side expressed GFP, n = 86). Reducing ODR-7 activity in the AWA neurons thus makes them susceptible to LIM-4 activity and reveals an underlying potential to produce AWC- or AWB-like cell fates.
Discussion
LIM-4 is a cell fate switch
LIM-4 plays a crucial role in determining the fate of the AWB olfactory neurons: It promotes the appropriate AWB cell fate while repressing an inappropriate AWC fate. In lim-4 mutants, AWB adopts many molecular, morphological, and functional characteristics of AWC. Conversely, ectopic expression of LIM-4 from the odr-3 promoter is sufficient to cause the AWC cells to take on several morphological and molecular aspects of the AWB fate.
The analysis of lim-4 mutants and LIM-4 ectopic expressors suggests that LIM-4 acts as a binary cell fate switch in AWB and AWC. Cells without functional LIM-4 take on an AWC fate, whereas cells with LIM-4 are repressed for the AWC fate and take on the AWB fate. This role as a cell fate switch is so far unique for a LIM homeobox gene in C. elegans. As measured by a variety of cell fate markers and anatomical criteria, the neurons that express ttx-3, lin-11, or lim-6 have not adopted different cell fates in the absence of the respective gene’s function, but are functionally defective (Hobert et al. 1997, 1998b, 1999). The neurite-sprouting defects of the SAA neurons in lim-4 mutant animals resemble the neurite-sprouting defects observed in other LIM homeobox gene mutant animals. LIM-4 may thus be performing different functions in different cells. In the SAA neurons it may be acting more like the LIM homeobox genes characterized previously, whereas in AWB, LIM-4 performs a novel function, repressing one cell fate while promoting another cell fate.
The only other C. elegans LIM homeobox mutant that is suspected of undergoing a cell fate transformation is mec-3, in which the ALM neurons display characteristics of the BDU neurons (Way and Chalfie 1988). However, mec-3 is not known to be sufficient for a BDU to ALM transformation. Because ALM and BDU are sister cells, a mutation in mec-3 transforms an asymmetric cell division into a symmetric division. This differs from the situation with LIM-4 because the AWB and AWC neurons are not related to each other by lineage yet are both responsive to the same transcription factor. LIM-4 does not appear to affect the AWB cell lineage as AWB’s sister cell ADF, and its lineal cousin ASE, develop normally in lim-4 mutants. Thus, whereas the principle by which MEC-3 distinguishes between BDU and ALM cell fates is dictated by lineage, LIM-4 distinguishes between AWC and AWB within a hierarchy of cell function.
Analogous roles for LIM homeobox genes as switches for at least one aspect of motor neuron fate, the axon trajectory, have been reported recently for vertebrate and fly LIM homeobox genes. Cell fates within the vertebrate spinal cord are likely specified by the unique combination of LIM homeobox genes they express (Tsuchida et al. 1994). Mutants in the two redundant LIM homeobox genes lhx3 and lhx4 together cause the misrouting of a subpopulation of spinal cord axons (Sharma et al. 1998). Ectopic expression of LHX3 causes the reciprocal transformation in spinal cord axon trajectory. A similar function has been shown for lim3, the Drosophila homolog of these spinal cord LIM homeobox genes (Thor et al. 1999). In a lim3 mutant, a subpopulation of motor neurons develop projections onto inappropriate muscles, and ectopic expression of LIM3 causes the reciprocal defects in motor neuron projection. Like LIM-4 these LIM homeobox genes act as binary cell fate switches, distinguishing between two alternative cell fates. The completeness of the AWB to AWC transformation in lim-4 establishes that multiple aspects of cell fate, not just the axon trajectory, can be transformed by this class of genes.
All LIM homeobox genes are expressed throughout the life of the animal, leading to the hypothesis that these genes act continuously to maintain proper cell identity (Hobert and Ruvkun 1998a). Our experiment with a temperature-sensitive allele of LIM-4 suggests that this is not true for at least one of its functions. LIM-4 is required acutely to repress str-2::GFP expression in AWB during a discrete period of early development and not later in the life of the animal. Thus, the repression of AWC fate by LIM-4 is an irreversible decision made soon after neuron birth. None of the lim-4 alleles were temperature-sensitive for their str-1::GFP expression phenotypes, so it was not possible to test whether LIM-4 is similarly required acutely to promote the AWB fate or continuously to maintain it. It is possible that LIM homeobox genes have different temporal requirements for different aspects of their function.
