Abstract
RNA polymerase II nascent transcripts are capped during pausing before elongation. Here we report that hSPT5, the human homolog of yeast elongation factor SPT5, interacts directly with the capping enzyme. hSPT5 stimulated capping enzyme guanylylation and mRNA capping by severalfold. Although RNA 5′-triphosphatase activity was unaffected, binding to this domain in the full-length enzyme is likely involved in the stimulation, as hSPT5 did not increase the activity of the guanylyltransferase fragment. Consistent with capping enzyme binding, TFIIH-phosphorylated CTD stimulated guanylylation, and this increase was not additive with hSPT5.
Keywords: Capping enzyme, hSPT5, P-CTD, transcription elongation
Metazoan capping enzymes are bifunctional proteins with both RNA 5′- triphosphatase (RTP) and RNA guanylyltransferase (RGT) activities. In concert with RNA (guanine-7-) methyltransferase, they selectively modify the 5′ ends of newly initiated RNA polymerase II (Pol II) transcripts by adding a 7-methylguanosine cap soon after transcription initiation (Shatkin 1976; Shuman 1995; Moss 1996). This modification facilitates transcript processing, transport, translation, and turnover (Furuichi and Shatkin 1999).
The mammalian carboxy-terminal RGT domain contains the highly conserved KXDG motif, which is required for formation of the capping enzyme intermediate by phosphoamide linkage of GMP to the ε-amino group of the motif Lys (Shuman 1995). The amino-terminal RTP domain also contains a conserved motif, (I/V)HCXXGXXR(S/T)G, which is characteristic of both RTP and protein tyrosine phosphatases (PTPs) that form a motif cysteinyl–phosphate intermediate, suggesting that RTP and PTPs share a common mechanism (Takagi et al. 1997; Ho et al. 1998a; Wen et al. 1998).
Caps are added to nascent Pol II transcripts when they reach a chain length of 25–30 nucleotides (Coppola et al. 1983; Hagler and Shuman 1992; Rasmussen and Lis 1993). This specificity is accomplished by the direct binding of the capping enzyme to the phosphorylated form of the Pol II largest subunit carboxy-terminal domain (P-CTD; Cho et al. 1997; McCracken et al. 1997; Yue et al. 1997). The CTD, which is unique to Pol II, consists of a tandemly repeated heptad motif, YSPTSPS (Dahmus 1996). During the transition from initiation to stable elongation complex, the CTD undergoes phosphorylation, including TFIIH- catalyzed phosphorylation of Ser-5 in the heptad repeats (Lu et al. 1992; Shiekhattar et al. 1995; Trigon et al. 1998). The finding that the capping enzyme is recruited to transcription complexes via binding to P-CTD explains the selective capping of Pol II transcripts and may couple capping to the switch from transcription initiation to elongation.
An important issue is whether other transcription elements are involved in linking capping to transcription, and if the capping enzyme is regulated in transcription complexes or itself influences Pol II transcription. The capping enzyme apparently does not associate with basal transcription initiation factors (Cho et al. 1997), but deciphering the interactions between the capping enzyme and elongation complexes during the promoter clearance step of transcription may provide new insights into the functional interplay between capping and transcription events.
Transcription elongation is an important target for gene regulation (Uptain et al. 1997). Many eukaryotic elongation factors function by either preventing or overcoming Pol II transcriptional pausing. For example, SPT5, SPT4, and SPT6 in Saccharomyces cerevisiae function together in an elongation-related transcriptional process that is essential for viability (Swanson and Winston 1992). Recent biochemical and genetic studies (Hartzog et al. 1998; Wada et al. 1998a) showed that the yeast SPT5–SPT4 complex and its human counterpart DSIF (DRB sensitivity inducing factor) are novel transcription elongation factors. Human SPT5 has been shown to play a role in Tat activation of HIV-1 gene expression at the level of promoter clearance and/or transcription elongation (Wu-Baer et al. 1998).
SPT5 contains a region similar in sequence to Escherichia coli transcription elongation factor NusG, which binds directly to the bacterial RNA polymerase (Li et al. 1992; Sullivan and Gottesman 1992). Moreover, SPT5 was shown to interact with Pol IIa containing unphosphorylated CTD and not with the phosphorylated Pol IIo (Wada et al. 1998b; Yamaguchi et al. 1999a,b), indicating that the phosphorylation state of CTD affects this interaction. The ability of SPT5–SPT4 complexes to inhibit transcription elongation is likely related to the phosphorylation state of CTD, as CTD phosphorylation resulted in the release of DSIF from Pol II and reversal of its negative effects on elongation (Wada et al. 1998b).