LIM-4 controls the behavioral output of olfactory neurons
Perhaps the most striking aspect of AWB’s transformation into AWC in lim-4 mutants is its change in neuronal function. In wild-type animals, odorant stimulation of AWB and AWC leads to opposite behavioral responses. When activated by an odorant, AWC directs worms to move toward it, whereas activation of AWB causes worms to move away from the odorant. Previous studies in C. elegans suggest that the nature of the behavioral response to an odorant is defined by the sensory neuron and not by olfactory receptors (Troemel et al. 1997). Specifically, the ODR-10 diacetyl receptor mediates attractive behaviors in its native neuron AWA and repulsive behaviors when placed in AWB (Sengupta et al. 1996; Troemel et al. 1997). We show here that ODR-10 is functional in AWC as well, in which it mediates attractive behaviors. Strikingly, ODR-10 can also function in AWB cells lacking lim-4 function, but these transformed neurons now mediate attractive responses, demonstrating that AWB in lim-4 forms active, functional connections.
All three pairs of olfactory neurons in C. elegans—AWA, AWB, and AWC—express candidate-seven transmembrane domain olfactory receptors and share components of their signal transduction pathways, notably the G-protein α subunit ODR-3 (Roayaie et al. 1998). AWB and AWC require many of the same components for odorant sensation, including a cGMP-gated channel (Coburn and Bargmann 1996; Komatsu et al. 1996). The differences in the responses generated by activation of these neurons might therefore lie in their patterns of connectivity to downstream interneurons. According to electron microscope reconstructions of the C. elegans nerve ring, AWB and AWC have nearly exclusive sets of downstream targets (White et al. 1986). AWC forms synapses primarily onto the interneurons AIY, AIB, and AIA, whereas AWB forms connections primarily onto AIZ and the sensory neuron ADF. LIM-4 might act upstream of the genes that are required for choosing appropriate synapses, repressing those that determine AWC’s pattern of connectivity and activating those that determine AWB’s connectivity.
A model for the specification of olfactory cell fates
ODR-7 is a predicted nuclear hormone receptor required for the proper specification of the AWA olfactory neurons (Sengupta et al. 1994). In an odr-7 mutant, AWA fails to express the diacetyl receptor ODR-10 (Sengupta et al. 1996), and we show here that it ectopically expresses the putative AWC receptor STR-2. odr-7 and lim-4 thus act analogously, as repressors of AWC cell fate and promoters of the AWA and AWB fates, respectively. In the acj6 POU-domain gene mutant of Drosophila, the odorant specificities of certain olfactory neurons have changed, suggesting an analogous alteration in cell fates (Clyne et al. 1999a,b).
Although the odr-3 promoter is expressed in the AWA, ADF, and ASH neurons, those cells were not affected by odr-3::LIM-4 expression in wild-type animals. Only the AWB and AWC neurons seem to be capable of responding to LIM-4’s AWB-promoting activity. The two AWC neurons can be thought of as two distinct cell types, as only one of the two AWC cells expresses str-2::GFP (E.R. Troemel and C.I. Bargmann, unpubl.). The AWB neurons in lim-4 mutants adopt only one of those two fates, or possibly both at once, as both express str-2::GFP. The further subdivision of AWC’s cell fate must therefore be determined by a separate factor acting in parallel to LIM-4, or by environmental cues present at AWC’s cell position. Thus, there seems to be a specific group of cells responsive to LIM-4, including AWB and the two AWCs. Because AWA neurons in the odr-7 mutant have become more AWC-like, we reasoned that they may now be part of this LIM-4-responsive group. We found that an odr-7-defective AWA neuron can express AWB markers if it also expresses LIM-4.