In this study, using the yeast two-hybrid screen to look for novel human proteins that interact with the capping enzyme, we isolated hSPT5, the human homolog of yeast SPT5. hSPT5 bound human capping enzyme (HCE) in vitro and, in addition, stimulated enzyme guanylylation and cap formation severalfold without affecting its RNA 5′-triphosphatase activity. CTD that was phosphorylated by TFIIH also dramatically stimulated guanylylation, an enhancement not augmented by the presence of hSPT5. Our studies indicate that 5′-capping and Pol II transcription are coupled by interactions between capping enzyme and hSPT5, as well as P-CTD, and that mRNA capping is stimulated by these interactions.
Results
Isolation of hSPT5 by association with HCE
To identify proteins that interact with HCE, a fusion protein of full-length HCE and GAL4 DNA-binding domain (DBD) was used as bait to screen a human fetal brain library fused to the GAL activation domain (AD). Twenty-one HIS+/lacZ+ candidates were obtained from 6.1 × 106 transformants, and none interacted with negative control proteins. DNA sequence and BLAST database searches revealed that 13 clones perfectly matched the nucleotide sequence of hSPT5, and three corresponded to Pol II CTD fragments containing 42 heptad repeats; the five others are being characterized. Two hSPT5 clones contained the entire ORF, and 9 of the 13 clones were in the translation reading frame. The full-length hSPT5 clone was then isolated from a human HeLa cDNA library, and its reading frame was confirmed using the coupled transcription/translation system.
HCE directly interacts with hSPT5 in vitro
To test for direct interaction between the mammalian capping enzyme and hSPT5, purified GST-fused hSPT5 was linked to glutathione–agarose and incubated with purified His-tagged HCE. About 10% of the input HCE bound to hSPT5 (Fig. 1A, lane 3) and not to GST (lane 2). Binding was relatively stable, and >60% of the complexes remained after washing with buffer containing 0.5 m NaCl (data not shown). In addition, HeLa cell extract incubated with monoclonal antibody to hSPT5 (and not anti-GST control monoclonal) immunoprecipitated the capping enzyme as measured by radiolabeling with [α-32P]GTP and SDS-PAGE (data not shown).
Figure 1.
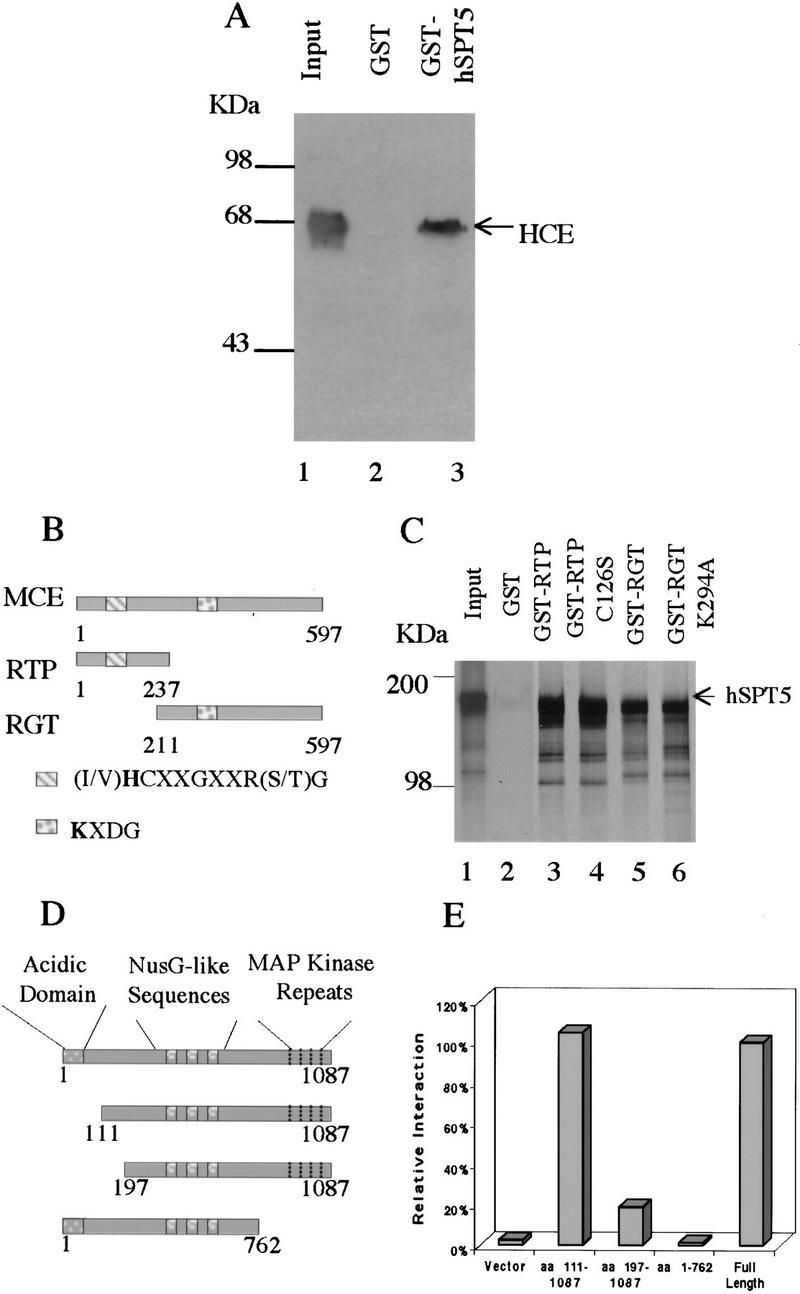