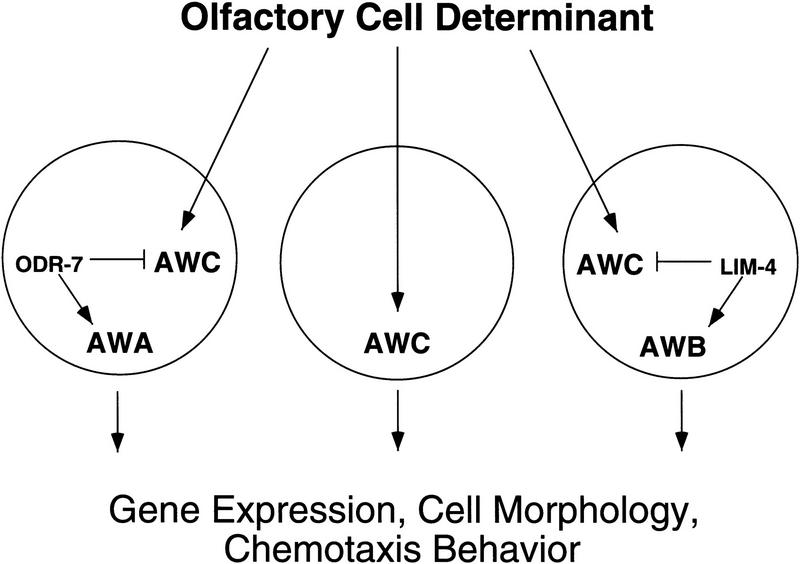
These results suggest a model for the specification of olfactory neuron identities in the C. elegans amphid (Fig. 8). lim-4 and odr-7 mutants reveal a common AWC-like developmental potential in the olfactory neurons AWA and AWB, which are not closely related to AWC or each other by lineage. This potential could be specified by an olfactory neuron fate determinant that is generated through the cell lineage, or induced by the embryonic environment, in three separate cells, AWA, AWB, and AWC. Without further modification, all of these cells can take on some of the characteristics of AWC. Modification of the AWC-like state in AWA and AWB is achieved by the transcription factors ODR-7 and LIM-4, either by themselves or in cooperation with other unidentified factors. It is these factors that allow the three olfactory neurons to establish their unique patterns of gene expression, their cell morphologies, and perhaps their synaptic connectivities, ultimately determining the behavioral outputs mediated by each cell. We speculate that the AWC-like fate may serve as an olfactory ground state on which the other neuronal fates can be elaborated. If this is true, the AWC-like fate could be a basic blueprint for making an olfactory cell, to which evolution can make alterations that diversify the animal’s repertoire of responses to volatile chemicals. Intriguingly, the head of the skin-penetrating nematode parasite Strongyloides stercoralis has been reconstructed from electron microscope sections and only one neuron has the structural features of an olfactory cell (Ashton et al. 1995). It may be interesting to determine whether this cell most closely resembles C. elegans AWC. A possible role for AWC as a cellular module is likely just one example of a general strategy used during evolution to transform a simple organ with a few cell types into a complex multifunctional organ.
Figure 8.
A model for olfactory cell fate specification in C. elegans. Analysis of lim-4 and odr-7 mutants suggests that a common cell fate specification pathway operates in AWA, AWB, and AWC, despite the fact that these neurons are not related by lineage. A common olfactory cell fate determinant may specify generic features of these three olfactory cells. In the absence of ODR-7 and LIM-4, these cells will develop like AWC, a potential olfactory default state. Modification of this fate in AWA and AWB is accomplished by ODR-7 and LIM-4, either by themselves or in cooperation with other factors. These factors direct the three olfactory neurons to adopt distinct gene expression patterns, cell morphologies, and behavioral functions.
Materials and methods
Strain maintenance
Wild-type strains were C. elegans variety Bristol, strain N2. Strains were maintained by standard methods (Brenner 1974).
Isolation of mutants, mapping, and cloning lim-4
str-2::GFP(kyIs140); rol-6(e187) worms were mutagenized with EMS according to standard protocols and screened for ectopic expression of GFP under a fluorescence dissecting scope in the F2 generation (Anderson 1995). The three lim-4 alleles were outcrossed once to a str-2::GFP strain and at least twice to N2. lim-4(ky403) was mapped with respect to Tc1 transposable element polymorphisms in the DP13 strain (Williams 1995). Mapping localized the mutant to the X chromosome between the stP40 and stP156 polymorphisms. Rescue of the str-2::GFP misexpression phenotype was observed in three independent lines injected with the cosmid ZC64 at 10 μg/ml. The lim-4 genomic-coding sequence in the three mutants was amplified by PCR in two pieces and PCR products were sequenced directly. Lesions were confirmed by sequencing a second independently amplified PCR product.