Mammalian capping enzymes interact with hSPT5 in vitro and in vivo. (A) Binding to HCE. Purified GST–hSPT5 was incubated with purified HCE (lane 1). Bound HCE was detected by SDS-PAGE followed by immunobloting with anti-MCE antisera (Yue et al. 1997; lanes 2,3) and densitometry. (Input lane) 50% of the amount of HCE used for the binding assays. (B) Schematic diagram of the MCE, RTP, and RGT domains. (C) Purified GST, GST–RTP, GST–RTP (C126S), GST–RGT, or GST–RGT (K294A) was incubated with [35S]Met-labeled hSPT5. Bound proteins were separated by SDS-PAGE followed by autoradiography. (Input lane) 10% of the amount of hSPT5 used for the assays. (D) Schematic diagram of hSPT5 protein constructs used to define sequence requirements for interaction with HCE. (E) Full-length hSPT5 and indicated truncation mutants were transformed into yeast Y190 harboring HCE. Yeast colonies that grew on SD/−Trp/−Leu were assayed for liquid β-gal activity.
Both RTP and RGT fragments of mammalian capping enzyme bind to hSPT5
HCE and mouse capping enzyme (MCE) are 97% identical (Yue et al. 1997) and consist of RTP and RGT domains (Fig. 1B). To investigate which domain(s) associate with hSPT5, [35S]Met-labeled hSPT5 was synthesized in vitro. The GST- fused MCE RTP fragment and RGT fragment were bound to glutathione–agarose and incubated with hSPT5. As shown in Figure 1C (lane 1) hSPT5 had an apparent molecular mass of ∼160 KDa by SDS-PAGE; the smaller products presumably are prematurely terminated products. As in the case of the full- length HCE (Fig. 1A), the RTP and RGT fragments both formed complexes with hSPT5 (Fig. 1C, lanes 3,5). About 10% of the input in each case bound to the GST- fused protein fragments and not to GST (Fig. 1C, lane 2). Mutations in motif active site Cys or Lys of RTP and RGT, respectively, which eliminate enzyme activity in each case (Yue et al. 1997; Wen et al. 1998), had no effect on binding (Fig, 1C, lanes 4,6). These findings indicate that hSPT5 binds not only to the full-length capping enzyme but also to the RTP and RGT fragments, suggesting that both domains are involved in the interaction.
Sequences in hSPT5 required for interaction with HCE
hSPT5 contains a very acidic amino-terminal domain and MAP kinase repeats near the carboxyl terminus. It also contains repeated elements that are homologous to a region of E. coli transcription elongation factor NusG (Fig. 1D). The two-hybrid system was used to map the domain(s) of hSPT5 that interacts with HCE. Yeast cells harboring HCE were transformed with full-length hSPT5; hSPT5(1–762); hSPT5(111–1087), or hSPT5(197–1087) (Fig. 1D). By β-gal liquid assay, hSPT5(1–762) completely lost any productive interaction with HCE, hSPT5(197–1087) retained only 21% interaction, and hSPT5(111–1087) was fully active compared with full-length hSPT5 (Fig. 1E). On the basis of these results, a carboxy-terminal region, including the MAP kinase repeats and a region within amino acids 111–197 is involved in the association of hSPT5 with HCE, whereas the very acidic domain (amino acids 1–106) is not required for binding.
hSPT5 enhances guanylylation and cap formation but not RTP activity
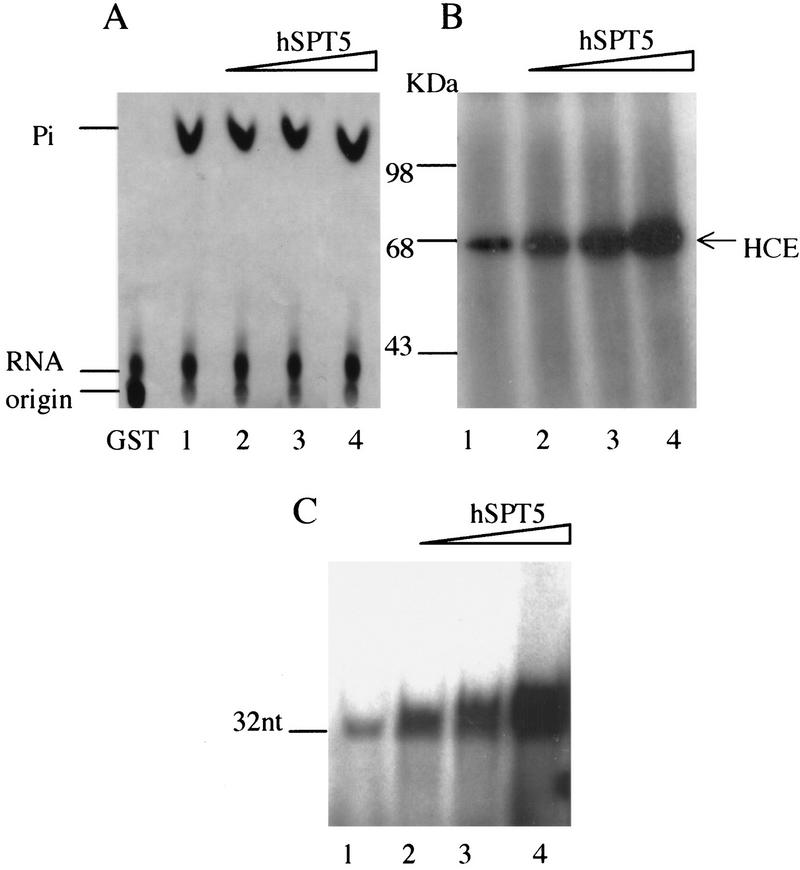
Because hSPT5 interacts with both the RTP and RGT domains of the mammalian capping enzyme, we examined its effects on the different steps of the capping reaction. First, purified hSPT5 was incubated with γ-32P labeled RNA and HCE. As shown in Figure 2A, RTP-mediated release of 32P by HCE was not changed by increasing levels of hSPT5. Next we investigated whether hSPT5 influences guanylylation in an assay containing [α- 32P]GTP and HCE. In contrast to RTP activity, hSPT5 stimulated capping enzyme guanylylation in a concentration-dependent manner (Fig. 2B). The stimulation was 2.1-fold when HCE and hSPT5 were present at a molar ratio of 1:2 (Fig. 2B, lane 2) and increased to 5.3-fold at a 1:8 ratio (Fig. 2B, lane 4). hSPT5 also markedly stimulated cap formation as measured by transfer of GMP to RNA 5′ ends (Fig. 2C). At a HCE/hSPT5 molar ratio of 1:8, capping was increased 5.1-fold (Fig. 2C, lane 4). These data suggest that hSPT5 enhances guanylylation and cap formation without affecting RTP activity, a notable finding in light of the rate-limiting nature of the guanylylation step in capping (Shuman 1995).
Figure 2.
hSPT5 increases RGT and capping but not RTP activity. HCE (3.5 pmoles) was assayed for RTP activity (A), guanylylation (B), and RNA capping (C) in the presence of increasing amounts of hSPT5 (0, 7, 14, 28 pmoles) as described in Materials and Methods. (A) 32Pi released from γ-32P-labeled runoff transcripts was detected by TLC. Guanylylated HCE (B) and capped RNA (C) were analyzed by PAGE and autoradiography.
Involvement of RTP domain/hSPT5 interaction in stimulation of capping
Although hSPT5 stimulated the activity of the full-length HCE (Figs. 2B,C), it had no apparent effect on the guanylylation of the RGT active fragment (Fig. 3A). Because hSPT5 binds RTP fragment, we tested whether its stimulation of the full-length capping enzyme is affected by the presence of this fragment. Purified MCE RTP fragment added to the guanylylation reaction mixture containing full-length HCE and hSPT5 (1:2 molar ratio) diminished the stimulatory effect of hSPT5 (Fig. 3B), and at a RTP/hSPT5 molar ratio of 4:1 the enhancement of guanylylation was almost abolished (Fig. 3B, lane 5), apparently because of the RTP fragment competing for hSPT5 binding of the corresponding region in the full-length capping enzyme. Based on these results, we speculate that hSPT5 interaction with the RTP domain has an allosteric effect on HCE that increases its RGT activity.
Figure 3.