Germ-line transformation
Transgenic strains were created according to methods described previously (Mello and Fire 1995). Transgenic animals used for behavioral assays were generated in a lin-15(n765) background and rescued with a lin-15 pJM23 coinjection marker at 50 μg/ml. These strains were maintained by picking non-Multivulva worms. Other transgenic lines were made with the dominant pRF4 rol-6(su1006) coinjection marker at 50 μg/ml, and maintained by picking Roller animals. The odr-3::LIM-4 transgene was injected at 100 μg/ml, and all other transgenes were injected at 50 μg/ml. In all cases in which phenotypes were quantified in extrachromosomal array-bearing lines, at least five lines exhibited the same qualitative phenotype and the strongest was chosen for quantitation.
Transgenes and cDNA
The str-2::GFP(kyIs140), str-1::GFP(kyIs104), and odr-3::GFP(kyIs126) transgenes were integrated strains described previously (Troemel et al. 1997; Dwyer et al. 1998; Roayaie et al. 1998).
Six GFP marker strains were crossed into lim-4(ky403) to determine the specificity of the mutant’s phenotype. The ASI, ADL, and ADF GFP marker transgenes were fusions of GFP to promoters of putative seven-transmembrane domain proteins (ASI = M7.13, ADL = F47C12.5, ADF = T08G3.3; Y. Zhang, E.R. Troemel, J. Hao, and C.I. Bargmann, unpubl.). The ASI and ADL strains were integrated and the ADF strain was extrachromosomal. For AWA, an integrated odr-10::GFP(kyIs37) transgene was used (Sengupta et al. 1996). For ASEL and ASER extrachromosomal fusions with promoters of the guanylyl cyclase, genes gcy-5 and gcy-6 were used (Yu et al. 1997). The ADF, ADL, ASEL, and ASER markers were crossed into lim-4(ky403) and hemizygous F1 males were scored. For the ASI and AWA markers, homozygous lim-4; GFP strains were generated.
lim-4 cDNA
The cDNA was isolated by PCR from a mixed-stage cDNA library with primers that match the amino and carboxyl terminus of the predicted ZC64.4 gene. The predicted gene structure of ZC64.4 (GenBank accession no. U39740) was confirmed with one exception. ZC64.4 was predicted to contain a 17-amino-acid insertion at amino acid position 289. This insertion most likely results from a misprediction of the gene structure because we could not detect it in cDNAs that were isolated. Moreover, the insertion is in the middle of the homeodomain, which would be highly unusual.
lim-4::GFP2(mgIs19)
A 4.5-kb fragment containing 3.6 kb 5′ to the predicted ATG start codon and the first three lim-4 exons was amplified from genomic DNA and cloned into the GFP reporter gene vector pPD95.75 generously provided by A. Fire (Carnegie Institute, Baltimore, MD). Transgenic lines were generated by injecting 50 μg/ml lim-4::GFP2 and 100 μg/ml pRF4 DNA into wild-type worms. The extrachromosomal DNA was integrated by use of a Stratalinker 1800 UV light source at 300 J/m2.
lim-4::GFP1
A total of 2682 bp containing the remaining introns and exons of the lim-4 gene were added to the lim-4::GFP2 fusion. GFP was fused to the last amino acid of the lim-4 coding sequence.
odr-3::LIM-4
The pD95.77 expression plasmid, generously provided by A. Fire, was digested with MscI and EcoRI to remove GFP, and the EcoRI overhang was blunted. The lim-4 cDNA was amplified by PCR with primers bearing blunt restriction site, digested, and ligated into the vector. The PCR product was sequenced to ensure that no errors were introduced. An EcoRV fragment of the odr-3 promoter (Roayaie et al. 1998) was cloned into the SmaI site in the vector.
str-2::ODR-10
Amplification of the str-2 coding region was done by PCR at 3.7 kb upstream. PstI and BamHI sites engineered into the PCR primers were used to insert the amplified product into a vector containing an odr-10 cDNA (Sengupta et al. 1996).