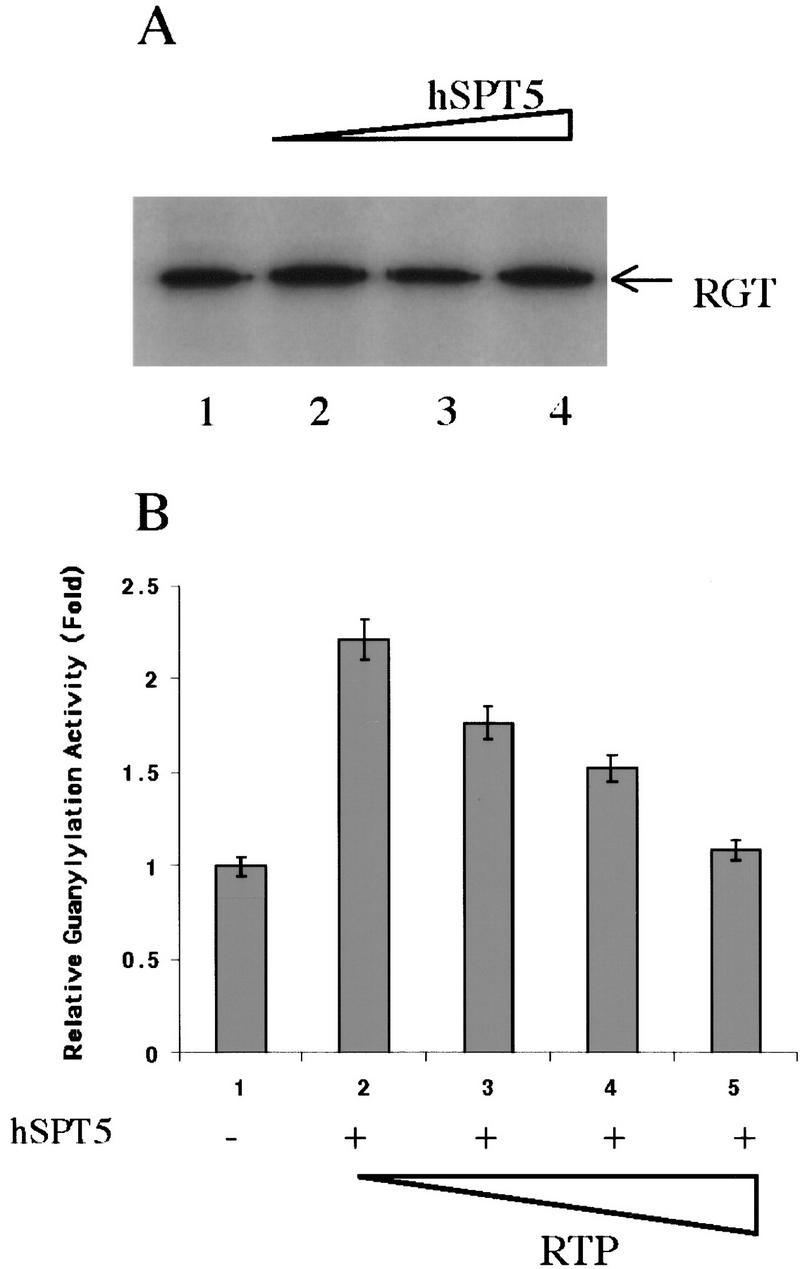
RTP–hSPT5 interactions can reverse guanylylation stimulation. (A) Purified RGT fragment (3.5 pmoles) was incubated with [α-32P]GTP in the presence of 0, 7, 14, and 28 pmoles of hSPT5 and assayed as in Fig. 2B. (B) HCE (3.5 pmoles, lanes 1–5) was assayed for guanylyltransferase activity in the presence of hSPT5 (7 pmoles, lanes 2–5) and 7, 14, or 28 pmoles of MCE–RTP fragment (lanes 3–5). Guanylylation of 32P-labeled HCE was quantitated by PhosphorImager after SDS-PAGE.
Stimulation of capping reaction by P-CTD is not increased by hSPT5 and enhancement of capping by hSPT5 is not affected by CTD
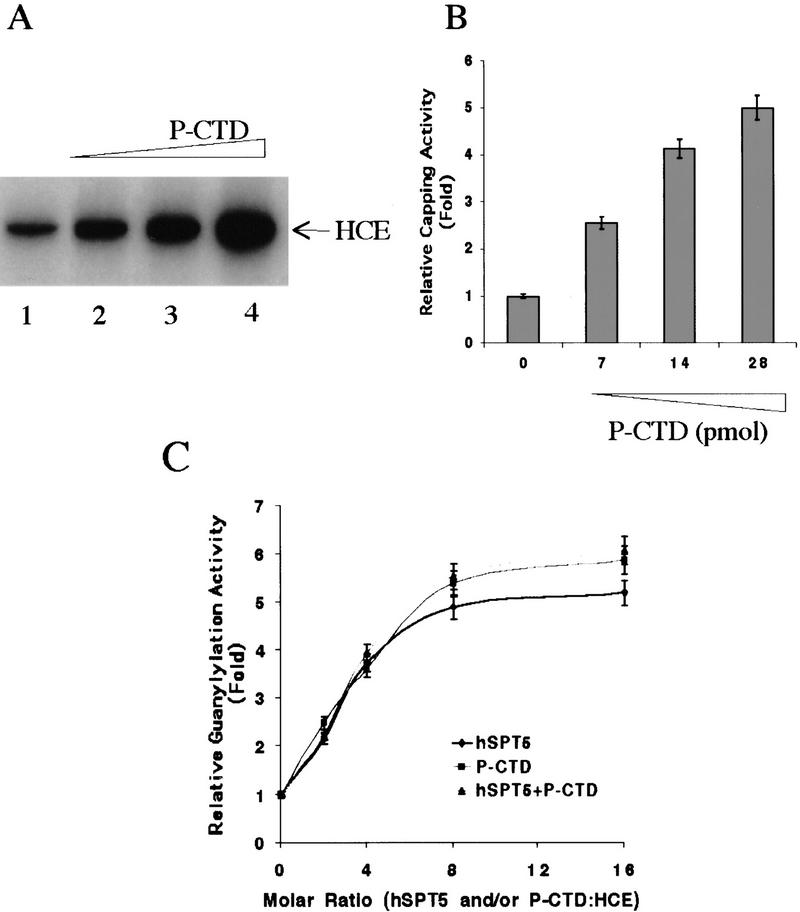
To test the possibility that P-CTD influences capping, a recombinant CTD with 42 repeats was phosphorylated by incubation with purified TFIIH. Addition of the P-CTD to HCE in the RGT reaction mixture dramatically increased the formation of the 32P-labeled capping enzyme–GMP intermediate (Fig. 4A). Similar to hSPT5, stimulation was 2.6-fold at a 1:2 molar ratio (Fig. 4A, lane 2), and increased to 5.4-fold at a molar ratio of 1:8 (Fig. 4A, lane 4). P-CTD also markedly stimulated cap formation (Fig. 4B). These findings indicate that like hSPT5, P-CTD can influence capping by direct interaction with HCE.
Figure 4.
Stimulation of capping reaction by P-CTD is not enhanced by hSPT5. (A) CTD was phosphorylated by incubation with TFIIH, and the effect of 0, 7, 14, or 28 pmoles on HCE guanylylation was assayed as in Fig. 2B. (B) TFIIH-phosphorylated CTD was incubated with HCE (3.5 pmoles), [α-32P]GTP and runoff transcripts and assayed for RNA capping as in Fig. 2C. (C) hSPT5, P-CTD or both proteins (0, 7, 14, 28, 56 pmoles) was incubated with 3.5 pmoles of HCE and [α-32P]GTP and assayed for guanylylation as in Fig. 2.
Because both hSPT5 and P-CTD activate the capping reaction, we tested for cooperative effects by the two proteins. HCE was incubated with equal molar amounts of hSPT5, P-CTD, or both proteins in the presence of [α-32P]GTP. Under these conditions, the formation of 32P-labeled guanylylated HCE was increased similarly by either hSPT5 or P-CTD, but the effects were not additive (Fig. 4C). Addition of eight-fold molar excess of CTD did not decrease the stimulation of HCE guanylylation by hSPT5 (data not shown).
Discussion
The selective capping of Pol II transcripts is explained by the interaction between the capping enzyme and the P-CTD of Pol II which, together with a recent report that phosphorylated RNA Pol II stimulates pre-mRNA splicing (Hirose et al. 1999), links RNA processing with transcription. Our studies provide evidence that the capping enzyme also interacts with and is activated by a transcription elongation factor component, hSPT5.
hSPT5 functions together with hSPT4 as transcription elongation factor DSIF. It also can associate with RNA Pol II containing unphosphorylated but not phosphorylated CTD, suggesting that hSPT5, by this interaction, inhibits the switch of transcription complexes from initiation to processive elongation. P-TEFb is a positive transcription elongation factor (Marshall and Price 1992, 1995) that has been shown to phosphorylate CTD (Marshall et al. 1996). With subsequent binding of the capping enzyme to the P-CTD, hSPT5 in transcription complexes may then interact functionally with the capping enzyme to up-regulate RGT activity and enhance capping.
Previous studies demonstrated that TFIIH selectively phosphorylates Ser-5 in the CTD repeats. We found that TFIIH-phosphorylated CTD stimulated the capping enzyme severalfold, in agreement with a recent report that a synthetic P-CTD consisting of two repeats with phosphorylated Ser-5 increased the yield of the guanylyltransferase–GMP intermediate (Ho and Shuman 1999). Thus, CTD in the phosphorylated form can directly activate the capping enzyme. hSPT5 and P-CTD were similarly active in stimulating guanylylation, but surprisingly, the effects were not additive. These studies indicate that P-CTD and hSPT5 can interact directly with the RGT domain of the capping enzyme, and they may use the same or overlapping binding site(s) to increase capping. This could result in a mutual block to capping enzyme access and thus limit stimulation. Further studies on the binding of hSPT5 to the capping enzyme and the capping enzyme to P-CTD should provide new insights into the regulation of capping by these protein–protein interactions.