Dye-filling
Worms were placed in 4 μl of a DiD solution (10 mg/ml DiD dissolved in DMSO) diluted in 1 ml of M9 buffer with food, incubated for 12 to 16 hr, and allowed to recover for 4 to 6 hr on a fresh seeded plate before examination with TRITC fluorescence filters. GFP markers for AWB and AWC cells were included to facilitate cell identifications.
Temperature shifts
To synchronize populations of mutant animals, L4 larval stage animals were placed at 15°C or 25°C and then disintegrated as gravid adults in a bleach solution (40% bleach, 0.4 m NaOH), leaving a synchronized population of eggs. Animals were shifted between temperatures at the L1 larval stage and assayed as young adults, 48 to 72 hr later.
Single worm chemotaxis assays and laser ablations
Single worm assays were performed as described previously (Troemel et al. 1997). Briefly, a healthy adult worm was placed at the center of a square agarose plate, two 1 μl point sources of the odorant (diacetyl diluted 1:1000 in ethanol) were spotted at one end of the plate and two 1 μl spots of the diluent (ethanol) were placed at the other end of the plate. The animal’s tracks were observed after 1 hr. Plates were divided into six zones with assigned values from −3, for the zone farthest from the odorant, to +3, for the zone closest to the odorant. Assays were scored by summing the scores for the zones through which the worm traveled. For example, a worm that moved directly toward the odorant passed through zones 1, 2, and 3 and received the maximum score of 6. Conversely, worms that were completely repulsed by the odorant received a score of −6. Worms unable to sense the odorant moved randomly and averaged a score of 0. Results were analyzed with a Mann Whitney rank sum test.
Laser ablations were performed in the L1 or L2 larval stages as described previously (Bargmann and Avery 1995). Animals used for laser ablation carried both the str-2::ODR-10 and the str-2::GFP transgenes. Only animals that expressed GFP in the appropriate cells were used in behavioral assays. The transgene-bearing AWC cell was identified for ablation by the presence of the str-2::GFP coinjection marker. To confirm that ablations were effective, animals were examined for str-2::GFP in the AWC cell after the assay. Only animals with no GFP expression in AWC were included.
Acknowledgments
We thank Shannon Grantner and Yongmei Zhang for excellent technical support; Joe Hao and Tim Yu for help with confocal microscopy; and Candace Chi, Gage Crump, Sue Kirch, Noelle L’Etoile, David Tobin, and Fan Wang for comments on the manuscript and discussions about experiments. We are grateful to Alan Coulson and the Sanger Centre for cosmids. This work was supported by grants from the Human Frontiers Science Program (to C.I.B.) and Hoechst AG to the Department of Molecular Biology, Massachusetts General Hospital (G.R.). A.S. is a Howard Hughes Medical Institute predoctoral fellow, O.H. was a Human Frontiers Science Program postdoctoral fellow, E.R.T. was a National Science Foundation predoctoral fellow, and C.I.B. is an Assistant Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cori@itsa.ucsf.edu; FAX (415) 476-3493.
References
- Anderson P. Mutagenesis. Methods Cell Biol. 1995;48:31–58. [PubMed] [Google Scholar]
- Ashton FT, Bhopale VM, Fine AE, Schad GA. Sensory neuroanatomy of a skin-penetrating nematode parasite: Strongyloides stercoralis. I. Amphidial neurons. J Comp Neurol. 1995;357:281–295. doi: 10.1002/cne.903570208. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Avery L. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 1995;48:225–250. doi: 10.1016/s0091-679x(08)61390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck LB. Information coding in the vertebrate olfactory system. Annu Rev Neurosci. 1996;19:517–544. doi: 10.1146/annurev.ne.19.030196.002505. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Certel SJ, de Bruyne M, Zaslavsky L, Johnson WA, Carlson JR. The odor specificities of a subset of olfactory receptor neurons are governed by Acj6, a POU-domain transcription factor. Neuron. 1999a;22:339–347. doi: 10.1016/s0896-6273(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron. 1999b;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Coburn CM, Bargmann CI. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron. 1996;17:695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- Colbert HA, Smith TL, Bargmann CI. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci. 1997;17:8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss J, Heilig JS. Arrowhead encodes a LIM homeodomain protein that distinguishes subsets of Drosophila imaginal cells. Dev Biol. 1997;190:129–141. doi: 10.1006/dbio.1997.8659. [DOI] [PubMed] [Google Scholar]
- Dwyer ND, Troemel ER, Sengupta P, Bargmann CI. Odorant receptor localization to olfactory cilia is mediated by ODR-4, a novel membrane-associated protein. Cell. 1998;93:455–466. doi: 10.1016/s0092-8674(00)81173-3. [DOI] [PubMed] [Google Scholar]
- Freyd G, Kim SK, Horvitz HR. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- Grigoriou M, Tucker AS, Sharpe PT, Pachnis V. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development. 1998;125:2063–2074. doi: 10.1242/dev.125.11.2063. [DOI] [PubMed] [Google Scholar]
- Hobert O, Mori I, Yamashita Y, Honda H, Ohshima Y, Liu Y, Ruvkun G. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron. 1997;19:345–357. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- Hobert O, Ruvkun G. A common theme for LIM homeobox gene function across phylogeny? Biol Bul. 1998a;195:377–381. doi: 10.2307/1543153. [DOI] [PubMed] [Google Scholar]
- Hobert O, D’Alberti T, Liu Y, Ruvkun G. Control of neural development and function in a thermoregulatory network by the LIM homeobox gene lin-11. J Neurosci. 1998b;18:2084–2096. doi: 10.1523/JNEUROSCI.18-06-02084.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Tessmar K, Ruvkun G. The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development. 1999;126:1547–1562. doi: 10.1242/dev.126.7.1547. [DOI] [PubMed] [Google Scholar]
- Komatsu H, Mori I, Rhee JS, Akaike N, Ohshima Y. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and themosensation in C. elegans. Neuron. 1996;17:707–718. doi: 10.1016/s0896-6273(00)80202-0. [DOI] [PubMed] [Google Scholar]
- Lundgren SE, Callahan CA, Thor S, Thomas JB. Control of neuronal pathway selection by the Drosophila LIM homeodomain gene apterous. Development. 1995;121:1769–1773. doi: 10.1242/dev.121.6.1769. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Tanaka T, Furuyama T, Kashihara Y, Ishii N, Tohyama M, Kitanaka J, Takemura M, Mori T, Wanaka A. Differential expression of LIM-homeodomain genes in the embryonic murine brain. Neurosci Lett. 1996;211:147–150. doi: 10.1016/0304-3940(96)12749-x. [DOI] [PubMed] [Google Scholar]
- McIntire SL, Jorgensen E, Kaplan J, Horvitz HR. The GABAergic nervous system of Caenorhabditis elegans. Nature. 1993;364:337–341. doi: 10.1038/364337a0. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Mitani S, Du H, Hall DH, Driscoll M, Chalfie M. Combinatorial control of touch receptor neuron expression in Caenorhabditis elegans. Development. 1993;119:773–783. doi: 10.1242/dev.119.3.773. [DOI] [PubMed] [Google Scholar]
- Mori I, Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature. 1995;376:344–348. doi: 10.1038/376344a0. [DOI] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, Bargmann CI. The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20:55–67. doi: 10.1016/s0896-6273(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Colbert HA, Bargmann CI. The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell. 1994;79:971–980. doi: 10.1016/0092-8674(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Chou JH, Bargmann CI. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996;84:899–909. doi: 10.1016/s0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95:817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, Bargmann CI. Reprogramming chemotaxis responses: Sensory neurons define olfactory preferences in C. elegans. Cell. 1997;91:161–169. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans R Soc Lond B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Williams BD. Genetic mapping with polymorphic sequence-tagged sites. Methods Cell Biol. 1995;48:31–58. doi: 10.1016/s0091-679x(08)61384-9. [DOI] [PubMed] [Google Scholar]
- Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific neurons: A new family of chemosensory receptors. Proc Natl Acad Sci. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chou JH, Bradley J, Bargmann CI, Zinn K. The Caenorhabditis elegans seven-transmembrane protein ODR-10 functions as an odorant receptor in mammalian cells. Proc Natl Acad Sci. 1997;94:12162–12167. doi: 10.1073/pnas.94.22.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]