The yeast capping enzyme consists of two separately encoded subunits, RTP (Cet1) and RGT (Ceg1). Cet1 and Ceg1 form heterodimers, an interaction that stimulates RGT activity (Ho et al. 1998b) or reverses the inhibition of RGT caused by P-CTD binding to Ceg1 (Cho et al. 1998). On the basis of these results and our findings, it is likely that RTP can regulate RGT allosterically in yeast. The RTP domain in metazoan bifunctional capping enzymes may be involved similarly in the regulation of RGT activity. We found that hSPT5 increased the RGT activity of the full-length capping enzyme but not that of the RGT fragment, and enhancement was prevented by the addition of the RTP fragment. RTP and RGT sequences in the capping enzyme may interact to limit activity, and association of hSPT5 with the RTP region may activate capping by opening the RGT catalytic domain. Alternatively, the RTP–hSPT5 association may induce a conformational change in the full-length protein, which increases binding between RGT and hSPT5 to promote capping.
Because the basal levels of guanylylation of the full-length capping enzyme and the RGT fragment are similar (data not shown), we favor the second possibility.
The capping enzyme associates with Pol II transcription complexes, apparently during the transition from initiation to elongation, and may also function as a regulator of transcription elongation by binding transcription elongation factors like DSIF. Further study of the effect of capping on transcription will likely uncover other aspects of the functional interplay between RNA processing and transcription.
Materials and methods
Yeast two-hybrid screen
HCE was cloned into the GAL4–DBD fusion vector pAS2-1. pAS2-1 HCE was transformed into yeast strain Y190, and the resulting transformants were tested for the absence of autoactivation of lacZ and HIS3 reporter genes. For the library selection, Y190 cells transformed with pAS2-1 HCE were transformed with a human fetal brain library constructed into a GAL4–AD fusion vector pACT2 (Matchmaker, Clontech). Transformants were plated on SD/−Trp/−Leu/−His + 35 mm 3-AT, and yeast plasmid DNAs of the HIS+ lacZ+ colonies were isolated and rescued into E.coli KC8. To verify the interaction, the library plasmids were cotransformed with pAS2-1, pAS2-1 lamin C or pAS2-1 HCE, and the activation of lacZ reporter genes was tested. The inserts of the library plasmids in positive clones were sequenced.
In vitro translation
Full-length hSPT5 was isolated from human HeLa Marathon-Ready cDNA (Clontech) with primers 5′-GAATCCATGTCGGACAGCGAGGACATCAAC-3′ and 5′-CTCGAGTCAGGCTTCCAGGAGCTTCCCCAG-3′, introducing the EcoRI site at the 5′ end and the XhoI site at the 3′ end. The PCR products were cloned into pCR2.1 (Invitrogen) and subcloned into His-tagged fusion vector pET28a (Novagen). pET28a hSPT5 was transcribed and translated in vitro by the TNT Quick Coupled Transcription/Translation System (Promega) with 20 μCi of [35S]Met (1000Ci/mmole, Amersham).
Expression and purification of hSPT5 recombinant proteins
His-tagged hSPT5 was expressed and purified on Ni–NTA agarose (Qiagen) from E. coli BL21 (DE3) transformed with pET28a hSPT5 as described (Wen et al. 1998). The hSPT5 fragment was subcloned into GST fusion vector pGEX-4T-1. GST-fused hSPT5 was expressed and purified on glutathione–agarose (Sigma) from E. coli BL21 (DE3) harboring pGEX-4T-1 hSPT5 as described (Wen et al. 1998).
Expression and purification of MCE and HCE recombinant proteins
GST–MCE–RTP and GST–MCE–RGT were expressed and purified as described (Wen et al. 1998). pHis (T)–hCAP1a was the kind gift of Dr. K. Mizumoto (Kitasato University, Tokyo, Japan). E. coli BL21 (DE3) containing pHis(T)–hCAP1a was grown at 17°C in LB medium and induced with 0.5 mm IPTG. Cells were harvested after 17 hr and protein purified as described (Wen et al. 1998).
Purification and phosphorylation of CTD fusion protein
The CTD EcoRI–XhoI fragment with 42 repeats isolated from the yeast two-hybrid screen was subcloned into vector pGEX-4T-1. GST–CTD fusion proteins were expressed and purified as above. The proteins, bound to glutathione–agarose, were incubated with 2 μl of TFIIH in 60 mm KCl, 50 mm Tris (pH 7.8), 10 mm MgCl2, 1 mm DTT, and 1 mm ATP at 30°C for 90 min. Bound proteins were washed by lysis/wash buffer (Wen et al. 1998) and eluted in 10 mm reduced glutathione, 25 mm Tris at pH 7.5, 5 mm MgCl2, and 0.5 mm DTT. Phosphorylation of CTD was confirmed by gel shift using SDS-PAGE and immunoblotting.
In vitro binding assay
Purified GST–MCE–RTP or GST–MCE–RGT bound to glutathione–agarose (Sigma) was incubated with [35S]Met-labeled hSPT5 in 20 mm Tris (pH 7.9) 0.1 m NaCl, 0.2 mm EDTA, 1 mm DTT, 1 mm PMSF, and 0.2% NP-40 overnight at 4°C and then washed with 20 mm Tris (pH 7.9), 0.5 m NaCl, 0.2 mm EDTA, 1 mm DTT, 1 mm PMSF, and 0.4% NP-40. The bound proteins were eluted from the beads by boiling in SDS sample buffer and separated by SDS-PAGE followed by autoradiography.
Purified GST–hSPT5 bound to glutathione–agarose was incubated with purified His-tagged HCE and washed in the same buffer as above. The bound proteins were separated by SDS-PAGE and analyzed by Western blot with anti-MCE antisera (Yue et al. 1997).
Liquid β-gal assay
hSPT5(111–1087), hSPT5(197–1087), or hSPT5(1–762) EcoRI-XhoI fragment was generated by PCR as above and cloned into vector pACT2. Yeast Y190 harboring pAS2-1 HCE with pACT2, pACT2 hSPT5, pACT2 hSPT5(111–1087), pACT2 hSPT5(197–1087), or pACT2 hSPT5(1–762) was grown to log phase and the OD600 was recorded. Harvested cells were disrupted by three freeze/thaw cycles in Z buffer. Reactions were started by adding ONDG as substrate and terminated by adding Na2CO3 when visible yellowing occurred. Cell debris was removed, and the OD420 was recorded. Each reaction was performed in triplicate.
Assays for RNA 5′-triphosphatase (RTP), RNA guanylyltransferase, and cap formation
These assays were carried out as described previously (Yue et al. 1997).
Acknowledgments
We thank G. Wang and Dr. J. Huibregtse for their generous gifts and helpful assistance with the yeast two-hybrid screen, T.K. Kim and Dr. D. Reinberg for TFIIH, Dr. K. Mizumoto for pHis (T)–hCAP1a, J. Bauman for purified HCE, and Drs. T. Wada and H. Handa for anti-p160 mAb. We also thank H. Chen for valuable suggestions and Dr. Z. Yue for encouragement. We are grateful to Dr. Reinberg for critical comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL shatkin@mbcl.rutgers.edu; FAX (732) 235-5318.
References
- Cho E-J, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E-J, Rodriguez CR, Takagi T, Buratowski S. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1998;12:3482–3487. doi: 10.1101/gad.12.22.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola JA, Field AS, Luse DS. Promoter-proximal pausing by RNA polymerase II in vitro: Transcripts shorter that 20 nucleotides are not capped. Proc Natl Acad Sci. 1983;80:1251–1255. doi: 10.1073/pnas.80.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Furuichi Y, Shatkin AJ. Encyclopedia of life sciences. London, UK: Macmillan; 1999. Caps on eukaryotic mRNAs. . (In press.) [Google Scholar]
- Hagler J, Shuman S. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science. 1992;255:983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes & Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- Ho C K, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998a;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- Ho CK, Schwer B, Shuman S. Genetic, physical, and functional interactions between the triphosphatase and guanylyltransferase components of the yeast mRNA capping apparatus. Mol Cell Biol. 1998b;18:5189–5198. doi: 10.1128/mcb.18.9.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Horwitz S, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N proteins of phage λ. J Biol Chem. 1992;267:6012–6019. [PubMed] [Google Scholar]
- Lu H, Zawel L, Fisher L, Egly JM, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Poxviridae: The viruses and their replication. In: Fields BN, Knipe D, Howley PM, editors. Fields virology. 3rd ed. Philadelphia, PA: Lippincott-Raven Publishers; 1996. pp. 2637–2671. [Google Scholar]
- Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin AJ. Capping of eukaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- Shiekhattar R, Mermelstein F, Fisher RP, Drapkin R, Dynlacht B, Wessling HC, Morgan DO, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;16:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Gottesman ME. Requirement of E. coli NusG protein in factor-dependent transcription termination. Cell. 1992;68:989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Winston F. SPT4, SPT5 and SPT6 interactions: Effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T, Moore CR, Diehn F, Buratowski S. A RNA 5′- triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- Trigon S, Serizawa H, Conaway JW, Conaway RC, Jackson SP, Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J Biol Chem. 1998;273:6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcriptional elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 1998a;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effects of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998b;24:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Yue Z, Shatkin AJ. Mammalian capping enzyme binds RNA and uses protein tyrosine phosphatase mechanism. Proc Natl Acad Sci. 1998;95:12226–12231. doi: 10.1073/pnas.95.21.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Baer F, Lane WS, Gaynor RB. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J Mol Biol. 1998;277:179–197. doi: 10.1006/jmbi.1997.1601. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999a;19:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999b;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]